Abstract
The Jun N-terminal kinases (JNKs) recently have been shown to be required for thymocyte apoptosis and T cell differentiation and/or proliferation. To investigate the molecular targets of JNK signaling in lymphoid cells, we used mice in which the serines phosphorylated by JNK in c-Jun were replaced by homologous recombination with alanines (junAA mice). Lymphocytes from these mice showed no phosphorylation of c-Jun in response to activation stimuli, whereas c-Jun was rapidly phosphorylated in wild-type cells. Despite the fact that c-jun is essential for early development, junAA mice develop normally; however, c-Jun N-terminal phosphorylation was required for efficient T cell receptor-induced and tumor necrosis factor-α-induced thymocyte apoptosis. In contrast, c-Jun phosphorylation by JNK is not required for T cell proliferation or differentiation. Because jnk2−/− T cells display a proliferation defect, we concluded that JNK2 must have other substrates required for lymphocyte function. Surprisingly, jnk2−/− T cells showed reduced NF-AT DNA-binding activity after activation. Furthermore, overexpression of JNK2 in Jurkat T cells strongly enhanced NF-AT-dependent transcription. These results demonstrate that JNK signaling differentially uses c-Jun and NF-AT as molecular effectors during thymocyte apoptosis and T cell proliferation.
Recent studies have shown that the Jun N-terminal kinase (JNK) signaling pathway regulates multiple biological processes in lymphocytes. The JNK pathway is activated in response to a variety of extracellular stresses and mitogenic factors including T cell antigen receptor (TCR) ligation (1). Knockout mice lacking individual jnk genes and transgenic mice overexpressing a dominant negative JNK protein have demonstrated a role for the JNK pathway in thymocyte apoptosis, T cell proliferation, and T cell differentiation (2–5). JNK signaling has been reported to be required for TCR-mediated apoptosis of thymocytes by CD3 crosslinking in the presence of anti-CD28 antibody, which provides an auxiliary signal (3, 4). Recent studies also have implicated the JNK mitogen-activated protein kinases in the control of T cell differentiation (2, 5). JNK2 was found to be required for T helper cell type 1 (Th1) differentiation (5), whereas jnk1−/− mice showed increased Th2 differentiation (2). These findings demonstrated that the JNK isoenzymes play distinct roles during T cell differentiation. Similarly, the analysis of JNK signaling in T cell proliferation using jnk mutant mice revealed opposite functions for JNK1 and JNK2. Peripheral jnk2−/− T cells show severely impaired T cell activation in a dose-dependent manner as measured by proliferation and IL-2 production (4), but the absence of jnk1 was reported to result in hyperproliferation, albeit IL-2 production was normal (2). However, the role of JNK2 is controversial. In one study, mice with JNK2 mutations showed defects in T cell differentiation but not activation, whereas in another they were found to have defects in activation but not differentiation (2, 4).
The JNKs phosphorylate and modify the activity of the AP-1 transcription factors c-Jun, JunD, JunB, ATF2, and ATFa but also may regulate the activity of other proteins including ELK-1, Sap-1, and NF-ATc3 (6–10). Some of the known JNK substrates, most notably the AP-1 and the nuclear factor of activated T cell (NF-AT) transcription factors, have been implicated in T cell function and are thus potential effector molecules of JNK signaling in T cells. c-Jun is a major component of the heterodimeric transcription factor AP-1, which can associate with other transcription factors including the NF-AT to regulate the transcription of T cell-specific target genes such as IL-2 (11–16). c-jun−/− lymphoid cells generated by RAG2 complementation showed incomplete restoration of thymocytes, decreased proliferation of mature B cells, and increased proliferation of T cells, but the function of c-Jun in thymocyte apoptosis and T cell differentiation has not been analyzed (17). The transactivation potential of c-Jun is regulated through phosphorylation of the serine residues 63 and 73 within its transactivation domain by the JNKs (18, 19). N-terminal phosphorylation of c-Jun is thought to increase transcription by stimulating the recruitment of the coactivator proteins CREB binding protein (CBP) and p300 to target gene promoters (20, 21).
The NF-ATc (where c indicates a cytoplasmic component) family of proteins (NF-ATc1 to NF-ATc4) translocate to the nucleus in response to a rise in intracellular calcium and combine with a nuclear component (NF-ATn) to form the NF-AT1 transcription complex. In lymphocytes, NF-ATn is controlled by the ras/protein kinase C pathway. Although NF-ATn has never been purified, it can be replaced with AP-1, c-Maf, and GATA4 (12, 22). Gene-targeting experiments have revealed that the NF-ATc transcription factors have multiple functions in various aspects of T cell biology (13). In particular, NF-ATc transcription factors appear to be both positive and negative regulators of T cell proliferation. NF-ATc1−/− T cells show a decrease in activation-induced proliferation (23), but absence of NF-ATc2 and NF-ATc3 results in hyperactivation of peripheral T cells (24, 25). Moreover, overexpression of a dominant negative NF-AT protein in T cells results in markedly reduced IL-2 production (26). Based on in vitro experiments, NF-ATc3 has been proposed to be a target of JNK-mediated phosphorylation, as JNK can negatively regulate NF-ATc3 translocation to the nucleus in transfected cells (9).
