Abstract
The phytohormone abscisic acid (ABA) functions through a family of fourteen PYR/PYL receptors, which were identified by resistance to pyrabactin, a synthetic inhibitor of seed germination. ABA activates these receptors to inhibit type 2C protein phosphatases, such as ABI1, yet it remains unclear whether these receptors can be antagonized. Here we demonstrate that pyrabactin is an agonist of PYR1 and PYL1, but unexpectedly an antagonist of PYL2. Crystal structures of the PYL2–pyrabactin and PYL1–pyrabactin–ABI1 complexes reveal the mechanism responsible for receptor-selective activation and inhibition, which enables us to design mutations that convert PYL1 to a pyrabactin-inhibited receptor and PYL2 to a pyrabactin-activated receptor, and to identify new pyrabactin-based ABA receptor agonists. Together, our results establish a new concept of ABA receptor antagonism, illustrate its underlying mechanisms, and provide a rational framework for discovering novel ABA receptor ligands.
The plant hormone abscisic acid (ABA) controls many important physiological processes, including seed germination, bud dormancy, and adaptive responses to environmental stresses such as drought and salinity. While downstream mediators of ABA signaling have been established, the protein receptors for ABA have eluded from identification for many years because of the high level of receptor redundancy. Through chemical genetics and yeast two-hybrid screening, a new class of START proteins has recently been described as ABA receptors in Arabidopsis thaliana1–3. These receptors are designated as pyrabactin resistance 1 (PYR1) and thirteen members of PYR1-like (PYL) receptors1 or as regulatory components of ABA receptors (RCAR)2. ABA binding to these receptors increases their ability to bind and inhibit type 2C protein phosphatases (PP2Cs), such as ABI1, ABI2, and HAB1. In the absence of ABA, these PP2Cs bind and inactivate subfamily 2 members of SNF1-related kinases (SnRK2 kinases) by dephosphorylating serine and threonine residues in their activation loop4–7. ABA-induced PP2C inhibition leads to SnRK2 activation by autophosphorylation and/or phosphorylation by upstream kinases. Activated SnRK2s in turn phosphorylate and activate downstream effectors such as the basic leucine-zipper transcription factors called ABFs/AREBs to switch on stress response programs7–15. The molecular mechanisms of ABA binding to the PYR/PYL receptors and PP2C inhibition have been revealed by a series of recent structural studies16–20. These structural studies highlight a conserved gate-latch-lock mechanism underlying ABA perception and signal transduction16–20. The apo-ABA receptor contains an open ligand binding pocket. ABA binding induces the closure of the ligand entry gate that allows the receptor to bind to and competitively inhibit PP2Cs. The interactions between PP2Cs and ABA receptors further induce conformational changes that lock the receptor in the closed conformation.
While the basic mechanisms of receptor activation from ABA binding to PP2C inhibition are illustrated by structures, fundamental questions pertaining to the functional regulation of the receptor activity remain. For example, the endogenous concentration of ABA in unstressed conditions is estimated to be 0.7–1.5 fg per guard cell pair, which corresponds to 0.7–1.5 μM21–23, a concentration range that is sufficient to bind and activate several different recombinant PYL–PP2C complexes1–3,16. ABA concentrations were also determined in vivo in guard cells by employing ABA-responsive reporter gene activation, indicating a cellular ABA level in unstressed conditions above a threshold of 0.3 μM24. Furthermore, several subtypes of ABA receptors (PYL5–14) show ABA-independent interactions with PP2Cs1,2, and yet the ABA response is inactivated in unstressed plants. These observations suggest that some or all ABA receptors need to be inhibited in unstressed conditions, yet the mechanism of how ABA receptors are inhibited is unknown.
While ABA is a pan-agonist of ABA receptors, pyrabactin was identified as selective agonist of PYR1 in a seed germination assay25 that promoted interaction of the HAB1 PP2C with a subset of PYR/PYL proteins1. Here we demonstrate that pyrabactin is surprisingly a PYL2 selective antagonist and identify the mechanism of receptor-selective activation and inhibition by pyrabactin. Our results demonstrate a new concept of ABA receptor antagonism, lay a theoretic foundation for future identification of physiological ABA receptor antagonists, and establish a structural framework to screen and design subtype-selective agonists and antagonists for unraveling ABA biology.
RESULTS
Subtype-selective activation and inhibition of ABA receptors by pyrabactin
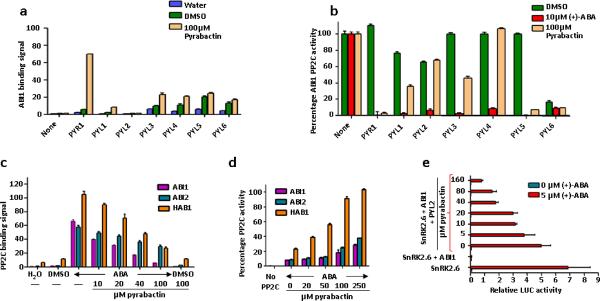
Pyrabactin selectively promoted the interaction of the HAB1 PP2C with PYR1, PYL1, and PYL3, but not PYL2 and PYL4, in yeast two hybrid assays1. To determine the biochemical basis of these observations, we used purified proteins of PYR1 and PYL1 to PYL6 (Fig. 1a and Supplementary Fig. 1a) to measure their interactions with the three PP2Cs, HAB1, ABI1, and ABI2, in vitro. Pyrabactin strongly promoted PYR1 to interact with all three PP2Cs, consistent with its original identification as a selective PYR1 agonist. Pyrabactin also promoted interaction of PYL1, PYL3, PYL6, and surprisingly PYL4 with the three PP2Cs. In contrast, pyrabactin did not promote PYL2 interaction with any of the three PP2Cs. In phosphatase assays, pyrabactin promoted PYR1, PYL1, PYL3, PYL5, and PYL6, but not PYL2 and PYL4, to inhibit all three PP2Cs (Fig. 1b and Supplementary Fig. 1b). Interestingly, in contrast to other receptors analyzed, PYL6 has high basal activity in both PP2C binding and inhibition. These results demonstrate different modes of PP2C binding and PP2C inhibition for different members of the PYR/PYL family.
