Abstract
Transcriptional silencing associated with aberrant promoter hypermethylation is a common mechanism of inactivation of tumor suppressor genes in cancer cells. To globally profile the genes silenced by hypermethylation in prostate cancer, we screened a whole genome expression microarray for genes reactivated in the LNCaP, DU-145, PC-3 and MDA2b prostate tumor cell lines after treatment with the demethylating drug 5-aza-2 deoxycytidine and histone deacetylation inhibiting drug trichostatin A. A total of 2997 genes showed at least 2-fold upregulation of expression after drug treatment in at least one prostate tumor cell line. For validation we examined the first 45 genes, ranked by upregulation of expression, that had a typical CpG island and were known to be expressed in the normal cell counterpart. Two important findings were firstly that several genes known to be frequently hypermethylated in prostate cancer were apparent. Secondly, validation studies revealed eight novel genes hypermethylated in the prostate tumor cell lines, four of which were unmethylated in normal prostate cells and hypermethylated in primary prostate tumors (SLC15A3 66%, KRT7 54%, TACSTD2 17%, GADD45b 3%). Thus, we established the utility of our screen for genes hypermethylated in prostate cancer cells. One of the novel genes was TACSTD2/TROP2 a marker of human prostate basal cells with stem cell characteristics. TACSTD2 was unmethylated in prostatic intraepithelial neoplasia and may have utility in emerging methylation-based detection of prostate cancer tests. Further study of the hypermethylome will provide insight into the biology of the disease and facilitate translational studies in prostate cancer.
Keywords: Prostate cancer, promoter hypermethylation, tumor suppressor genes, expression microarray
Introduction
Aberrant DNA hypermethylation of CpG islands in the promoter region of genes is well established as a common mechanism for the transcriptional silencing of tumor suppressor genes in cancer cells and, thus, as an alternative mechanism of functional inactivation (1). The GSTP1, p16INK4a, CDH1, APC and RASSF1A tumor suppressor genes as well as a number of other cancer genes have been identified as hypermethylated with associated loss of expression in prostate cancer (2). By definition, a candidate gene approach has resulted in the examination of only a limited number of genes for epigenetic alteration. Many other tumor suppressor and cancer genes important in prostate tumorigenesis likely remain to be identified. A global approach to the identification of epigenetically silenced genes in prostate tumor cells could provide methylation signatures for early detection and for predictive classification studies, identify novel targets for therapy, and lead to further elucidation of the biology of this disease.
One global approach to the identification of epigenetically silenced genes in tumor cells is based on the reversal of epigenetic silencing by drugs such as 5-aza-2 deoxycytidine (5Aza-dC) resulting in re-expression analyzed by well-annotated gene expression arrays. This approach can preferentially identify re-expression of epigenetically silenced genes over methylated CpG islands that do not affect transcription. A proportion of the reexpressed genes will have been silenced by promoter hypermethylation in the untreated tumor cell lines (3-6).
In the present study, we examined the global reactivation of epigenetically silenced genes in prostate cancer by analysis of a gene expression microarray with RNA isolated from 4 prostate tumor cell lines after treatment with 5Aza-dC and trichostatin A (TSA). Through intuitive selection of upregulated genes followed by validation, we have evidence that at least 20 of 45 genes examined are hypermethylated in prostate cancer, and thus, our screen preferentially selected for epigenetically silenced genes. We report here 4 genes newly identified as hypermethylated in primary prostate tumor specimens. Informed analysis of function combined with a pathway and network database analysis supports the relevance of these genes to prostate cancer.
Materials and Methods
Cell lines and Drug Treatment
Four prostate cancer cell lines LNCaP, DU-145, PC-3 and MDA2b were obtained directly from the American Type Culture Collection (ATCC) and were cultured according to ATCC recommendations. The 4 prostate cancer cell lines were treated with 5-aza-2 deoxycytidine (5Aza-dC, Sigma, St.Louis, MO) and trichostatin A (TSA, Wako, Richmond, VA) in a combined treatment. 5Aza-dC was dissolved in phosphate buffered saline (PBS) as a 5mM stock solution, and stored in aliquots at -80°C. TSA was dissolved in absolute ethanol as a 330μM stock solution, and stored at -20°C. Cells were exposed to 5Aza-dC to a final concentration of 5μM at 0, 24 and 72 hours, over two cell divisions by counting of the cells, and then, treated with TSA to a final concentration of 500nM during the 24 hours before RNA extraction. Mock (untreated) cells were cultured with the equivalent volume of PBS alone and, for the 24 hours before RNA extraction, with the equivalent volume of EtOH.
