Abstract
Secondary active transporters catalyze concentrative transport of substrates across lipid membranes by harnessing the energy of electrochemical ion gradients. These transporters bind their ligands on one side of the membrane, and undergo a global conformational change to release them on the other side of the membrane. Over the last few years, crystal structures have captured several bacterial secondary transporters in different states along their transport cycle, providing insight into possible molecular mechanisms. In this review, we will summarize recent findings focusing on the emerging structural and mechanistic similarities between evolutionary diverse transporters. We will also discuss the structural basis of substrate binding, ion coupling and inhibition viewed from the perspective of these similarities.
Introduction
Channels and transporters are membrane proteins that catalyze passage of solutes across lipid membranes. Channels form pores selective for ions and small molecules such as water or urea, which permeate at rates approaching the diffusion limit. The turnover cycle of transporters is significantly slower and involves isomerization between states, in which substrate binding sites are accessible from one side of the membrane or the other (Box 1). They accept a broad range of solutes, including ions, neurotransmitters, nutrients, and numerous drugs. Primary active transporters couple substrate movements to a source of chemical energy, such as ATP-hydrolysis, reduction-oxidation reactions or light. Passive and secondary active transporters are driven only by the electrochemical gradients of the transported species. These transporters facilitate diffusion of a single solute, dissipating its concentration gradient, or couple thermodynamically unfavorable concentrative transport of a solute to energetically favorable dissipative movements of others.
Box 1.
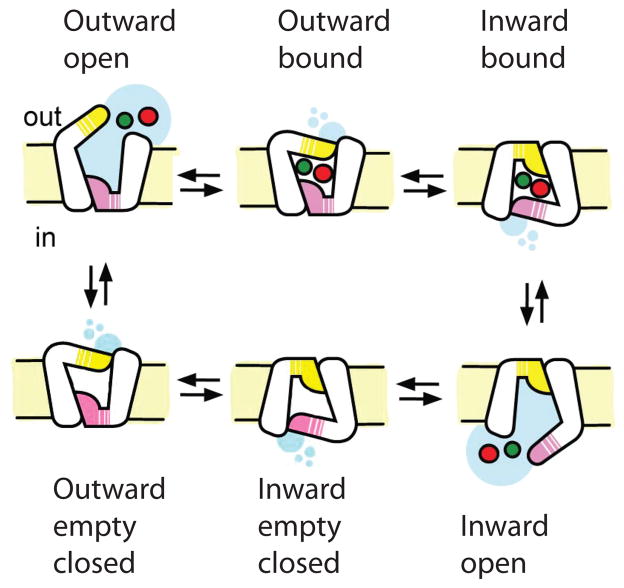
In early 1950s, a saturable catalytic carrier mechanism was proposed to explain the experimentally measured rates of trans-membrane solute transport which were inconsistent with simple diffusion [78, 79]. As the carriers were unlikely to diffuse across membranes, the alternating access mechanism was formulated, according to which the carrier or transporter isomerized between an outward facing state with a centrally located substrate binding site accessible from the external solution, and an inward facing state with the site accessible from the cytoplasm [80, 81]. Shortly after, coupled active sugar/protons and sugar/sodium transport was reported [82, 83]. Two conceptual models prevailed – the “rocker-switch” and the “gated pore”. The former focused on the rocking movements of protein domains pivoting at the substrate-binding site. The latter emphasized local motions of outside and inside gates, flanking the substrate-binding site and undergoing alternating openings. Recent studies suggest that the transport mechanisms of secondary transporters likely integrate both conformational modes, therefore including an isomerization between the outward and inward facing states, as well as local opening and closure of the gates that partially or completely occlude bound substrates (Box 1, Figure 1). Additional intermediates may occur between inward and outward facing states either in the substrate-bound or substrate-free brunch of the cycle. Importantly, thermodynamics dictates that transport is driven only by the difference of the electrochemical potentials of the solutes on two sides of the membrane, and is independent of the path. Hence, the number and relative stability of the involved states may vary as long as none of the intermediates has a free energy that is too high or too low, otherwise yielding high kinetic barriers. Supporting this view, various transporters crystallized in different states, which provide structural snapshots along the transport cycle.
Secondary transporters have been classified into distinct families based on their sequence and functional similarities [1, 2]. However, many of these families are evolutionarily and structurally related [3–5], suggesting that a few membrane protein folds have successfully evolved to provide widely divergent functionalities [6, 7]. For example, several transporter families with little or no detectable sequence similarity assume the same structural fold, typified by the sodium-coupled leucine transporter LeuT, a bacterial homologue of the neurotransmitter sodium symporters (NSS) [8–15]. The major facilitator superfamily (MFS) represents another common fold, encompassing up to 25 % of all transporters [16–18]. By contrast, other folds, like those of GltPh [19] and NhaA [20], the prokaryotic homologues of mammalian glutamate transporters and sodium/hydrogen antiporters, respectively, seem to represent relatively small groups of families [4, 21, 22].
In this review, we discuss structural data on secondary transporters. We focus primarily on transporters with MFS, LeuT and GltPh folds because they have been captured by crystallography in distinct states, providing snapshots of the long-standing alternating access mechanism. We emphasize architectural and mechanistic similarities, as well as the structural basis of substrate specificity, ionic coupling and transport inhibition.
