Abstract
Helicases are essential enzymes that utilize the energy of nucleotide hydrolysis to drive unwinding of nucleic acid duplexes. Helicases play roles in all aspects of DNA metabolism including DNA repair, DNA replication and transcription. The subcellular locations and functions of several helicases have been studied in detail; however, the roles of specific helicases in mitochondrial biology remain poorly characterized. This review presents important recent advances in identifying and characterizing mitochondrial helicases, some of which also operate in the nucleus.
Keywords: mitochondrial DNA, helicases, BER, aging, mtDNA deletions
Introduction
This article is part of a series of articles on the biological importance of the mitochondrial genome for the aging process and the mechanisms that maintain and ensure its stability. Recent studies demonstrate that mitochondrial DNA (mtDNA) mutations and deletions accumulate with age in several model organisms, suggesting that defects in mtDNA metabolism and/or decreased stability of mtDNA play a causative role in aging.
All DNA metabolic processes involve helicase activity, and thus helicases are omnipresent and crucial enzymes in genome stability. While several mitochondrial helicases have been described and characterized, they remain relatively poorly characterized. This review describes recent evidence that mitochondrial helicases play a critical role in the maintenance of mtDNA stability, possibly preventing age-associated accumulation of mtDNA mutations.
DNA helicases, which unwind DNA duplexes as well as unusual three- and four-stranded DNA structures, play critical roles in DNA replication and DNA repair. Most experimental evidence suggests that the two mtDNA strands replicate asynchronously, starting at the origin of the heavy strand and proceeding unidirectionally to the origin of the light strand, after which replication of the light strand is initiated (Falkenberg et al., 2007). However, Holt and colleagues suggest that the two mtDNA strands may also replicate in a synchronous, bidirectional manner, characterized by coordinated replication of leading and lagging strands (Bowmaker et al., 2003; Yang et al., 2002). Helicase activity is required as a cofactor for DNA polymerase γ (Polγ), which is inactive on duplex DNA in vitro in the absence of a helicase and a single-stranded DNA (ssDNA) binding protein (Korhonen et al., 2004). The fidelity and efficiency of DNA replication influences the stability of the mitochondrial genome.
MtDNA stability also depends on the relative rates of DNA damage and repair. Under physiological conditions, mtDNA is uniquely susceptible to oxidative DNA damage, because of its physical proximity to reactive oxygen species (ROS) generated during oxidative phosphorylation in the inner mitochondrial membrane. Oxidative lesions in the mtDNA are repaired by base excision repair (BER) enzymes, all of which are thought to play dual roles in the nuclear and mitochondrial compartments.
BER is a well-characterized multi-step pathway, initiated by a DNA glycosylase that removes a damaged base. Other BER enzymes or activities that act downstream of the glycosylase include an AP endonuclease that cleaves the abasic site and/or trims DNA ends, 5′deoxyribose phosphate lyase (dRPase) and DNA synthesis catalyzed by a DNA polymerase (DNA polymerase γ in mitochondria), and DNA ligase. In short patch BER (SP-BER), the DNA synthesis tract is 1-2 nucleotides. When the 5′ end is blocked, repair proceeds via long-patch BER (LP-BER; reviewed in (Akbari et al., 2008; Hegde et al., 2008; Liu et al., 2008; Maynard et al., 2009; Szczesny et al., 2008)). In LP-BER, strand-displacement DNA synthesis generates a 2-7 nt long flap, which is removed by a structure-specific endonuclease (Hegde et al., 2008), and results in a 2-7 nt DNA synthesis tract. LP-BER occurs in mitochondria (Akbari et al., 2008; Liu et al., 2008; Szczesny et al., 2008) in a FEN-1 dependent or FEN-1 independent manner (Akbari et al., 2008; Liu et al., 2008; Szczesny et al., 2008). Nuclear helicases, such as the Werner syndrome helicase (WRN, are thought to participate in LP-BER through interactions with FEN-1 (Bohr, 2008). Human Dna2 also interacts with FEN-1, and this interaction may be important in DNA replication and DNA repair (Budd and Campbell, 1997; Zheng et al., 2008).
The RecQ helicases are an important group of mammalian DNA helicases that play roles in DNA repair and DNA replication. Mutations in human genes encoding RecQ helicases cause premature aging (Werner syndrome, Rothmund-Thomsons syndrome) or premature cancer (Bloom syndrome) (Bohr, 2008). The possible roles of mammalian RecQ helicases in mitochondria are of great interest, but are not yet well understood.
Until recently, mitochondrial helicases had only been identified and characterized in yeast. Twinkle, a helicase believed to participate in replication of the mtDNA in human mitochondria, was the first mammalian mitochondrial helicase identified, and its mutations were associated with autosomal ophtalmoplegia (Spelbrink et al., 2001). Other mammalian DNA helicases recently identified include hPif1 (Futami et al., 2007)), hSuv3 (Dmochowska et al., 1999) and hDna2 (Zheng et al., 2008). All three are homologues of known yeast mitochondrial helicases. Preliminary studies suggest that the roles of the mammalian DNA helicases may differ from the roles of their yeast counterparts (Copeland and Longley, 2008).
This review focuses on the biochemical properties of mtDNA helicases, and their possible roles in maintaining stability and/or preventing age-associated instability of mtDNA.
