Abstract
We report here nine unusual cases of Kala-azar, of which parasites were isolated and found by 18S rRNA gene sequencing to be most similar to Leptomonas species. One of these isolates was used to inoculate Balb/c mice; organs were collected and directly submitted to a genus-specific rDNA-ITS1 PCR analysis: this revealed the presence of both Leptomonas sp. and Leishmania donovani. Therefore, we conclude that there was a mixed infection of Leptomonas sp. and L. donovani in this isolate. We consider that mixed infection may be present in the patients themselves, Leptomonas persisting in them because of the immuno-suppression associated with Kala-azar.
Keywords: Leptomonas, Leishmania donovani, Visceral leishmaniasis, hsp70 PCR-RFLP, rDNA-ITS1 PCR, Kala-azar
1. INTRODUCTION
Kala-azar (or visceral leishmaniasis, VL) is a vector-borne disease caused by two species of the Leishmania donovani complex: L. donovani and L. infantum. Ninety percent of the new cases of VL occurring annually originate from five countries: Bangladesh, Brazil, India, Nepal and Sudan (Desjeux, 1992). In India, VL is essentially encountered in Bihar and Eastern Uttar Pradesh and L. donovani is the only etiological agent reported so far. A recent study with multilocus microsatellite typing revealed that L. donovani parasites from Bangladesh, Bihar (India) and Nepal formed a very homogeneous population regardless of geographical origin, clinical manifestation, and independently of their grade of susceptibility to antimonial drugs (Alam et al., 2009). The same authors suggested that the circulation of a single homogeneous population of L. donovani in Bihar (India), Bangladesh and Nepal was, most probably, related to the epidemic spread of visceral leishmaniasis in this area. All these features of Indian kala-azar imply that in the practice, most of the parasites isolated from VL cases are not typed, assuming that they are all L. donovani. This contrasts with the situation in other leishmaniasis endemic regions where typing is more systematically done: f.i. in Latin America, where different Leishmania species can be endemic (Reithinger and Dujardin, 2007). In the present study, we aimed to verify the identity of 120 VL isolates collected from Bihar and Uttar Pradesh between March 2005 and October 2008. This was done by restriction fragment length polymorphism analysis of an hsp70 PCR-amplified gene fragment (PCR-RFLP), a simple and powerful method we are currently aiming to promote worldwide for Leishmania species identification (Montalvo et al, 2010).
2. MATERIALS AND METHODS
2.1 Cultured isolates and patients
One hundred and twenty individuals (all from India, 107 from the high endemic zone of Bihar and 13 from the low endemic zone of Uttar Pradesh), having symptoms of Kala-azar, were subjected to splenic aspiration at the Kala-Azar Medical Research Centre (KAMRC) and Banaras Hindu University (BHU) with prior consent, between March 2005 and October 2008. Splenic aspirates were collected in NNN tubes containing biphasic media (Blood-agar and c-RPMI) in sterile conditions and cultured at 26°C for a maximum of 3 passages before DNA isolation. Ethical clearance was obtained from the ethical committee of the Institute of Medical Sciences, BHU, Varanasi.
2.2 Growth curve of parasites
This was done for each strain by inoculating 100 μL of parasite culture containing 1 × 106 parasites in 5 mL of c-RPMI and maintaining it for 7 days. The parasites were counted daily using a hemocytometer under a light microscope to evaluate parasite growth. The data were log transformed and linear regression and calculation of the slopes was done using GraphPad Prism 5.0 software (San Diego, CA).
2.3 Molecular analyses
DNA was extracted from cultures and mice spleen with a standard phenol-chloroform method. Hsp70 and 18S rRNA genes were amplified from culture DNA as described elsewhere (Montalvo et al, 2010; Meredith et al., 1993), using primers R221 and R332 for the 18S rRNA gene (Van Eys et al., 1992). An rDNA-ITS1 fragment (approximately 320 bp in L. donovani) was directly amplified from spleen isolated DNA as described elsewhere (el Tai et al., 2000). Amplified DNA from the hsp70 gene was subjected to restriction digestion with HaeIII and the strain MHOM/NP/02/BPK043/0 (Rijal et al., 2007) was used as reference for L. donovani identification. We sequenced DNA from hsp70 and 18S rRNA genes.