We and others have previously shown that c-Jun is required for embryonic development (27, 28). However, the absence of c-Jun N-terminal phosphorylation (JNP) does not affect fetal development (29). Thus, JNP is not essential for all functions of c-Jun, although it does play a role in fibroblast proliferation and neuronal apoptosis (29). To define the relevance of c-Jun as a target of JNK signaling in lymphoid cells, we have used B and T cells isolated from mice in which a mutant allele of c-jun, lacking the JNK phosphoacceptor sites (junAA), has replaced the endogenous c-jun gene. Here, we provide genetic evidence that N-terminal phosphorylation of c-Jun is involved in thymocyte apoptosis but surprisingly is not required for B and T cell proliferation and differentiation. This finding demonstrates that JNK function must be exerted on a substrate other than c-Jun. We found that the DNA binding and transactivation activity of the NF-AT transcription factor complex is regulated by JNK2 upon peripheral T cell stimulation, suggesting that NF-AT, but not c-Jun, is an essential mediator of JNK signaling in T cell proliferation.
Materials and Methods
Mice.
Mice carrying a mutant c-jun allele, which has the JNK phosphoacceptor serines 63 and 73 mutated to alanines, were generated by using homologous recombination in embryonic stem cells as described (29). The mice used in this study were in a 129×C57/B6×CBA genetic background. The jnk2 mutant mice were in a 129×C57/B6 background (4). For all experiments, sex-matched littermates were used.
Flow Cytometric Analysis.
Single cell suspensions of thymus, spleen, and bone marrow were prepared and stained by standard procedures and analyzed on a FACScan flow cytometer (Becton Dickinson) using cellquest software.
In Vitro Apoptosis Assays.
Freshly isolated thymocytes were plated at 5 × 105 cells/ml in each well of a 24-well dish. For anti-CD3ɛ-induced cell death, wells were precoated with various concentrations of the antibody for 1 h at 37°C, and thymocytes subsequently were cultured for the indicated periods. Apoptosis also was induced by addition of various concentrations of anti-Fas (clone Jo-2, PharMingen), dexamethasone (Fluka), or by UVC irradiation using a Stratagene crosslinker. Apoptotic cells were double-positive for Annexin-V and PI. After incubation for the indicated periods at 37°C, the cells were harvested and stained with FITC-conjugated Annexin-V (PharMingen) and propidium iodide and analyzed with a FACScan cytometer as described above. Thymocytes cultured in medium alone served as controls to measure spontaneous rate of cell death. Viability of thymocytes is expressed as percentage of viable thymocytes treated with apoptotic stimuli over viability of untreated thymocytes. The results are representative of four independent experiments, each using three pairs of mice. All experiments were performed in duplicate.
In Vivo Thymocyte Death Assay.
Twelve-week-old wild-type and junAA mice were injected with either PBS or various doses of purified anti-CD3 antibody (PharMingen) intraperitoneally. After 45 h, mice were killed and thymi were removed. Single cell suspensions were prepared and subjected to flow cytometric analysis after staining with appropriate antibodies. The results are representative of two independent experiments, each using five pairs of mice for each dose of CD3 antibody.
T Cell Differentiation.
Total splenocytes or purified CD4+ T cells were differentiated in vitro into effector T cells by activation with Con A in combination with IL-12 and anti-IL-4 antibody (Th1 differentiation) or IL-4 and anti-IL-12 antibody (Th2 differentiation) as described (24).
Proliferation Assays and ELISA.
T cells from spleen of wild-type and junAA mice were purified by FACS sorting the CD4+ and CD8+ populations. Purified cells were plated in 96-well plates precoated with anti-CD3ɛ for 2 h at 37°C in the absence or presence of soluble anti-CD28. After 60 h, cultures were pulsed for 10–12 h with 1 μCi of [3H]thymidine/well, and cells subsequently were harvested and analyzed by standard procedures. T lymphocytes stimulated with anti-CD3ɛ for 24 h were harvested, and the supernatants were used for measurement of IL-2 levels by ELISA (R & D systems). B cell proliferation was analyzed by stimulation of B220+ spleen cells with IL-4 (50 units/ml), soluble anti-CD40 (1 μg/ml), or lipopolysaccharide (10 μg/ml). The magnitude of stimulated [3H]thymidine incorporation was used as an indicator of cell proliferation. The results are representative of three independent experiments, each using three pairs of mice. All experiments were performed in triplicate.
Western Blot Analysis.
Western blot analysis was performed with 50 μg of nuclear cell extracts from primary splenocytes as described (4) and probed with anti-phospho-c-Jun (serine 63) (from M. Yaniv, Pasteur Institute, Paris), anti-c-Jun (Transduction Laboratories, Lexington, KY), anti-JNK2 (SC-7345, Santa Cruz Biotechnology), and anti-NF-ATc1 antibodies. Proteins were visualized by ECL (Amersham Pharmacia).
Cell Culture and Transfections.
Jurkat TAg (simian virus 40 T antigen) cells (30) were grown in RPMI 1640 supplemented with 10% (vol/vol) FCS, 20 mM L-glutamine, 100 units/ml penicillin G, and 100 mg/ml streptomycin in a 5% CO2, 95% air-humidified atmosphere. For transfections, 107 Jurkat-TAg cells were electroporated in 0.3 ml of complete medium (Bio-Rad gene pulser; 960 mF, 250 V, 0.4-cm gap width) with 6 μg of hemagglutinin-tagged JNK-2 mammalian expression construct and 2 μg of NF–AT-luciferase reporter plasmid (31).
Reporter Gene Assays.
Cells were harvested 24 h after transfection and aliquoted in duplicate into 24-well flat-bottom plates and stimulated with ionomycin (1 μM) and phorbol 12-myristate 13-acetate (PMA, 25 ng/ml). Luciferase activity was measured 18 h after stimulation. To monitor protein expression, cells were lysed 36 h after transfection in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS), and Western analysis was performed (32).