Dna2
Yeast Dna2
Yeast Dna2 (yDna2) is an essential, 170 kDa protein with an N-terminal nuclease RecB homology domain, and a C-terminal helicase domain that includes canonical helicase motifs I, II, III, V and VI. Purified yDna2 has 5′ – 3′ DNA-dependent DNA helicase and ATPase activity on forked DNA molecules and an endonuclease activity (Bae and Seo, 2000; Budd and Campbell, 1995; Budd et al., 2000; Lee et al., 2000). Purified yDna2 preferentially cleaves 5′- protruding ssDNA ends in vitro, and this reaction is stimulated by the presence of RNA in the 5′ end of the DNA substrate (Bae and Seo, 2000). Yeast Dna2 mutants are temperature-sensitive, and hypersensitive to DNA damaging agents including bleomycin (Choe et al., 2002), X-rays (Budd and Campbell, 2000), hydroxyurea (HU) and methyl methanesulfonate (MMS) (Imamura and Campbell, 2003). Thus, yeast Dna2 plays a role in DNA repair.
YDna2 also interacts with yeast FEN-1 (yFEN-1), and plays a likely role in maturing Okazaki fragments during DNA replication. In particular, it has been proposed that yDna2 removes the primer from the lagging DNA strand and leaves a short flap, which is cleaved by yFEN-1, resulting in a product that can be ligated (Bae and Seo, 2000; Budd and Campbell, 1997; Pike et al., 2009). yDna2 may also play a role in double strand break (DSB) repair, promoting 5′-strand resection, and in concert with yeast Sgs1, a WRN and BLM ortholog, producing long 3′ ssDNA overhangs (Zhu et al., 2008).
Human Dna2
Human Dna2 (hDna2), which is less well characterized than yDna2, complements the phenotype of yeast Dna2 mutants (Imamura and Campbell, 2003), indicating functional conservation of these two proteins. Interestingly, yDna2 has three nuclear localization signals while hDna2 has none, and in some cell types, hDna2 localizes primarily to the mitochondria, with none or only a small amounts present in the nucleus. Thus, although yDna2 may largely function in nuclear replication, telomere maintenance and DSB repair, hDna2 may play important functions in mitochondrial DNA metabolism. Because hDna2 depletion results in G2/M arrest, aneuploidy and internuclear chromatin bridges, hDna2 must also play some roles in nuclear DNA metabolism (Duxin et al., 2009; Zheng et al., 2008).
Human Dna2 ATPase and helicase activities are stimulated by ssDNA (Masuda-Sasa et al., 2006). The protein also has an associated endonuclease activity, which cleaves 5′-flaps, 3′-flaps, 3′-overhangs and 5′-overhangs, but does not cleave ssDNA flanked by dsDNA (Kim et al., 2006; Masuda-Sasa et al., 2006). In vitro, hDna2 stimulates ligation of a flap substrate in the presence of T4 DNA ligase and hFEN-1, suggesting that hDna2, like yDna2, may promote Okazaki fragment maturation (Kim et al., 2006) or possibly LP-BER, as discussed below.
Human Dna2 co-localizes with mtDNA and Twinkle in the nucleoids in a subset of mitochondria. hDna2 co-localization with Twinkle increased drastically in cells expressing some but not all mutant forms of Twinkle (Duxin et al., 2009), even though all mutant forms of Twinkle proteins caused replication fork stalling. Mutations that altered subcellular localization of Twinkle were not associated with defects in ssDNA binding or helicase activity (Duxin et al., 2009; Goffart et al., 2009; Wanrooij et al., 2007). Thus, the mechanism of re-distribution of hDna2 mutants is not understood at present.
Human Dna2 also interacts with Polγ in the absence of DNA or RNA. This interaction stimulates primer elongation by Polγ on a D-loop plasmid substrate that resembles the mtDNA D-loop, but does not stimulate elongation by Polγ on oligo-primed ssDNA. In mitochondrial extracts, hDna2 partially removes all but 1-10 nucleotides of a 5′ DNA or RNA flap, as observed with yDna2. Subsequently, hDna2 stimulates cleavage of remaining short 5′ flaps, as well as gap-filling, primer removal and ligation by Polγ, FEN-1 and Lig IIIα. These data suggest that hDna2 may be involved in RNA primer and flap removal during mitochondrial DNA replication (Zheng et al., 2008). Cells that have been depleted for hDna2 by siRNA knockdown show a modest decrease in replication intermediates, supporting a role for hDna2 in mitochondrial replication in vivo (Duxin et al., 2009).
In vitro and in vivo experiments support a role for hDna2 in mitochondrial LP-BER. Purified hDna2 and mitochondrial extracts cleave and produce identical products from nicked or flap DNA substrates containing the AP-site analogue tetrahydrofuran (THF). In an in vitro reconstituted LP-BER assay, hDna2 and hFEN-1 synergistically stimulate formation of ligated products. Furthermore, immunodepletion of either hDna2 or hFEN-1 from mitochondrial extracts resultes in decreased LP-BER activity (Zheng et al., 2008). In vivo, hDna2 knock down decreases repair of hydrogen peroxide-induced oxidative damage in mtDNA (Duxin et al., 2009; Zheng et al., 2008).
In summary, these results suggest that hDna2 plays a different biological role than yDna2. YDna2 is thought to play roles in DSB processing, Okazaki fragment processing, telomere maintenance, and possibly mtDNA stability, while the primary role of hDna2 is likely to be processing flap intermediates during LP-BER in mitochondria.