Structural repeats
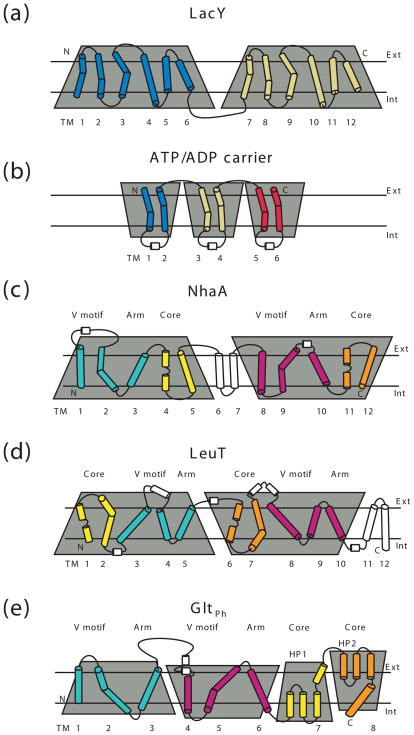
A common feature shared by all secondary transporters is the presence of the structural repeats. Each repeat typically comprises several contiguous trans-membrane segments (TMs), which assume a specific three-dimensional fold, repeated two or three times (Figure 2). Structural repeats are thought to arise from gene duplication followed by fusion and sequence divergence [23], which make such repeats often impossible to detect until the three-dimensional structures are determined. Interestingly, carriers, such as the zinc transporter YiiP [24] and multidrug resistance protein EmrE [25, 26], may represent ancient functional homodimers, preceding gene duplication. A glimpse at possible evolutionary molecular events is provided by studies on EmrE, whose protomers can assemble in parallel or antiparallel orientations [27, 28].
Figure 2. Membrane topology of transporters with parallel and inverted structural repeats.
The topologies are shown for the MFS transporter LacY (a), ATP/ADP carrier (b), NhaA (c), LeuT (d) and GltPh (e). The structural repeats are highlighted by the shaded trapezoids, which emphasize their relative orientation. The N- and C-terminal TM segments comprising the transporter cores in GltPh, NhaA and LeuT are colored yellow and orange, respectively. The remaining N- and C-terminal portions of the repeats are colored cyan and magenta, respectively. The segments that are not part of the symmetrical repeats are white. Non-helical regions in the middle of the core TMs are shown as lines.
Structural repeats with parallel orientations are related by a pseudo-symmetry axis perpendicular to the membrane plain. In MFS transporters, such repeats form two 6 TM bundles, each containing two 3 TM anti-parallel repeats, suggesting that MFS transporters evolved from a 3 TM ancestral unit [29]. In the mitochondrial ATP/ADP carrier a 2 TM unit is repeated three times, yielding a three-fold pseudo-symmetric structure [30]. So-called “inverted” structural repeats with anti-parallel orientation and a pseudo-symmetry axis parallel to the membrane have been found in transporters with LeuT fold, NhaA and GltPh (reviewed in 31, 32). The crystal structures of these apparently unrelated proteins reveal a striking similarity. LeuT and NhaA contain 5 TM repeats, and GltPh contains a 3 TM repeat followed by a repeat of an unusual motif: a reentrant helical hairpin (HP) and a TM (Figure 2). The first two TMs within each repeat in LeuT, and the last two TMs of each repeat in NhaA form 4 TM transporter cores (Figures 3 and 4). Notably, the first TM of each repeat in the core domain contains an unwound region in the middle of the membrane, which plays a critical role in ligand binding and transport. In GltPh the core consists of two HP-TM motifs that are central to substrate and ion binding. Intriguingly, in all these transporters, the remaining three adjacent TMs of each repeat form similar structural motifs. The first two TMs of each repeat give rise to interlocking V-shaped motifs (Figure 3). The third “arm” TM of each repeat extends from the V-motif encasing the core domain. This architectural similarity suggests a possible common ancestry and mechanistic parallels. Of note, other transporters and channels, including chloride/proton antiporters [33], aquaporins [34], ammonia [35] and urea channels [36], also contain inverted structural repeats. However, the TMs of one repeat do not interlace with the TMs of the other, but instead the TMs in each repeat coalesce yielding pseudo-symmetric halves.
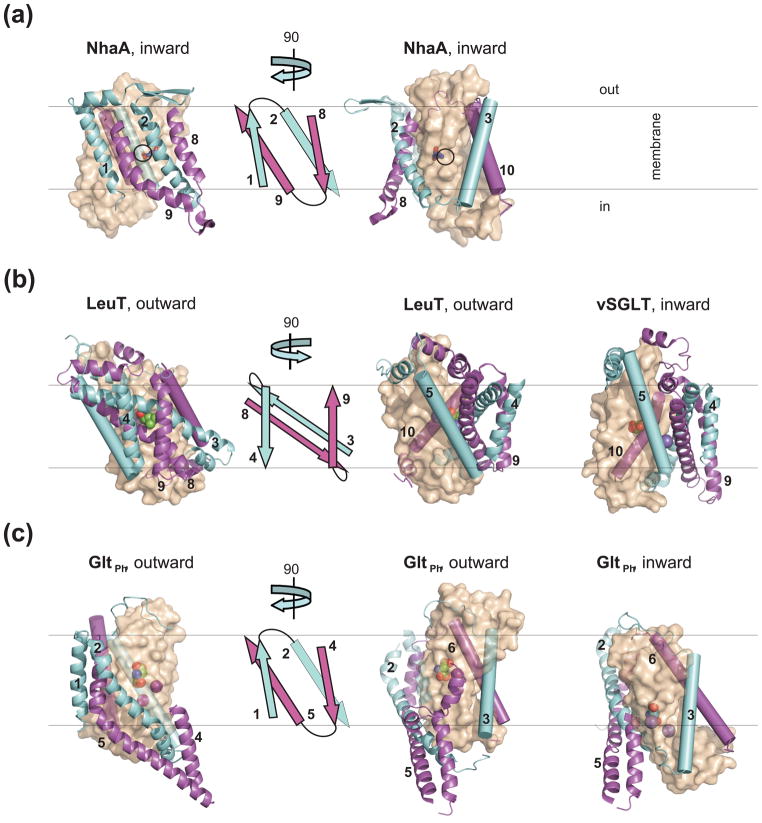
Figure 3. Architecture of the transporters with inverted structural repeats.