Nuclear Extracts and Electrophoretic Mobility Shift Assay.
Nuclear extracts were prepared from total lymph node cells as described (22). The double-stranded oligonucleotide used in the mobility shift assay to detect NF-AT DNA binding was from the distal NF-AT binding site from the human IL-2 promoter (5′-gatcggaggaaaaactgtttcatacagaaggcgt-3′), and the AP-1 oligonucleotide sequence was 5′-ggggtgactcagggg-3′.
Results
The Hematopoietic System Is Not Altered in the Absence of JNP.
To examine if JNP has a role in the development of the hematopoietic system, we first determined the distribution of the different thymic subpopulations. No differences in the percentages and ratios of immature double negative CD4−CD8−, double positive CD4+CD8+, or mature single positive CD4+ and CD8+ cells between junAA and wild-type mice were observed. In addition, the expression of other differentiation markers (TCR, CD25, HSA) that characterize the maturation stages of each population was normal in the absence of JNP (data not shown). Normal staining patterns also were found for c-kit, B220, and Mac1/GR1 in the bone marrow, and the ratio of CD4+ and CD8+ mature T cells and the percentage of B220+ B cells in the spleen of junAA mice were comparable to wild-type mice. The analysis of whole blood from junAA mice by clinical chemistry revealed no obvious abnormalities in the numbers of platelets, white blood cells and red blood cells, hemoglobin content, or hematocrit (data not shown).
junAA Thymocytes Are Protected from Tumor Necrosis Factor α (TNFα)- and Anti-CD3-Induced Cell Death.
To investigate the role of c-Jun in JNK-mediated thymocyte apoptosis, we compared the susceptibility of wild-type and junAA thymocytes to cell death induced by incubation with anti-CD3 antibody and other stimuli. Although the rate of spontaneous apoptosis was similar between wild-type and junAA thymocytes in vitro, c-Jun N-terminal phosphorylation was required for maximal TCR-induced apoptosis. Thymocytes lacking JNP were consistently more resistant than their wild-type counterparts to incubation with low doses (1 and 10 μg/ml) of anti-CD3 (100% vs. 96.1% and 93.5% vs. 78.1% viability at 24 h) (Fig. 1A). At a higher anti-CD3 dose (100 μg/ml), 83.6% of junAA thymocytes were viable at 24 h compared with 72.9% of wild-type thymocytes (Fig. 1A). After 48 h, junAA thymocytes were still more resistant than wild-type thymocyte CD3 (45.8% vs. 25.9%) (Fig. 1B). This phenotype is similar to, but less pronounced than, the apoptosis defect observed in jnk2−/− thymocytes, suggesting that c-Jun is a downstream target of the JNK signaling cascade during TCR-mediated cell death in vitro.
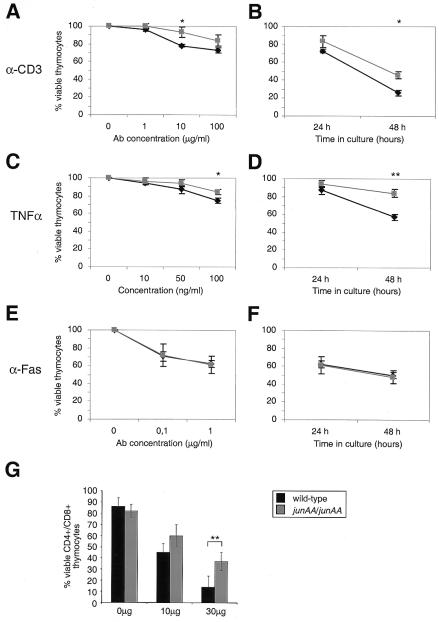
Figure 1.
Resistance of junAA-deficient thymocytes to death caused by apoptotic stimuli in vitro and in vivo. (A and B) Thymocytes from wild-type (black symbols) and junAA (gray symbols) mice were cultured on 24-well plates with various concentrations of immobilized anti-CD3ɛ antibody for 24 h (A) or for the indicated periods (B) in the presence of 100 μg/ml anti-CD3ɛ antibody, and their viability was determined. (C and D) Cell death was induced by addition of various concentrations of TNFα to the medium, and cell viability was determined after 24 h (C) or after 24 and 48 h postaddition of 50 ng/ml TNFα (D). (E and F) Thymocyte viability was determined after treatment with various concentrations of anti-Fas antibody for 24 h (E) or with 1 μg/ml anti-Fas antibody after 24 and 48 h (F). One representative result of four independent experiments, each using three pairs of mice, is shown. (G) Death of CD4+CD8+ double positive thymocytes was induced by i.p. injection of the indicated doses of anti-CD3ɛ antibody in wild-type (black symbols) and junAA (white symbols) mice, and the number of CD4+CD8+ double positive thymocytes was determined after 45 h. Statistical significance was analyzed by Student's t test. *, P < 0.05; **, P < 0.01.
