Abstract
Communication between organelles is an important feature of all eukaryotic cells. To uncover components involved in mitochondria/endoplasmic reticulum (ER) junctions, we screened for mutants that could be complemented by a synthetic protein designed to artificially tether the two organelles. We identified the Mmm1/Mdm10/Mdm12/Mdm34 complex as a molecular tether between ER and mitochondria. The tethering complex was composed of proteins resident of both ER and mitochondria. With the use of genome-wide mapping of genetic interactions, we showed that the components of the tethering complex were functionally connected to phospholipid biosynthesis and calcium-signaling genes. In mutant cells, phospholipid biosynthesis was impaired. The tethering complex localized to discrete foci, suggesting that discrete sites of close apposition between ER and mitochondria facilitate interorganelle calcium and phospholipid exchange.
Eukaryotic cells evolved segregation of functions into separate organelles. Compartmentalization increases the efficiency of biochemical reactions by creating tailored chemical microenvironments, but also creates a need for communication and routes of metabolite exchange. Membrane lipids, for example, are primarily synthesized in the endoplasmic reticulum (ER) and distributed to other organelles. Many organelles exchange phospholipids with the ER via vesicular transport. In contrast, mitochondria are not connected to vesicular trafficking pathways, and many lipids of the inner and outer mitochondrial membranes (IMM and OMM) cannot be synthesized within mitochondria but are imported by unclear mechanisms. Phospholipids may transfer from the ER to the OMM at spatially restricted sites, which are frequently observed by electron microscopy and have been enriched by cell fractionation (1–3).
Other work has implicated ER-mitochondrial contact sites in Ca++ transport between the ER and mitochondria (4–6), suggesting a mechanism that may exploit the formation of an encapsulated space at the contact sites, akin to that formed at neuronal or immunological synapses. Such a connection between the ER and the mitochondria might buffer and control cytosolic and mitochondrial Ca++ concentrations (7). Several proteins have been implicated to participate in ER-mitochondria contacts, including the ER resident Ca++ channel IP3 receptor, the mitochondrial voltage-dependent anion channel, the chaperones grp75 and sigma-1R, the sorting protein PACS-2, and the mitofusin Mfn2 (8–11).
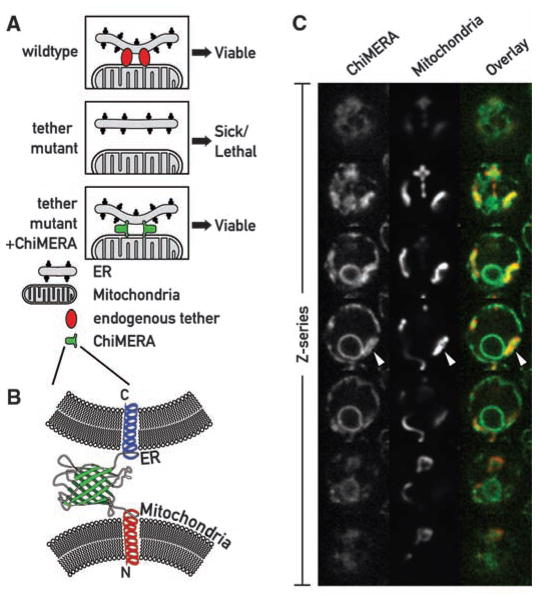
To explore a role for ER-mitochondrial junctions, we sought mutants in the yeast Saccharo-myces cerevisiae, in which tethering between the two organelles was impaired. We reasoned that, if such contacts are important, defects in proteins that establish these interactions would be detrimental, yet perhaps could be suppressed by artificially tethering ER and mitochondria (Fig. 1A). We designed a synthetic ER-mitochondria tether (“ChiMERA” for construct helping in mitochondria-ER association) (Fig. 1B) consisting of an N-terminal mitochondrial signal sequence and transmembrane domain derived from Tom70, a central module composed of green fluorescent protein (GFP), and a C-terminal ER tail-anchor derived from Ubc6. The design was based on a similar chimeric protein that strengthened mitochondria-ER interactions (12). The GFP moiety allowed us to visualize