Abstract
Nonmalignant human mammary epithelial cells (HMEC) seeded in laminin-rich extracellular matrix (lrECM) form polarized acini and, in doing so, transit from a disorganized proliferating state to an organized growth-arrested state. We hypothesized that the gene expression pattern of organized and growth-arrested HMECs would share similarities with breast tumors with good prognoses. Using Affymetrix HG-U133A microarrays, we analyzed the expression of 22,283 gene transcripts in 184 (finite life span) and HMT3522 S1 (immortal nonmalignant) HMECs on successive days after seeding in a lrECM assay. Both HMECs underwent growth arrest in G0–G1 and differentiated into polarized acini between days 5 and 7. We identified gene expression changes with the same temporal pattern in both lines and examined the expression of these genes in a previously published panel of microarray data for 295 breast cancer samples. We show that genes that are significantly lower in the organized, growth-arrested HMEC than in their proliferating counterparts can be used to classify breast cancer patients into poor and good prognosis groups with high accuracy. This study represents a novel unsupervised approach to identifying breast cancer markers that may be of use clinically.
Introduction
Loss of growth control and disruption of tissue architecture are among the earliest hallmarks of cancer. We hypothesized that the gene expression changes that occur during the organization and growth arrest of cultured mammary acini would share similarities with breast tumors that had a good prognosis. To test this hypothesis, we examined temporal changes in gene expression in two nonmalignant human mammary epithelial cells (HMEC) grown in a three-dimensional laminin-rich extracellular matrix (lrECM) assay (1). In this model, single nonmalignant breast epithelial cells form polarized growth-arrested multicellular structures that resemble acini over a period of several days. This transition from the unpolarized actively dividing state to a polarized nondividing state is the reverse of what happens in the early stages of tumorigenesis.
We used two nonmalignant HMECs, a nonimmortalized HMEC strain, 184 (2, 3), and a spontaneously immortalized cell line, HMT3522 S1 (4), and monitored the changes in gene and protein expression as the cells formed acinar structures in three-dimensional lrECM cultures. During the process of self-organization and withdrawal from the cell cycle, we noted a progressive hypo-phosphorylation of the retinoblastoma (Rb) gene and an induction of the cyclin-dependent kinase (CDK) inhibitor p27kip1. Analysis of the changes in gene expression in both cell lines allowed the identification of sets of commonly regulated genes.
Although established breast cancer prognostic markers, such as tumor size, grade, nodal, and hormone receptor status, are useful in predicting survival in large populations (5–7), there is a pressing need to develop better prognostic signatures to predict recurrence and overall survival. A particular benefit would be the identification of patients with good prognoses, whose tumors are highly unlikely to recur and who nevertheless are being treated with cytotoxic chemotherapies (8). The advent of gene expression technologies has greatly aided the identification of molecular signatures with value for tumor classification and prognosis prediction (9–14). van de Vijver et al. have developed a 70-gene signature that effectively stratifies patients into good and poor prognosis groups (15, 16). Paik et al. (17) have proposed a 21-gene signature with which to calculate a “recurrence score” that predicts the likelihood of recurrence in estrogen receptor (ER)-positive lymph node–negative patients. In each of these studies, the predictive signatures have been derived by using a training set of patients of known outcome followed by testing these signatures in a validation set of patients. In contrast, our approach has been to identify directly genes, the expression of which changes as cultured mammary epithelial cells transition from a disorganized to an organized state, and to then test the proof of principle and the possible use of these genes as prognostic markers in a validation set of patients.
Materials and Methods
Cell culture
Finite life span 184 HMECs were obtained from reduction mammoplasty tissue and grown in a serum-free MCDB 170 medium (mammary epithelial growth medium, Clonetics Division of BioWhittaker, Walkersville, MD) as described previously (2, 18). HMT3522 S1 mammary epithelial cells were cultured in a H14 medium (DMEM/F12 containing 250 ng/mL insulin, 10 μg/mL transferrin, 2.6 ng/mL sodium selenite, 10−10 mol/L estradiol, 1.4 × 10−6 mol/L hydrocortisone, 10 ng/mL epidermal growth factor, and 5 μg/mL prolactin). The cells were cultured in a three-dimensional lrECM (Matrigel, BD Biosciences, Franklin Lakes, NJ) as described (19). The colonies were isolated from the Matrigel in ice-cold PBS/5 mmol/L EDTA after 3, 5, 7, and 10 days. For Western blot analysis, the colonies were lysed in 150 mmol/L NaCl, 1% NP40, and 50 mmol/L Tris (pH 8.0). RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Indirect immunofluorescence and image acquisition
Acinar structures were fixed on glass slides in methanol-acetone (1:1) at −20°C for 10 minutes and air dried. A primary block was done in an immunofluorescence buffer (130 mmol/L NaCl, 7 mmol/L Na2HPO4, 3.5 mmol/L NaH2PO4, 7.7 mmol/L NaN3, 0.1% bovine serum albumin, 0.2% Triton X-100, 0.05% Tween 20) plus 10% goat serum for 1 hour at room temperature. A secondary block was done in immunofluorescence buffer plus 10% goat serum plus 20 μg/mL goat anti-mouse F(ab′)2 fragment for 30 minutes. The primary antibody rat anti-α6 integrin (Chemicon International, Temecula, CA) was diluted 1:50 in the latter blocking buffer and incubated overnight (15–18 hours) at 4°C. Secondary antibodies conjugated with fluorescent dye FITC or Texas red were diluted 1:100 in blocking buffer and incubated 1 hour at room temperature. Immunofluorescent images were acquired using an inverted microscope equipped with a digital camera and SPOT software. Confocal analysis was done using a Zeiss 410 confocal microscopy system (Zeiss, Thornwood, NY). The images presented are representative of two or more independent experiments. All the images were converted to TIFF format and arranged using Adobe Photoshop 5.0.