Oligonucleotide Array Hybridization
Total RNA used for microarray analysis was isolated from treated and mock cultured cells using TRIZOL reagent (Invitrogen, Carlsbad, CA) and purified using an RNAeasy Mini Kit (Qiagen, Valencia, CA), combined with DNase treatment. RNA quality was confirmed by the ratio of 28S and 18S ribosomal RNA after agarose gel electrophoresis. Total RNA was reverse-transcribed using oligo(dT)24 primer and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) for 1.5 hours at 42°C. cDNA was labeled with Cy3 or Cy5 (Amersham Biosciences, Piscataway, NJ) and then hybridized, according to the manufacturer's instructions for 18 hours at 42 °C, to slides with oligonucleotides from 14,802 genes, which were processed and spotted in the DNA Microarray facility at Fox Chase Cancer Center from 15k human oligonucleotide microarray (MWG-Biotech Inc, High Point, NC). The Gene list, Gene ID and Template files for human 15k oligo A can be viewed at http://www.ocimumbio.com/web/arrays/download.asp
Each cell line was hybridized twice, with a dye-flip replicate using reversed labeling. The hybridized slides were scanned using a GMS 428 Scanner (Affymetrix, Santa Clara, CA) to generate high-resolution images for both Cy3 and Cy5 channels. Image analysis was performed using the ImaGene software (BioDiscovery, Inc, El Segundo, CA). The spots were identified using an optimized segmentation algorithm. Spots of poor quality, as well as spots with signal levels indistinguishable from the background, were flagged as bad spots. The image data were extracted and analyzed using the Functional Genomics Data Pipeline (7).
Data Analysis
Normalization and background correction were performed using LOWESS with a width of 0.7 and local background correction. The PC-3 flip array data was removed as quality control measures and its MA plot suggested a poor quality array. Our goal was to identify genes that changed expression due to methylation in the maximum number of cell lines, since this provided the greatest probability of in vivo methylation. We therefore categorized the genes based on their fold change, determining a cut-off of 2-fold (i.e., ratio = 2, log-ratio = 1) between treated and untreated cell lines using the mean value of the dye-flips. We did not correct for disparate values between the dye-flip readings, as we took one large value as indicative of potential methylation in that cell line, and our goal was to count the number of cell lines with such potential methylation. We ranked the genes based on number of cell lines with potential methylation. We noted several well-known methylation targets, which we took as partial validation of our approach, and pursued the novel genes further.
Specimen Collection and DNA Extraction
After IRB approval and informed consent, primary prostate tumor tissues were obtained from surgical specimens resected at Fox Chase Cancer Center followed by pathological review and dissection of tumor cell-enriched areas. In this study we used histologically normal tissue from a prostate with no evidence of cancer obtained from cystoprostatectomy of age-matched bladder cancer patients. DNA was extracted from fresh-frozen tissues or paraffin blocks using a standard technique of digestion with proteinase K followed by phenol-chloroform extraction (8).
Bisulfite Modification
Genomic DNA (1μg) in a volume of 50μl was denatured by NaOH (0.2M) for 10 min at 37°C and then modified by hydroquinone and sodium bisulfite treatment at 50°C for 17 hours under a mineral oil layer. Modified DNA was purified using Wizard DNA Clean-Up system (Promega, Madison, WI). Modification was completed by NaOH (0.3M) treatment for 5 min at room temperature, followed by glycogen, ammonium acetate and ethanol precipitation (9). Pellets were resuspended in water and stored at -20°C for immediate use or −80°C for longer-term storage.
Genomic Sequencing
A typical 200-400bp size fragment containing the promoter CpG island was PCR amplified with bisulfite modified prostate cancer cell line DNA, normal prostate tissue DNA, and normal lymphocyte DNA for each gene analyzed. The PCR product was loaded into a 1.5% agarose gel, cut out and cleaned (Qiagen, Valencia, CA). We used a single PCR amplification for each of the KRT7, TACSTD2, SLC15A3, GADD45b, IFI30, ANXA2 and AQP3 genes. The primers used for each gene analyzed are given in Supplementary Table 2.
Quantitative Real-Time Methylation Specific PCR
Primer sequences to methylated DNA sequence were designed together with an internal Taqman probe labeled with FAM and MGBNFQ for SLC15A3, KRT7, GADD45b, IFI30 or TACSTD2. In vitro methylated normal human genomic DNA, confirmed by bisulfite sequencing to show methylation for the gene to be analyzed, was used as a positive control. The concentration of this DNA was determined and a series of dilutions made for the standard curve. Unmethylated sequence of the ACTINB gene was used as a normalizing control. The percentage of methylated alleles was calculated for a gene based on the standard curve. An Applied Biosystems 7500 Real-Time PCR machine was used for PCR and data analyzed with SDS 1.3.1 software. AQP3 and ANXA2 were assessed by conventional gel-based MSP. Primer and probe sequences are given in Supplementary Table 2.