Twinkle
Twinkle is a mammalian mitochondrial member of the RecA/DnaB superfamily, with structural similarity to phage T7 gene 4 primase/helicase and other hexameric ring helicases. Human Twinkle has an N-terminal primase-like domain; however, no primase activity has been identified. The C-terminal helicase domain includes Walker A and B motifs, and the two domains are connected by a short linker region. Twinkle monomers form hexamers in vitro, even in absence of Mg2+ and nucleotide (Farge et al., 2008; Goffart et al., 2009; Spelbrink et al., 2001). The C-terminal domain is required for oligomerization, and the N-terminal domain contributes to the stability of the hexamers (Farge et al., 2008). Purified recombinant hTwinkle has an NTP-dependent 5′ - 3′ helicase activity that is supported by hydrolysis of UTP, GTP or ATP (Goffart et al., 2009; Korhonen et al., 2003). Twinkle's preferred DNA substrate is forked DNA, requiring a 5′ ssDNA tail for loading and a short 3′ tail for initiating the helicase activity (Korhonen et al., 2003).
Mitochondrial single-stranded DNA binding protein (mtSSB) strongly stimulates the unwinding activity of hTwinkle (Korhonen et al., 2003), but even in the presence of mtSSB, hTwinkle unwinds relatively short stretches (approximately 20 bp) of DNA. However, in the presence of Polγ, Twinkle can unwind longer stretches of dsDNA, to such extend that it promotes Polγ DNA synthesis tracts up to 2000 nt, even though Polγ cannot elongate through dsDNA. In vitro, hTwinkle, Polγ and mtSSB constitute the minimal replisome for the mtDNA, and can produce a 15 Kb product, which is nearly the size of the mitochondrial genome (16 Kb) (Korhonen et al., 2004).
Mutations in the gene encoding hTwinkle can cause autosomal dominant progressive external ophthalmoplegia (adPEO), a disease that manifests in older adults with large deletions in mtDNA deletions in post-mitotic tissues. Overexpression of wild type mTwinkle in mice increases mtDNA copy number, and Twinkle knockdown in human osteosarcoma cells decreases mtDNA copy number (Tyynismaa et al., 2004). In a mouse model for PEO, older mice expressing a PEO-associated Twinkle mutant accumulate mtDNA deletions in muscle and brain, and have a cellular phenotype similar to PEO patients, including aberrant mitochondrial structures. These mice also exhibit mtDNA depletion in brain, but not in muscle and heart (Tyynismaa et al., 2005). Muscle cells from six week old mutant Twinkle mice accumulate mtDNA replication intermediates, and when the mutated Twinkle was overexpressed in human cells, mtDNA depletion and accumulation of replication intermediates was also observed. When overexpressed in human cells, most Twinkle mutants induce changes in structure of the mitochondrial nucleoid. These results support the notion that Twinkle is the replicative helicase in mammalian mitochondria, and that Twinkle mutations result in replication stalling, both in mouse cells in vivo and in cultured cells (Goffart et al., 2009; Wanrooij et al., 2007).
In vitro, hTwinkle mutant and wild-type monomers form mixed hexamers, which may partially explain the dominant negative character of adPEO mutations. The severity of the adPEO phenotype of the patients correlates inversely with the amount of residual helicase activity in Twinkle mutants, suggesting that the helicase activity is essential for the biological function of Twinkle (Goffart et al., 2009). Moreover, in vitro studies in HEK293 cells expressing mutant Twinkle suggest that Twinkle mutants inhibit transcription of the mitochondrial genome independent of its effect on mtDNA copy number (Goffart et al., 2009). Therefore, Twinkle is well established as the mammalian mitochondrial replication helicase, and mutations in this protein can lead to mtDNA deletions. It is not yet known whether Twinkle plays a role in mitochondrial DNA repair.
Suv3
The yeast Suv3 (ySUV3) protein belongs to the Ski2 family of DExH-box RNA helicases. It is an NTP-dependent RNA helicase in vitro that localizes to mitochondria in yeast cells (Dziembowski et al., 1998; Margossian et al., 1996; Stepien et al., 1992). Yeast SUV3-1 was originally identified as a dominant nuclear suppressor of deletions of the mitochondrial var1 gene in Saccharomyces cerevisiae (Butow et al., 1989). Subsequently, it was found that ySuv3 plays a major role in post-transcriptional and translational processes (Conrad-Webb et al., 1990). The product of the ySUV3 gene forms, together with the Dss1p RNase, a well-characterized mitochondrial degradosome complex responsible for degradation of mitochondrial transcripts and RNA surveillance (Dziembowski et al., 1998).
The human SUV3 (hSUV3) gene was identified and cloned based on a high degree of sequence homology to the yeast ortholog (Dmochowska et al., 1999). Similar to ySuv3, hSuv3 primarily localizes to the mitochondria, although a small fraction can be found in the nucleus (Minczuk et al., 2002; Szczesny et al., 2007). Like the mitochondrial helicase Twinkle, hSuv3 associates with the mitochondrial nucleoid (Wang and Bogenhagen, 2006). In vitro, the recombinant enzyme is an ATP-dependent 5′-3′ dsDNA helicase with broad DNA substrate specificity (Minczuk et al., 2002; Shu et al., 2004). SUV3 knockout mice die in utero (Szczesny et al., 2007), and conditional Cre recombinase-mediated mSUV3 knockdown mice show delayed growth and symptoms of accelerated aging (Paul et al., 2009). This observation suggests that mSuv3 may promote mtDNA stability and prevent aging-associated mtDNA instability.