Figure 1.
Pyrabactin functions both as selective ABA receptor agonist and antagonist. (a) Stimulation of PYR/PYL–ABI1 interaction by pyrabactin. PP2C interaction in the presence and absence of 100 μM pyrabactin was determined by AlphaScreen luminescence proximity assays as detailed in Methods (n=3, error bars=s.d.). (b) Pyrabactin-dependent inhibition of ABI1 phosphatase activities by PYR/PYL proteins (n=3, error bars=s.d.). (c) Pyrabactin inhibits the ABA-stimulated interaction between PYL2 and PP2Cs as determined by Alphascreen assays in the presence or absence of 10 μM (+)-ABA and the indicated concentrations of pyrabactin (n=3, error bars=s.d.). (d) Pyrabactin relieves the ABA-stimulated inhibition of PP2C phosphatase activity by PYL2. Reactions contained 10 μM (+)-ABA and the indicated concentrations of pyrabactin (n=3, error bars=s.d.). (e) Pyrabactin inhibits ABA-stimulated reporter gene activity in a PYL2-reconstituted ABA signaling pathway in Arabidopsis mesophyll protoplasts. Expression plasmids for PYL2, ABI1, SnRK2.6, and the transcription factor ABF2 were cotransfected into protoplasts together with a luciferase reporter plasmid driven from the ABA-responsive RD29B gene (n=3, error bars=s.d.).
Surprisingly, not only did pyrabactin fail to activate PYL2, but it also inhibited ABA-dependent PYL2 interaction with all three PP2Cs in a concentration-dependent manner, suggesting pyrabactin may be a PYL2-selective antagonist (Fig. 1c). Consistent with this observation, pyrabactin did not promote PYL2 to inhibit the three PP2Cs (Fig. 1b), but high concentrations of pyrabactin reversed ABA-dependent inhibition of PP2Cs by PYL2 (Fig. 1d). In addition, pyrabactin promoted PYR1 in a concentration-dependent manner to induce the expression of RD29B-LUC, an ABA-inducible reporter (Supplementary Fig. 1c), but inhibited PYL2 and ABA-dependent induction in a reconstituted ABA signaling pathway in Arabidopsis protoplasts26 (Fig. 1e). These results collectively establish that pyrabactin is a PYL2-selective antagonist.
Crystal structures of the PYL1–pyrabactin–ABI1 agonist and the PYL2–pyrabactin antagonist complexes
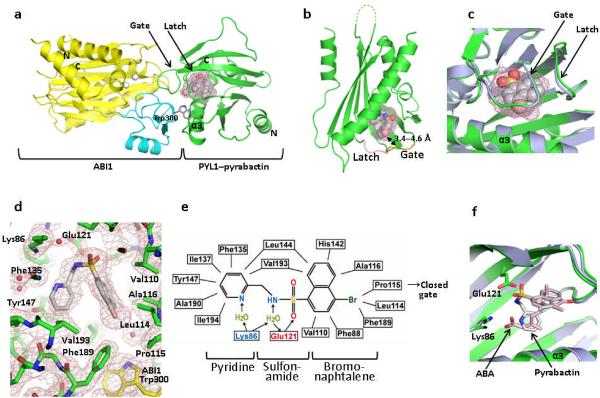
To understand the molecular basis of pyrabactin as a subtype-selective agonist and antagonist of ABA receptors, we determined the crystal structures of a PYL1–pyrabactin–ABI1 ternary complex and a PYL2–pyrabactin binary complex at resolutions of 2.15 Å and 1.85 Å, respectively. These structures were solved by molecular replacement starting from the PYL1–ABA–ABI1 and apo-PYL2 structures16,17,20, with the statistics of data and structures summarized in Table 1. For the PYL1–ABA–ABI1 complex, there are two ternary complexes in the P1 unit cell. Consistent with the agonist property of pyrabactin for PYL1, the overall arrangement of the PYL1–pyrabactin–ABI1 complex resembles the agonist structure of the PYL1–ABA–ABI1 complex, with the gate and latch loops of PYL1 (residues 112–116 and 142–144) adopting the closed conformation that is further stabilized by the insertion of the locking residue, Trp300 from ABI1 (see Fig. 2a–c and Supplementary Fig. 2a for an overlay of the ABA-bound and pyrabactin-bound structures). In the complex, the gate–latch interface is tightly packed against the active site of ABI1, therefore providing a mechanism of phosphatase inhibition.
Table 1.