As we have observed that JNK signaling was essential for efficient thymocyte apoptosis in a stimulus-dependent manner (4), we tested survival of thymocytes in response to various apoptotic stimuli. The proinflammatory cytokine TNFα is a potent inducer of both JNK and AP-1 activity. In transformed cell lines, JNK activation was shown to occur through a noncytotoxic pathway (33–35). Interestingly, JNP is required in thymocytes for TNFα-induced apoptosis as increasing doses of TNFα (50 and 100 ng/ml) failed to efficiently kill junAA cells (94.4% vs. 86.9% and 84.4% vs. 74.8% viability at 24 h) (Fig. 1C). The resistance of junAA thymocytes to TNFα-induced apoptosis became most apparent after 48 h of incubation (Fig. 1D) when 43.1% of wild-type cells had died, whereas only 16.1% of junAA thymocytes were dead, suggesting that JNK-mediated phosphorylation of c-Jun plays a proapoptotic role in TNFα-induced cell death in thymocytes. In contrast, junAA thymocytes were not significantly more resistant than wild-type thymocytes to apoptosis induced by incubation with agonistic anti-Fas antibody, the synthetic glucocorticoid dexamethasone, and UV-C irradiation (Fig. 1 E and F; data not shown). To confirm a relevant function of JNP in CD3-induced cell death in vivo, we challenged junAA and control mice with anti-CD3 antibody injections. At a low dose of anti-CD3 (10 μg), no statistically significant differences between wild-type and junAA mutant mice could be observed (43% vs. 58% CD4+CD8+ double positive thymocytes) (Fig. 1G). However, the junAA mutation resulted in markedly decreased thymocyte death in response to a higher dose of anti-CD3 (30 μg), as after 45 h only 14% of wild-type CD4+CD8+ double positive thymocytes were present compared with 37% in mice lacking JNP (Fig. 1G). Mice lacking jnk2 are also resistant to CD3-induced thymocyte apoptosis in vivo (4), suggesting that c-Jun may be a molecular target of JNK2 in this biological process. Therefore, JNP appears to contribute to anti-CD3- and TNFα-induced thymocyte death.
T Cell Differentiation Without JNP.
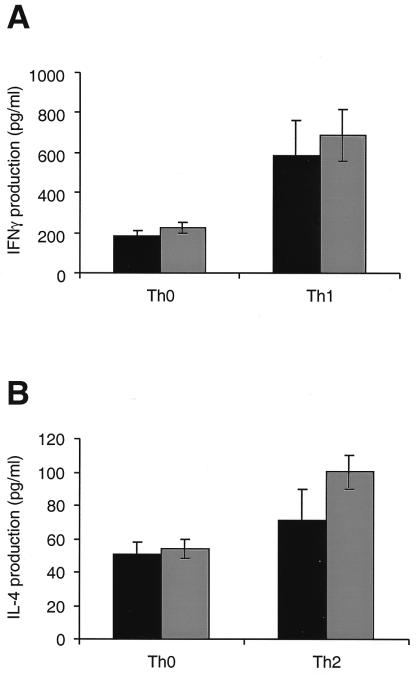
Naive CD4+ Th cells can differentiate into two subsets of effector T cells that have been defined on the basis of their distinct cytokine secretion patterns and their immunomodulatory effects. Th1 cells produce IFNγ, which is required for cell-mediated inflammatory reactions; Th2 cells secrete IL-4, IL-5, IL-6, IL-10, and IL-13, which mediate B cell activation and differentiation (36). Moreover, AP-1 activity is strongly induced during Th2 differentiation, and c-Jun is present in Th2 cells (37). To test whether c-Jun is required for JNK-mediated T cell differentiation, naive junAA and wild-type control splenocytes (or CD4+ purified T cells, data not shown) were differentiated in vitro into effector cells by activation with Con A in combination with IL-12 and anti-IL-4 antibody (Th1 differentiation) or IL-4 and anti-IL-12 antibody (Th2 differentiation). After 4 days of differentiation, the cells were washed and counted, and equal numbers of cells were restimulated with Con A in the absence of exogenous cytokines. Supernatants were harvested after 24 h and analyzed for cytokine secretion. No significant difference in the production of IFNγ in effector Th1 cells was observed (585 pg/ml in wild-type and 688 pg/ml in junAA cells) (Fig. 2A). Similarly, IL-4 production was normal in effector Th2 cells from junAA mice (100 pg/ml) compared with wild-type littermate controls (71 pg/ml) (Fig. 2B). Similar results were obtained after restimulation with anti-CD3 antibody (data not shown). The reported increase in Th2 differentiation in jnk1−/− mice is much more dramatic than in JNP-deficient mice, suggesting that c-Jun is only a minor, if at all, target of JNK signaling in this biological process. In the absence of a polarizing cytokine, CD4+ T cells develop into a Th0-like population that produces low levels of both IL-4 and IFNγ. To determine the Th phenotype of CD4+ T cells in junAA mice under neutral conditions, splenocytes were differentiated in the absence of added cytokines. After restimulation with Con A (or anti-CD3 antibody, data not shown), the production of both IL-4 and IFNγ was comparable between wild-type and junAA Th0 cells (Fig. 2). These data suggest that c-Jun is not an important substrate of neither JNK1- nor JNK2-mediated T cell differentiation.
Figure 2.
Analysis of cytokine production in effector Th1 and Th2 cells from wild-type and junAA mice. (A) IFNγ production by Th0 and Th1 cells differentiated from wild-type (filled bar) and junAA (empty bar) T cells was determined by ELISA. (B) IL-4 production by Th0 and Th2 cells differentiated from wild-type (filled bar) and junAA (empty bar) T cells was determined by ELISA. One representative result of three independent experiments, each using three pairs of mice, is shown.
JNP Is Not Required for B and T Cell Proliferation.