(a) NhaA (PDB code 1zcd); (b) LeuT in the outward-facing state (PDB code 2a65) and vSGLT in the inward-facing state (PDB code 3dh4); (c) GltPh in the outward- (PDB code 2nwx) and inward- (PDB code 3kbc) facing states. The leftmost panels show a single protomer viewed in the membrane plane from the side of the V-motifs with cartoon topologies of the V-motifs shown immediately to the right. In the right panels the structures are rotated by about 90 °, as indicated by the arrows. The V-motifs and the arms TM-s are colored as in Figure 2 and shown in cartoon representation and as cylinders, respectively. The cores are shown in transparent surface representation and colored wheat; the bound substrates and ions are shown as spheres. In NhaA, D164 is shown as sticks and black circles indicate expected site of ion binding. This and other figures were prepared in PyMol [84].
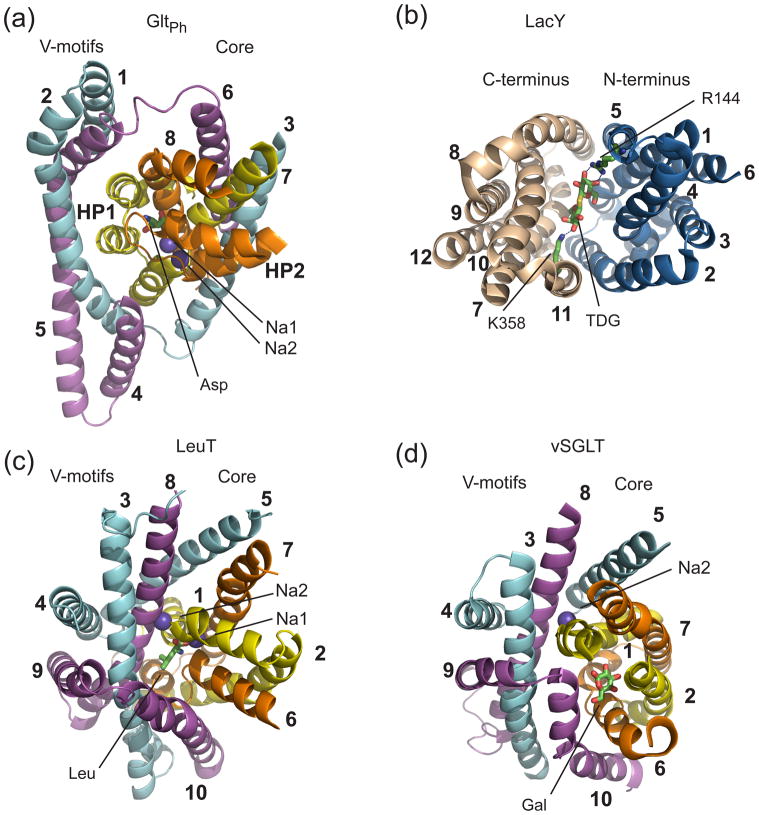
Figure 4. Binding sites.
(a) GltPh; (b) LacY (PDB code 1pv7); (c) LeuT; (d) vSGLT. Transporters are shown in cartoon representation and colored as in Figure 2. LeuT and vSGLT are viewed from the extracellular space and LacY is viewed from the cytoplasm; GltPh is tilted to reveal the binding site. The TMs that are not part of the symmetrical repeats, extracellular loops, and parts of TM helices are removed for clarity. The substrates and two coordinating residues in LacY are shown stick representation and bound Na+ ions are shown as purple balls.
Molecular basis of the alternating access mechanism
The molecular mechanisms of secondary transporters appear to be deeply rooted in the symmetry of the structural repeats. The mechanism of MFS transporters is best understood in the context of the bacterial lactose permease LacY, and the glycrol-3-phosphate transporter GlpT [16,17]. Their structures reveal large aqueous vestibules formed between the symmetrical 6 TM bundles, open to the cytoplasm, with the substrate-binding sites located at the apex, approximately halfway across the membrane. It has been postulated that the rocking motions of the bundles, pivoting around the bound substrate, provide alternating access to the binding site [37, 38]. Structures of two MFS transporters, the multidrug resistance transporter EmrD [18] and oxalate transporter OxlT [29], capture considerably more compact states, in which the substrate-binding sites are occluded from both sides of the membrane, suggesting that transitions between outward and inward facing states may not be two-state processes only and may proceed via intermediate states [29].
Aqueous vestibules leading to the substrate-binding sites, either from the extracellular or intracellular sides of the membrane, are also observed in LeuT fold, GltPh and NhaA (Figures 3 and 4). However, these are not formed between the pseudo-symmetrical structural components, but between the V-motifs and the cores. While the symmetry-related V-motifs are similarly oriented relative to the membrane, the core repeats are oriented differently with respect to the membrane and the V-motifs. In LeuT, the N-terminus of TM1 (LeuT TM numbering is used from hereafter for all LeuT fold transporters) and the C-terminus of the symmetry-related TM6 form extensive interactions with the V-motifs at the cytoplasmic side, while their respective C- and N-termini lean away from the V-motifs, yielding an extracellular vestibule (Figure 3). This remarkable asymmetry led Forrest and colleagues to propose that isomerization of the transporter into an inward facing state would occur upon the repeats swapping their conformations [39]. The swap results in the extracellular termini of TM1 and TM6 packing against the V-motifs, and the intracellular termini leaning away, yielding the closure of the extracellular vestibule and opening of a cytoplasmic vestibule. Consistently, the structure of the bacterial LeuT fold sodium/galactose symporter vSGLT, captured in the inward facing, state shows a conformation similar to the prediction (Figure 3) [9]. Since then, structures of other transporters with LeuT fold have been determined. The agmatine/arginine exchanger AdiC [11, 13] and nucleobase/cation symporter Mhp1 [10] were crystallized in outward facing states, while the L-carnitine/γ-butyrobetaine antiporter CaiT was in the inward facing state [15]. The sodium/betaine symporter BetP [14] and proton-dependent amino acid transporter ApcT [12] were captured in intermediate states, although closer to the inward facing state. Remarkably, Mhp1 has also been crystallized in the inward facing state, providing particularly detailed information on the conformational transitions [40]. Overall, these structures reveal variable orientations of the core relative to the V-motifs, and thus provide evidence for a rocking-like movement of domains pivoting roughly around the substrate-binding site.