Results and Discussion
Selection of Reactivated Genes for Validation
We analyzed differential expression on a 14,802 human gene microarray between RNA from 4 mock (untreated) prostate cancer cell lines and the same cell lines treated with 5Aza-dC over two cell doubling times and a single dose of TSA 24 hours before harvesting. We combined the 5Aza-dC treatment with TSA since there is a reported synergistic effect on demethylation of DNA (10). A total of 2997 genes were upregulated at least 2-fold in one or more of the four prostate cancer cell lines analyzed compared to the mock cells. The list of genes was then ranked in descending order from upregulation in all 4 cell lines to upregulation in 1 cell line only (Supplementary Table 1). Further data can be found at http://www.ncbi.nlm.nih.gov/geo/ upon publication. The list of ranked upregulated genes was then prioritized for validation by examination of expression status in the normal cell counterpart compared to the tumor cell and the presence and location of a CpG island in the promoter region (11). In addition, known imprinted genes e.g. IGF2, poorly annotated genes, or genes previously identified by us as upregulated but not to have cancer specific methylation e.g. TGM2 and GAGE7 (11) were excluded. Accordingly, we selected the first 45 genes that had higher or equal expression in normal prostate compared to prostate cancer cells according to the Cancer Genome Anatomy Project (CGAP) Serial Analysis Gene Expression (SAGE) database and that contained a CpG island within the promoter by the criteria of Takai and Jones (12) through WebGene analysis of the genomic sequence. Genes that showed no expression in normal cells or did not have a CpG island were excluded from immediate study (Table 1). Importantly, we noted that three of the most frequently methylated genes identified to date in prostate cancer i.e. GSTP1 (13), PDLIM4 (14) and IGFBP3 (15) were included in this 45 gene list.
Table 1. Upregulated genes identified by statistical analysis and database interrogation.
The table shows 45 genes that showed at least 2-fold upregulation in at least one of the four cell lines. Gene names in bold indicate genes identified as hypermethylated in prostate cancer cells in this study. Gene names in bold italics indicate genes previously identified as under epigenetic regulation in prostate cancer cells. Location is chromosomal map location. The tumor cell lines identified as hypermethylated by bisulfite sequencing are listed. Numbered refererences indicate published report of DNA methylation or other epigenetic regulation. Gene function was obtained from Entrez Gene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene).
| Gene Name | Location | Function | Methylation |
|---|---|---|---|
| NM_000228_1-- LAMB3 | 1q32.2 | Cell Communication | (40) |
| NM_005319_1-- H1F2 | 6p22.2 | Nucleosome condensation | Unmethylated |
| NM_000598_1-- IGFBP3 | 7p13 | Cell growth and differentiation, apoptosis | (15) |
| NM_002353_1-- TACSTD2 | 1p32.1 | Receptor activity | Methylated (LNCaP) |
| NM_000852_1-- GSTP1 | 11q13.2 | Transferase activity | (13) |
| NM_019554_1-- S100A4 | 1q21.3 | Calcium ion binding | (41) |
| NM_006332_1-- IFI30 | 19p13.11 | Immune response | Methylated (LNCaP, PC-3) |
| NM_004165_1-- RRAD | 16q22.1 | Nucleotide binding, GTPase activity | (42) |
| NM_000574_1-- DAF | 1q32.2 | Complement activation | Unmethylated |
| XM_027365_1-- ARL6IP | 16p12.3 | Protein transport, cell signaling | Unmethylated |
| NM_001647_1-- APOD | 3q29 | Lipid metabolic process | (31) |
| NM_014164_1-- FXYD5 | 19q13.12 | Cell adhesion | Unmethylated |
| NM_002306_1-- LGALS3 | 14q22.3 | Extracellular matrix organization, biogenesis | (43) |
| NM_001924_1-- GADD45a | 1p31.2 | Apoptosis, DNA repair | (31) |
| NM_001909_1—CTSD | 11p15.5 | Proteolysis | Unmethylated |
| NM_005556_1-- KRT7 | 12q13.13 | Cytoskeleton organization and biogenesis | Methylated (LNCaP, MDA2b) |
| NM_003246_1—THBS1 | 15q14 | Cell motility and adhesion, inflammation | (44) |