2.4 Sequence analysis
Sequences from 18S rRNA and hsp70 genes were aligned to available GenBank accessions from several trypanosomatid species. Based on these alignments, sequences were clustered using the Neighbor-Joining method from calculated p-distances, whereby 2000 bootstrap replicates were analyzed(Saitou and Nei, 1987). The software package MEGA 4.0 was used for aligning the sequences and constructing the dendrograms (Tamura et al., 2007). The 18S rRNA genes were analyzed in two steps: a first tree included an extended set of organisms, and was based on a selection of 368 conserved alignment positions. In a second step, the entire alignment (665 sites) was used to build a tree only including the organisms most closely related to BHU151 and BHU154, as among these also the more variable positions could be reliably aligned. The hsp70 alignment included a geographically and genetically representative set of all known Leishmania species or complexes, and consisted of 1234 sites, which were all used in the tree building process.
2.5 In vivo studies
Six Balb/c mice (30 ± 5g, IMS, BHU, India) were used for in vivo infections with 108 parasites in stationary growth phase. BHU151 and BHU154 clinical isolates were each inoculated in two mice, while two other mice were injected with isolate BHU193. Inoculation was done intracardially, and after 45 days the surviving mice were euthanized and their splenic and liver tissue was collected for DNA isolation (Manandhar et al., 2008).
3. RESULTS
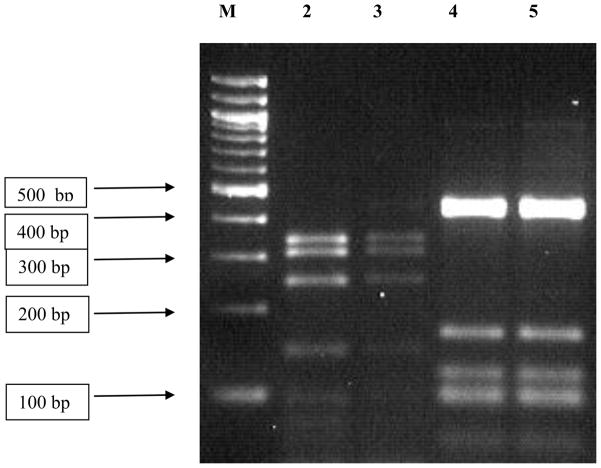
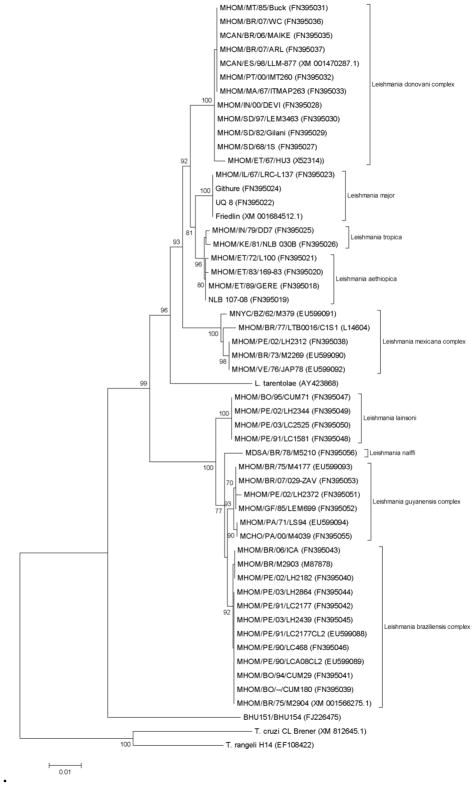
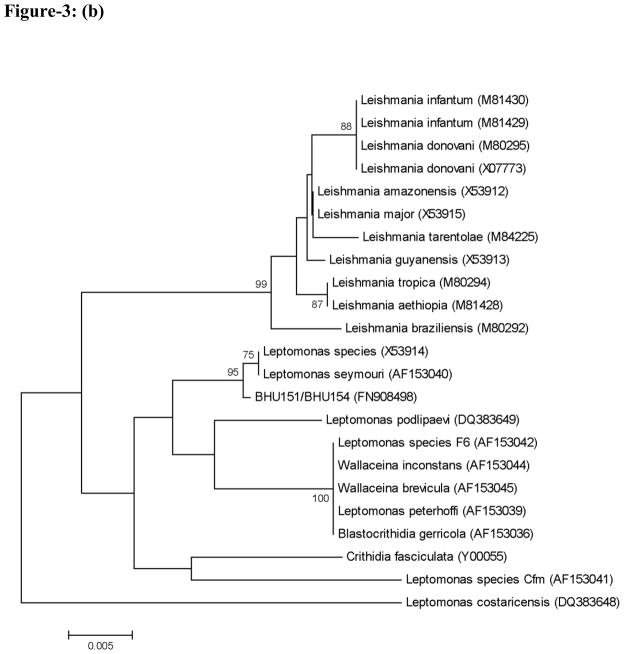
Two patterns were observed from hsp70 PCR-RFLP among the 120 isolates here studied (Figure 1): (i) 111 isolates showed the same pattern as the one encountered in the L. donovani reference strain MHOM/NP/02/BPK043/0, and (ii) 9 isolates (6 from Bihar and 3 from Uttar Pradesh) showed an identical restriction pattern different from L. donovani. Analysis of the hsp70 gene sequence in two of these (GenBank accession no. FJ226475) revealed that they do not correspond to any Leishmania or Sauroleishmania (Figure 2). The nucleotide dissimilarity (p-distance) to the most closely related Leishmania sequence is 7.9 %, while the variation seen between any 2 Leishmania sequences is maximum 5.3%. In order to identify the organism from which this aberrant hsp70 gene was amplified, 18S rRNA gene sequences were used for clustering analysis (Figure 3). According to Figure 3a, constructed from the conserved regions in the 18S rRNA gene fragment studied, BHU151 and BHU154 are most closely related to Leptomonas accessions AF153040 and X53914. This was confirmed with a bootstrap support of 95% in Figure 3b, which is based upon the entire 18S rRNA gene fragment. We thus further refer to the 9 Indian isolates as Leptomonas sp. BHU, being conscient that a future revision of this status is warranted along with the revision of the genus Leptomonas itself(Yurchenko et al., 2006).