its tethering activity directly (Fig. 1C and fig. S1A). ChiMERA localized mainly to ER membranes and stained both peripheral and perinuclear ER. In addition, ChiMERA localized to patches of contact with mitochondria (Fig. 1C). Mitochondria in untransformed wild-type (WT) cells showed the characteristic morphology of a uniformly tubular network (Fig. 2B and fig. S1B) and adopted a pronounced patchy structure at the artificial contact sites in cells expressing ChiMERA (Fig. 1C and fig. S1A; see also Fig. 2B). Thus, ChiMERA tethers mitochondria and ER as intended. The dominant ER staining probably reflects a partial dominance of the ER targeting sequence over the mitochondrial one. We also generated a second version of the artificial tether in which the yeast Tom70 presequence and transmembrane domain was replaced by the mouse AKAP1 presequence (12). This construct also localized predominantly to the ER but accumulated in only one to three discrete foci per cell (fig. S2). These foci colocalized with both ER (fig. S2A) and mitochondria (fig. S2B). This construct, however, did not create distortions in mitochondrial shape. It is likely that this synthetic protein does not exert a substantial tethering force and instead labels preexisting ER-mitochondrial interfaces. We refer to this construct as ChiMERA-ra (for ChiMERA with reduced affinity).
Fig. 1.
A synthetic biology screen to uncover mutants of the ER-mitochondria connection. (A) Rationale of the screen. (Top) In WT cells a yet unknown endogenous complex tethers the ER to the mitochondria. (Middle) Mutations causing the loss of the endogenous complex are detrimental and cause slow growth or cell death. (Bottom) Artificial ER-mitochondria tethering by ChiMERA can suppress the defects associated with the loss of the endogenous tether. (B) Outline of the ChiMERA. A central GFP molecule (green) is flanked by the mitochondria-directed N-terminal Tom70 presequence and transmembrane sequence (red) and the ER-directed C-terminal Ubc6 tail anchor sequence (blue). (C) Confocal Z-series across a yeast cell expressing the ChiMERA and a mitochondrial marker (mt-dsRed). ChiMERA displays a characteristic ER staining with additional thicker structures (arrowheads), which colocalize with mitochondria and represent sites of artificial tethering.
Fig. 2.
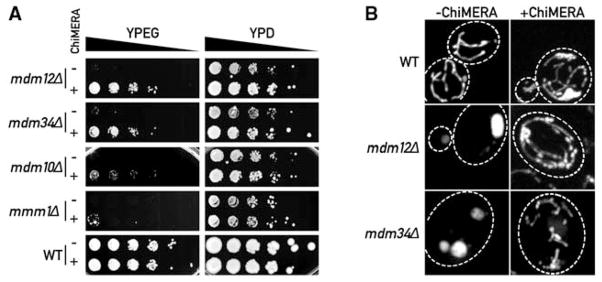
ChiMERA expression suppresses phenotypes associated with deletions of ERMES complex components. (A) 10-fold serial dilutions of a saturated culture were spotted on YP + ethanol/glycerol medium (YPEG) that supports growth by respiration or YP + dextrose (YPD) that allows fermentation. ChiMERA expression rescues growth of mdm12Δ, mdm34Δ, mdm10Δ, and mmm1Δ strains to different extents on respiration medium. (B) The mitochondrial shape defect of mdm12Δ and mdm34Δ strains is partially suppressed by expression of the ChiMERA. Cells bearing a mitochondria-targeted dsRed were imaged with the use of confocal fluorescence microscopy. Z projections across the whole volume of the cells are shown. A broken white line outlines the perimeter of the cell. ChiMERA expression has no effect in WT cells, other than formation of artificial tethering structures. mdm12Δ and mdm34Δ strains display rounded and enlarged mitochondria. The mitochondrial tubular shape is substantially restored upon ChiMERA expression. A quantitation of this effect is shown in fig. S4.