| AF373867_1-- TBX1C | 22q11.21 | Transcription factor | Unmethylated |
| NM_004039_1-- ANXA2 | 15q22.2 | Calcium ion binding | Methylated (LNCaP) |
| NM_002167_1-- ID3 | 1p36.12 | Negative regulation of transcription | (31) |
| NM_002165_1-- ID1 | 20q11.21 | Negative regulation of transcription | Unmethylated |
| NM_004925_1—AQP3 | 9p13.3 | Transport | Methylated (LNCaP) |
| NM_000610_1-- CD44 | 11p13 | Cell adhesion | (45) |
| NM_003687_1—PDLIM4 | 5q23.3 | Protein, metal ion binding | (14) |
| NM_002084-- GPX3 | 5q33.1 | Glutathione peroxidase activity | (31) |
| NM_002229_1-- JUNB | 19p13.13 | Transcription factor | (31) |
| NM_006005_1—WFS1 | 4p16.1 | Putative transmembrane protein | Not done |
| NM_001953_1-- ECGF1 | 22q13.33 | Growth factor | Unmethylated |
| NM_004417_1-- DUSP1 | 5q35.1 | Protein binding | (31, 34) |
| NM_006111_1-- ACAA2 | 18q21.1 | Acetyl-CoA c-acyltransferase activity | Unmethylated |
| NM_005822_1-- DSCR1L1 | 6p12.3 | Central nervous system development | Not done |
| NM_002204_1-- ITGA3 | 17q21.33 | Receptor | Unmethylated |
| NM_006496_1-- GNAI3 | 17q24.1 | Nucleotide binding | Unmethylated |
| NM_006509_1-- RELB | 19q13.32 | Transcription factor | Unmethylated |
| NM_005536_1-- IMPA1 | 8q21.13 | Phosphatidylinositol biosynthetic process | Not done |
| NM_006317_1-- BASP1 | 5q15.1 | Brain acid-soluble protein | Methylated in all 4 lines |
| AL050044_1-- GADD45b (hypothetical protein dkfzp566b133) | 19p13.3 | Apoptosis, Regulation of MAPKK activity | Methylated (DU-145, LNCaP) |
| NM_005737_1-- ARL7 | 2q37.1 | Binds and exchanges GTP and GDP | Unmethylated |
| NM_005919_1-- MEF2B | 19p13.11 | Transcription factor | Not done |
| NM_001673_1-- ASNS | 7q21.3 | Aspartate and asparagine activity | Unmethylated |
| NM_005253_1-- FOSL2 | 2p23.2 | Transcription factor | Unmethylated |
| NM_016582_1-- SLC15A3 (PHT2) | 11q12.2 | Transporter activity | Methylated in all 4 lines |
| NM_021967_1-- SERF1A | 5q13.2 | Nervous system development | Not done |
| NM_001386_1-- DPYSL2 | 8p21.2 | Dihydropyrimidinase activity | Not done |
| NM_002083_1-- GPX2 | 14q23.3 | Response to oxidative stress | Not done |
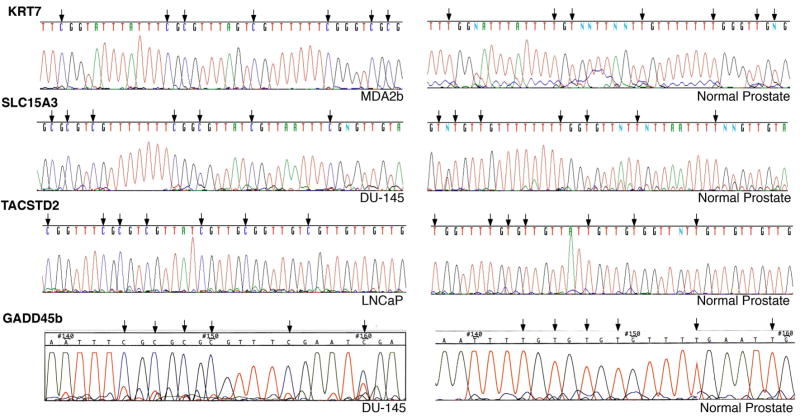
We then examined the published literature through the GeneCard (www.genecards.org) and PubMed databases which revealed that 15 genes were previously reported to be hypermethylated in prostate cancer (Table 1). These 15 genes were excluded from immediate analysis. We therefore prioritized the remaining genes for study and the promoter methylation status of 23 of this set of genes was first validated by direct bisulfite sequencing of the untreated prostate tumor cell line DNA. Eight genes showed extensive methylation, i.e. in a majority of CG dinucleotides (Figure 1 and Supplementary Figure 1), in at least one prostate tumor cell line (Table 1). To examine if hypermethylation was specific to neoplastic prostate cells, we performed direct bisulfite sequencing of a normal prostate tissue DNA obtained from the cystoprostatectomy (for bladder cancer) of an age-matched male with no clinical or histopathological evidence of prostate cancer and also a normal lymphocyte DNA. With the exception of BASP1, all genes were unmethylated in the normal prostate tissue DNA (Figure 1 and Supplementary Figure 1) and unmethylated in the normal lymphocyte DNA.
Figure 1. Bisulfite sequencing of the promoter CpG island of the KRT7, SLC15A3, TACSTD2 and GADD45b genes in prostate tumor DNA and normal DNA.
Representative sequencing of prostate tumor cell line DNA and histologically normal prostate tissue (from an age-matched cystoprostatectomy patient) DNA after bisulfite modification. Unmethylated cytosines (C) are converted to Uracil (T). The presence of C preceding a G in the sites indicated by black arrows demonstrates that these cytosines were methylated in the tumor cell line DNA. The presence of T instead of C in the same positions demonstrates that these cytosines were unmethylated in the normal prostate tissue DNA.