The first structure of GltPh in complex with aspartate revealed the transporter in the outward facing state (Figure 3) [19]. The inward facing state was then trapped by intra-molecular cross-linking a double cysteine mutant [41, 42]. Its structure shows that the V-motifs remain largely unaltered while the core together with two arm TMs 3 and 6 traverse ~15 Å of the lipid bilayer toward the cytoplasm (Figure 3). In the outward facing state, the first core repeat unit (HP1-TM7) interacts with the V-motifs, while the second (HP2-TM8) lines the extracellular vestibule. As in the LeuT-fold transporters, the inward facing state forms upon the two core repeats swapping their conformations, with HP2-TM8 packing against the V-motifs, and HP1-TM7 lining the intracellular vestibule. Consistently, modeling based on these symmetry considerations has predicted similar conformational changes [43].
A common mechanistic aspect of GltPh and LeuT fold transporters is the movement of the core relative to the V-motifs. It is tempting to hypothesize that similar conformational changes also underlie the mechanism of NhaA. Indeed, residues important for pH-dependent activation of NhaA map to the interface between the V-motifs and the core [44], suggesting that relative movements of these domains are involved in activation and/or transport. Interestingly, in GltPh and NhaA, the V-motifs participate in subunit oligomerization [44–46]. These oligomeric interactions may stabilize the V-motifs within the membrane, and suggest that, upon conformational transitions, their positions remain fixed, while the cores re-orient, which is consistent with the observation that protomers can function independently [47, 48]. Conversely, BetP and CaiT oligomerize via peripheral helices adjacent to the core, suggesting that the V-motifs could be the moving component. For monomeric transporters like vSGLT, Mhp1 and ApcT, it is possible that both the core and the V-motifs re-orient relative to the membrane during transport.
Substrate-binding sites
Do conformational transitions between outward and inward facing states affect the ligand binding sites? Evidence thus far is mixed. In GltPh the substrate-binding site forms entirely within the core (Figures 3 and 4), therefore trans-membrane movements of the core do not affect the network of hydrogen bonds and ionic interactions underlying the selectivity and affinity of GltPh for its substrate. Similarly, sites D163 and D164, which are critical for proton and Na+ binding in NhaA, are buried within the core (Figure 3). In contrast, in transporters with MFS and LeuT folds, the substrate-binding sites are located at the interfaces between structural components, which probably move during transport.
In LacY, the substrate-binding site is positioned between the two pseudo-symmetrical bundles with N-terminal bundle coordinating one galactopyranosyl moiety of the substrate, β-D-galactopyranosyl-thio-β-D-galactopyranoside (TDG), and the C-terminal bundle coordinating the other (Figure 4) [16]. Two positively charged residues, R144 and K358, located in the symmetrical TMs 5 and 11, are hydrogen-bonded to disaccharide hydroxyls. Despite this apparent symmetry, the N-terminal bundle plays a greater role in establishing the transporter affinity and specificity [37]. K358 can be mutated to a neutral amino acid or the coordinated galactose ring replaced with glucose without significant effects on substrate affinity or transport. In contrast, R144 and the spatially adjacent E126 and W151 are among the few amino acids essential for transport and specificity [37]. It is possible that as the bundles undergo a rocking motion, the N-terminal part remains unaffected, while the C-terminal part undergoes changes without detrimental effects on substrate binding.
The substrate-binding sites in the outward facing LeuT, AdiC and Mhp1 also have a bipartite nature, with half-sites provided by the cores and the V-motifs (Figure 4). The substrates bind near the unwound regions of the core TMs 1 and 6, which form a pocket. In LeuT, the main chain amides and carbonyl oxygen atoms of these non-helical regions coordinate the carboxyl- and amino-groups of the substrate, respectively, and the V-motifs present a patch of hydrophobic residues selective for the apolar side chains [8, 49]. Strikingly, AdiC binds arginine in exactly the same manner, except that the guanidinium group forms cation-π interactions with the essential V-motif W293 [50]. In Mhp1, L-5-benzyl-hydantoin binding site also overlaps closely with the location of the substrate-binding site in LeuT, with the core W220 and the V-motif W117 forming π-stacking interactions with the benzyl and hydantoin moieties, respectively [10]. However, how do these binding sites rearrange upon transition between outward and inward facing states is not known. Remarkably, in the inward facing vSGLT and CaiT, and in the intermediate BetP, the position and architecture of the substrate-binding sites are distinct from what is observed in the outward facing LeuT, AdiC and Mhp1 [9, 15, 14]. The binding pockets form deeper within the core, so that not only TMs 1 and 6, but also the peripheral TMs 2 and 7 are involved in substrate coordination (Figure 4). Furthermore, when viewed from the extracellular side, the cores are rotated counterclockwise, so that the V-motifs no longer contribute to substrate coordination, and the substrates are laterally shifted compared to the leucine position in LeuT. In vSGLT, the extent of this rotation is the greatest, and the bound sugar faces the arm TM10, and is about 8 Å away from the V-motifs (Figure 4).