Figure 1.
Agarose gel analysis of hsp70 PCR-RFLP from different parasite isolates. The PCR products of 1420 bp were digested with the restriction enzyme HaeIII. Digested products were separated on 3% Small Fragment Agarose (Eurogentec, Belgium). Lane M is a 100 bp Ladder (New England Biolabs,UK). Lanes 2 and 3: L. donovani BHU193 and MHOM/NP/02/BPK043/0 respectively, a pattern found in 111 out of the 120 studied isolates. Lanes 4 and 5: BHU250 and BHU234, respectively, a pattern found in 9 out of the 120 studied isolates.
Figure 2.
Neighbour-Joining tree based on p-distances of the hsp70 sequences of Leishmania (Fraga et al., 2010), and BHU151/BHU154 (both of which have the same sequence); outgroup: Trypanosoma cruzi and T. rangeli. The analysis is based on an alignment of 1234 nucleotides. GenBank accession numbers are given between brackets. Distances are measured along the horizontal branches, according to the scale shown. Bootstrap values above 70% are indicated at the internodes.
Figure 3.
Neighbour-Joining trees based on p-distances of partial 18S rRNA gene sequences. GenBank accession numbers are given between brackets. Distances are measured along the horizontal branches, according to the scale shown. Bootstrap values above 70% are indicated at the internodes, and the root was placed arbitrarily. (A) Tree based on conserved alignment positions including a more extended set of taxa. (B) Tree based on the entire sequenced gene fragment.
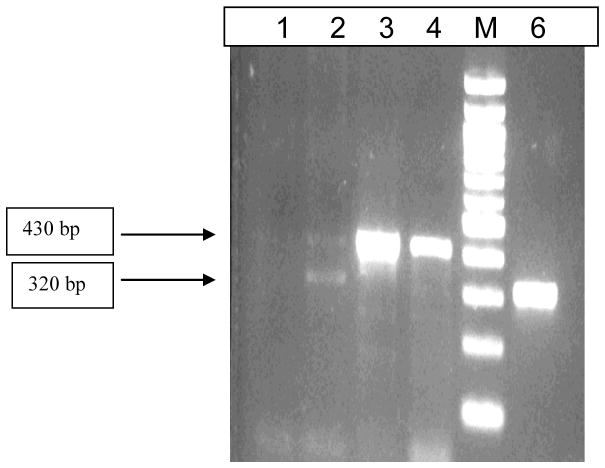
To evaluate whether Leishmania is still present in the cultures from these isolates, one L. donovani isolate (BHU193) and two Leptomonas sp. BHU isolates (BHU151 and BHU154) were injected in Balb/c mice. All the mice inoculated with isolate BHU154 died after 20 days. Spleen dabbed smears were found positive for amastigote parasites by microscopy in the mice that were infected with BHU151. At day 45 after infection, mice were sacrificed and DNA was isolated from their splenic tissue. Given the low sensitivity of hsp70 PCR-RFLP for direct application on tissue samples, rDNA-ITS1 PCR was applied (Figure 4). This revealed that the parasites present in the mice 45 days after infection showed the signals of both Leptomonas sp. BHU (approximately 430 bp) and L. donovani (approximately 320 bp).