Data collection and refinement statistics
| PYL2–Pyrabactin | PYL1–Pyrabactin–ABI1 | PYL2 A93F–Pyrabactin | PYL2 A93F–Pyrabactin–HAB1 | PYL2 A93F–Pyrabactin–ABI2 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C2221 | P1 | C2221 | P212121 | P212121 |
| Cell dimensions | |||||
| a, b, c(Å) | 62.27, 105.07, 185.08 | 59.98, 66.71, 72.60 | 62.08, 105.57, 182.90 | 62.08, 105.57, 182.90 | 62.13, 97.59, 134.50 |
| α,β,γ(°) | 90, 90, 90 | 115.8, 95.4, 105.6 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 30–1.85 (1.92–1.85)* | 30–2.15 (2.23–2.15) | 30–2.10 (2.18–2.10) | 30–2.55 (2.64–2.55) | 30–2.10 (2.18–2.10) |
| Rsym or Rmerge | 0.100 (0.706) | 0.098 (0.752) | 0.093(0.725) | 0.164(0.793) | 0.102(0.764) |
| I/σI | 32.2 (3.90) | 49.1 (4.5) | 27.6 (2.9) | 15.9 (2.4) | 32.4 (3.7) |
| Completeness (%) | 96.9 (92.8) | 97.6 (96.2) | 99.9 (99.9) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 12.7(12.8) | 13.4 (2.3) | 8.2 (8.4) | 8.4 (8.6) | 14.7 (14.6) |
| Refinement | |||||
| Resolution (Å) | 30–1.85 | 30–2.15 | 30–2.10 | 30–2.55 | 30–2.10 |
| No. reflections | 46913 | 46283 | 32811 | 18181 | 25445 |
| Rwork / Rfree (%) | 19.4/23.4 | 21.2% | 23.0% | 22.3% | 19.4% |
| No. atoms | |||||
| Protein | 4259 | 6956 | 4259 | 3706 | 3574 |
| Ligand/ion | 66 | 48 | 66 | 34 | 96 |
| Water | 327 | 180 | 223 | 128 | 171 |
| B-factors | |||||
| Protein | 39.49 | 48.59 | 55.39 | 49.61 | 46.35 |
| Ligand/ion | 67.16 | 42.23 | 101.40 | 84.16 | 40.79 |
| Water | 44.46 | 49.19 | 50.00 | 40.24 | 47.29 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.016 | 0.021 | 0.022 | 0.018 | 0.023 |
| Bond angles (°) | 1.493 | 1.73 | 1.66 | 1.44 | 1.74 |
Values in parentheses are for highest-resolution shell. One crystal was used for each structure.
Figure 2.
Structure of the PYL1–pyrabactin–ABI1 complex. (a) Structure overview of the complex. The N- and C- termini of PYL1 and ABI1, the locking residue Trp300, and the PYL1 α3-helix are indicated. Pyrabactin is shown as ball representation with the surrounding ligand binding pocket as mesh. (b) Structure of pyrabactin as ball model in the PYL1 pocket are shown as mesh. The gate and latch are shown in yellow and magenta with the gate–pyrabactin distance indicated. (c) Close view of the pyrabactin-bound PYL1 ligand binding pocket (green) overlaid with the PYL1 structure in the PYL1–ABA–ABI1 complex (blue-grey). (d) 2 F0−Fc electron density map of bound pyrabactin and its surrounding residues contoured at 1.0 σ. (e) Schematic representation of the interactions between pyrabactin and PYL1 binding pocket residues. Charged interactions and hydrogen bonds are indicated by arrows, hydrophobic interactions by solid lines with hydrogen bond donors in blue and acceptors in red. The position of pyrabactin relative to the closed gate is indicated. (f) Overlay of pyrabactin (grey) with (+)-ABA (pink) in the PYL1 binding pocket.
Pyrabactin, which is clearly defined by a high resolution electron density map, adopts a л-shape configuration in the PYL1 pocket (Fig. 2d and Supplementary Fig. 2b). The inter-molecular interactions between pyrabactin and PYL1 are diagrammed in Fig. 2e. The binding mode of pyrabactin mimics that of ABA with the naphthalene double ring of pyrabactin overlapping extensively with the cyclohexene ring of ABA (Fig. 2f). The bromide group from the naphthalene ring forms several Van der Waals interactions with Leu-Pro-Ala, three residues from the ligand entry gate (Fig. 2d,e), and these interactions are important to keep the gate in the closed conformation as the equivalent P88S mutation in PYR1 abolishes its responses to pyrabactin1. The nitrogen from the pyridine ring of pyrabactin functionally mimics the acidic group of ABA and forms a water-mediated hydrogen bond with Lys86. These interactions between pyrabactin and PYL1 provide a basis for understanding the structure-functione relationship of pyrabactin derivatives, and help to explain why the nitrogen group of pyridine is required for agonist activity of pyrabactin1.
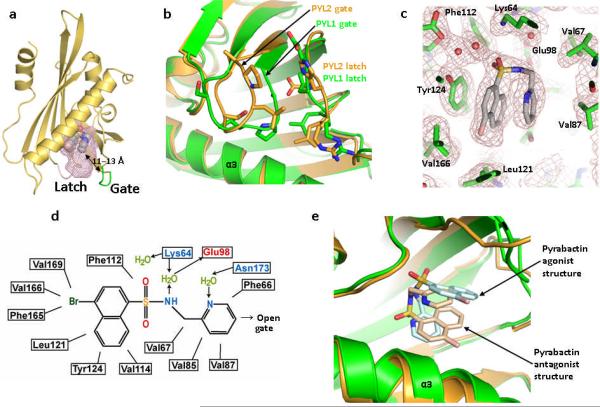
The PYL2–pyrabactin crystals were obtained under the same conditions as for the ABA-bound PYL2, but were formed in the same space group as the apo-PYL2. In contrast to the closed conformation of PYL1, pyrabactin-bound PYL2 adopts an open conformation where its ligand entry gate assumes a position similar to the apo-PYL2 structure (Fig. 3a,b). There are three PYL2–pyrabactin complexes in each asymmetric unit and pyrabactin assumes a similar л configuration in each complex. Interestingly, the binding orientation of pyrabactin in PYL2 is flipped 180° from that of PYL1. In this conformation, pyrabactin forms extensive interactions with PYL2 (Fig. 3c,d). The sulfonamide functional group mimics the acidic group of ABA, forming two water-mediated hydrogen bonds with Lys64 (Lys89 equivalent in PYL1) and one water mediated hydrogen bond with Glu98. The naphthalene ring of pyrabactin forms parallel packing interactions with the phenol ring of Tyr124 and the pyridine ring of pyrabactin (Fig. 3c). We also determined the crystal structure of pyrabactin itself, which adopts a more extended conformation than that in the ligand binding pockets of PYL1 and PYL2 (Supplementary Fig. 2c), indicating that pyrabactin undergoes an induced fit to accommodate the shape of the ligand binding pockets.