Flow cytometry
To analyze the DNA content, acini were suspended in 300 μL 0.25% trypsin and incubated at 37°C for exactly 10 minutes. The dispersed cells were washed thrice in PBS and fixed in 40% ethanol at 4°C overnight. They were then incubated with 500 μg/mL RNase A in PBS at 37°C for 30 minutes and stained with 69 μmol/L propidium iodide solution in PBS at room temperature for 30 minutes. The DNA content was determined by flow cytometry using a FACScan (Becton Dickinson, Hialeah, FL), and the data were analyzed with CellQuest software (Becton Dickinson). Statistics for the reduction in S-phase kinetics were based on the means of repeated measurements: S1, day 3, n = 4; S1, day 5, n = 5; S1, day 7, n = 6; 184, day 3, n = 2; 184, day 5, n = 1; and 184, day 7, n = 2.
Immunoblotting
Whole-cell lysates (20–100 μg protein) were separated by SDS-PAGE and transferred electrophoretically to polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking in 10% nonfat dry milk for 90 minutes at room temperature, they were incubated with the following antibodies at 1:500 to1:1,000 dilution in 2% nonfat dry milk for 2 hours at room temperature or overnight at 4°C: rabbit anti-phosphorylated Rb at S807/811 and S795 (Cell Signaling Technology, Beverly, MA), rabbit anti-phosphorylated Rb at S612 and T821 (Biosource, Camarillo, CA), and mouse anti-p27kip1 (BD Transduction Laboratories, Franklin Lakes, NJ). The blots were then incubated with horseradish peroxidase–conjugated sheep anti-mouse IgG or anti-rabbit IgG at 1:2,000 in 2% nonfat dry milk for 90 minutes at room temperature. The blots were developed using SuperSignal West Pico Chemilumiscent Substrate (Pierce, Rockford, IL).
Survival analysis
A database consisting of the microarray profiles of 295 human breast tumors with the associated clinical data (15) was obtained from Rosetta Inpharmatics (http://www.rii.com/publications/2002/nejm.html). For survival analysis of the 19 individual marker genes, the patients were stratified into quartiles for expression of each marker, and the survival curves were computed using the method of Kaplan-Meier. Statistical significance was determined using the log-rank test. Statistical analyses were done using GraphPad Prism. For survival analysis of the set of 249 marker genes, the patients were stratified into two groups using GeneSpring software by hierarchical cluster analysis with a distance metric of the expression pattern of all 249 genes. Kaplan-Meier survival curves, log-rank statistics, and the estimated hazard ratio for these two groups were computed using the Excel add-in EcStat.
Microarray hybridization and analysis
Cell samples were harvested in duplicate at three time points, 3, 5, and 7 days, after seeding in lrECM. Purified total cellular RNA was biotin labeled and hybridized to human oligonucleotide microarrays (Affymetrix HG-U133A; Affymetrix, Santa Clara, CA) as described previously (20). Experiments with Affymetrix-present P-call rates of >30% were included in the analysis. Signal values from each of the 22,283 probe sets were calculated by means of robust multiarray analysis (21) using Bioconductor (http://www.bioconductor.org) in the R computing environment. The signal values were inverse log2 transformed and then imported into GeneSpring software (Silicon Genetics, Palo Alto, CA), and each array was normalized to its median signal intensity. The genes were normalized to the mean of the 3-day time point for each cell type independently. For method 1 of selecting significantly differential, temporally coregulated genes, significantly up-regulated genes in each cell specimen were identified by first selecting the genes induced at least 1.5-fold in at least one of the six conditions and then doing an ANOVA analysis as a function of time. Variances were calculated using the cross-gene error model (GeneSpring), Ps cutoff 0.05 (multiple testing correction: Benjamini-Hochberg false discovery rate). About 5% of the identified genes in each set would be expected to pass this restriction by chance. Significantly down-regulated genes were identified in the same manner after normalizing to the 7-day time point. Genes that were up-regulated or down-regulated early in each cell line were selected from the significantly up-regulated or down-regulated gene lists. The early genes were defined as those with a mean expression at 5 days of at least 50% of their mean expression at 7 days. For method 2 of selecting significantly differential, temporally coregulated genes, we first selected all genes that were at least 1.5-fold differential (up or down) in at least one of the four samples from the two later time points (days 5 and 7) for either cell specimen. We then did an ANOVA as a function of time. Variances were not assumed equal (Welch ANOVA), Ps cutoff 0.05 (multiple testing correction: Benjamini-Hochberg false discovery rate). From this list of genes that were significantly differential in either cell line, we then identified genes that were up-regulated or down-regulated early (mean expression at 5 days of at least 50% of their mean expression at 7 days) or late in each cell line. We then identified those genes that were coordinately regulated in both cell lines.