Frequency of Hypermethylation of Studied Genes
Since there is evidence that transformed tumor cell lines can have more gene methylation than the patient tumor specimen counterparts (16), we next examined the frequency and timing of hypermethylation of the KRT7, TACSTD2, SLC15A3, GADD45b and IFI30 genes by quantitative real-time MSP of DNAs from a set of 19 PIN and 35 prostate tumor tissue specimens from patients (Table 2 and Figure 2). The tumors consisted of 18 low and intermediate grade (≤ Gleason 7), 16 high grade (≥ Gleason 8) and one of unknown grade. Thirteen tumors were stage I or II and 22 tumors were stage III or IV. Hypermethylation of the ANXA2 and AQP3 genes was examined by conventional gel-based MSP and verified by bisulfite direct sequencing in a subset of 12 PIN and 20 prostate tumor tissue specimen DNA described above.
Table 2. Frequency of hypermethylation of KRT7, SLC15A3 and TACSTD2 by lesion and histological grade and stage.
Fisher's exact test was used to explore whether the hypermethylation of a given gene was related to tumor grade or stage. Results were declared statistically significant at the 5% significance level. M = number of patients with methylated gene. U = number of patients with non-methylated gene.
| KRT7 | SLC15A3 | TACSTD2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| M | U | M | U | M | U | ||||
| PIN (19) | 10 (53%) | 9 (47%) | p-value 1.00 | 7 (37%) | 12 (63%) | p-value 0.051 | 0 (0%) | 19 (100%) | p-value 0.08 |
| Prostate tumors (35) | 19 (54%) | 16 (46%) | 23 (66%) | 12 (34%) | 6 (17%) | 29 (83%) | |||
| Gleason ≤ 7 | 13 (72%) | 5 (28%) |
p-value 0.037 |
12 (67%) | 6 (33%) |
p-value 1.00 |
2 (11%) | 16 (89%) |
p-value 0.39 |
| Gleason ≥ 8 | 5 (31%) | 11 (69%) | 11 (69%) | 5 (31%) | 4 (25%) | 12 (75%) | |||
| Stage I-II | 9 (69%) | 4 (31%) |
p-value 0.29 |
9 (69%) | 4 (31%) |
p-value 1.00 |
1 (7.7%) | 12 (92.3%) |
p-value 0.38 |
| Stage III-IV | 10 (45%) | 12 (55%) | 14 (64%) | 8 (36%) | 5 (23%) | 17 (77%) | |||
Figure 2. Distribution plots for methylation levels of the KRT7, SLC15A3 and TACSTD2 genes. (a) by Gleason score. (b) by pathological stage.
Quantitative methylation specific PCR of 19 PIN and 35 primary prostate tumor specimen DNA. The percentage of methylated alleles (PMA) was calculated as the ratio between the PCR amplification product of the gene of interest and the ACTINB reference gene multiplied by 100. The PMA is given as a log value Ln (% + 1). The horizontal solid bar indicates the median and the hatched bar indicates a PMA of 5% which we chose as a cut-off point for scoring as methylated.
The keratin 7 (KRT7) gene was hypermethylated in 10 of 19 (53%) PIN and 19/35 (54%) tumors. There was a significant difference (p = 0.037) in frequency between Gleason ≤7 13/18 and 5/16 Gleason ≥8, but not between stage I-II and III-IV prostate tumors (Table 2). Cytokeratins are a subfamily of intermediate filament proteins characterized by biochemical diversity being represented in human epithelial tissues by at least 20 different polypeptides. Expression of particular cytokeratins is often cell-type specific. KRT7 is expressed in the ductal epithelium of the genitourinary tract. Expression also varies by course of terminal differentiation (17). Hypermethylation of KRT7 might indicate lineage differentiation or provide a growth advantage through cell-cell adhesion. A recent study reported the subgroup of clear cell renal cell carcinoma with KRT7 expression to be associated with genetic stability, a distinct global expression signature and a more indolent clinical course (18). A network of molecular interactions and canonical pathways for KRT7 formed by Ingenuity Pathways Analysis (IPA) was extensive. Consequently, an example of interaction with one cancer relevant pathway (Wnt signaling) only is shown in Supplementary Figure 2.
SLC15A3, also known as PHT2 or PTR3, is a member of the SLC15 family of electrogenic transporters that utilize the proton-motive force for uphill transport of short chain peptides and peptido-mimetics into a variety of cells (19). While the function of SLC15A3 in cancer cells is unclear, IPA identified interaction of SLC15A3 with the NFkB signaling pathway and p38 MAPK signaling pathway as well as other cancer-related pathways (Supplementary Figure 2). We found SLC15A3 to be methylated in 7 of 19 (37%) PIN and 23 of 35 (66%) of prostate tumors (Table 2 and Figure 2).