Figure 3.
PYL2–pyrabactin antagonist structure. (a) Overview of the PYL2–pyrabactin structure with the pocket shown as mesh. The gate and latch are shown in green and magenta with the gate-pyrabactin distance indicated. (b) Overlay of the gate and latch regions of the PYL2 antagonist (brown) and PYL1 agonist (green) structures. (c) 2 F0−Fc electron density map of bound pyrabactin and its surrounding residues contoured at 1.0 σ. (d) Schematic representation of the interactions between pyrabactin and PYL2 binding pocket residues. Charged interactions and hydrogen bonds are indicated by arrows, hydrophobic interactions by solid lines with hydrogen bond donors in blue and acceptors in red. The position of pyrabactin relative to the open gate is indicated. (e) Overlay of pyrabactin in the PYL2 antagonist (brown/ pale brown) and PYL1 agonist (green/ pale cyan) structures.
I137V converts PYL1 from a pyrabactin-activated to a pyrabactin-inhibited receptor
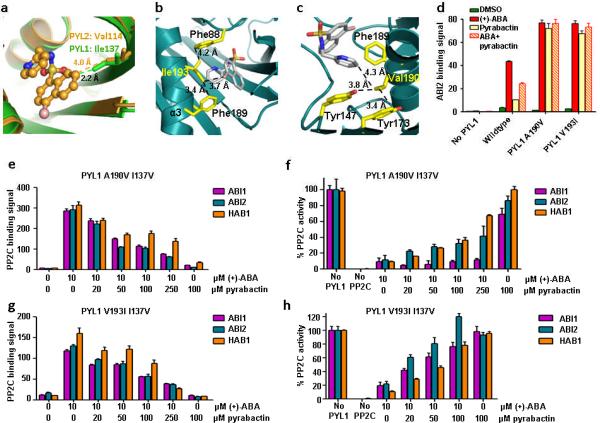
One key question is why pyrabactin adopts different conformations between PYL1 and PYL2. The ligand binding pockets of PYL2 and PYL1 only differ in three residues (PYL2/PYL1: Val114/Ile137, Val166/Ala190, and Val170/Ile194) (Supplementary Fig. 3). Structural modeling indicated that Val166/Ala190 and Val170/Ile194 changes would not interfere with the binding of pyrabactin in either the PYL1 or the PYL2 conformation. In contrast, the Val114/Ile137 change in PYL1 collides with the naphthalene ring of pyrabactin in the PYL2 conformation (C–C distance of 2.2 Å) (Fig. 4a) and forces pyrabactin to flip by 180°. Mutation of PYL1 Ile137 to valine, which was predicted to allow pyrabactin to adopt the PYL2-bound conformation, converted PYL1 to a pyrabactin-inhibited receptor. Due to the very weak pyrabactin agonist activity in wildtype PYL1, this change is most clearly seen in the context of two mutant PYL1 receptors, in which residues Ala190 and Val193 from helix 3 were exchanged against larger side chains that stabilize the binding of pyrabactin, as modeled in Fig. 4b and 4c. A190V and V193I mutations strongly increased PYL1 agonist activity (Fig. 4d and Supplementary Fig. 4). Introduction of the I137V mutation into either PYL1 A190V or PYL1 V193I strongly shifted the equilibrium from a predominantly pyrabactin-activated receptor to a predominantly pyrabactin-antagonized receptor, for which pyrabactin inhibited ABA-promoted activation in a concentration-dependent manner (Fig. 4e–h). From these results we conclude that removal of a single methyl group (isoleucine vs. valine) is sufficient to convert PYL1 from a pyrabactin-activated receptor into a pyrabactin-repressed receptor.
Figure 4.
I137V converts PYL1 into a pyrabactin-inhibited receptor. (a) PYL1 Ile137 clashes with the pyrabactin antagonist structure. Overlay of pyrabactin in the PYL2 ligand binding pocket (brown) with the PYL1 ligand binding pocket (green). Ile137 and the corresponding residue in PYL2, Val114, are represented as stick models. Their distances to pyrabactin in the PYL2 antagonist structure are indicated. (b,c) V193I and A190V mutations stabilize pyrabactin in the PYL1 pocket by forming hydrophobic interaction networks with pyrabactin and ligand binding pocket residues. (b) Model of Ile193 in the PYL1 pocket interacting with pyrabactin, Phe189, and Phe88, (c) Model of Val190 in the PYL1 pocket interacting with pyrabactin, Tyr173, Tyr147, and Phe189. (d) PYL1 A190V and V193I mutations increase the pyrabactin-mediated PYL1–ABI2 interaction. AlphaScreen interaction in the presence or absence of 10 μM (+)-ABA, 100 μM pyrabactin, or 5 μM (+)-ABA + 100 μM pyrabactin (n=3, error bars=s.d.). (e+g) Pyrabactin inhibits the ABA-stimulated interaction between PYL1 I137V and PP2Cs. Alphascreen interactions with PYL1 A190V I137V (e) or V193I I137V (g) (n=3, error bars=s.d.). (f+h) Pyrabactin relieves the ABA-stimulated inhibition of PP2C phosphatase activity by PYL1 A190V I137V (f) or V193I I137V (h) (n=3, error bars=s.d.).