Results
Temporal analysis of gene expression in two nonmalignant human breast epithelial cells grown in three-dimensional lrECM cultures
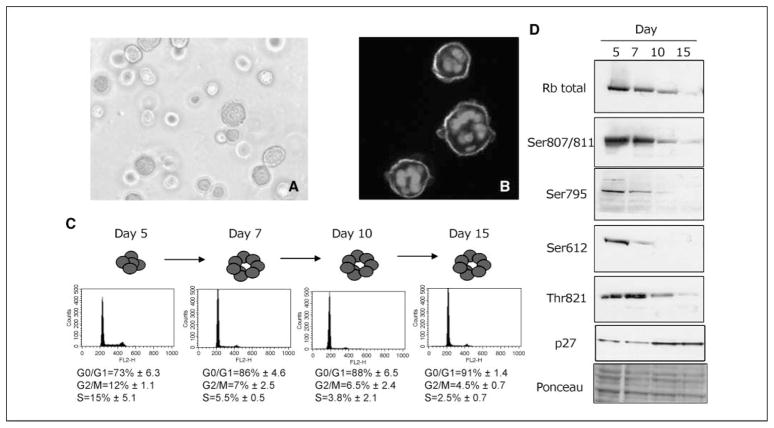
To identify consistent changes in gene and regulatory protein expression levels, we characterized two independently derived nonmalignant HMECs: one finite life span strain (184; ref. 2) and one spontaneously immortalized line (HMT3522 S1; ref. 4). Both cells formed acinus-like structures with similar morphology and basal polarity when cultured from single cells in lrECM (Fig. 1A and B; see also ref. 1). We did temporal studies to determine when, and in what order, changes in critical cell cycle regulatory molecules occurred in nonmalignant HMECs in three-dimensional lrECM cultures. Flow cytometric analyses indicated that, after undergoing a limited number of cell divisions, the majority of nonmalignant S1 cells accumulated in the G0–G1 phase of the cell cycle by day 7 (Fig. 1C). The product of the Rb susceptibility gene, a central player in the G1-S transition, is inactivated by phosphorylation, allowing cell cycle progression. Rb inactivation occurs through the sequential actions of cyclin D/CDK4 and CDK6 and cyclin E-CDK2 complexes (22). We analyzed cell cycle regulators known to affect G1 checkpoints in a time-dependent manner in S1 cells in three-dimensional cultures. Phosphorylation of several sites on Rb was found to gradually decrease between days 5 and 10 in these nonmalignant cells, consistent with the growth-suppressive role of the hypophosphorylated form (Fig. 1D). Cyclins D1, E, and A, as well as their binding partners, CDK4, CDK6, and CDK2, also decreased during this period (data not shown). In contrast, the CDK inhibitor p27kip1 increased between days 5 and 10 (Fig. 1D). The pronounced down-modulation of Rb phosphorylation and the elevation of p27kip1 protein levels were changes observed in both the S1 and 184 cells (data not shown). Although other studies have used different HMECs (MCF10A) to determine how cells can escape normal growth control (23–26), the mechanisms by which mammary cells actually initiate and maintain growth arrest during the process of acini formation in the context of three-dimensional lrECM had remained to be determined.
Figure 1.
HMEC cultured in lrECM form polarized structures and arrest growth in G0–G1. A, phase-contrast image of typical acinar structures formed by 184 cells at day 7 in lrECM. Structures reach dimensions of 20 to 40 μm diameter. Bar, 25 μm. B, indirect immunofluorescent image showing basal polarity of α6 integrin (green ) in 184 cells at day 7 in lrECM. Red, cell nuclei were stained with 4′,6-diamidino-2-phenylindole. C, flow cytometric analyses of propidium iodide–stained S1 cells indicated that the majority of the cells accumulated in the G0–G1 phase of the cell cycle by day 7 in lrECM. D, Western blot analyses of cell cycle regulatory molecules in S1 cells at days 5, 7, 10, and 15 after suspension in lrECM indicated that total as well as specific phosphorylated forms (Ser807/Ser811, Ser795, Ser612, and Thr821) of Rb decreased whereas p27 increased over the time course. Ponceau staining of major proteins indicated that equivalent amounts of total protein were loaded in each lane.
Global gene expression analysis of the time course of HMECs in three dimensions
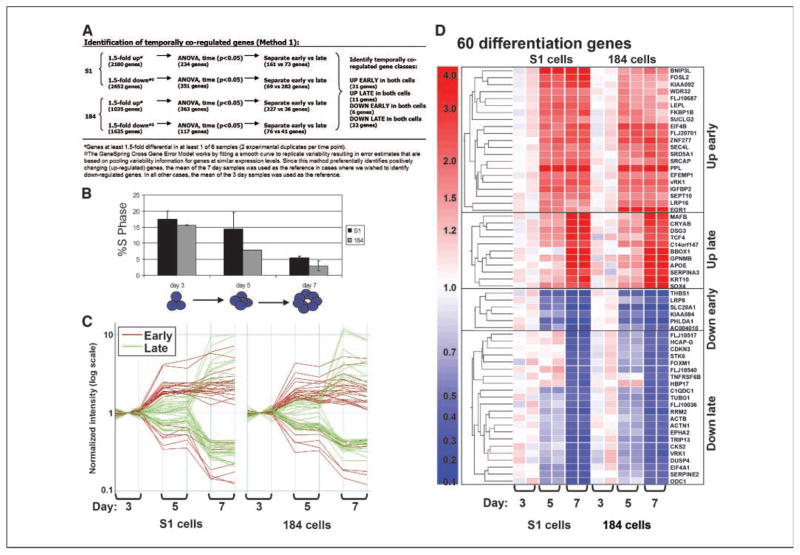
To probe systematically the molecular changes that accompany acinus formation, we analyzed the expression profiles of 22,283 transcripts using Affymetrix HG-U133A microarrays. Microarray experiments were done with biological duplicates using RNA samples harvested from S1 and 184 cells after 3, 5, and 7 days of culture in lrECM. Comparison of the decrease in the percentages of cells in S phase showed that growth arrest occurred with kinetics that were significantly correlated (>95% confidence level) in both cell types (correlation coefficient, 0.89; Fig. 2A). Further, the S-phase decrease was significant (P = 0.05, ANOVA). Hence, we reasoned that the gene expression changes important for these processes would follow a common temporal pattern in both cell lines and that changes that were cell type-specific could be disregarded.