We next used a construct expressing ChiMERA from a centromeric expression plasmid in a classical sectoring screen (13). We screened ~100,000 ethylmethanesulfonate-treated colonies and retrieved two mutants that could not grow in the absence of the plasmid. Genetic complementation (14) indicated that the mutations are two different alleles of MDM12. Accordingly, a strain bearing a complete deletion of MDM12 grew on respiration plates when it expressed the ChiMERA, but the strain failed to grow without it (Fig. 2A). Mdm12 is a peripheral OMM protein (15) (fig. S3). Mdm12 can be isolated from cell extracts as a complex with Mmm1 and Mdm10 and localizes in a few punctate structures along the mitochondrial network, together with a fourth protein, Mdm34 (16, 17) (Fig. 3A). All four proteins are required for respiratory growth and for the maintenance of the proper tubular morphology of mitochondria. We deleted MMM1, MDM10, and MDM34 and assessed the extent to which ChiMERA could rescue their phenotypes. ChiMERA expression partially suppressed growth defects, albeit to highly different extents (Fig. 2A). mdm12Δ, mmm1Δ, mdm10Δ, and mdm34Δ strains displayed enlarged and spherical mitochondria as compared with the tubular WT mitochondria (18). ChiMERA expression also restored the mitochondrial morphology of mdm12Δ and mdm34Δ strains (Fig. 2B and fig. S4), although it failed to do so in mmm1Δ and mdm10Δ strains, consistent with the lesser rescue of growth on respiration medium (Fig. 2A). An Mdm34-mCherry fusion protein colocalized with both the punctate structures labeled by ChiMERA-ra and the broad ER-mitochondria interfaces created by ChiMERA (fig. S5).
Fig. 3.
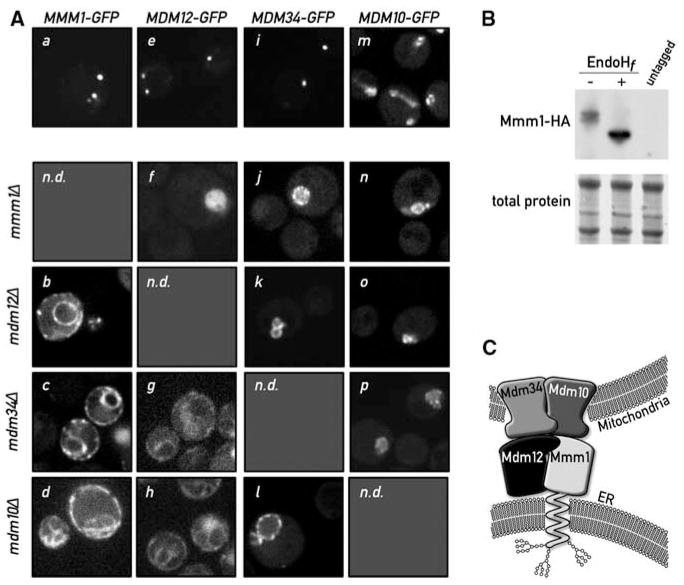
Mmm1 is an integral ER protein. (A) Disruption of a single ERMES component causes disassembly of the complex. Mmm1-GFP is localized in punctate structures in WT cells (a). Upon deletion of MDM12 (b), MDM34 (c), or MDM10 (d), Mmm1-GFP relocalizes to the ER. Mdm12-GFP displays a punctate pattern in WT cells (e). In the absence of Mmm1 (f), Mdm12-GFP displays a uniform mitochondrial localization (note the rounded swollen mitochondrial shape in mmm1Δ strains), but in the absence of MDM34 (g) or MDM10 (h), Mdm12-GFP relocalizes to the ER. Mdm34-GFP is also in punctate structures in WT cells (i). Upon mutation of any other complex member, Mdm34 relocalizes more uniformly to the mitochondrial membrane (j, k, l). Mdm10-GFP localizes to the whole surface of the mitochondria (m) and localizes to the rounded mitochondria after deletion of any other ERMES component (n, o, p). n.d., not determined. (B) Mmm1 is N-glycosylated. Whole-cell extract from a strain bearing a functional HA-tagged MMM1 gene was subjected to SDS–polyacrylamide gel electrophoresis with or without pretreatment with the glycosidase EndoHf. Detection was performed by Western blotting with an anti-HA antibody. The shift in electrophoretic mobility upon glycosidase treatment is indicative that Mmm1 is N-linked glycosylated. (C) Model of ERMES-mediated ER-mitochondria tethering. Mmm1 is an integral ER protein glycosylated on its N-terminal side. Mmm1 interacts with Mdm10, a OMM β-barrel protein. Mdm34 and Mdm12 promote this association, most probably via direct association.