Human Suv3 is expressed in all tissues, with the highest levels detected in liver (Dmochowska et al., 1999). Transient silencing of the hSUV3 gene in HeLa cells induces apoptosis and an increased frequency of sister chromatid exchange during mitotic cell division (Pereira et al., 2007). Lee and coworkers (Khidr et al., 2008; Wang et al., 2009) suggested that hSuv3 may maintain mitochondrial homeostasis in human cells by serving as a component of an RNA degradosome. Thus, an inducible shRNA-mediated knock-down of hSuv3 in U2OS cells resulted in the accumulation of shortened polyadenylated mtRNA species and showed impaired mitochondrial protein synthesis, which in turn led to mitochondrial dysfunction and cellular senescence or death. More recently, it was demonstrated that expression of dominant-negative form of hSuv3 lacking ATPase and helicase activities lead to disruption in mtRNA, resulting in accumulation of molecules with extended poly(A) tails and 3′-end truncated degradation intermediates (Szczesny et al.). Downregulation of hSUV3 also leads to reduced mtDNA copy number, and some data have implicated hSuv3 in the maintenance of mitochondrial DNA and cell viability. Human Suv3 also interacts with the RecQ helicases WRN and BLM (Pereira et al., 2007) and with the cofactor of survivin HBXIP (Minczuk et al., 2005).
Overall, Suv3 seems to play an important role in mitochondrial homeostasis, most likely by regulating mtRNA metabolism in yeast and mammalian mitochondria. Although no human ortholog of the Dss1p RNase has been found to date, a recent publication indicates that polynucleotide phosphorylase (PNPase) is a hSuv3-interacting component of a putative human mitochondrial degradosome (Wang et al., 2009).
Pif1
S. cerevisiae Pif1 protein (yPif1) belongs to the SF1 super-family of helicases, which is similar to the RecD subfamily. Yeast Pif1 exhibits ssDNA-stimulated ATPase and 5′ – 3′ helicase activity and localizes to the nucleus and to mitochondria (Lahaye et al., 1991; Zhang et al., 2006; Zhou et al., 2000). yPif1 may play roles in telomere maintenance by inhibiting telomerase activity (Zhou et al., 2000), rDNA replication (Ivessa et al., 2000), preventing gross-chromosomal rearrangements (reviewed in (Cheng et al., 2007)), and Okazaki fragment processing (Budd et al., 2006; Pike et al., 2009; Rossi et al., 2008).
The yPif1 gene product is necessary for recombination of mtDNA between rho+ and rho- cells with tandemly arrayed repeats. For example, yPif1 mutants exhibit increased formation of petites with or without exposure to UV light (Foury and Dyck, 1985; Foury and Kolodynski, 1983; Lahaye et al., 1991; Wagner et al., 2006), mtDNA loss and fragmentation after ethidium bromide treatment (Cheng et al., 2007), and mild sensitivity towards MMS and hydroxyurea (Wagner et al., 2006). These results strongly suggest that yPif1 plays a role in mtDNA repair/recombination.
Recently, yPif1 has been shown to associate with mtDNA in vivo (Cheng et al., 2007). However, overexpression of yPif1 causes growth defects in wild type yeast and yPif1 mutants are viable only on non-fermentable carbon sources (Lahaye et al., 1991; Wagner et al., 2006). Furthermore, overexpression of a helicase-dead yPif1 mutant in wild type cells caused increased formation of petite colonies, indicating that the helicase mutant had a toxic effect on mitochondria, even in the presence of wild type yPif1 (Wagner et al., 2006), and suggesting a possible dominant-negative effect.
Recent results also suggest that yPif1 may prevent accumulation of oxidative DNA damage in mtDNA. Pif1Δ yeast have a 29-fold higher spontaneous frequency of mtDNA mutations than wild type and a 7-fold higher level of yNtg1-sensitive sites (likely oxidized pyrimidines) than wild type yeast (Doudican et al., 2005; O'Rourke et al., 2002).Yeast Pif1 and Ntg1 showed synergistic interactions, including synergistic increases in DNA damage-induced yNtg1-sensitive lesions in pif1Δ ntg1Δ double mutants and synergistic defects in mitochondrial respiration. The latter effect was not seen in ntg1Δ strains with mutations in other recombination genes, suggesting that yPif1 participates in the repair of oxidative lesions by a recombination-independent mechanism (O'Rourke et al., 2002). Furthermore, yPif1 may compensate for defects in yNtg1. Based on these results, it has been suggested that yPif1 might inhibit replication fork progression to allow time for repair of mtDNA, or change the mtDNA structure to make it more accessible for repair (Doudican et al., 2005; O'Rourke et al., 2002). In mammalian cells, oxidative lesions accumulate significantly with age in the mtDNA (Hudson et al., 1998). Thus, if there is functional conservation among yeats and human Pif1, these results implicate hPif1 as an important factor in maintaining mtDNA stability during the aging process.
Yeast pif1Δ cells are defective in growth on non-fermentable media after exposure to ethidium bromide, likely due to rapid fragmentation and depletion of mtDNA and inhibition of mtDNA replication (Cheng et al., 2007; Cheng et al., 2009). These results also argue that yPif1 plays a direct role in mtDNA replication. Similar effects on respiration are not observed in pif1-m2 mutants (mutated only in nuclear pif1), but are observed in pif1-m2 Dna2 double mutants, indicating that Dna2 also is essential for mitochondrial function and/or mtDNA stability (Budd et al., 2006).