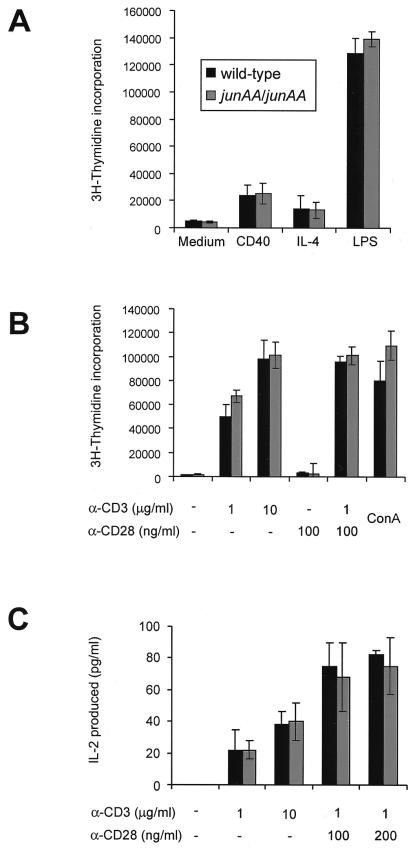
To investigate the requirement of JNP for lymphocyte activation, the proliferation of JNP-deficient lymphocytes in response to treatment with various stimuli was measured. Proliferation of splenic B cells in response to lipopolysaccharide, CD40 antibody, and IL-4 stimulation was not affected by the absence of JNP (Fig. 3A), similar to that of B cells lacking the JNKs (4, 53). Splenic T cells were sorted for CD4+ and CD8+, and T cell activation was monitored by cell proliferation and IL-2 production. TCR ligation by anti-CD3 in combination with anti-CD28 antibody and Con A treatment resulted in comparable rates of T cell proliferation and similar levels of IL-2 production in both wild-type and junAA T cells (Fig. 3 B and C). There was no significant difference in proliferation rates and IL-2 production at limiting doses of anti-CD3 stimulation as was observed with jnk2−/− T cells. Similar results were obtained with total splenocytes (data not shown). These findings indicate that JNP is not essential for the proliferation of both B and T cells.
Figure 3.
Normal proliferation of junAA B and T cells. (A) Purified B cells from spleens of 8-to 10-wk-old mice were cultured in media or in the presence of plate-bound anti-CD40 antibody, IL-4, or lipopolysaccharide. (B) Purified T cells from spleens of 8-to 10-wk-old mice were cultured in media or in the presence of various concentrations of plate-bound anti-CD3ɛ antibody and soluble anti-CD28 antibody, or in the presence of Con A. (C) IL-2 production was measured by ELISA after 24-h stimulation of purified T cells as described above. One representative result of four independent experiments, each using three pairs of mice, is shown.
JNK2 Regulates NF-AT Activity in T Cells.
Phosphorylation regulates both the stability of c-Jun and autoregulation of c-jun transcription (38, 39). The absence of a proliferation defect in JNP-deficient T cells could be explained by altered stability of the JunAA protein and subsequent higher JunAA protein levels, which might functionally compensate the absence of JNP. Examination of JNP in vivo revealed that c-Jun was N-terminally phosphorylated upon T cell activation in wild-type T cells but not in activated junAA T cells (Fig. 4A). Accordingly, the levels of c-Jun were increased in wild-type T cells in response to TCR ligation (Fig. 4A). The levels of JunAA protein in JNP-deficient T cells also increased, albeit the protein levels appeared to be slightly reduced compared with wild type (Fig. 4A). Therefore, although JNP and the JNP-mediated increase in c-Jun protein levels were affected in junAA T cells, these cells do not exhibit a proliferation defect. This finding argues against JNP or JNP-mediated increase in c-Jun as critical events regulating T cell activation.
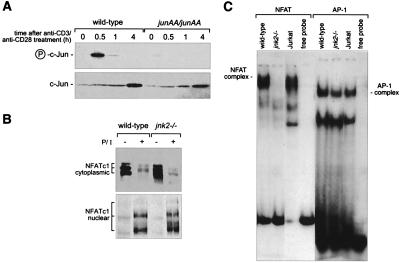
Figure 4.
Reduced NF-AT DNA-binding activity jnk2-deficient T cells. (A) JNP and c-Jun protein levels were measured by using nuclear extracts from untreated or anti-CD3/anti-CD28 stimulated splenocytes. (B) Cytoplasmic and nuclear extracts from untreated (−) and PMA/ionomycin-stimulated (for 3 h, indicated by +) wild-type and jnk2−/− lymph node T cells were prepared, and NF-ATc1 protein was detected by Western analysis. (C) Nuclear extracts from PMA/ionomycin-stimulated (for 3 h) wild-type and jnk2−/− lymph node T cells were prepared 3 h after stimulation, and NF-AT and AP-1 DNA binding was detected by electrophoretic mobility shift assay.
In an attempt to identify factors, which might be responsible for T cell signal transduction via the JNK pathway, we examined whether NF-AT DNA-binding activity is altered in the absence of JNK2. The NF-AT transcription complex was absent in nonstimulated nuclear extracts of wild-type and jnk2−/− T cells but appeared upon T cell stimulation. NF-ATc1 was expressed at wild-type levels and efficiently translocated from the cytoplasm into the nucleus in both jnk2−/− and wild-type T cells (Fig. 4B). NF-ATc2 (also called NFATp or NF-AT1) also was expressed at equivalent levels and translocated to the nucleus in both wild-type and jnk2−/− T cells (data not shown). However, NF-AT DNA binding using the distal IL-2 site (14) could not be detected in jnk2−/− T cells stimulated with either PMA and ionomycin or anti-CD3 (Fig. 4C). In contrast, AP-1 DNA-binding activity was not reduced in the absence of jnk2 (Fig. 4C).