In addition to these primary sites, secondary substrate binding sites have been postulated for LeuT and CaiT. Simulations suggested that an additional substrate binding site is located in the extracellular vestibule of LeuT [51]. It was termed the “symport-effector” site, and it was hypothesized that substrate binding there prompts vestibule closure and release of the substrate from the primary site into the cytoplasm [51]. Interestingly, in the inward facing CaiT, carnitine binds not only at the primary but also at a secondary site located in the intracellular vestibule [12]. Strikingly, mutations near these secondary sites in the extracellular and intracellular vestibules of LeuT and CaiT, respectively, abolished transport [12, 51].
To complete the transport cycle, empty transporters must return to the outward facing state (Figure 1). Interestingly, in the ApcT structure, the postulated substrate-binding site is occluded from solvent, but contains water molecules instead of the substrate [12]. Whether transport of water during the backstroke will turn out to be a common mechanism is not known, however it would provide a universal means to compensate thermodynamically for the lack of bound polar ligands.
Figure 1. Alternating access mechanism for an ion/substrate symport.
The cycle starts with an empty outward facing transporter, which binds the substrate and coupled ion (red and green spheres, respectively) from the extracellular solution in a reaction coupled to the closure of the gate (yellow). The transporter undergoes a conformational transition into an inward facing state in which the extracellular gate can no longer open while opening of the intracellular gate (pink) is enabled. The release of the substrate and ion into the cytoplasm yields an empty inward facing state, and isomerization back into the outward facing state completes the cycle.
Gates
In addition to global structural transitions between outward and inward facing states, transporters also undergo local conformational changes in response to ligand binding. These transitions are sometimes conceptualized in terms of opening and closure of “gates”, which completely or partially occlude bound ligands. In GltPh, HP2 occludes aspartate from the extracellular solution, and is captured in the open state in the crystal structure of GltPh in complex with the blocker L-threo-β-benzyloxyaspartate (TBOA) [52]. Its dynamic nature and role as a gate have also been postulated based on molecular dynamics studies [53, 54]. In transporters with LeuT fold, substrates are occluded from the extracellular solution by bulky hydrophobic residues in the extracellular vestibule. For instance, a pair of aromatic residues in LeuT and one tryptophan in AdiC serve as gates [8, 50]. The wider openings of the vestibule, observed in apo AdiC and Mhp1 and a blocker-bound LeuT, is due to the extracellular helical segments of core TM1 and TM6 leaning further away from the V-motifs, and the reciprocal outward bending of the arm TM10. The extent of these movements varies significantly between these transporters. The recent structure of the inward facing state of Mhp1 captures the transporter in an apo state [40]. The substrate-binding site is significantly perturbed, suggesting that substrate binding to this state could also induce a significant conformational change. It seems also possible that the substrate-bound inward facing state is intrinsically unstable and relaxes to the outward facing occluded conformation observed in the earlier structure. By contrast, in LacY, only subtle conformational changes occur upon binding of the ligand, which remains largely exposed to the cytoplasm. Nevertheless, binding still occurs via an induced fit mechanism, as side chain interactions, including a salt bridge between critical coordinating residues, R144 and E269, are broken [55].
Ion coupling
To couple transport of substrates and ions, and prevent dissipative leaks, transporters should only isomerize between outward and inward facing states when both substrate and ion binding sites are loaded or un-occupied (Box 1, Figure 1). Hence, a strict thermodynamic coupling must exist between ligand binding and the formation of a transport-competent conformation.
In GltPh, two Na+-binding (Na) sites have been structurally characterized, while a third site, which has been recently suggested by a 1:3 aspartate to Na+ uptake stoichiometry [56], remains to be located. Two Na sites have been observed in LeuT, and were hypothesized to occur at equivalent positions in BetP. Only one Na site was proposed in vSGLT and Mhp1. Although Na+ and substrate bind in a highly cooperative manner [10, 52, 57], only Na+ ions at the Na1 site of LeuT and the proposed BetP site are directly coordinated by substrates. Other Na+ ions bind at sites distanced by 7–10 Å and therefore must be coupled to substrates via allosteric interactions. The structure of Mhp1 bound to both substrate and Na+ reveals a much greater closure of the extracellular vestibule compared to the transporter bound only to Na+, demonstrating that binding of both species is needed for complete closure of the extracellular gate [10]. In GltPh, the Na2 and substrate-binding sites are positioned between HP2 and the rest of the core, suggesting a structural basis for the strict coupling of Na+ and aspartate binding to the extracellular gate closure [52].
Ion binding within secondary transporters is mediated mostly by the unwound TM regions [58]. The two GltPh Na sites and the Na1 LeuT site are located entirely within the core, and are likely to remain largely unperturbed upon transition of the transporters between outward and inward facing states. In contrast, LeuT Na2 site, expected to be conserved also in BetP, vSGLT and Mhp1, is located at the interface between the core and V-motifs. This site forms between two hydroxyl moieties originating from the V-motif TM8 (T354–S355 in LeuT, T467–S468 in BetP, S364–S365 in vSGLT, S312–T313 in Mhp1) and carbonyl oxygen atoms of the C-terminal end of the cytoplasmic TM1 helix. In the outward facing LeuT and Mhp1, this helix packs against the V-motif forming an occluded Na site with appropriate ion coordination geometry. However, in the inward facing vSGLT and Mhp1, these helices lean away from each other, exposing the Na site to the cytoplasm and disrupting coordination (Figure 3). Molecular dynamic simulations suggest that Na+ is unstable at these positions and that the crystal structures represent ion-releasing states of these transporters [40, 59].