Human Pif1
In human cells, there are two isoforms of Pif1 (hPif1), produced by alternative splicing: nuclear hPif1α (74kDa) and mitochondrial hPif1β (80kDa). These enzymes have a common central helicase domain, but different C-terminal domains. The helicase domain of hPif1 is homologous to yPif1, but the C and the N terminal domains show no marked homology to yPif1 (Futami et al., 2007), suggesting that there could be important functional differences between yPif1 and hPif1. However, like yPif1, hPif1α inhibits telomerase (Zhang et al., 2006) and hPif1 siRNA-mediated knock down slowed cell cycle progression in S-phase, suggesting a role for hPif1 in replication (Futami et al., 2007). Purified N-terminally truncated hPif1α exhibited ssDNA-stimulated ATPase activity, and 5′ to 3′ DNA/DNA and DNA/RNA helicase activity in vitro (Futami et al., 2007; Zhang et al., 2006). hPif1α also has ATP-independent ssDNA annealing activity, which competes with the helicase activity. The helicase domain unwinds 40 bp duplexes 50% less efficiently than 20 bp duplexes, and fails to unwind 100bp DNA substrates. A minimum of approximately 30 nt was required for binding to ssDNA, ATPase activity and helicase activity (George et al., 2009). Interestingly, the helicase domain of hPif1 preferentially binds and unwinds DNA structures that mimic stalled DNA replication forks (George et al., 2009). hPif1β is poorly characterized, due to difficultly in purifying and/or overexpressing the recombinant protein (unpublished results). However, the high homology between the yeast and the human proteins suggests that their in vivo functions may be somewhat conserved. Thus, it will be important to investigate a possible role for hPif1 in repair of oxidative DNA damage and in preventing aging-associated loss of mtDNA stability.
YB-1
The multifunctional Y-box binding protein 1 (YB-1) is one of the most evolutionarily conserved DNA binding proteins characterized to date. Human YB-1 plays roles in transcriptional regulation, translational regulation, DNA repair, drug resistance and responses to extracellular signals (Kohno et al., 2003). Human YB-1 was first identified as a protein that binds to the Y-box (a cis-acting element with a CCAAT motif) in the class II major histocompatibility complex (Didier et al., 1988). hYB-1 is a transcriptional regulatory protein and translational repressor (Evdokimova et al., 2006).
Mutations in hYB-1 have been identified in several human cancers (Kuwano et al., 2003), and hYB-1 may suppress carcinogenesis (Bader et al., 2003) through its role as a translational suppressor (Bader and Vogt, 2005), mediated by Akt phosphorylation (Bader and Vogt, 2008). In addition, hYB-1 interacts with p53 and modulates p53-induced cell cycle arrest and apoptosis (Braithwaite et al., 2006).
There is evidence that hYB-1 also plays a role in DNA repair. For example, recombinant YB-1 binds abasic site-containing DNA with higher affinity than undamaged DNA (Hasegawa et al., 1991). YB-1 also binds to DNA containing cisplatin adducts (Ise et al., 1999) and mismatched DNA (Gaudreault et al., 2004). Moreover, hYB-1 interacts physically and functionally with the oxidized pyrimidine DNA glycosylase NTH1 (Marenstein et al., 2001), increasing NTH1 glycosylase and AP-lyase activity by altering the steady state equilibrium of the hNTH1-AP site Schiff base intermediate. Human YB-1 also interacts with the major BER abasic endonuclease, APE1 (Chattopadhyay et al., 2008), and with the NEIL2 DNA glycosylase, stimulating its glycosylase activity several-fold. In this study, it was demonstrated that hYB-1 interacts with the other NEIL2-associated BER proteins, namely, DNA ligase III alpha and DNA polymerase beta, and thus could form a large multiprotein complex (Das et al., 2007). Moreover, hYB-1 translocates to the cytosol after oxidative stress, and NEIL2-initiated BER is significantly reduced in YB-1 depleted cells. These results, together with the observation that hYB-1 interacts with other DNA repair proteins, including MSH2, DNA polymerase delta, Ku80 and WRN proteins (Gaudreault et al., 2004), demonstrate that hYB-1 is an important modulator of BER in the nucleus, and may be involved in the general response to DNA damage.
YB-1 was initially characterized as a dsDNA binding protein. However, YB-1 may also generate and stabilize ssDNA in promoter regions of MHC genes (MacDonald et al., 1995), suggesting that YB-1 could participate in other DNA metabolizing processes involving the double- to single-stranded DNA transition. In fact, Gaudreault and colleagues demonstrated that recombinant hYB-1 can unwind DNA duplexes containing blunt ends, 5′ or 3′ recessed ends, or forked structures. This unwinding activity increases on cisplatin-modified or heteroduplex DNA containing mismatches (Gaudreault et al., 2004). Moreover, hYB-1 also promotes efficient strand annealing of both DNA-DNA and DNA-RNA duplexes, and the coordination of the two activities, melting and annealing, was regulated in a dose-dependent manner (Skabkin et al., 2001).
We have recently identified a new role for hYB-1 in mitochondrial mismatch repair. Mismatch repair (MMR) is an essential repair pathway in preventing mutagenesis and microsatellite instability (Skabkin et al., 2001). MMR proteins had been identified in yeast and coral mitochondria, but MMR had not yet been reported or characterized in human mitochondria, despite deletions of the highly repetitive sequences and microsatellite instability in mtDNA of cells with intact nuclear MMR. We showed that human mitochondria have a robust mismatch-repair activity, which is distinct from nuclear MMR and depends on the mitochondrial polymerase, Polγ (de Souza-Pinto et al., 2009). We identified a mismatch-binding activity, which was independent of key nuclear MMR factors and was detected in mitochondrial extracts from cells lacking MSH2, suggesting distinctive pathways for nuclear and mitochondrial MMR. Surprisingly, hYB-1 is one of the components of the recently identified mitochondrial mismatch-binding complex. We found a subfraction of hYB-1 protein was detected in mitochondria in HeLa cells, and this subfraction increased in cells cultured in presence of high concentrations of thymidine, which induced mtDNA mutagenesis. More importantly, mismatch-binding activity decreased significantly in mitochondrial extracts from cells depleted for YB-1 by siRNA or immunodepletion. Moreover, hYB-1 depletion caused an increase in mtDNA mutagenesis. These results have shown, for the first time, that human mitochondria contain a functional MMR repair pathway, and implicates hYB-1 in the mismatch-binding and recognition steps of this pathway (de Souza-Pinto et al., 2009). Because hYB-1 plays several roles in nuclear DNA metabolism, it seems likely that YB-1 also plays other yet-to-be identified roles in mtDNA metabolism.