A93F converts PYL2 from a pyrabactin-inhibited to a pyrabactin-activated recptor
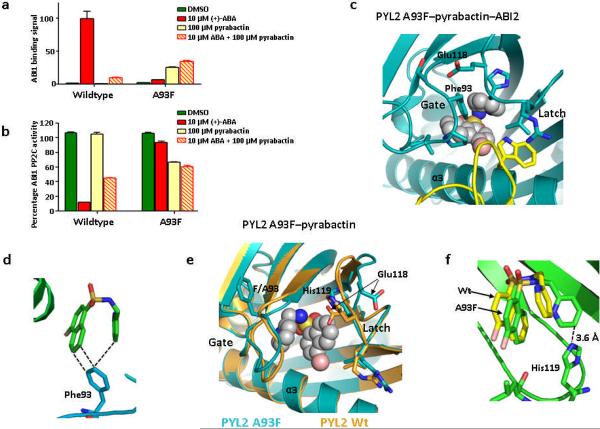
Another key question is why pyrabactin binding does not activate PYL2. Structural analysis indicates that pyrabactin in the context of PYL2 does not form any direct interactions with residues from the ligand entry gate. The closest distance between the pyridine ring of pyrabactin and the Leu-Pro-Ala gating residues is 4.8 Å–7.0 Å when the Leu-Pro-Ala loop assumes the ABA-bound closed conformation, but is 11 Å –13 Å when the Leu-Pro-Ala gate is in open conformation as seen in the pyrabactin-bound PYL2 structure (Fig. 3a). In contrast, the distance of the naphthalene double ring of pyrabactin or the cyclohexene ring of ABA to the three side chains of the Leu-Pro-Ala gate in the PYL1 structure is 3.4 Å –4.6 Å (Fig. 2b), a range for strong Van der Waals interactions, thus helping to keep the Leu-Pro-Ala gate in the closed conformation. These observations suggest that the antagonism of pyrabactin to PYL2 is because the pyridine group is positioned too far away to make any direct contacts with the Leu-Pro-Ala loop that would help keep the gate closed. To validate this hypothesis, we changed the alanine in the Leu-Pro-Ala loop to a larger residue. Indeed, mutation of A93F, designed to close the distance between the gate loop and the pyridine ring, converted PYL2 from a pyrabactin-repressed to a pyrabactin-activated receptor, as determined by PP2C binding and phosphatase assays (Fig. 5a,b). As expected from structural modeling, replacement of the gate residue Ala93 with the bulky phenylalanine ring also abrogated ABA to promote PYL2 activation (Fig. 5a,b).
Figure 5.
A93F converts PYL2 into a pyrabactin-activated receptor. (a+b) PYL1 A93F is activated by pyrabactin to bind and inhibit ABI1 as determined by AlphaScreen assay (a) and by phosphatase assay (b); (n=3, error bars=s.d.). (c) Close view of pyrabactin in the PYL2 A93F–pyrabactin–ABI2 trimeric complex. ABI2 with Trp290 is shown in yellow. (d) Phe93 interacts with both ring systems of pyrabactin in the PYL2 A93F–pyrabactin–ABI2 structure. (e) Structure of pyrabactin in the PYL2 A93F ligand binding pocket (cyan) overlaid with the PYL2 wildtype structure (brown). (f) His119 is flipped into the ligand binding pocket to allow direct interaction with the pyridine ring of pyrabactin in the PYL2 A93F–pyrabactin structure.
Crystal structures of PYL2 A93F–pyrabactin agonist complexes
To gain further understanding on how the A93F mutation converts PYL2 into a pyrabactin-activated, ABA-insensitive receptor, we solved the crystal structures of a dimeric complex of PYL2 A93F bound to pyrabactin at a resolutions of 2.10 Å and two trimeric complexes of PYL2 A93F–pyrabactin bound to ABI2 and HAB1 at a resolution of 2.10 Å and 2.55 Å, respectively. The two trimeric structures closely resemble the active PYL2–ABA–HAB116 and PYL1–ABA–ABI117,20 structures, with the gate and latch loops in the closed conformation, and the PP2C locking residue (Trp290 in ABI2 and Trp385 in HAB1) inserted between the gate and latch to make water-mediated contacts with the bound pyrabactin (Fig. 5c and Supplementary Fig. 5a,b). Pyrabactin adopts an intermediate conformation between the agonist and antagonist conformation (Fig. 5c) to allow formation of Van der Waals interactions with Phe93, which contacts both ring systems of pyrabactin (Fig. 5d), thereby stabilizing the gate in the closed conformation as predicted.