Tumor-associated calcium signal transducer-2 (TACSTD2) or TROP-2 is a cell surface glycoprotein. Its function remains largely unknown; however, it is phosphorylated by protein kinase C and cross-linking TACSTD2 with antibodies causes a transient increase in intracellular calcium levels, implying that it has a role in signal transduction (20). A recent report identified two subpopulations of basal cells based on TROP2 expression and reported TROP2 to be a marker of human prostate basal cells with stem cell characteristics (21). Prostate tumors with TACSTD2/TROP2 hypermethylation and associated loss of expression may identify a distinct subgroup in terms of tumor lineage or tumor behavior. IPA of TACSTD2 is shown in Supplementary Figure 2 and interestingly, TACSTD2 interacts with SMARCA4/BRG1 reported to be mutated in tumor cell lines including DU-145 (22) and a component of the SWI-SNF chromatin remodeling complex. In our study we found TACSTD2 to be unmethylated in 19 PIN but hypermethylated in 6/35 tumors (p = 0.08) (Table 2 and Figure 2). TACSTD2 has been reported to be hypermethylated in glioblastomas (23). Current opinion is that patients diagnosed with PIN are not at a higher risk of prostate cancer than are patients diagnosed as benign (24). Thus, a gene methylated in prostate tumors but not PIN might be a useful marker for the differentiation of PIN from other more aggressive lesions e.g. intraductal carcinoma. TACSTD2 may therefore have utility in emerging methylation-based detection of prostate cancer tests (25-28).
Promoter hypermethylation of the growth arrest and DNA-damage-inducible, beta (GADD45b also referred to as hypothetical protein dkfzp566b133) gene was found by qMSP in 1 prostate tumor only (Gleason 9, stage IV). Direct bisulfite sequencing of the tumor DNA confirmed this result (data not shown). GADD45 is involved in the regulation of cell cycle arrest and apoptosis and inhibition of cell growth. GADD45 responds to environmental stresses by mediating activation of the p38/JNK pathway (29). A selective IPA of the ten pathways most strongly associated with GADD45b is shown in Supplementary Figure 2. GADD45b has been reported to be hypermethylated in hepatocellular tumors (30). Another GADD45 gene, GADD45a, was previously reported as hypermethylated in prostate tumors (31).
The IFI30, ANXA2 and AQP3 genes were unmethylated in all PIN and prostate tumor DNA examined. It is possible that analysis of a larger number, or a particular subtype, of primary prostate tumor specimens may yet reveal methylation of these genes. Of the four prostate tumor cell lines, 3 lines were established from distant metastases and the other, LNCaP, from a lymph node metastasis (32). The source of the cell lines taken together with our observation that these genes were unmethylated in primary specimens highlights that the prostate tumor cell lines may be unrepresentative of the disease found in the population. It will be interesting to see if any of these three genes are subsequently identified as aberrantly methylated in other types of cancer.
Evidence for Epigenetic Reactivation of Additional Genes
Evidence for the specificity and potential of our screen was provided by the inclusion of the GSTP1 gene known to be hypermethylated in the LNCaP and PC3 cell lines (13) as well as in the majority of primary prostate tumors, and two other genes: PDLIM4 (14) and IGFBP3 (15) reported to be frequently methylated in primary prostate cancer in the 45 selected genes. Since an oligonucleotide probe on an array may not discriminate between alternative splice forms of the same gene, genes where expression of not all isoforms is lost with promoter hypermethylation, e.g. RASSF1A, will likely not appear upregulated by a global reactivation approach (33). There was evidence from the literature for epigenetic regulation of several other genes in the 45 studied. The references for such genes are given in Table 1. In our list (Table 1), there were also additional genes reported by several groups to be upregulated after epigenetic reactivation but with no evidence of methylation in prostate tumor cells e.g. DUSP1 (31, 34). Such genes may be upregulated by demethylation and reactivation of another gene or may result from a stress response to the drug treatment. In general, our study compares well to the “hit rate” i.e. the ratio of genes hypermethylated among genes upregulated after demethylation reported in other global reactivation studies (5, 6, 31). We did not place emphasis upon the higher fold of reactivation since we considered this to be arbitrary given the current limited understanding of the degree of reexpression necessary to restore the normal function of a particular gene.
The Prostate Cancer Cell Hypermethylome
The average total number of genes methylated with functional significance in the prostate tumor cell is unknown but might be reasonably estimated as several hundred (35, 36). Several prostate cancer global methylation studies have been reported. Yu et al compared the results from PC-3, DU-145 and LNCaP lines on an oligonucleotide-based methylation array to an expression array (37) and reported a number of genes methylated by array analysis, some of which were replicated by non-quantitative conventional MSP analysis but not direct bisulfite sequencing. Lodygin et al performed a global epigenetic reactivation of the same 3 prostate tumor cell lines and identified a number of known, as well as novel, genes methylated in prostate cancer from the subset of 50 genes examined (31). Several of the genes in the ranked selection by Lodygin were also in our selected list. Their study did not perform quantitative MSP or examine normal cells for imprinted or tissue-specific methylated genes. Hoque et al also studied epigenetic reactivation in the same 3 lines as well as the 22Rv1 line. They selected 45 reactivated genes of which 9 were previously reported to be hypermethylated in cancer cells, 16 of the remaining 36 were methylated in the cell lines, 8 in primary prostate tumors of which 3 did not have methylation in normal prostate cells (38). Chung et al used methylated CpG island amplification (MCA) coupled with representational difference analysis (RDA) of PC-3, DU-145 and LNCaP and validation by combined bisulfite restriction analysis (COBRA) and pyrosequencing identified 6 novel methylated genes in primary prostate tumors that had significantly higher methylation than in adjacent normal prostate tissue (39).