Thus, one core molecular function of the Mmm1/Mdm10/Mdm12/Mdm34 complex is to connect ER and mitochondria, which can be bypassed by expressing a synthetic tether. Henceforth, we refer to the complex as ERMES (ER-mitochondria encounter structure).
ERMES localizes into a few discrete foci (16, 17), and mutations in any single component cause the puncta to disappear (16, 17). We reasoned that ERMES components may be respectively localized to the ER and mitochondria, and they might serve to zipper the organelles together. ERMES disruption would then cause some of the components to partition with ER, whereas some others would partition with mitochondria.
To address this possibility, we replaced the gene encoding each ERMES component with that of a functional GFP fusion protein and imaged both WT and mutant cells (Fig. 3A). In WT cells, ERMES components displayed the expected punctate pattern, with the exception of Mdm10, which displayed a more widespread localization throughout the mitochondrial network. This is consistent with the observation that Mdm10 is not only a component of ERMES but is also a component of the sorting and assembly machinery (SAM) complex in the OMM (19). As expected from its assignment as an OMM protein, Mdm34-GFP relocalized uniformly to mitochondria in the absence of Mdm10, Mdm12, and Mmm1 (Fig. 3A; note the swollen morphology of mitochondria in the mutants). Likewise, Mdm10-GFP remained colocalized with mitochondria in all strains tested. In contrast, and contrary to expectation, Mmm1-GFP relocalized to the ER, showing a characteristic peripheral and perinuclear staining in the absence of Mdm10, Mdm12, or Mdm34. Mdm12-GFP, a membrane peripheral protein (fig. S3), relocalized to the ER in the absence of Mdm10 or Mdm34, and the mitochondria in the absence of Mmm1, suggesting that its localization depends on the availability of interaction partners at either organelle.
Mmm1 harbors a classical helical transmembrane domain, previously proposed to be inserted into either the outer or inner mitochondrial membrane (20), yet our localization data suggested that Mmm1 is instead inserted into the ER. Many integral ER membrane proteins are N-glycosylated within their lumenal domains, and four potential N-glycosylation sites lie in the N terminus of Mmm1 preceding the transmembrane domain. To address whether these sites were glycosylated, we replaced the MMM1 gene with fully functional hemagglutinin (HA)–tagged MMM1 and assessed the electrophoretic mobility of HA-Mmm1 from cell extracts with or without treatment with the glycosidase EndoHf, which removes N-linked glycans. Treatment with EndoHf caused HA-Mmm1 to migrate faster (Fig. 3B and fig. S6A), indicating that Mmm1 is N-glycosylated and therefore is an integral ER membrane protein that was misannotated as a mitochondrial protein. The misannotation was partly due to the interpretation of a tobacco etch virus (TEV) protease cleavage accessibility assay (20). We used the same assay with an ER-targeted TEV protease and confirmed Mmm1 ER-membrane insertion (fig. S6B).
Taken together, our results indicate that ERMES functions as a molecular zipper bridging between ER and mitochondria. ERMES consists of the ER-resident membrane protein Mmm1 and the OMM-resident β-barrel protein Mdm10 (21). The well-characterized interaction of these two proteins (16, 19) requires Mdm12 and Mdm34. Defects in these latter proteins can be compensated by expressing ChiMERA, a synthetic protein that bridges ER and mitochondrial membranes, strongly suggesting that one primary physiological role of ERMES is that of a mechanical tether. Mutation of a single ERMES component causes complex disassembly but results in different growth phenotypes. Deletions of MDM12 and MDM34 are easily compensated for by ChiMERA expression, whereas deletions of MMM1 and MDM10 are less efficiently compensated. The variability in phenotypes indicates that Mmm1 and Mdm10 still provide useful activity in the absence of a functional complex, provided that an artificial tether is present. Mdm10 is a component of both ERMES and of the SAM complex (19) that imports β-barrel proteins in the OMM, and therefore, this latter activity would not be expected to be compensated for by ChiMERA expression.