Figure 2.
Temporally coregulated genes in HMEC grown in three-dimensional culture. A, temporal pattern of growth arrest in S1 and HMEC184 cells. The percentage of cells in S phase decreased significantly between days 3 and 7. B, scheme is provided outlining method 1 used to select sets of temporally coregulated genes. Sixty genes were determined by method 1 to show significant changes in expression in both HMEC specimens during the time course of incubation in lrECM. C, expression levels of the 60 genes that were coordinately expressed in both cell types. The genes were grouped into categories based on whether they showed coordinate up-regulation early by our definition (3–5 days), up-regulation late (5–7 days), down-regulation early, or down-regulation late during the time course (see text for details). Note that the scale is logarithmic. D, changes in the expression levels of the 60 individual genes are plotted and organized by hierarchical cluster analysis. Red, up-regulated genes; blue, down-regulated genes. Complete gene names and Genbank IDs are available (see Supplementary Data).
We first identified genes that showed at least 1.5-fold changes during the time course in the individual cell specimens (P < 0.05, ANOVA; within this window, 363 genes were up-regulated and 117 genes down-regulated in 184 cells and 234 genes were up-regulated and 351 genes down-regulated in S1 cells). We then divided these lists into genes, the expression of which changed ‘early’ by our definition (between days 3 and 5) or ‘late’ (between days 5 and 7) in S1 and 184. Finally, we identified the genes from each temporal group that were common to both cell types (Fig. 2B). A total of 60 genes with common temporal patterns were identified, including 21 genes that were up-regulated early, 11 genes that were up-regulated late, 6 genes that were down-regulated early, and 22 genes that were down-regulated late (Fig. 2C and D; Supplementary Data). The magnitude of the expression changes of the 22 down-regulated genes in HMEC ranged from 2.5-fold (ACTB) to 5.4-fold (TRIP13).
Correlation of the differentially expressed genes with survival of breast cancer patients
To relate the process of acinar development in three-dimensional lrECM cell cultures to the changes that occur in breast cancer, we examined the expression levels of the differentially regulated genes identified using our model using previously published microarray data for a panel of 295 breast cancer samples from the fresh-frozen tissue bank of the Netherlands Cancer Institute (Amsterdam, the Netherlands), including 151 lymph node–negative disease and 144 lymph node–positive disease patient samples (15). Fifty-five of the 60 genes selected in our three-dimensional culture analysis were included on these microarrays. We looked at 5- and 10-year survival data and applied Student’s t test to determine how many of the genes modulated in three-dimensional cultures showed survival-associated expression changes. Student’s t tests were done to determine whether the difference in the expression level of a given gene in two groups (e.g., patients who survived 5 years versus patients who did not) was large enough that it was not likely to be due to chance. The numbers and percentages of genes exhibiting significantly different expression in the tumors of patients with differential survival (P < 0.05) were tabulated for (a) all the genes represented on the microarrays, (b) genes selected based on differential expression during the three-dimensional lrECM time course, or (c) randomly generated gene lists (Table 1). The percentage of genes with survival-associated expression changes was highest for those genes down-regulated late (between days 5 and 7) in the time course. The percentage for this gene list exceeded those of the unfiltered list of all 25,773 genes represented on the arrays, 5 random gene lists, and all other three-dimensional lrECM gene lists. The list of genes that were down-modulated late in the lrECM time course showed a marked enrichment in genes, the expression level of which correlated to 5-year (68%) and 10-year (53%) survival. The levels of the majority of the late down-regulated genes were higher in patients who died within 5 or 10 years (ACTB, VRK1, ODC1, CKS2, FLJ10036, FLJ10540, FOXM1, RRM2, TRIP13, CDKN3, STK6, FLJ10517, TUBG1, ACTN1, TNFRSF6B, and EPH2), whereas the levels of three genes (DUSP4, HBP17, and EIF4A1) were lower in these patients. The magnitude of the expression changes of the down-regulated genes in the 250 tumor samples ranged from 1.5-fold (FLJ10036) to 9.3-fold (HPV17).
Table 1.