The dimeric PYL2 A93F–pyrabactin structure shows mixed conformations of the activated and repressed receptors for the ligand entry gate and latch loops (Fig. 5e and Supplementary Fig. 5c). Although the gate adopts an open conformation with Phe93 facing away from the ligand binding pocket as Ala93 in wildtype apo PYL2 (Fig. 5e), the latch is deposited into the active conformation. The latch residue His119 is flipped into the ligand binding pocket mimicking its conformation in the active trimeric complex (Fig. 5c,e) and forms direct contacts with pyrabactin (Fig. 5f). Glu118, the residue immediately preceding the His-Arg-Leu latch, is flipped outside of the pocket, as in ABA-activated PYL216, to allow closure of the gate onto the latch upon PP2C binding (Fig. 5e). In this state, the mutated receptor is poised to be activated upon the binding of pyrabactin and a PP2C, therefore providing the basis of pyrabactin agonism by the mutated receptor.
Identification of pyrabactin-based ABA receptor agonists
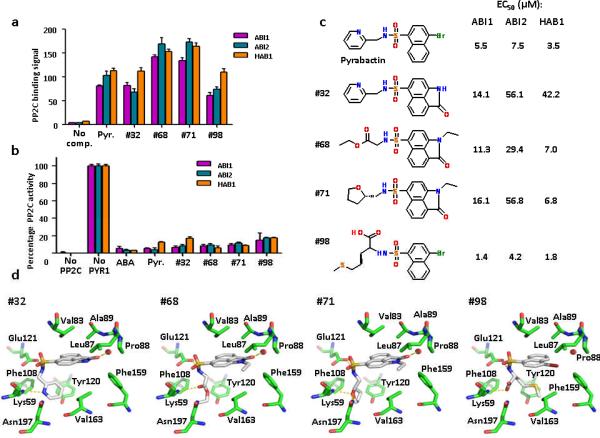
The ability to switch the activation/repression response of PYL1 and PYL2 to pyrabactin by single point mutations demonstrates the detailed levels of our mechanistic understanding of ABA receptor activation and repression, and provides a rational model to screen for novel ABA receptor activators and inhibitors. To demonstrate the validity of this approach, we explored the structural information of pyrabactin bound to PYL1 and PYL2. In both the PYL1 and PYL2 structures, the sulfonamide group of pyrabactin forms extensive interactions with the receptor by mimicking the carboxylate group of ABA. We therefore searched virtual library servers (Chembridge: www.hit2lead.com and ZINC8: http://zinc.docking.org/) representing more than 10 million commercially available compounds for molecules containing a naphthalene-1-sulfonamide group. These compounds were then computationally docked into the PYL1 ligand binding pocket (Supplementary Fig. 6). To validate our docking approach, we tested the top 64 docking matches for their ability to promote PYR1–PP2C interaction and PYR1-dependent inhibition of PP2C activity in vitro. At least 4 of the 64 compounds efficiently activated PYR1 with efficacies (Fig. 6a,b and Supplementary Fig. 7) and EC50 values (Fig. 6c and Supplementary Fig. 8) similar to that of pyrabactin. The success of this virtual docking exercise demonstrates the proof of concept for future screening and design of potent ABA receptor ligands to explore ABA biology and for agriculture applications.
Figure 6.
Identification of pyrabactin-based ABA-receptor agonists. (a+b) Novel ABA agonists induce PYR1–PP2C interaction as determined by AlphaScreen assays (a) and induce PYR1-mediated PP2C inhibition as determined by phosphatase assay (b); Pyr=pyrabactin, (+)-ABA was used at 10 μM, all other compounds at 100 μM (n=3, error bars=s.d.). (c) 2D-structures of pyrabactin and pyrabactin-based ABA-receptor agonists. Listed next to each structure are the EC50 values for the stimulation of the PYR1 interaction with ABI1, ABI2, and HAB1 as determined by the binding curves shown in Supplementary Fig. 8. The EC50 values for pyrabactin are the average values from all binding curves in Supplementary Fig. 8. (d) Docking models for ABA agonists in the PYR1 ligand binding pocket. Hydrogen bonds are shown as dotted yellow lines, a water molecule between Leu87 and Pro88 as red sphere.
DISCUSSION
In this paper, we demonstrate that pyrabactin has both receptor-selective agonist and antagonist properties by receptor–PP2C interaction assays, PP2C inhibition assays, and protoplast reporter assays. Crystal structures of the PYL2–pyrabactin antagonist and the PYL1–pyrabactin–ABI1 agonist complexes reveal that pyrabactin adopts two different pseudo-symmetrical configurations. In the PYL1 structure, pyrabactin mimics ABA. The bromo-naphtalene group of pyrabactin interacts with the same three gate residues (Leu114-Pro115-Ala116) as the ABA cyclohexene ring to pull the gate into the closed position, therefore allowing the formation of the gate-latch interface that docks into the active site of ABI1. In the antagonist conformation, pyrabactin occupies the ABA-binding pocket but does not interact with the gate residues, lacking the interacting energy to pull the gate into the closed position and therefore leaving the gate in the inactive open conformation. These results further highlight opening and closing of the ligand entry gate as the critical mechanism that determines activation and inhibition of ABA receptors.