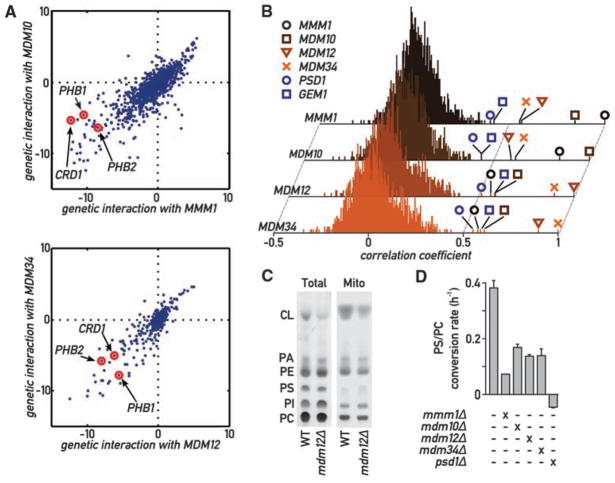
From these results, it appeared plausible that impairing ER-mitochondrial tethering provided the molecular basis for the phenotypes associated with ERMES defects, thus offering a unique opportunity to investigate the physiology of ER-mitochondria communication. To evaluate systematically the phenotypic consequences of loss of ERMES, we analyzed a recently generated epistasis mini-array profile (E-MAP) (see supplement) (22). The E-MAP methodology identifies functionally related genes by comparing their profiles of genetic interaction. In the map, a set of 1493 null or hypomorphic alleles, including mmm1Δ, mdm10Δ, mdm12Δ, and mdm34Δ strains, was crossed to a set of 484 query null or hypomorphic strains selected primarily for their association with mitochondrial and ER biology. The phenotype of the ~700,000 double mutants was assessed by colony size and used to calculate quantitative genetic interactions. This analysis confirmed previously characterized genetic interactions. For instance, prohibitins (PHB1 and PHB2), which have been shown to be synthetic lethal to ERMES component deletions (23), display some of the strongest synthetic phenotypes (Fig. 4A).
Fig. 4.
Global analysis of ERMES genetic interactions. (A) Genetic interactions of MMM1 and MDM10 (top) and of MDM12 and MDM34 (bottom). Positive values indicate epistatic or suppressive interactions (i.e., double mutant grows better than expected from the combination of the phenotypes of each single mutant); negative values indicate a synthetic sick/lethal genetic interaction (i.e., double mutant grows worse than expected). (B) Histograms of correlation coefficients generated by comparing the profiles of genetic interaction for each ERMES component to all other 1493 profiles in the E-MAP analysis. ERMES components display strongest correlation to each other and to GEM1 and PSD1. (C) Thin-layer chromatography (TLC) analysis of steady-state total and mitochondrial phospholipids in WT and mdm12Δ strains. PA, phosphatidic acid; PI, phosphatidylinositol. (D) The aminoglycerophospholipid biosynthesis pathway is slowed down in ERMES mutants. Cultures of the indicated genotypes were labeled with 14C-serine, then chased with an excess cold serine. The variation of the PC/PS ratio was then measured over time by quantitative TLC, and its slope was estimated with linear regression. Error bars represent the SE of the linear regression.
We used the interaction patterns of the 1493 genes in this analysis to calculate pairwise correlation coefficients. In this analysis, genes encoding ERMES components consistently showed strong correlation with each other (Fig. 4B), confirming that these genes are functionally related and that this analysis has the power to uncover this relation. Two genes, GEM1 and PSD1, showed strong correlation to every ERMES gene. Gem1 is a calcium-binding rho-like GTPase inserted in the OMM and involved in calcium-dependent mitochondrial movement and inheritance (24, 25). Psd1 is a phosphatidylserine (PS) decarboxylase involved in the de novo biosynthesis of aminoglycerophospholipids. ER-mitochondria connections have been previously shown to be important for interorganelle calcium and phospholipid exchange, further supporting a role for ERMES in mediating ER-mitochondrial junctions.