The type of coupled ions and transport stoichiometries vary between evolutionarily related transporters [60] and likely correlate with the nature of the available ion gradients, and the magnitude of the electrochemical gradients of the substrates, respectively. The structure of the proton-dependent amino acid transporter ApcT provides an interesting example of how a Na+-binding site can be converted to a protonation site. In a related mammalian sodium-coupled neutral amino acid transporter 2 (SNAT2), Na+ shown to bind to a site equivalent to Na2 in LeuT [61]. However in ApcT, this site is occupied by the amine moiety of K158, which has an anomalously shifted ionization constant, and likely serves as a proton binding site [12]. Similarly, comparison of the related sodium-coupled BetP and ion-independent CaiT reveals that CaiT harbors an arginine, R262, at a position similar to K158 in ApcT, suggesting that the guanidinium group could replace Na+ at the Na2 site. Comparative studies of LeuT and mammalian NSS illustrate how fine-tuning of ion-coordinating residues and local electrostatics can yield new ion-binding sites. Unlike LeuT, serotonin and GABA transporters (SERT and GAT-1, respectively) couple substrate uptake to symport of chloride ions in addition to Na+. Two research groups have suggested that E290 in LeuT, which is proximal to Na1, favors Na+ binding by contributing to the local negative electrostatic potential [62, 63]. In GAT-1 and SERT, which harbor serines at the equivalent positions, a chloride ion could be coordinated by the hydroxyl group and provide the favorable negative charge. Consistently, the serine to glutamate mutation in the mammalian transporters leads to the loss of chloride-stimulated transport [62, 63], and a reciprocal E290S mutation in LeuT results in leucine binding that is chloride-dependent [63].
Inhibition
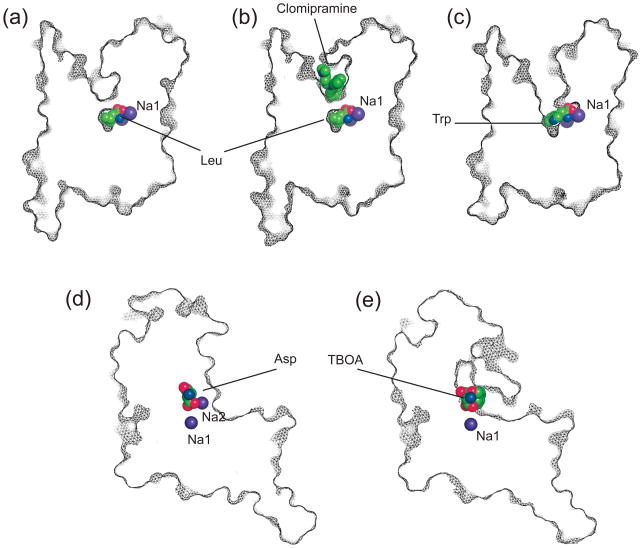
A complete transporter cycle involves ligand binding events and multiple conformational transitions, and inhibition of any of these steps is expected to abolish transport. Substrate-binding sites are the targets of competitive blockers, which usually contain chemical moieties similar to substrates, as well as additional bulky, typically aromatic, groups. Indeed, systematic studies suggest that a size boundary separates transported substrates and non-transportable blockers [49, 64–66]. Many antidepressants and illicit drugs, such as cocaine, are competitive inhibitors of NSS, and bind to the outward facing transporters in Na+- and chloride-dependent manners [67–69], suggesting that their binding sites overlap with that of the substrate. The structural basis of competitive inhibition has been revealed for TBOA, an inhibitor of mammalian glutamate transporters and GltPh, as well as for tryptophan, which inhibits LeuT. These blockers bind within the substrate-binding pockets, and are coordinated similarly to the substrates, but are however too large to allow closure of the extracellular gates (Figure 5).
Figure 5. Competitive blockers and non-competitive inhibitors.
(a) LeuT bound to leucine; (b) LeuT bound to leucine and non-competitive inhibitor clomipramine (PDB code 2qei); (c) LeuT bound to competitive blocker tryptophan (PDB code 3f3a); (d) GltPh bound to aspartate; (e) GltPh bound to competitive blocker TBOA (PDB code 2nww). Shown are the thin slices through the transporter protomers in surface representation and viewed in the membrane plain. Bound ligands are emphasized as balls.
Non-competitive inhibitors do not interfere with substrate binding, but instead reduce the turnover rate of transporters. Such an inhibitive mode has been reported for NSS inhibitors acting on LeuT [70]. The structures of LeuT in complexes with these compounds, although unlikely to be relevant to their high affinity competitive inhibition of NSS transporters, reveal the mechanism of non-competitive inhibition. Tricyclic antidepressants (TCA) and SERT-specific reuptake inhibitors (SSRI) bind within the extracellular vestibule of LeuT, above the substrate-binding site, which itself remains unperturbed and contains leucine (Figure 5) [70–72]. The locations of the binding sites suggest that these compounds may interfere with the closure of the extracellular vestibule that is essential for isomerization into the inward facing state. LeuT extracellular vestibule appears to be a “hot spot” for binding, as other hydrophobic compounds, including tryptophan and detergent molecules, have been observed there crystallographically [49, 73]. Highlighting the potential significance of non-competitive inhibition, low affinity allosteric binding sites have been postulated for TCA and SSRI inhibitors in NSS (their location may not coincide with the LeuT site), which prolong the residence of the inhibitors in the primary sites [74–76]. Also, the SERT inhibitor ibogaine inhibits transport non-competitively by stabilizing the transporter in its inward facing state [77].
Summary
Crystal structures of secondary transporters have captured diverse functional states and revealed the presence of structural domains with substrate and inhibitor binding sites lying at or near the interfaces. In several active secondary transporters, a conformation swap between symmetrical structural elements within these domains underlies the transitions between the outward and inward facing states. The most striking feature emerges from comparing the structures of GltPh, NhaA and transporters with the LeuT fold. Specifically, these transporters seem to assemble from similar structural modules, which yield similar three-dimensional arrangements, even though they occur in different order within primary sequences. Future crystal structures will reveal whether different combinations of these modules produce distinct transporter folds as well as whether the modular assembly is common among other transporter families.