Discussion
This review describes recent advances in identifying and characterizing mammalian DNA and DNA/RNA helicases, with an emphasis on the impact of these enzymes on mitochondrial DNA metabolism and/or stability. Strong in vitro and in vivo evidence indicates that Twinkle associates with Polγ and is the primary mitochondrial replicative helicase and/or primase. The role of helicases in mtDNA damage processing is best understood in yeast, where yPif1 plays an essential role in repair of oxidative DNA damage. By analogy, it is likely that mammalian DNA helicases also facilitate repair of oxidative DNA damage in mtDNA, thereby preventing the accumulation of age-associated oxidative base damage and deletions in the mtDNA.
In the case of hDna2, recently published results demonstrated direct participation in processing BER intermediates (Duxin et al., 2009; Zheng et al., 2008), and a role in long-patch BER (Copeland and Longley, 2008; Zheng et al., 2008). However, hDna2 is expressed in the nucleus and in mitochondria, and its specific role in each compartment is not yet known. While one group suggests that hDna2 is exclusively mitochondrial (Zheng et al., 2008), some experiments using hDna2 knockdown cells provide evidence of a nuclear role as well (Duxin, 2008). Additional studies are required to clarify the possible roles of Dna2 in DNA repair or other aspects of DNA metabolism in the mitochondria or nucleus.
Recent studies in mice expressing error-prone Polγ suggest that DNA deletions and clonal mutations drive the aging process (Vermulst et al., 2008). MtDNA deletions may be the result of defects in DNA repair pathways including DSB repair (Krishnan et al., 2008) and replication fork stalling and/or collapse due to the inability to unwind the double helix ahead of the fork. Helicases active in the mitochondrial compartment prevent such mtDNA deletions, as suggested for Twinkle and hDna2 (Zheng et al., 2008).
Interactions have been demonstrated between hSuv3 and BLM or WRN, and ySuv3 and Sgs1. Interestingly, hSuv3 and RecQ helicases have opposite polarities (Pereira et al., 2007), which may indicate the requirement for bidirectional DNA unwinding in some DNA transactions. While neither WRN nor BLM have been demonstrated in mitochondria, preliminary studies suggest that RecQ4 may be present in human mitochondria (unpublished data). In the nucleus, the RecQ helicases are involved in several DNA repair pathways, including BER. Thus, hSuv3 may cooperate with other helicases during BER in mitochondria. It is interesting to note that human BLM complements the HU- and MMS-sensitivity of yDna2 mutants, indicating a functional overlap between different helicases in vivo. Similarly, the possible role of yPif1 in counteracting Sgs1 function in topoIII mutants in yeast (Wagner et al., 2006), could suggest an interplay between hPif1β and a RecQ helicase in mammalian mitochondria.
The observed redistribution of mitochondrial hDna2 to the nucleoids in cells expressing mutated Twinkle proteins (Duxin et al., 2009) also suggests a functional link between the two mitochondrial helicases. However, the correlation between the enzymatic activities of the different Twinkle mutants and their ability to cause hDna2 redistribution is unclear, and the molecular events leading to hDna2 redistribution remain to be determined. These results indicate that a functional interplay between these two helicases may be important in resolving structures that arise during DNA replication.
Mutation of yPif1 suppressed the lethality of a yDna2 mutant, suggesting that yPif1 may generate intermediates that are processed by yDna2. Campbell and co-workers (Budd et al., 2006) suggested that yPif1 generates long 5′ flaps which are removed by yDna2. This is supported by in vitro studies showing that Pif1 stimulates flap displacement in Okazaki fragment processing, resulting in longer 5′ flaps which seem to inhibit the formation of the final ligation product in the absence of Dna2 (Pike et al., 2009; Rossi et al., 2008). Since hDna2 is largely mitochondrial (Zheng et al., 2008), it is possible that hPif1β and hDna2 cooperate during mtDNA replication or repair, and the loss of these activities could lead to accumulation of intermediates that would, ultimately lead to mtDNA deletion.
In summary, multiple mitochondrial helicases play likely roles in mtDNA maintenance and replication, and their functions may be coordinated with each other or with other mitochondrial enzymes by an as yet unknown mechanism. This model is summarized graphically in Figure 1. Additional studies are needed to refine our understanding of the compartment-specific and unique roles of these enzymes, especially those that localize to both the nuclear and mitochondrial compartments. It will also be important to identify biologically-relevant substrates for this diverse group of enzymes, and the degree to which their biological roles are unique or overlapping. Even with large gaps in our knowledge about mitochondrial helicases remaining, it is reasonable to conclude that as a class of enzymes, mitochondrial helicases are critical and essential for maintaining the integrity of the mitochondrial genome, and in turn, for maintaining mitochondrial function, especially in aging cells and/or organisms.