The phenotypic similarity between ERMES and PSD1 was particularly notable because Psd1 is the only aminoglycerophospholipid bio-synthetic enzyme in mitochondria (1) (fig. S7), implying that its substrate has to be transported from the ER and its product back to the ER. To assess a role of ER-mitochondrial junctions in lipid biosynthesis, we first analyzed mitochondrial phospholipid content in ERMES-disrupted (mdm12Δ) cells at steady-state conditions. All phospholipid classes were represented in ratios comparable to those of WT cells, with the notable exception of cardiolipin (CL), which was much reduced in its relative abundance (Fig. 4C). CL is a mitochondria-specific phospholipid essential for respiration. Deletion of any ERMES component, as well as of PSD1, is synthetically lethal with a deletion of CRD1 (Fig. 4A) (26), which encodes the CL synthase (27). Moreover, psd1Δ cells also display reduced CL levels (28). Thus, similar to psd1Δ cells, ERMES mutant strains suffer from mitochondrial phospholipid abnormalities and cannot tolerate the absence of a functional CL biosynthesis pathway, even when grown in fermentable medium. In addition, an extensive cross talk has been observed between aminoglycerophospholipid and CL synthesis, along with a reduction in CL levels in mmm1Δ, mdm10Δ, and mdm34Δ strains (23).
The maintenance of a balance between most phospholipid species besides CL in mdm12Δ mutant cells suggests that ER-mitochondria connections might affect the rate of phospholipid biosynthesis but have little effect on steady-state levels because of compensatory mechanisms (28). To assess this possibility, we monitored the conversion of PS to phosphatidylcholine (PC) in a 14C-serine pulse-chase. This experiment was conducted in a psd2Δ background to exclude the contribution of the vacuolar PS decarboxylase Psd2 (29). An otherwise WT strain displayed a high rate of PS-to-PC conversion (Fig. 4E), whereas a psd1Δ psd2Δ strain failed to convert PS to PC. Mutants of ERMES showed a two- to fivefold reduction in PS-to-PC conversion rate (Fig. 4D), consistent with the notion that disruption of ERMES impairs phospholipid exchange between ER and mitochondria. This defect was partially reversed by ChiMERA expression in mdm12Δ psd2Δ and mdm34Δ psd2Δ strains (fig. S8). Contrary to psd1Δ psd2Δ strains, however, none of the psd2Δ ERMES mutants were auxo-trophic for ethanolamine, showing that, consistent with the pulse-chase labeling data, Psd1 still displayed activity even in the absence of tethering.
Thus, ERMES-mediated ER-mitochondria connections are necessary for efficient inter-organellar phospholipid exchange. A reduced rate of lipid exchange in turn slows down amino-glycerophospholipid turnover, which results in impaired CL synthesis (28). It is likely that part of the pronounced defects in ERMES mutant cells results from problems in mitochondrial membrane maintenance (23).
Our strategy of discovering unknown genes by encoding their function into artificial constructs may be applicable to other situations in which a synthetic protein can substitute for an endogenous gene(s).
Mmm1 and Mdm12 belong to the SMP-domain protein family that has multiple members across all eukaryotes (30), making it likely that complexes containing one or more of these SMP-domain–containing proteins perform similar functions in metazoan cells.
The organization of the ERMES complex into 2 to 10 foci per cell suggests that the number and extent of interorganelle exchange surfaces have been vastly overestimated (2), or that eukaryotes may have evolved multiple specialized and spatially restricted structures that perhaps are dedicated to particular tasks. Each ERMES punctum represents a large macromolecular assembly that is expected to contain, for example, ~250 molecules of Mmm1 (31). The mechanical tethering provided by ERMES might organize specialized membrane domains that serve as platforms to recruit other molecules, which, in turn, carry out the physiological roles of the junctions, such as lipid transport or calcium exchange. ERMES foci localize adjacent to actively replicating mitochondrial nucleoids, as part of a poorly characterized assembly spanning both OMM and IMM (32). Such structures would span a third membrane and thus could strategically connect the ER lumen and the mitochondrial genome. The purpose of such a link is unclear, but we can speculate that it might help coordinate mitochondrial growth by coupling mitochondrial genome replication and membrane upkeep. In this light, the presence of Mdm10 in both SAM and ERMES complexes provides the intriguing possibility that such a link includes regulation of mitochondrial protein import.