Acknowledgments
The authors would like to thank Nicolas Reyes for help with figures, helpful discussions and comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saier MH, Jr, et al. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hediger MA, et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 3.Lolkema JS, Slotboom DJ. Classification of 29 families of secondary transport proteins into a single structural class using hydropathy profile analysis. J Mol Biol. 2003;327:901–909. doi: 10.1016/s0022-2836(03)00214-6. [DOI] [PubMed] [Google Scholar]
- 4.Brett CL, et al. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 5.Lolkema JS, Slotboom DJ. The major amino acid transporter superfamily has a similar core structure as Na+-galactose and Na+-leucine transporters. Mol Membr Biol. 2008;25:567–570. doi: 10.1080/09687680802541177. [DOI] [PubMed] [Google Scholar]
- 6.Oberai A, et al. A limited universe of membrane protein families and folds. Protein Sci. 2006;15:1723–1734. doi: 10.1110/ps.062109706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theobald DL, Miller C. Membrane transport proteins: surprises in structural sameness. Nat Struct Mol Biol. 2010;17:2–3. doi: 10.1038/nsmb0110-2. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita A, et al. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 9.Faham S, et al. The Crystal Structure of a Sodium Galactose Transporter Reveals Mechanistic Insights into Na+/Sugar Symport. Science. 2008 doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao X, et al. Structure and mechanism of an amino acid antiporter. Science. 2009;324:1565–1568. doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]
- 12.Shaffer PL, et al. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325:1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460:1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ressl S, et al. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature. 2009;458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 15.Tang L, et al. Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol. 2010;17:492–496. doi: 10.1038/nsmb.1788. [DOI] [PubMed] [Google Scholar]
- 16.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, et al. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 18.Yin Y, et al. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yernool D, et al. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 20.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 21.Schushan M, et al. Model-guided mutagenesis drives functional studies of human NHA2, implicated in hypertension. J Mol Biol. 2010;396:1181–1196. doi: 10.1016/j.jmb.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lolkema JS, Slotboom DJ. Sequence and hydropathy profile analysis of two classes of secondary transporters. Mol Membr Biol. 2005;22:177–189. doi: 10.1080/09687860500063324. [DOI] [PubMed] [Google Scholar]
- 23.Lolkema JS, et al. Evolution of antiparallel two-domain membrane proteins: tracing multiple gene duplication events in the DUF606 family. J Mol Biol. 2008;378:596–606. doi: 10.1016/j.jmb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317:1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- 25.Steiner-Mordoch S, et al. Parallel topology of genetically fused EmrE homodimers. Embo J. 2008;27:17–26. doi: 10.1038/sj.emboj.7601951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YJ, et al. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci U S A. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nasie I, et al. Topologically Random Insertion of Emre Supports a Pathway for Evolution of Inverted Repeats in Ion-Coupled Transporters. J Biol Chem. 2010;285:15234–15244. doi: 10.1074/jbc.M110.108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rapp M, et al. Emulating membrane protein evolution by rational design. Science. 2007;315:1282–1284. doi: 10.1126/science.1135406. [DOI] [PubMed] [Google Scholar]
- 29.Hirai T, Subramaniam S. Structure and transport mechanism of the bacterial oxalate transporter OxlT. Biophys J. 2004;87:3600–3607. doi: 10.1529/biophysj.104.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pebay-Peyroula E, et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 31.Forrest LR, Rudnick G. The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24:377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramson J, Wright EM. Structure and function of Na(+)-symporters with inverted repeats. Curr Opin Struct Biol. 2009;19:425–432. doi: 10.1016/j.sbi.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutzler R, et al. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 34.Fu D, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 35.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 36.Levin EJ, et al. Crystal structure of a bacterial homologue of the kidney urea transporter. Nature. 2009;462:757–761. doi: 10.1038/nature08558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law CJ, et al. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forrest LR, et al. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci U S A. 2008;105:10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimamura T, et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328:470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan RM, et al. The chloride permeation pathway of a glutamate transporter and its proximity to the glutamate translocation pathway. J Biol Chem. 2004;279:20742–20751. doi: 10.1074/jbc.M304433200. [DOI] [PubMed] [Google Scholar]
- 42.Reyes N, et al. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462:880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crisman TJ, et al. Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0908570106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Padan E, et al. NhaA crystal structure: functional-structural insights. J Exp Biol. 2009;212:1593–1603. doi: 10.1242/jeb.026708. [DOI] [PubMed] [Google Scholar]
- 45.Appel M, et al. Conformations of NhaA, the Na+/H+ exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J Mol Biol. 2009;388:659–672. doi: 10.1016/j.jmb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Herz K, et al. Beta-sheet-dependent dimerization is essential for the stability of NhaA Na+/H+ antiporter. J Biol Chem. 2009;284:6337–6347. doi: 10.1074/jbc.M807720200. [DOI] [PubMed] [Google Scholar]
- 47.Rimon A, et al. Monomers of the NhaA Na+/H+ antiporter of Escherichia coli are fully functional yet dimers are beneficial under extreme stress conditions at alkaline pH in the presence of Na+ or Li+ J Biol Chem. 2007;282:26810–26821. doi: 10.1074/jbc.M704469200. [DOI] [PubMed] [Google Scholar]