Figure 4.
rDNA-ITS1 PCR products from splenic tissue of mice, showing fragments from Leptomonas sp. BHU and L. donovani. Lane M is 100 bp Ladder, Lane 1: mouse liver injected with Leptomonas sp. BHU isolate BHU151; Lane2: mouse spleen injected with Leptomonas sp. BHU isolate BHU151; Lanes 3 and 4: Leptomonas sp. BHU cultures BHU151 and BHU154; Lane 6: L. donovani reference isolate BHU193.
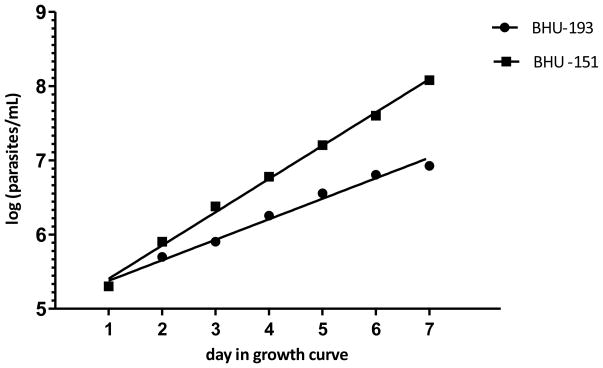
In order to compare the growth behavior of L. donovani and Leptomonas sp. BHU, we established the growth curve of isolates BHU193 and BHU151 (Figure 5). We found that isolate BHU151 showed unique features of multiplication. After 5 days, BHU151 shows a maximum density of 1.6 × 107 parasites/mL whereas L. donovani BHU193 showed 3.6 × 106 parasites/mL. After log transforming the growth data and applying linear regression, Leptomonas sp. BHU showed a slope of 0.01868 (95% CI: 0.01733 to 0.02004), which is significantly higher than the slope for L. donovani of 0.01151 (95% CI: 0.009921 to 0.01311). Therefore it is clearly evidenced by the growth curves that Leptomonas sp. BHU grows faster in culture than L. donovani.
Figure 5.
Growth Curve of Leptomonas sp. BHU (BHU151) and L. donovani (BHU193). The concentration of parasites per mL is log transformed. The slope of BHU193 and BHU151 is respectively 0.01151 (95% CI: 0.009921 to 0.01311) and 0.01868 (95% CI: 0.01733 to 0.02004).
4. DISCUSSION
The molecular characterization of 9/120 VL isolates (7.5%) originating from Bihar and Uttar Pradesh and maintained in vitro for 3 passages, revealed that they correspond to parasites different from L. donovani and phylogenetically clustered more closely to monoxenous parasites of the Leptomonas genus (further called Leptomonas sp. BHU). The identification of the parasites here described should be considered with some care. Indeed, the taxonomy of the insect trypanosomatids requires new criteria other than morphology and 18S rRNA gene based phylogenies to re-evaluate the generic affinities of known isolates (Podlipaev et al., 2004). Surprisingly, after in vivo mice infection with one of these isolates, Leptomonas sp. BHU parasites were encountered in the spleen of the animals together with L. donovani. Assuming that sterility was guaranteed all along the chain from the isolation from patients, the in vitro maintenance and the harvesting for DNA extraction and inoculation, we consider that mixed infection was present in these isolates, and hence also in the patients from whom these were derived.