Number of potential prognostic markers for breast cancer among the genes that were differential in the three-dimensional time course (method 1)
| Total no. genes | No. significantly* differential genes |
||
|---|---|---|---|
| Survival (5 yrs), n (%) | Survival (10 yrs), n (%) | ||
| All genes | 25,773 | 2,418 (9.4) | 67 (0.26) |
| Early up | 20 | 7 (35) | 5 (25) |
| Early down | 5 | 1 (20) | 1 (20) |
| Late up | 11 | 3 (27) | 1 (9) |
| Late down | 19 | 13 (68)† | 10 (53)† |
| Random list 1 | 20 | 0 | 1 (5) |
| Random list 2 | 20 | 5 (25) | 3 (15) |
| Random list 3 | 20 | 4 (20) | 0 |
| Random list 4 | 20 | 0 | 0 |
| Random list 5 | 20 | 0 | 0 |
| Random list 6 | 55 | 23 (42) | 17 (31) |
| Random list 7 | 71 | 11 (15) | 7 (9.8) |
NOTE: Total number reflects genes included on the Rosetta micro-arrays; hence, values in some cases are less than the total number included on the Affymetrix microarrays.
P < 0.05, Student’s t test.
Greater than 50%.
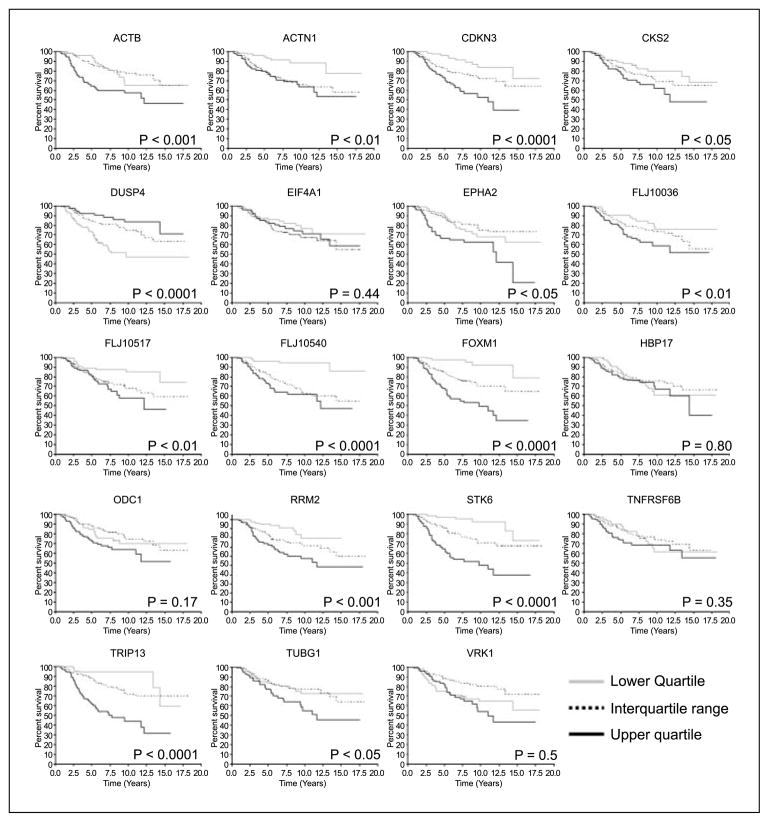
We identified 22 genes in all that were down-regulated in both HMECs between days 5 and 7 of lrECM culture. Of these, 19 genes were represented in the published data set for the 295 patient tumor samples. We stratified the 295 tumors into quartiles based on the relative expression level of each of the genes in the selected set and further analyzed the relationship of the expression level of each individual gene to survival (Fig. 3). The resulting Kaplan-Meier curves showed that gene expression levels correlated significantly with outcome for 14 of the 19 selected markers. For 13 of the 14 markers, gene expression was lower in tumors from patients with better outcomes, whereas in one case (DUSP4) gene expression was lower in tumors from patients with poorer outcomes.
Figure 3.
Fourteen of the 19 genes down-regulated late during acinar morphogenesis showed significant correlations with patient survival. Two hundred ninety-five patients were grouped into quartiles based on the relative expression of each selected gene in their corresponding tumors. Survival of each quartile was plotted according to the method of Kaplan-Meier. Ps, outcomes of the log-rank tests between the upper and lower quartiles.
To test whether expression levels of the 19 selected marker genes correlated with lymph node status, we calculated Pearson product-moment correlation coefficients. Although no genes were correlated at the 95% confidence level, expression levels of 4 of the 19 genes showed a trend toward a correlation with lymph node number or status (80% confidence level), including DUSP4, HBP17, TNFRSF6B, and TUBG1.
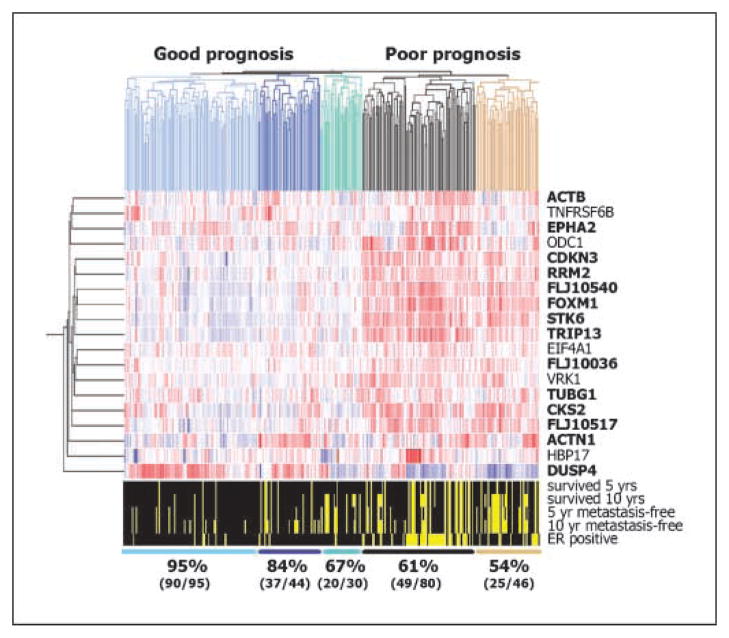
Collective gene signatures have the potential to discriminate among clinical end points more accurately than markers used individually. Hence, we tested the ability of our set of 19 genes to classify breast cancer patients into prognostic groups. We used hierarchical cluster analysis to separate the patients into groups and then determined the overall 10-year survival rates for these groups (Fig. 4). The cluster analysis separated the patients into five groups, three of which had tumors that expressed comparatively lower levels of most of the 19 genes and two of which expressed higher levels. The 10-year survival rates for these five groups were 95%, 84%, 67%, 61%, and 54%, respectively.