Summary
In summary, we report for the first time 4 novel genes with promoter hypermethylation in primary prostate tumor specimens but that are unmethylated in normal cells. One of these genes, TACSTD2/TROP2, is a potential marker for detection or diagnosis of prostate cancer rather than for PIN only, as well as a prostate stem cell marker (21). The specificity of our screen for hypermethylated genes is further supported by the reactivation of GSTP1 and other genes known to be frequently methylated in prostate cancer in our selection. Further mining of the data provided here (Supplementary Table 1) as well as emerging technologies will increase our knowledge of the prostate hypermethylome. Such studies should lead to further understanding of the biology of prostate tumorigenesis and the identification of further hypermethylated genes as candidate markers for the diagnosis and prognosis of prostate cancer.
Supplementary Material
After bisulfite modification, unmethylated cytosines (C) are converted to Uracil (T). The presence of C preceding a G in the sites indicated by black arrows demonstrates that these cytosines were methylated in the tumor cell line DNA, while the presence of T instead of C in the same positions demonstrates that these cytosines were unmethylated in the normal prostate tissue DNA.
The IPA (Ingenuity Systems, www.ingenuity.com) for TACSTD2 and SLC15A3 are shown as generated. The IPA for KRT7 and GADD45b include only interactions and pathways selected for cancer relevance as the original IPA generated an extremely large number of interactions. A direct interaction is indicated by a solid line and an indirect interaction by a broken line. CP: canonical pathway. Gene names enclosed in square = growth factor, diamond = enzyme, anvil = transporter, vertical oval = transmembrane receptor, inverted triangle = kinase, circles = other.
Footnotes
Conflict of Interest: P.C. is a paid consultant to Oncomethylome Sciences. The terms of this arrangement are being managed by Fox Chase Cancer Center in accordance with its conflict of interest policies.
Supplementary information is available at Cancer Prevention Research's website
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2002;8:514–9. [PubMed] [Google Scholar]
- 3.Cairns P. 5′-azacytidine expression arrays. Methods in molecular biology (Clifton, NJ. 2009;507:165–74. doi: 10.1007/978-1-59745-522-0_13. [DOI] [PubMed] [Google Scholar]
- 4.Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:961–6. [PubMed] [Google Scholar]
- 5.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–9. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–95. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 7.Grant JD, Somers LA, Zhang Y, Manion FJ, Bidaut G, Ochs MF. FGDP: functional genomics data pipeline for automated, multiple microarray data analyses. Bioinformatics. 2004;20:282–3. doi: 10.1093/bioinformatics/btg407. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook J, Russell DW. A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning. [Google Scholar]
- 9.Herman JG, Graff JR, Myöhänen S, BD N, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 11.Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res. 2006;66:5021–8. doi: 10.1158/0008-5472.CAN-05-3365. [DOI] [PubMed] [Google Scholar]
- 12.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the π-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci USA. 1994;91:11733–7. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanaja DK, Ballman KV, Morlan BW, et al. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin Cancer Res. 2006;12:1128–36. doi: 10.1158/1078-0432.CCR-05-2072. [DOI] [PubMed] [Google Scholar]
- 15.Perry AS, Loftus B, Moroose R, et al. In silico mining identifies IGFBP3 as a novel target of methylation in prostate cancer. Br J Cancer. 2007;96:1587–94. doi: 10.1038/sj.bjc.6603767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smiraglia DJ, Rush LJ, Fruhwald MC, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10:1413–9. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 17.Omary MB, Ku NO, Strnad P, Hanada S. Toward unraveling the complexity of simple epithelial keratins in human disease. The Journal of clinical investigation. 2009;119:1794–805. doi: 10.1172/JCI37762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertz KD, Demichelis F, Sboner A, et al. Association of cytokeratin 7 and 19 expression with genomic stability and favorable prognosis in clear cell renal cell cancer. International journal of cancer. 2008;123:569–76. doi: 10.1002/ijc.23565. [DOI] [PubMed] [Google Scholar]
- 19.Daniel H, Kottra G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch. 2004;447:610–8. doi: 10.1007/s00424-003-1101-4. [DOI] [PubMed] [Google Scholar]
- 20.Fornaro M, Dell'Arciprete R, Stella M, et al. Cloning of the gene encoding Trop-2, a cell-surface glycoprotein expressed by human carcinomas. International journal of cancer. 1995;62:610–8. doi: 10.1002/ijc.2910620520. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci U S A. 2008;105:20882–7. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong AK, Shanahan F, Chen Y, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–7. [PubMed] [Google Scholar]