The conceptually most exciting aspect of these studies is the identification of ABA receptor antagonism. Our works provide comprehensive evidence for the phenomenon of ABA receptor antagonism and its underlying mechanisms through combinatorial approaches of structural, biochemical, mutagenesis and chemical screening studies. The antagonism concept of ABA receptors mirrors the mechanism of the receptor activation and has profound implications in the regulation of ABA receptor physiology. It is well documented that the concentration of endogenous ABA in unstressed plant tissues is in a range that is sufficient to bind and activate several different recombinant PYL–PP2C complexes1–3,16,21,24,27,28. In addition, some ABA receptors, including PYL6, have high levels of constitutive activity in inhibition of all three PP2Cs (Fig 1b and Supplementary Fig. 1b) and are co-expressed with these PP2Cs in several organs, including seeds and guard cells (Supplementary Fig. 9), and yet the ABA response is silent in normal unstressed plants. This raises the important question of how the basal activity of ABA receptors is inhibited. Our data lead to the intriguing hypothesis of the existence of physiological antagonists that can inhibit the basal activation of ABA receptors. We speculate that likely candidates are catabolic derivatives or storage forms of ABA, as ABA metabolic products are known to be involved in ABA responses28. On the other hand, PYR/PYL receptors are prone to the binding of diverse ligands due to their large hydrophobic pockets, a characteristic shared with other START proteins, which can bind chemically distinct sets of lipids, hormones, and antibiotics29. As demonstrated by this study, PYR/PYL receptors can bind to pyrabactin and many other sulfonamide derivatives, with chemical structures completely different from ABA. Thus, derivatives from other hormone pathways that functionally antagonize ABA pathways could also serve as potential ABA receptor antagonists. Identification of endogenous ABA receptor antagonists will be an exciting direction for future research.
The mechanisms of ABA receptor antagonism presented here also complement the activation mechanism of ABA receptors elucidated by earlier structural studies16–20, and provide a full picture for up and down regulation of ABA receptors. This detailed mechanistic understanding of the receptor regulation has allowed us to manipulate the receptor activation and repression properties as well as ligand specificity, which will be important new tools for metabolic engineering and to unravel the biology of individual receptors in the context of high receptor redundancy. Furthermore, the structure information of the ABA receptor agonism and antagonism provides a solid framework for computational screening of virtual chemical libraries, which have allowed us to identify four novel ABA receptor activators. ABA signaling plays the central role in plant resistance to environmental stresses such as drought conditions. The ability of identifying agonists of ABA receptors opens a new avenue for making other small molecules of ABA mimics, which should have practical applications for improving crop yield under stress conditions.
ONLINE METHODS
Protein Preparation
PYL1 (residues 36–211), PYL2 (residues 14–188), and HAB1 (residues 172–511) were prepared as recombinant proteins in E. coli as described16. ABI1 (residues 117–434) and ABI2 (residues 101–423) were expressed in E. coli BL21 (DE3) as H6Sumo fusion proteins and purified following the same general method as for PYL116. PYL–pyrabactin complexes we prepared by incubating purified PYL1 and PYL2 with pyrabactin at a 1:5 molar ratio for 30 minutes on ice prior to crystallization trials. For ternary complexes, we added pyrabactin and purified PYL proteins to purified PP2Cs at a 5:1:1 molar ratio in the presence of 5mM MgCl2.
Small scale purifications of H6GST-tagged PYL/PYR proteins for binding studies of wildtype and mutant proteins were performed by standard glutathione sepharose chromatography. Biotinylated PP2C proteins were prepared as recombinant fusion proteins with in vivo biotinylated 14 amino acid avitags as previously described for HAB116.
Crystallization
PYL2–pyrabactin crystals were grown at room temperature in hanging drops containing 2.4 μl of the purified PYL2 protein and 1.6 μl of well solution (2 M ammonium sulfate, 0.1 M HEPES pH 7.5 and 10 % (v/v) glycerol). Crystals appeared within 1–2 days and grew to a dimension of approximately 250 μm in length on the 6th day. Crystals were transferred to well buffer with 20 % (v/v) glycerol prior to flash freezing in liquid nitrogen.
The PYL1–pyrabactin–ABI1 complex crystals were grown at room temperature in hanging drops containing 1.0 μl of the above protein–ligand–PP2C solution and 1.0 μl of well solution (0.2 M ammonium sulfate, 0.1 M BisTris pH 5.5, and 22 % (w/v) PEG 3350). Crystals appeared within 1–2 days and grew to a dimension of about 120 μm in length on the 4th day. Crystals were serially transferred to well buffer with increasing PEG 3350 concentration (35 % w/v final) before flash freezing in liquid nitrogen.
The PYL2 A93F–pyrabactin crystals were grown at room temperature in hanging drops containing 1.0 μl of the purified PYL2 protein and 1.0 μl of well solution (2 M ammonium acetate pH 8.1 and 22 % (w/v) PEG 3350). Crystals appeared within 1–2 days and grew to a dimension of about 150–200 μm in length after 2 weeks. Crystals were serially transferred to well buffer with increasing PEG 3350 concentration (40 % v/v final) prior to flash freezing in liquid nitrogen.
The PYL2 A93F–pyrabactin–ABI2 complex crystals were grown at 4 °C in hanging drops containing 1.6 μl of the protein–ligand–PP2C solutions and 2.4 μl of well solution (0.1 M HEPES pH 7.5, 10 % (w/v) PEG 8000 and 10% (w/v) sucrose). Crystals appeared within 1–2 days and grew to a dimension of about 200 μm in length on the 4th day. Crystals were serially transferred to well buffer with increasing sucrose concentration (35 % w/v final) before flash freezing in liquid nitrogen.
The PYL2 A93F–pyrabactin–HAB1 complex crystals were grown at room temperature in hanging drops containing 1.0 μl of the protein–ligand–PP2C solution and 1.0 μl of well solution (0.2 M ammonium sulfate, 0.1 M Tris pH 7.5, 10 % (w/v) ethylene glycol and 23 % (w/v) PEG 3350). Crystals were grown to a dimension of about 250 μm in length and flash-frozen in liquid nitrogen on the 3rd day.