The occurrence of insect trypanosomatids in humans is exceptional, but reports are worldwide available, involving protozoan parasites genetically related with several different species such as Herpetomonas sp. in Texas (McGhee and Cosgrove, 1980), Herpetomonas samuelpessoai in the south of France (Morio et al., 2008), Leptomonas pulexsimulantis in Brazil (Pacheco et al., 1998), or a yet unidentified trypanosomatid in Spain (Jimenez et al., 1996). Also in other mammals similar observations were described, e.g. Herpetomonas ztiplika-like parasites in rats from Egypt (Podlipaev et al., 2004). Interestingly, most of the human cases in which insect trypanosomatids were encountered were HIV positive (Chicharro and Alvar, 2003), even though a symptomatic infection with such trypanosomatids was also seen in immunocompetent patients (Boisseau-Garsaud et al., 2000).
Our report is exceptional in the sense that it concerns 9 different patients from different regions of India. Furthermore, none of them showed HIV/AIDS, but VL-diagnosis was confirmed by the rK39 strip test (Sundar et al., 2006) and clinical data such as complete blood count, platelet count, and splenic smear microscopy. VL is known to be associated with a strong immuno-suppression and as such it might allow non-human trypanosomatids to install in the body. After isolation from this type of samples, the insect parasites would rapidly overgrow L. donovani in culture, because of their high multiplication rate (Figure 5). Back in a mammal host (mice in this study), L. donovani would reappear because of higher fitness in that host.
How the patients got infected with these insect parasites is still unknown, but sandflies themselves might be involved, as suggested by the recent description of L. donovani as well as other -yet unidentified- trypanosomatids in Nepalese specimens of Phlebotomus argentipes, the main vector of VL in the Indian sub-continent (Bhattarai et al., 2009). Further studies are needed to get a more extensive perception of this phenomenon. However, considering our results, we strongly recommend to undertake a precise species identification of any in vitro maintained isolate from a VL patient if it is to be used for further studies of L. donovani, even in regions where only L. donovani is reported to be endemic.
Acknowledgments
This work was supported by NIAID, NIH Grant Number 1P50AI074321-01, the EC-funded project Kaladrug-R (FP7, Contract 222895) and by the Directorate-General for Development Cooperation of the Belgian Government (third framework agreement with ITMA). Authors sincerely acknowledge Prof. Rajeev Raman, Cytogenetics laboratory, Department of Zoology, Banaras Hindu University, Varanasi for providing the Genetic Analyzer facility. VKP is thankful to ICMR (New Delhi, India) for providing financial assistance. MV has a fellowship of the Agency for Innovation by Science and Technology in Flanders (IWT)
Footnotes
ETHICAL APPROVAL
Ethical clearance to conduct the project was obtained from the ethical committee of the Institute of Medical Sciences, Banaras Hindu University.
CONFLICT OF INTEREST:
No conflict
References
- Alam MZ, Kuhls K, Schweynoch C, Sundar S, Rijal S, Shamsuzzaman AK, Raju BV, Salotra P, Dujardin JC, Schonian G. Multilocus microsatellite typing (MLMT) reveals genetic homogeneity of Leishmania donovani strains in the Indian subcontinent. Infect Genet Evol. 2009;9:24–31. doi: 10.1016/j.meegid.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Bhattarai NR, Das ML, Rijal S, Van der Auwera G, Picado A, Khanal B, Roy L, Speybroeck N, Berkvens D, Davies CR, Coosemans M, Boelaert M, Dujardin JC. Natural infection of Phlebotomus argentipes with Leishmania and other trypanosomatids in a visceral leishmaniasis endemic region of Nepal. Trans R Soc Trop Med Hyg. 2009;103:1087–1092. doi: 10.1016/j.trstmh.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Boisseau-Garsaud AM, Cales-Quist D, Desbois N, Jouannelle J, Jouannelle A, Pratlong F, Dedet JP. A new case of cutaneous infection by a presumed monoxenous trypanosomatid in the island of Martinique (French West Indies) Trans R Soc Trop Med Hyg. 2000;94:51–52. doi: 10.1016/s0035-9203(00)90435-8. [DOI] [PubMed] [Google Scholar]