Figure 1.
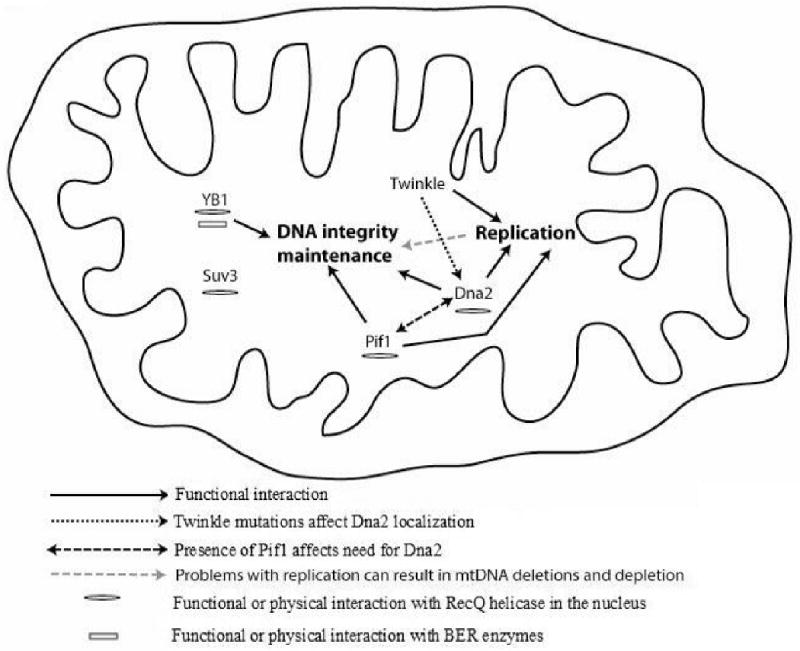
Interplay between mitochondrial helicases, mtDNA maintenance and replication Evidence suggests roles for mitochondrial helicases in both replication and DNA repair. Results also suggest a functional interaction between some of the mitochondrial helicases and it has been suggested that mtDNA repair play a major role in mtDNA deletions. Hence the role of mitochondrial helicases in replication and mtDNA integrity maintenance is possibly interconnected.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–16. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Bader AG, Felts KA, Jiang N, Chang HW, Vogt PK. Y box-binding protein 1 induces resistance to oncogenic transformation by the phosphatidylinositol 3-kinase pathway. Proc Natl Acad Sci U S A. 2003;100:12384–9. doi: 10.1073/pnas.2135336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Vogt PK. Inhibition of protein synthesis by Y box-binding protein 1 blocks oncogenic cell transformation. Mol Cell Biol. 2005;25:2095–106. doi: 10.1128/MCB.25.6.2095-2106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Vogt PK. Phosphorylation by Akt disables the anti-oncogenic activity of YB-1. Oncogene. 2008;27:1179–82. doi: 10.1038/sj.onc.1210719. [DOI] [PubMed] [Google Scholar]
- Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol Chem. 2000;275:38022–31. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–20. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278:50961–9. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- Braithwaite AW, Del Sal G, Lu X. Some p53-binding proteins that can function as arbiters of life and death. Cell Death Differ. 2006;13:984–93. doi: 10.1038/sj.cdd.4401924. [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci U S A. 1995;92:7642–6. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–42. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat Res. 2000;459:173–86. doi: 10.1016/s0921-8777(99)00072-5. [DOI] [PubMed] [Google Scholar]
- Budd ME, Choe W, Campbell JL. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J Biol Chem. 2000;275:16518–29. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Zhu H, Perlman P, Conrad-Webb H. The role of a conserved dodecamer sequence in yeast mitochondrial gene expression. Genome. 1989;31:757–60. doi: 10.1139/g89-134. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, Bhakat KK. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–80. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7:211–22. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Cheng X, Qin Y, Ivessa AS. Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p. Mol Genet Genomics. 2009;281:635–45. doi: 10.1007/s00438-009-0438-6. [DOI] [PubMed] [Google Scholar]
- Choe W, Budd M, Imamura O, Hoopes L, Campbell JL. Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol Cell Biol. 2002;22:4202–17. doi: 10.1128/MCB.22.12.4202-4217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad-Webb H, Perlman PS, Zhu H, Butow RA. The nuclear SUV3-1 mutation affects a variety of post-transcriptional processes in yeast mitochondria. Nucleic Acids Res. 1990;18:1369–76. doi: 10.1093/nar/18.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC, Longley MJ. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol Cell. 2008;32:457–8. doi: 10.1016/j.molcel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chattopadhyay R, Bhakat KK, Boldogh I, Kohno K, Prasad R, Wilson SH, Hazra TK. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem. 2007;282:28474–84. doi: 10.1074/jbc.M704672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988;85:7322–6. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmochowska A, Kalita K, Krawczyk M, Golik P, Mroczek K, Lazowska J, Stepien PP, Bartnik E. A human putative Suv3-like RNA helicase is conserved between Rhodobacter and all eukaryotes. Acta Biochim Pol. 1999;46:155–62. [PubMed] [Google Scholar]
- Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:5196–204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–82. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Malewicz M, Minczuk M, Golik P, Dmochowska A, Stepien PP. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol Gen Genet. 1998;260:108–14. doi: 10.1007/s004380050876. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ovchinnikov LP, Sorensen PH. Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle. 2006;5:1143–7. doi: 10.4161/cc.5.11.2784. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–99. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Farge G, Holmlund T, Khvorostova J, Rofougaran R, Hofer A, Falkenberg M. The N-terminal domain of TWINKLE contributes to single-stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 2008;36:393–403. doi: 10.1093/nar/gkm1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F, Dyck EV. A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. Embo J. 1985;4:3525–3530. doi: 10.1002/j.1460-2075.1985.tb04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983;80:5345–9. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull. 2007;30:1685–92. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- Gaudreault I, Guay D, Lebel M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004;32:316–27. doi: 10.1093/nar/gkh170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George T, Wen Q, Griffiths R, Ganesh A, Meuth M, Sanders CM. Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. Nucleic Acids Res. 2009;37:6491–502. doi: 10.1093/nar/gkp671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet. 2009;18:328–40. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa SL, Doetsch PW, Hamilton KK, Martin AM, Okenquist SA, Lenz J, Boss JM. DNA binding properties of YB-1 and dbpA: binding to double-stranded, single-stranded, and abasic site containing DNAs. Nucleic Acids Res. 1991;19:4915–20. doi: 10.1093/nar/19.18.