Figure 4.
The set of 19 genes down-regulated late during acinar morphogenesis can be used to accurately cluster 295 breast cancer samples into prognostic groups. Rows, relative expression levels of the genes (right); columns, individual patients. Scale of relative expression levels is the same as that in Fig. 2D. Genes and tumor samples were arranged by a hierarchical cluster analysis using a Pearson metric. Dendrograms, top, left, degree of relatedness of the samples and genes. Dendrogram branch colors, top, different prognostic groups. Bold font, genes were significantly associated with survival (P < 0.05, Kaplan-Meier analysis; Fig. 3). Bottom, clinical variables for the 295 patient samples. Black regions, a given variable applies to that patient.
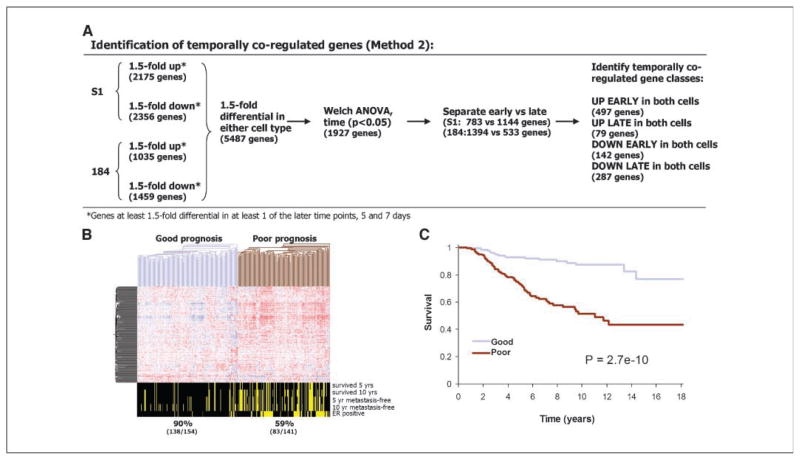
To test whether other sets of genes down-regulated late in the lrECM time course identified by using other selection strategies would also include useful breast cancer markers, we applied a second selection strategy (Fig. 5A) and tested the ability of the resulting gene set to predict breast cancer prognosis. This second method was less restrictive than the first and resulted in the identification of 287 genes that were significantly down-regulated late in the three-dimensional time course of both HMEC specimens (see Supplementary Data for complete gene list and gene expression information). Seventeen of the 22 genes selected using method 1 were also included in the 287 genes selected using method 2. We tested the ability of these genes to predict breast cancer prognosis by using hierarchical cluster analysis in the same set of previously published microarray data from 295 breast cancers (15). A large majority, 249 of the 287 genes, was included on these microarrays. Of the 17 overlapping (methods 1 and 2) genes on the Affymetrix chips, 15 were present in the Rossetta data set: ACTB, ACTN1, CDK3, CKS2, DUSP4, EPHA2, HBP17, FOXM1, ODC1, RRM2, STK6, TNFRSF6B, TRIP13, TUBG1, and VRK1.
Figure 5.
A second set of 249 genes down-regulated late in the time course of HMECs in three-dimensional lrECM cultures classifies good versus poor prognosis in breast cancer patients. A, scheme outlining method 2 used to select sets of temporally coregulated genes. B, set of 249 down-regulated genes identified by method 2 clustered the 295 breast cancer samples into two prognostic groups. Genes are arranged by a hierarchical cluster analysis using a Pearson metric, and samples are arranged using a distance metric. Bottom, clinical variables. C, overall survival in the two groups was plotted by the method of Kaplan-Meier. P, outcome of the log-rank tests between the good and poor prognosis groups.
Hierarchical cluster analysis using the 249-gene signature classified the samples into two groups of approximately equal numbers of tumors (Fig. 5B). Overall 10-year survival rates were 90% (138 of 154) for the good prognosis group and 59% (83 of 141) for the poor prognosis group. To assess the significance of these predictions and take into account patients that could not be followed the entire length of the study, we did a Kaplan-Meier analysis. The results show that the 249-gene profile was highly informative in identifying patients with poor outcome (P = 2.7 × 10−10, log-rank test; Fig. 5C). The estimated hazard ratio for poor outcome (failure to survive) in the group with the poor prognosis signature compared with the good prognosis signature was 4.7 [95% confidence interval (95% CI), 2.8–7.9].