Supplementary Material
Acknowledgments
We thank all members of the Walter lab for their support; N. Krogan, S. Wang, and N. Bajwa for their work creating the genetic interaction maps; and J. Shaw and K. Okamoto for their kind gift of plasmids and discussions. This work was supported by NIH. B.K. is a fellow of the Swiss National Science Foundation; M.S. is supported by a Human Frontiers Science Program Career Development Award; and P.W and J.S.W. are investigators at the Howard Hughes Medical Institute.
Footnotes
References and Notes
- 1.Voelker DR. J Lipid Res. 2003;44:441. doi: 10.1194/jlr.R200020-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Achleitner G, et al. Eur J Biochem. 1999;264:545. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 3.Vance JE. J Biol Chem. 1991;266:89. [PubMed] [Google Scholar]
- 4.Rizzuto R, Simpson AW, Brini M, Pozzan T. Nature. 1992;358:325. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 5.Rizzuto R, Brini M, Murgia M, Pozzan T. Science. 1993;262:744. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 6.Rizzuto R, et al. Science. 1998;280:1763. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 7.Pinton P, et al. Oncogene. 2008;27:6407. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmen T, et al. EMBO J. 2005;24:717. doi: 10.1038/sj.emboj.7600559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szabadkai G, et al. J Cell Biol. 2006;175:901. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi T, Su T. Cell. 2007;131:596. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 11.de Brito OM, Scorrano L. Nature. 2008;456:605. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 12.Csordás G, et al. J Cell Biol. 2006;174:915. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hieter P, Mann C, Snyder M, Davis RW. Cell. 1985;40:381. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 14.Rose MD, Broach JR. Methods Enzymol. 1991;194:195. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- 15.Berger KH, Sogo LF, Yaffe MP. J Cell Biol. 1997;136:545. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boldogh IR, et al. Mol Biol Cell. 2003;14:4618. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youngman MJ, et al. J Cell Biol. 2004;164:677. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McConnell SJ, Stewart LC, Talin A, Yaffe MP. J Cell Biol. 1990;111:967. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meisinger C, et al. EMBO J. 2007;26:2229. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo-Okamoto N, Shaw JM, Okamoto K. J Biol Chem. 2003;278:48997. doi: 10.1074/jbc.M308436200. [DOI] [PubMed] [Google Scholar]
- 21.Kutik S, et al. Cell. 2008;132:1011. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 22.Schuldiner M, et al. Cell. 2005;123:507. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 23.Osman C, et al. J Cell Biol. 2009;184:583. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Schwarz TL. Cell. 2009;136:163. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frederick RL, et al. J Cell Biol. 2004;167:87. doi: 10.1083/jcb.200405100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gohil VM, Thompson MN, Greenberg ML. J Biol Chem. 2005;280:35410. doi: 10.1074/jbc.M505478200. [DOI] [PubMed] [Google Scholar]
- 27.Chang SC, et al. J Biol Chem. 1998;273:14933. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- 28.Birner R, Bürgermeister M, Schneiter R, Daum G. Mol Biol Cell. 2001;12:997. doi: 10.1091/mbc.12.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trotter PJ, Voelker DR. J Biol Chem. 1995;270:6062. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- 30.Lee I, Hong W. FASEB J. 2006;20:202. doi: 10.1096/fj.05-4581hyp. [DOI] [PubMed] [Google Scholar]
- 31.Ghaemmaghami S, et al. Nature. 2003;425:737. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 32.Meeusen S, Nunnari J. J Cell Biol. 2003;163:503. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.