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29:573–9. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- Imamura O, Campbell JL. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc Natl Acad Sci U S A. 2003;100:8193–8. doi: 10.1073/pnas.1431624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, Izumi H, Ohmori H, Okamoto T, Ohga T, Uchiumi T, Kuwano M, Kohno K. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999;59:342–6. [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–89. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- Khidr L, Wu G, Davila A, Procaccio V, Wallace D, Lee WH. Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J Biol Chem. 2008;283:27064–73. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim HD, Ryu GH, Kim DH, Hurwitz J, Seo YS. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 2006;34:1854–64. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–8. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–32. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. Embo J. 2004;23:2423–9. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–9. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- Kuwano M, Uchiumi T, Hayakawa H, Ono M, Wada M, Izumi H, Kohno K. The basic and clinical implications of ABC transporters, Y-box-binding protein-1 (YB-1) and angiogenesis-related factors in human malignancies. Cancer Sci. 2003;94:9–14. doi: 10.1111/j.1349-7006.2003.tb01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. Embo J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Kim DW, Bae SH, Kim JA, Ryu GH, Kwon YN, Kim KA, Koo HS, Seo YS. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 2000;28:2873–81. doi: 10.1093/nar/28.15.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–87. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald GH, Itoh-Lindstrom Y, Ting JP. The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J Biol Chem. 1995;270:3527–33. doi: 10.1074/jbc.270.8.3527. [DOI] [PubMed] [Google Scholar]
- Marenstein DR, Ocampo MT, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J Biol Chem. 2001;276:21242–9. doi: 10.1074/jbc.M101594200. [DOI] [PubMed] [Google Scholar]
- Margossian SP, Li H, Zassenhaus HP, Butow RA. The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- Masuda-Sasa T, Imamura O, Campbell JL. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34:1865–75. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, Mroczek S, Pawlak SD, Stepien PP. Human ATP-dependent RNA/DNA helicase hSuv3p interacts with the cofactor of survivin HBXIP. Febs J. 2005;272:5008–19. doi: 10.1111/j.1742-4658.2005.04910.x. [DOI] [PubMed] [Google Scholar]
- Minczuk M, Piwowarski J, Papworth MA, Awiszus K, Schalinski S, Dziembowski A, Dmochowska A, Bartnik E, Tokatlidis K, Stepien PP, Borowski P. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–86. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol Cell Biol. 2002;22:4086–93. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul E, Cronan R, Weston PJ, Boekelheide K, Sedivy JM, Lee SY, Wiest DL, Resnick MB, Klysik JE. Disruption of Supv3L1 damages the skin and causes sarcopenia, loss of fat, and death. Mamm Genome. 2009;20:92–108. doi: 10.1007/s00335-008-9168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M, Mason P, Szczesny RJ, Maddukuri L, Dziwura S, Jedrzejczak R, Paul E, Wojcik A, Dybczynska L, Tudek B, Bartnik E, Klysik J, Bohr VA, Stepien PP. Interaction of human SUV3 RNA/DNA helicase with BLM helicase; loss of the SUV3 gene results in mouse embryonic lethality. Mech Ageing Dev. 2007;128:609–17. doi: 10.1016/j.mad.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–80. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J Biol Chem. 2008;283:27483–93. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Z, Vijayakumar S, Chen CF, Chen PL, Lee WH. Purified human SUV3p exhibits multiple-substrate unwinding activity upon conformational change. Biochemistry. 2004;43:4781–90. doi: 10.1021/bi0356449. [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Evdokimova V, Thomas AA, Ovchinnikov LP. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J Biol Chem. 2001;276:44841–7. doi: 10.1074/jbc.M107581200. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–31. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Stepien PP, Margossian SP, Landsman D, Butow RA. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc Natl Acad Sci U S A. 1992;89:6813–7. doi: 10.1073/pnas.89.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–56. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–98. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny RJ, Obriot H, Paczkowska A, Jedrzejczak R, Dmochowska A, Bartnik E, Formstecher P, Polakowska R, Stepien PP. Down-regulation of human RNA/DNA helicase SUV3 induces apoptosis by a caspase- and AIF-dependent pathway. Biol Cell. 2007;99:323–32. doi: 10.1042/BC20060108. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102:17687–92. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet. 2004;13:3219–27. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–4. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- Wagner M, Price G, Rothstein R. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics. 2006;174:555–73. doi: 10.1534/genetics.104.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J Biol Chem. 2009;284:20812–21. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem. 2006;281:25791–802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–51. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–36. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–4. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]