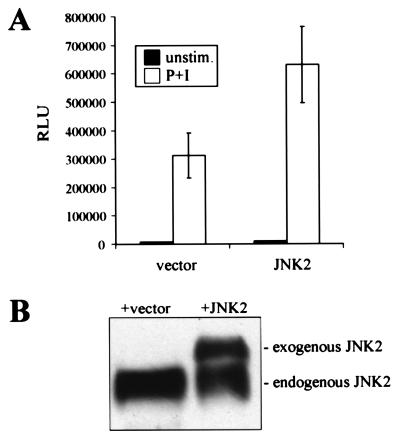
To further investigate the regulation of NF-AT by JNK2, a JNK2 expression construct was transfected into Jurkat T cells, and the levels of NF-AT transcriptional activity were measured by using a luciferase reporter gene. PMA/ionomycin treatment of vector-transfected cells resulted in elevated NF-AT reporter gene activity (Fig. 5A). PMA/ionomycin-stimulated Jurkat cells overexpressing JNK2 showed a 2-fold increase in NF-AT activity compared with vector-transfected cells (Fig. 5A). Ectopic expression of JNK2 in Jurkat cells led to an approximate doubling of total cellular JNK2 protein (Fig. 5B). The increase in JNK2 protein levels by transfection correlates well with the elevation of NF-AT reporter gene activity (Fig. 5), strongly suggesting that the NF-AT transcription factor complex is a physiological target of JNK2 and that JNK2 is a positive regulator of NF-AT complex formation and transcriptional activity.
Figure 5.
Overexpression of JNK2 results in elevated NF-AT transcriptional activity. (A) NF-AT activity assay of Jurkat T cells transfected with control vector and a JNK expression construct together with NF-AT luciferase reporter construct. Protein extracts were prepared 24 h after transfection, and luciferase activity was measured in duplicate. (B) JNK2 protein levels in Jurkat T cells transfected with control vector and a JNK expression construct were determined by Western analysis.
Discussion
Protein phosphorylation is an important posttranslational mechanism to regulate the subcellular localization and the activity of transcription factors (1, 12). Although the signaling events leading to the activation of the various groups of mitogen-activated protein kinases are relatively well understood, little is known about the substrates used by these kinases in different biological processes (1). The JNK family of mitogen-activated protein kinases was identified as the c-Jun phosphorylating kinases, and only recent results indicate that several other proteins can be phosphorylated by the JNKs in vitro (6–10). The JNK signaling pathway regulates several aspects of neural and T cell development (2, 4, 5, 40). Embryos lacking both jnk1 and jnk2 are characterized by a defect in neural closure and exhibit exencephaly due to an apoptosis defect (40, 41). c-Jun is not a substrate of the JNKs in this developmental process because neurogenesis proceeds normally in junAA mice (29). In contrast, c-Jun is the essential substrate downstream of JNK3 during kainate-induced neuronal apoptosis (29, 42). Because there are c-Jun-dependent and -independent biological functions of JNK signaling, other molecular effectors in addition to c-Jun may be used in neuronal cells.
In this study, mice lacking JNP and mice lacking jnk2 were used to investigate the downstream targets of the JNK pathway in lymphoid cells. Our data suggest that c-Jun is likely to be a target of the JNKs in thymocyte apoptosis. jnk2−/− and junAA mice are both more resistant than wild-type to CD3-induced thymocyte death in vivo and in vitro, providing evidence that JNK death signaling is partially mediated by c-Jun N-terminal phosphorylation. Surprisingly, junAA thymocytes are less susceptible to TNFα-induced death but are normally killed by the FAS pathway. The death machinery used by both pathways is believed to be similar, but importantly, TNF signaling, in contrast to FAS signaling, has both cytoprotective and cytotoxic effector arms (43). As c-jun can function both as an activator and as a repressor of transcription (44), it is possible that JNP is required for the efficient expression of a proapoptotic target gene. Alternatively, JNP may repress a target gene involved in cell survival, which would be consistent with previously published work showing JNK activation by TNF receptors through a noncytotoxic pathway (33–35).
The role of JNK2 in lymphocyte activation and differentiation is controversial. Other investigators have observed normal T cell proliferation in response to PMA/ionomycin stimulation but defective differentiation in mice lacking jnk2 (2). In contrast, we see no bias in differentiation but a defect in activation of IL-2 and proliferation in JNK2-deficient mice (4). These differences could be due to strain differences, which are known to profoundly effect T cell differentiation responses. In addition, whereas other investigators have seen delayed activation of JNK after T cell activation (45), we find that JNK activity in vivo is seen within 30 min of T cell activation. We have assayed JNK activity in vivo by using the difference between the phosphorylation of endogenous wild-type and endogenous c-Jun containing mutations of the phosphorylation sites (Fig. 4A). The other studies used an in vitro kinase assay that does not identify the actual residues phosphorylated in c-Jun. Hence, the apparent delay in JNK activation might reflect the activity of other kinases.