- 23.Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- 24.Epstein JI, Herawi M. Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: implications for patient care. J Urol. 2006;175:820–34. doi: 10.1016/S0022-5347(05)00337-X. [DOI] [PubMed] [Google Scholar]
- 25.Cairns P, Esteller M, Herman JG, et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7:2727–30. [PubMed] [Google Scholar]
- 26.Harden SV, Sanderson H, Goodman SN, et al. Quantitative GSTP1 methylation and the detection of prostate adenocarcinoma in sextant biopsies. J Natl Cancer Inst. 2003;95:1634–7. doi: 10.1093/jnci/djg082. [DOI] [PubMed] [Google Scholar]
- 27.Hoque MO, Topaloglu O, Begum S, et al. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J Clin Oncol. 2005;23:6569–75. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Tokumaru Y, Harden SV, Sun DI, Yamashita K, Epstein JI, Sidransky D. Optimal use of a panel of methylation markers with GSTP1 hypermethylation in the diagnosis of prostate adenocarcinoma. Clin Cancer Res. 2004;10:5518–22. doi: 10.1158/1078-0432.CCR-04-0108. [DOI] [PubMed] [Google Scholar]
- 29.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–30. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 30.Qiu W, Zhou B, Zou H, et al. Hypermethylation of growth arrest DNA damage-inducible gene 45 beta promoter in human hepatocellular carcinoma. Am J Pathol. 2004;165:1689–99. doi: 10.1016/s0002-9440(10)63425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–27. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- 32.van Bokhoven A, Varella-Garcia M, Korch C, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205–25. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- 33.Shames DS, Girard L, Gao B, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauhala HE, Porkka KP, Tolonen TT, Martikainen PM, Tammela TL, Visakorpi T. Dual-specificity phosphatase 1 and serum/glucocorticoid-regulated kinase are downregulated in prostate cancer. International journal of cancer. 2005;117:738–45. doi: 10.1002/ijc.21270. [DOI] [PubMed] [Google Scholar]
- 35.Costello JF, Frühwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type–specific patterns. Nature Genet. 2000;25:132–8. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 36.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 37.Yu YP, Paranjpe S, Nelson J, et al. High throughput screening of methylation status of genes in prostate cancer using an oligonucleotide methylation array. Carcinogenesis. 2005;26:471–9. doi: 10.1093/carcin/bgh310. [DOI] [PubMed] [Google Scholar]
- 38.Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–70. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung W, Kwabi-Addo B, Ittmann M, et al. Identification of novel tumor markers in prostate, colon and breast cancer by unbiased methylation profiling. PLoS ONE. 2008;3:e2079. doi: 10.1371/journal.pone.0002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sathyanarayana UG, Padar A, Suzuki M, et al. Aberrant promoter methylation of laminin-5-encoding genes in prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2003;9:6395–400. [PubMed] [Google Scholar]
- 41.Rehman I, Goodarzi A, Cross SS, et al. DNA methylation and immunohistochemical analysis of the S100A4 calcium binding protein in human prostate cancer. Prostate. 2007;67:341–7. doi: 10.1002/pros.20401. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki M, Shigematsu H, Shivapurkar N, et al. Methylation of apoptosis related genes in the pathogenesis and prognosis of prostate cancer. Cancer Lett. 2006;242:222–30. doi: 10.1016/j.canlet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Ahmed H, Banerjee PP, Vasta GR. Differential expression of galectins in normal, benign and malignant prostate epithelial cells: silencing of galectin-3 expression in prostate cancer by its promoter methylation. Biochemical and biophysical research communications. 2007;358:241–6. doi: 10.1016/j.bbrc.2007.04.114. [DOI] [PubMed] [Google Scholar]
- 44.Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202:233–40. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 45.Verkaik NS, van Steenbrugge GJ, van Weerden WM, Bussemakers MJ, van der Kwast TH. Silencing of CD44 expression in prostate cancer by hypermethylation of the CD44 promoter region. Lab Invest. 2000;80:1291–8. doi: 10.1038/labinvest.3780137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
After bisulfite modification, unmethylated cytosines (C) are converted to Uracil (T). The presence of C preceding a G in the sites indicated by black arrows demonstrates that these cytosines were methylated in the tumor cell line DNA, while the presence of T instead of C in the same positions demonstrates that these cytosines were unmethylated in the normal prostate tissue DNA.
The IPA (Ingenuity Systems, www.ingenuity.com) for TACSTD2 and SLC15A3 are shown as generated. The IPA for KRT7 and GADD45b include only interactions and pathways selected for cancer relevance as the original IPA generated an extremely large number of interactions. A direct interaction is indicated by a solid line and an indirect interaction by a broken line. CP: canonical pathway. Gene names enclosed in square = growth factor, diamond = enzyme, anvil = transporter, vertical oval = transmembrane receptor, inverted triangle = kinase, circles = other.