- Chicharro C, Alvar J. Lower trypanosomatids in HIV/AIDS patients. Ann Trop Med Parasitol. 2003;97(Suppl 1):75–78. doi: 10.1179/000349803225002552. [DOI] [PubMed] [Google Scholar]
- Desjeux P. Human leishmaniases: epidemiology and public health aspects. World Health Stat Q. 1992;45:267–275. [PubMed] [Google Scholar]
- el Tai NO, Osman OF, el Fari M, Presber W, Schonian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg. 2000;94:575–579. doi: 10.1016/s0035-9203(00)90093-2. [DOI] [PubMed] [Google Scholar]
- Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol. 10:238–245. doi: 10.1016/j.meegid.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Jimenez MI, Lopez-Velez R, Molina R, Canavate C, Alvar J. HIV co-infection with a currently non-pathogenic flagellate. Lancet. 1996;347:264–265. doi: 10.1016/s0140-6736(96)90441-9. [DOI] [PubMed] [Google Scholar]
- Manandhar KD, Yadav TP, Prajapati VK, Kumar S, Rai M, Dube A, Srivastava ON, Sundar S. Antileishmanial activity of nano-amphotericin B deoxycholate. J Antimicrob Chemother. 2008;62:376–380. doi: 10.1093/jac/dkn189. [DOI] [PubMed] [Google Scholar]
- McGhee RB, Cosgrove WB. Biology and physiology of the lower Trypanosomatidae. Microbiol Rev. 1980;44:140–173. doi: 10.1128/mr.44.1.140-173.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith SE, Zijlstra EE, Schoone GJ, Kroon CC, van Eys GJ, Schaeffer KU, el-Hassan AM, Lawyer PG. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch Inst Pasteur Tunis. 1993;70:419–431. [PubMed] [Google Scholar]
- Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, Van der Auwera G. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology. 2010;137:1159–1168. doi: 10.1017/S0031182010000089. [DOI] [PubMed] [Google Scholar]
- Morio F, Reynes J, Dollet M, Pratlong F, Dedet JP, Ravel C. Isolation of a protozoan parasite genetically related to the insect trypanosomatid Herpetomonas samuelpessoai from a human immunodeficiency virus-positive patient. J Clin Microbiol. 2008;46:3845–3847. doi: 10.1128/JCM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco RS, Marzochi MC, Pires MQ, Brito CM, Madeira Mde F, Barbosa-Santos EG. Parasite genotypically related to a monoxenous trypanosomatid of dog's flea causing opportunistic infection in an HIV positive patient. Mem Inst Oswaldo Cruz. 1998;93:531–537. doi: 10.1590/s0074-02761998000400021. [DOI] [PubMed] [Google Scholar]
- Podlipaev SA, Sturm NR, Fiala I, Fernandes O, Westenberger SJ, Dollet M, Campbell DA, Lukes J. Diversity of insect trypanosomatids assessed from the spliced leader RNA and 5S rRNA genes and intergenic regions. J Eukaryot Microbiol. 2004;51:283–290. doi: 10.1111/j.1550-7408.2004.tb00568.x. [DOI] [PubMed] [Google Scholar]
- Reithinger R, Dujardin JC. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol. 2007;45:21–25. doi: 10.1128/JCM.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal S, Yardley V, Chappuis F, Decuypere S, Khanal B, Singh R, Boelaert M, De Doncker S, Croft S, Dujardin JC. Antimonial treatment of visceral leishmaniasis: are current in vitro susceptibility assays adequate for prognosis of in vivo therapy outcome? Microbes Infect. 2007;9:529–535. doi: 10.1016/j.micinf.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sundar S, Singh RK, Maurya R, Kumar B, Chhabra A, Singh V, Rai M. Serological diagnosis of Indian visceral leishmaniasis: direct agglutination test versus rK39 strip test. Trans R Soc Trop Med Hyg. 2006;100:533–537. doi: 10.1016/j.trstmh.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Van Eys GJ, Schoone GJ, Kroon NC, Ebeling SB. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol Biochem Parasitol. 1992;51:133–142. doi: 10.1016/0166-6851(92)90208-2. [DOI] [PubMed] [Google Scholar]
- Yurchenko VY, Lukes J, Jirku M, Zeledon R, Maslov DA. Leptomonas costaricensis sp. n (Kinetoplastea: Trypanosomatidae), a member of the novel phylogenetic group of insect trypanosomatids closely related to the genus Leishmania. Parasitology. 2006;133:537–546. doi: 10.1017/S0031182006000746. [DOI] [PubMed] [Google Scholar]