Based on our findings, it is unexpected that JNP is not required for T cell proliferation mediated by JNK2, because lack of JNP results in a proliferation defect in other cell types such as mouse embryonic fibroblasts (29). However, the NF-AT transcription factor could be responsible for the T cell proliferation defect observed in jnk2−/− mice (4). NF-ATc1−/− and jnk2−/− T cells both showed reduced proliferation, indicating that the NF-AT transcription factor may be a physiological effector of JNK in this biological process (4, 23). The NF-AT transcription factor complex is composed of a nuclear component (NF-ATn) and a cytoplasmic component (NF-ATc), which translocates to the nucleus in response to various signals including TCR ligation (12, 22). Transcription factors like AP-1, c-Maf, and GATA4 as well as other unidentified components can function as NF-ATn and cooperatively bind to DNA with members of the NF-ATc family (12, 46, 47). The DNA binding activity of NF-AT is greatly reduced in activated jnk2−/− T cells, indicating that JNK2 is required for the biological function of the NF-ATc and/or the NF-ATn component of the transcription factor complex. The absence of NF-AT DNA binding could be explained by a subtle defect in T cell development in jnk2−/− mice. However, ectopic overexpression of JNK2 in Jurkat T cells resulted in elevated NF-AT-dependent transcription, suggesting a direct effect of JNK signaling on NF-AT. JNK2 does not appear to affect the protein levels and the stability of nuclear NF-ATc1 and NF-ATc2. Similarly, NF-ATc1 and NF-ATc2 translocation to the nucleus is normal in jnk2−/− peripheral T cells, excluding JNK2 as a regulator of this process. In agreement with our findings, Werlen et al. (48) have found using Jurkat T cells that stimulation of JNK activity resulted in elevated NF-AT-dependent transcription. However, our data are in contrast to other published observations, which suggested an inhibition of NF-AT activity by JNK signaling. JNK-mediated phosphorylation of NF-AT was reported to negatively regulate nuclear translocation in transfection studies in cell lines (9, 49). These differences could be due to the different cellular and experimental systems used, i.e., transient transfection in cell lines vs. primary knockout lymphocytes, because in different cellular systems distinct signaling pathways may regulate different NF-ATc transcription factors. Dong et al. (2, 50) found hyperproliferation and increased NF-AT activity in jnk1 mutant T cells, suggesting that JNK1 negatively regulates NF-AT. However, both of these phenotypes are not present in our strain of jnk1-deficient mice (53). It can only be speculated that differences in the design of the knockout strategy or genetic background of the mutant mouse strains are responsible for the observed discrepancies. As we have no evidence for a direct effect of JNK signaling on NF-ATc transcription factors, JNK2 may regulate the DNA-binding affinity of the NF-AT complex by controlling the activity of NF-ATn. Our data are consistent with a large body of experimental data that indicate that in primary lymphocytes, neurons, and other cell types and cell lines, signals that activate JNK synergize with Ca2+ signals to activate NF–AT-dependent transcription (15, 16, 22, 32, 51, 52).
Taken together, these data suggest that JNK signaling may use distinct substrates for different aspects of T cell biology. C-Jun is one target of JNK2 during thymocyte apoptosis, whereas the DNA binding activity of the NF-AT transcription factor complex is regulated by JNK2 during T cell proliferation. One of the major challenges will be to understand at the molecular level how distinct JNK isoenzymes, which can play nonoverlapping and even opposite roles in response to the same stimulus, are capable of recruiting various sets of target proteins to control a multitude of biological effects.
Acknowledgments
We are grateful to Svetlana Pekez for maintaining our mouse colony. We thank Karin Paiha and Peter Steinlein for FACS analysis; Moshe Yaniv for antibodies; Michael Karin for hemagglutinin-tagged JNK-2 construct; Manuela Baccarini, Thomas Decker, Peter Steinlein, and Georg Stingl for critical reading of the manuscript; and Hannes Tkadletz for help with preparing the illustrations. This work was supported in part by the Austrian Federal Ministry of Science, Transport, and the Arts and the Austrian Industrial Research Promotion Fund.
Abbreviations
- JNK
Jun N-terminal kinase
- TCR
T cell antigen receptor
- TNF
tumor necrosis factor
- PMA
phorbol 12-myristate 13-acetate
- NF-AT
nuclear factor of activated T cell
- NF-ATn
NF-AT with a nuclear component
- NF-ATc
NF-AT with a cytoplasmic component
- JNP
c-Jun N-terminal phosphorylation
- Th
T helper
References
- 1.Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 2.Dong C, Yang D D, Wysk M, Whitmarsh A J, Davis R J, Flavell R A. Science. 1998;282:2092–2095. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 3.Rincon M, Whitmarsh A, Yang D D, Weiss L, Derijard B, Jayaraj P, Davis R J, Flavell R A. J Exp Med. 1998;188:1817–1830. doi: 10.1084/jem.188.10.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabapathy K, Hu Y, Kallunki T, Schreiber M, David J P, Jochum W, Wagner E F, Karin M. Curr Biol. 1999;9:116–125. doi: 10.1016/s0960-9822(99)80065-7. [DOI] [PubMed] [Google Scholar]
- 5.Yang D D, Conze D, Whitmarsh A J, Barrett T, Davis R J, Rincon M, Flavell R A. Immunity. 1998;9:575–585. doi: 10.1016/s1074-7613(00)80640-8. [DOI] [PubMed] [Google Scholar]
- 6.Kallunki T, Deng T, Hibi M, Karin M. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 7.Gille H, Strahl T, Shaw P E. Curr Biol. 1995;5:1191–1200. doi: 10.1016/s0960-9822(95)00235-1. [DOI] [PubMed] [Google Scholar]