- 48.Groeneveld M, Slotboom DJ. Rigidity of the subunit interfaces of the trimeric glutamate transporter GltT during translocation. J Mol Biol. 2007;372:565–570. doi: 10.1016/j.jmb.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 49.Singh SK, et al. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010;463:828–832. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 51.Shi L, et al. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boudker O, et al. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–393. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 53.Shrivastava IH, et al. Time-resolved mechanism of extracellular gate opening and substrate binding in a glutamate transporter. J Biol Chem. 2008;283:28680–28690. doi: 10.1074/jbc.M800889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Z, Tajkhorshid E. Dynamics of the extracellular gate and ion-substrate coupling in the glutamate transporter. Biophys J. 2008;95:2292–2300. doi: 10.1529/biophysj.108.133421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirza O, et al. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. Embo J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groeneveld M, Slotboom DJ. Na+: aspartate coupling stoichiometry in the glutamate transporter homologue GltPh. Biochemistry. 2010;49:3511–3513. doi: 10.1021/bi100430s. [DOI] [PubMed] [Google Scholar]
- 57.Turk E, et al. A reinvestigation of the secondary structure of functionally active vSGLT, the vibrio sodium/galactose cotransporter. Biochemistry. 2006;45:1470–1479. doi: 10.1021/bi052160z. [DOI] [PubMed] [Google Scholar]
- 58.Screpanti E, Hunte C. Discontinuous membrane helices in transport proteins and their correlation with function. J Struct Biol. 2007;159:261–267. doi: 10.1016/j.jsb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Li J, Tajkhorshid E. Ion-releasing state of a secondary membrane transporter. Biophys J. 2009;97:L29–31. doi: 10.1016/j.bpj.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krishnamurthy H, et al. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, et al. A conserved Na(+) binding site of the sodium-coupled neutral amino acid transporter 2 (SNAT2) J Biol Chem. 2009;284:25314–25323. doi: 10.1074/jbc.M109.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forrest LR, et al. Identification of a chloride ion binding site in Na+/Cl -dependent transporters. Proc Natl Acad Sci U S A. 2007;104:12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zomot E, et al. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature. 2007;449:726–730. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
- 64.Washburn WN. Evolution of sodium glucose co-transporter 2 inhibitors as anti-diabetic agents. Expert Opin Ther Pat. 2009;19:1485–1499. doi: 10.1517/13543770903337828. [DOI] [PubMed] [Google Scholar]
- 65.Loo DD, et al. How drugs interact with transporters: SGLT1 as a model. J Membr Biol. 2008;223:87–106. doi: 10.1007/s00232-008-9116-6. [DOI] [PubMed] [Google Scholar]
- 66.Hirayama BA, et al. Common mechanisms of inhibition for the Na+/glucose (hSGLT1) and Na+/Cl-/GABA (hGAT1) cotransporters. Br J Pharmacol. 2001;134:484–495. doi: 10.1038/sj.bjp.0704274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beuming T, et al. The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat Neurosci. 2008;11:780–789. doi: 10.1038/nn.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersen J, et al. Location of the antidepressant binding site in the serotonin transporter: importance of Ser-438 in recognition of citalopram and tricyclic antidepressants. J Biol Chem. 2009;284:10276–10284. doi: 10.1074/jbc.M806907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tavoulari S, et al. Fluoxetine (Prozac) binding to serotonin transporter is modulated by chloride and conformational changes. J Neurosci. 2009;29:9635–9643. doi: 10.1523/JNEUROSCI.0440-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh SK, et al. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Z, et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–1393. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Z, et al. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nat Struct Mol Biol. 2009;16:652–657. doi: 10.1038/nsmb.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quick M, et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc Natl Acad Sci U S A. 2009;106:5563–5568. doi: 10.1073/pnas.0811322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plenge P, et al. Allosteric effects of R- and S-citalopram on the human 5-HT transporter: evidence for distinct high- and low-affinity binding sites. Eur J Pharmacol. 2007;567:1–9. doi: 10.1016/j.ejphar.2007.03.055. [DOI] [PubMed] [Google Scholar]
- 75.Zhong H, et al. An allosteric binding site at the human serotonin transporter mediates the inhibition of escitalopram by R-citalopram: kinetic binding studies with the ALI/VFL-SI/TT mutant. Neurosci Lett. 2009;462:207–212. doi: 10.1016/j.neulet.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 76.Neubauer HA, et al. Dissection of an allosteric mechanism on the serotonin transporter: a cross-species study. Mol Pharmacol. 2006;69:1242–1250. doi: 10.1124/mol.105.018507. [DOI] [PubMed] [Google Scholar]
- 77.Jacobs MT, et al. Ibogaine, a noncompetitive inhibitor of serotonin transport, acts by stabilizing the cytoplasm-facing state of the transporter. J Biol Chem. 2007;282:29441–29447. doi: 10.1074/jbc.M704456200. [DOI] [PubMed] [Google Scholar]
- 78.Widdas WF. Inability of diffusion to account for placental glucose transfer in the sheep and consideration of the kinetics of a possible carrier transfer. J Physiol. 1952;118:23–39. doi: 10.1113/jphysiol.1952.sp004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitchell P. Transport of phosphate across the surface of Micrococcus pyogenes; nature of the cell inorganic phosphate. J Gen Microbiol. 1953;9:273–287. doi: 10.1099/00221287-9-2-273. [DOI] [PubMed] [Google Scholar]
- 80.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180:134–136. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- 81.Patlak CS. Contributions to the theory of active transport: II. The gate type non-carrier mechanism and generalizations concerning tracer flow, efficiency, and measurement of energy expenditure. Bulletin of Mathematical Biology. 1957;19:209–235. [Google Scholar]
- 82.Crane RK. Hypothesis for mechanism of intestinal active transport of sugars. Fed Proc. 1962;21:891–895. [PubMed] [Google Scholar]
- 83.Mitchel P. Moleculae, Group and Electron Translocation through Natural Membranes. Biochem Soc Symp. 1963;22:142–169. [Google Scholar]
- 84.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. [Google Scholar]