Discussion
Three-dimensional lrECM cultures permit nonmalignant cells to exhibit self-organizing properties. Such cultures provide models that allow the study of processes that are aberrant in breast cancer (1, 19, 27, 28). To understand how breast epithelial cells transition from a proliferating, unorganized state to a resting, organized state and to relate this process to the opposing changes that occur in clinical breast cancer, we did genome-wide gene expression profiling for two nonmalignant HMECs in three-dimensional cultures and used published data from a panel of 295 breast cancer samples. We show that genes that are down-regulated as HMEC transition from a proliferative to an acinar phenotype can be used collectively as signatures to predict clinical breast cancer prognosis.
Our approach represents a new way to identify genome-wide cancer prognostic markers. We based all the marker selection steps on HMEC cultured in three-dimensional lrECM. The three-dimensional model system provided a means to focus on epithelial cells themselves and a defined, highly relevant biological process, the formation of breast acini. Whereas the stroma is absent, the three-dimensional lrECM assays seem to substitute for myoepithelial cells and other signals that are needed to form organized acini (29). Differentially expressed genes identified using this model system are likely to be functionally linked to the transformation relevant process. Further, we have applied an unsupervised method (hierarchical cluster analysis) to classify the patient samples using selected markers. Hence, neither our method of marker selection nor our sample classification method relies on any clinical information.
Gene expression profiling of tumors using DNA microarrays is a promising method for predicting prognosis and treatment response in cancer patients (17, 30–32). Two studies have recently used genome-wide microarray analysis to identify gene signatures that predict prognosis in breast cancer. The profiles studied by both groups of researchers were reported to be more powerful predictors of the outcome of disease than standard systems based on clinical and histologic criteria. The study by van’t Veer et al. (16) used the supervised classification of a primary data set of 78 tumor samples to identify a 70-gene signature that divided the samples into classes of poor and good prognosis. Validation of the classifier by a second overlapping set of 295 tumors showed that it accurately predicted 10-year survival in breast cancer patients (15). Overall, the 10-year survival rates were 54.6 ± 4.4% and 94.5 ± 2.6% for the poor and good prognosis groups, respectively. Their estimated hazard ratio for poor outcome (distant metastases) in the group with a poor prognosis signature compared with the group with the good prognosis signature was 5.1 (95% CI, 2.9–9.0; P < 0.001). In a similar study, Wang et al. (33) also used supervised classification in a training set of 115 breast tumor samples. They identified a 76-gene signature, which was verified in an independent set of 171 breast tumors. Their estimated hazard ratio for poor outcome (distant metastases within 5 years) in the group with the poor prognosis signature compared with the group with the good prognosis signature was 5.7 (95% CI, 2.6–12.4). Our 249-gene signature predicted 10-year survival rates of 59% and 90% for poor and good prognosis groups, respectively. The estimated hazard ratio for poor outcome (failure to survive) in the group with the poor prognosis signature compared with the good prognosis signature was 4.7 (95% CI, 2.8–7.9).
Our 249-gene signature overlapped by 11 genes with the 70-gene signature of van’t Veer et al. (including NUSAP1, UCHL5, RAMP, DC13, PRC1, COL4A2, PITRM1, CENPA, MELK, KNTC2, and MCM6; ref. 16) and by 7 genes with the 76-gene signature of Wang et al. (including MTB, POLQ, SUPT16H, FEN1, DUSP4, PLK1, and SMC4L1; ref. 33). The van’t Veer and Wang signatures overlapped by a single gene (cyclin E2). Our 19-gene signature had no genes in common with either of the previously published signatures.
Our 19-gene signature included several genes encoding proteins with roles in the cell cycle and in cell division. The cell cycle genes were previously identified as important markers of prognosis in ER-positive younger patients (34). In this earlier study, 50 cell cycle–related genes divided 83 ER-positive younger patients into two groups of good versus poor prognosis. The overall 10-year survival rates were 46% and 96% for the poor and good prognosis groups, respectively. Similarly, we found that a core group of predominantly cell cycle and mitotic organizing center genes (CDKN3, RRM3, FLJ10540, FOXM1, STK6, TRIP13, EIF4A1, FLJ10036, VRK1, TUBG1, CKS2, and FLJ10517) made a strong contribution to stratifying tumors into good versus poor prognostic groups. One gene from our 19-gene signature, STK6, which encodes Aurora-A (35, 36), was also included in the 50-gene signature of Dai et al. (34), whereas 15 genes from our 249-gene signature were included in the Dai et al. signature (including BM039, DKFZp762E1312, LOC51203, LOC51659, ID-GAP, KNSL6, PRC1, STK6, CDC45L, SNRPA1, H2AFZ, CENPA, CDC6, BIRC5, and BLM). In addition to cell cycle genes, our prognostic genes also encoded products with other functions, including genes involved in cytoskeletal regulation (ACTB and ACTN1; ref. 37), cell survival (TNFRSF6B; refs. 38, 39), polyamine biosynthesis (ODC1; ref. 40), and cell-cell interactions (EPHA2; ref. 41). The genes in these additional functional groups were important in subdividing the patients into subgroups with differing survival rates.
In conclusion, we report that the gene expression changes that commonly occur in nonmalignant HMEC grown in three-dimensional lrECM cultures provide gene expression signatures that effectively stratify patients into prognostic groups according to overall survival rates. Our 249-gene signature achieved a hazard ratio of 4.7, which is comparable with hazard ratios achieved by large-scale supervised breast cancer microarray studies. Our results underscore the relevance of three-dimensional lrECM cultures for studies of malignant transformation and suggest potentially valuable new biomarkers for further clinical evaluation.