- 8.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Dérijard B, Davis R J. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 9.Chow C W, Rincon M, Cavanagh J, Dickens M, Davis R J. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 10.Cavigelli M, Dolfi F, Claret F X, Karin M. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angel P, Karin M. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 12.Crabtree G R. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 13.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 14.Shaw J P, Utz P J, Durand D B, Toole J J, Emmel E A, Crabtree G R. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 15.Jain J, McCaffrey P G, Valge-Archer V E, Rao A. Nature (London) 1992;356:801–804. doi: 10.1038/356801a0. [DOI] [PubMed] [Google Scholar]
- 16.Crabtree G R. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Stewart V, Spyrou G, Hilberg F, Wagner E F, Alt F W. Immunity. 1994;1:65–72. doi: 10.1016/1074-7613(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 18.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 19.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 20.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 21.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 22.Flanagan W M, Corthesy B, Bram R J, Crabtree G R. Nature (London) 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida H, Nishina H, Takimoto H, Marengere L E, Wakeham A C, Bouchard D, Kong Y Y, Ohteki T, Shahinian A, Bachmann M, et al. Immunity. 1998;8:115–124. doi: 10.1016/s1074-7613(00)80464-1. [DOI] [PubMed] [Google Scholar]
- 24.Hodge M R, Ranger A M, Charles de la Brousse F, Hoey T, Grusby M J, Glimcher L H. Immunity. 1996;4:397–405. doi: 10.1016/s1074-7613(00)80253-8. [DOI] [PubMed] [Google Scholar]
- 25.Oukka M, Ho I C, de la Brousse F C, Hoey T, Grusby M J, Glimcher L H. Immunity. 1998;9:295–304. doi: 10.1016/s1074-7613(00)80612-3. [DOI] [PubMed] [Google Scholar]
- 26.Chow C W, Rincon M, Davis R J. Mol Cell Biol. 1999;19:2300–2307. doi: 10.1128/mcb.19.3.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilberg F, Aguzzi A, Howells N, Wagner E F. Nature (London) 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 28.Johnson R S, van Lingen B, Papaioannou V E, Spiegelmann B M. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 29.Behrens A, Sibilia M, Wagner E F. Nat Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 30.Northrop J P, Ullman K S, Crabtree G R. J Biol Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 31.Bram R J, Hung D T, Martin P K, Schreiber S L, Crabtree G R. Mol Cell Biol. 1993;13:4760–4769. doi: 10.1128/mcb.13.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graef I A, Mermelstein P G, Stankunas K, Neilson J R, Deisseroth K, Tsien R W, Crabtree G R. Nature (London) 1999;401:703–708. doi: 10.1038/44378. [DOI] [PubMed] [Google Scholar]
- 33.Reinhard C, Shamoon B, Shyamala V, Williams L T. EMBO J. 1997;16:1080–1092. doi: 10.1093/emboj/16.5.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natoli G, Costanzo A, Ianni A, Templeton D J, Woodgett J R, Balsano C, Levrero M. Science. 1997;275:200–203. doi: 10.1126/science.275.5297.200. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z G, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 36.Flavell R A, Rincon M, Zheng W P, Li B, Enslen H, Raingeaud J, Davis R J. Eur Cytokine Network. 1998;9:26–32. [PubMed] [Google Scholar]
- 37.Rincon M, Derijard B, Chow C W, Davis R J, Flavell R A. Genes Funct. 1997;1:51–68. doi: 10.1046/j.1365-4624.1997.00007.x. [DOI] [PubMed] [Google Scholar]
- 38.Angel P, Hattori K, Smeal T, Karin M. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 39.Musti A M, Treier M, Bohmann D. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 40.Sabapathy K, Jochum W, Hochedlinger K, Chang L, Karin M, Wagner E F. Mech Dev. 1999;89:115–124. doi: 10.1016/s0925-4773(99)00213-0. [DOI] [PubMed] [Google Scholar]
- 41.Kuan C Y, Yang D D, Samanta Roy D R, Davis R J, Rakic P, Flavell R A. Neuron. 1999;22:667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 42.Yang D D, Kuan C Y, Whitmarsh A J, Rincón M, Zheng T S, Davis R J, Rakic P, Flavell R A. Nature (London) 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- 43.Wallach D, Varfolomeev E E, Malinin N L, Goltsev Y V, Kovalenko A V, Boldin M P. Annu Rev Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner E F. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss L, Whitmarsh A J, Yang D D, Rincon M, Davis R J, Flavell R A. J Exp Med. 2000;191:139–146. doi: 10.1084/jem.191.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe S A, Zhou P, Dotsch V, Chen L, You A, Ho S N, Crabtree G R, Wagner G, Verdine G L. Nature (London) 1997;385:172–176. doi: 10.1038/385172a0. [DOI] [PubMed] [Google Scholar]
- 47.Zhou P, Sun L J, Dotsch V, Wagner G, Verdine G L. Cell. 1998;92:687–696. doi: 10.1016/s0092-8674(00)81136-8. [DOI] [PubMed] [Google Scholar]
- 48.Werlen G, Jacinto E, Xia Y, Karin M. EMBO J. 1998;17:3101–3111. doi: 10.1093/emboj/17.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chow C W, Dong C, Flavell R A, Davis R J. Mol Cell Biol. 2000;20:5227–5234. doi: 10.1128/mcb.20.14.5227-5234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong C, Yang D D, Tournier C, Whitmarsh A J, Xu J, Davis R J, Flavell R A. Nature (London) 2000;405:91–94. doi: 10.1038/35011091. [DOI] [PubMed] [Google Scholar]
- 51.Woodrow M, Clipstone N A, Cantrell D. J Exp Med. 1993;178:1517–1522. doi: 10.1084/jem.178.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holsinger L J, Graef I A, Swat W, Chi T, Bautista D M, Davidson L, Lewis R S, Alt F W, Crabtree G R. Curr Biol. 1998;8:563–572. doi: 10.1016/s0960-9822(98)70225-8. [DOI] [PubMed] [Google Scholar]
- 53.Sabapathy K, Kallurki T, David J-P, Graef I, Karin M, Wagner E F. J Exp Med. 2001;19:1–13. doi: 10.1084/jem.193.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]