Supplementary Material
Acknowledgments
Grant support: Grant DE-AC03 SF0098 (M.V. Fournier, P. Yaswen, and M.J. Bissell); U.S. Department of Energy, Office of Biological and Environmental Research Distinguished Fellowship Award, National Cancer Institute grant 2 R01 CA064786-09, and U.S. Department of Defense Breast Cancer Research Program Innovator Award grant BC012005 (M.J. Bissell); National Institute of Allergy and Infectious Diseases grant U19-AI057319 (K.J. Martin, K. Xhaja, and I. Bosch); and Postdoctoral Training Fellowship grant DOD BCRP DAMD17-00-1-0224 (P.A. Kenny).
We thank Martha Stampfer for kindly providing the 184 HMEC and her and Saira Mian for critical reading of the article and other members of our laboratories for fruitful discussions.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–8. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond SL, Ham RG, Stampfer MR. Serum-free growth of human mammary epithelial cells: rapid clonal growth in defined medium and extended serial passage with pituitary extract. Proc Natl Acad Sci U S A. 1984;81:5435–9. doi: 10.1073/pnas.81.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stampfer MR, Yaswen P. Culture systems for study of human mammary epithelial cell proliferation, differentiation, and transformation. Cancer Surv. 1993;18:7–34. [PubMed] [Google Scholar]
- 4.Briand P, Petersen OW, Van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol. 1987;23:181–8. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 5.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6:375–92. doi: 10.1023/a:1014778713034. [DOI] [PubMed] [Google Scholar]
- 6.Clark GM. Prognostic and predictive factors for breast cancer. Breast Cancer. 1995;2:79–89. doi: 10.1007/BF02966945. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgibbons PL, Page DL, Weaver D, et al. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:966–78. doi: 10.5858/2000-124-0966-PFIBC. [DOI] [PubMed] [Google Scholar]
- 8.Goldhirsch A, Glick JH, Gelber RD, Coates AS, Senn HJ. Meeting highlights: International Consensus Panel on the Treatment of Primary Breast Cancer. Seventh International Conference on Adjuvant Therapy of Primary Breast Cancer. J Clin Oncol. 2001;19:3817–27. doi: 10.1200/JCO.2001.19.18.3817. [DOI] [PubMed] [Google Scholar]
- 9.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–71. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 10.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–7. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 11.Martin KJ, Graner E, Li Y, et al. High-sensitivity array analysis of gene expression for the early detection of disseminated breast tumor cells in peripheral blood. Proc Natl Acad Sci U S A. 2001;98:2646–51. doi: 10.1073/pnas.041622398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nacht M, Ferguson AT, Zhang W, et al. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999;59:5464–70. [PubMed] [Google Scholar]
- 13.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 14.Ramaswamy S, Golub TR. DNA microarrays in clinical oncology. J Clin Oncol. 2002;20:1932–41. doi: 10.1200/JCO.2002.20.7.1932. [DOI] [PubMed] [Google Scholar]
- 15.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 16.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 17.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 18.Stampfer MR, Bartley JC. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc Natl Acad Sci U S A. 1985;82:2394–8. doi: 10.1073/pnas.82.8.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weaver VM, Petersen OW, Wang F, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–45. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warke RV, Xhaja K, Martin KJ, et al. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol. 2003;77:11822–32. doi: 10.1128/JVI.77.21.11822-11832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligo-nucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 23.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–92. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 25.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 26.Debnath J, Walker SJ, Brugge JS. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J Cell Biol. 2003;163:315–26. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–35. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003;116:2377–88. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002;16:693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weigelt B, Hu Z, He X, et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65:9155–8. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 31.Braun S, Vogl FD, Naume B, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 32.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 34.Dai H, van’t Veer L, Lamb J, et al. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005;65:4059–66. doi: 10.1158/0008-5472.CAN-04-3953. [DOI] [PubMed] [Google Scholar]
- 35.Kimura M, Matsuda Y, Eki T, et al. Assignment of STK6 to human chromosome 20q13.2→q13.3 and a pseudogene STK6P to 1q41→q42. Cytogenet Cell Genet. 1997;79:201–3. doi: 10.1159/000134721. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M, Kotani S, Hattori T, et al. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosphila and yeast Ipl1. J Biol Chem. 1997;272:13766–71. doi: 10.1074/jbc.272.21.13766. [DOI] [PubMed] [Google Scholar]
- 37.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–34. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 38.Pitti RM, Marsters SA, Lawrence DA, et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396:699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- 39.Yu KY, Kwon B, Ni J, Zhai Y, Ebner R, Kwon BS. A newly identified member of tumor necrosis factor receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J Biol Chem. 1999;274:13733–6. doi: 10.1074/jbc.274.20.13733. [DOI] [PubMed] [Google Scholar]
- 40.Kay JE, Cooke A. Ornithine decarboxylase and ribosomal RNA synthesis during the stimulation of lymphocytes by phytohaemagglutinin. FEBS Lett. 1971;16:9–12. doi: 10.1016/0014-5793(71)80671-3. [DOI] [PubMed] [Google Scholar]
- 41.Lindberg RA, Hunter T. cDNA cloning and characterization of eck, an epithelial cell receptor protein-tyrosine kinase in the eph/elk family of protein kinases. Mol Cell Biol. 1990;10:6316–24. doi: 10.1128/mcb.10.12.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.