Pyrabactin crystals were grown at room temperature in hanging drops containing 1 μl of 40 mM pyrabactin in methanol and 1 μl of well solution (50 % 2-methyl-2,4-pentanediol). Crystals were grown to a dimension of 100 μm in length and flash-frozen in liquid nitrogen on the 2nd day.
Data Collection and Structure Determination
All diffraction data sets were collected at 100 K using an X-ray beam at 1 Å wavelength with MAR300 and MAR225 CCD detectors (MAR Research) at the ID-D and ID-F beamlines of sector-21(LS-CAT) at the Advanced Photon Source at Argonne National Laboratory. The observed reflections were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package30.
Molecular replacement was performed by using the Collaborative Computational Project 4 (CCP4) program Phaser31. Programs O and Coot were used to manually fit the protein model32,33. Model refinement was performed with CNS and the CCP4 program Refmac534,35. Ramachandran statistics: residues in most favored regions/residues in additional allowed regions/residues in generously allowed regions/residues in disallowed regions were 89.8 %/9.6 %/0.6 %/0 0.0 % for PYL2–pyrabactin; 88.9 %/11.1 %/0.0 %/0.0 % for PYL1–pyrabactin–ABI; 89.1 %/10.2 %/0.2 %/0.0 % for PYL2 A93F; 83.6 %/16.4 %/ 0.0 %/0.0 % for PYL2 A93F–pyrabactin–HAB1; and 91.4 %/8.6 %/0.0 %/0.0 % for PYL2 A93F–pyrabactin–ABI2.
Molecular modeling
Starting with the pyrabactin structure, substructure searching was performed using Canvas (version 1.2, Schrödinger, LLC, New York) on Chembridge (www.hit2lead.com, 800,000 screening compounds) and ZINC8 (http://zinc.docking.org/, 13 million compounds) databases. Identified compounds were further subjected to cluster analysis by using ICM/molsoft (version 3.5-1n, Molsoft LLC) and molecular docking using Glide/Schrodinger (version 5.5, Schrödinger, LLC, New York). 64 of the top scoring 100 compounds were purchased from Chembridge, Otava, and Princeton BioMolecular Research. A schematic representation of the hit discovery strategy is shown as Supplementary Fig. 6.
Assays for the interactions between PYR/PYL and PP2Cs
Interactions between PYR/PYL and PP2Cs were assessed by luminescence-proximity AlphaScreen technology as described previously16. All reactions contained 100 nM recombinant H6GST-PYR/PYL proteins bound to Ni-acceptor beads and 100 nM recombinant biotin-PP2C bound to streptavidin acceptor beads in the presence and absence of the indicated amounts of (+)-ABA, pyrabactin, and docking compounds. Binding signals represent photon counts × 1000.
Assays of PP2C phosphatase activity
Phosphatase assays were performed by colorimetric determination of phosphate release from phospho-S175 of 100 μM of a SnRK2.6 activation loop phosphopeptide, as described previously16. Reactions contained 200 nM recombinant PP2Cs and 600 nM (Fig. 1b and Supplementary Fig. 1b: 2 μM) recombinant PYR/PYL proteins.
Mutagenesis
Site-directed mutagenesis was carried out using the QuikChange method (Stratagene). Mutations and all plasmid constructs were confirmed by sequencing.
Protoplast transient assays
SnRK2.6, ABI1, and PYL2 expression vectors were cotransfected with an ABA-inducible luciferase reporter in Arabidopsis mesophyll protoplasts as described previously16.
Supplementary Material
Acknowledgments
We thank the staff of LS-CAT for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the Advanced Photon Source was supported by the Office of Science of the U. S. Department of Energy. This work was supported by the Jay and Betty Van Andel Foundation (H.E.X.); National Institutes of Health (H.E.X., and J-K.Z.). L.-M.N. and F.-F.S. were supported by an overseas PhD scholarship from the NUS Graduate School for Integrative Sciences & Engineering (NGS).
Footnotes
Data deposition The structure factors and atomic coordinates discussed in this work have been deposited in the Protein Data Bank. The accession codes are: 3NMH for PYL2–pyrabactin, 3NMN for PYL1–pyrabactin–ABI1, 3NMP for PYL2 A93F–pyrabactin, 3NMT for PYL2 A93F–pyrabactin–HAB13, and NMV for PYL2 A93F–pyrabactin-ABI2.
Competing interest statement The authors declare no competing financial interests.
References
- 1.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–71. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–8. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 3.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 4.Belin C, et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–27. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–94. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:17588–93. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida R, et al. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281:5310–8. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 8.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci U S A. 2006;103:1988–93. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi Y, et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–49. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura N, et al. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50:935–49. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 11.Viswanathan C, Zhu JK. Molecular genetic analysis of cold-regulated gene transcription. Philos Trans R Soc Lond B Biol Sci. 2002;357:877–86. doi: 10.1098/rstb.2002.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–83. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida T, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–26. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–73. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–8. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–14. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–9. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–8. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 20.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–6. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 21.McCourt P, Creelman R. The ABA receptors -- we report you decide. Curr Opin Plant Biol. 2008;11:474–8. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Harris MJ, Outlaw WH, Mertens R, Weiler EW. Water-stress-induced changes in the abscisic acid content of guard cells and other cells of Vicia faba L. leaves as determined by enzyme-amplified immunoassay. Proc Natl Acad Sci U S A. 1988;85:2584–2588. doi: 10.1073/pnas.85.8.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang SQ, Outlaw WH, Jr., Aghoram K. Relationship between changes in the guard cell abscisic-acid content and other stress-related physiological parameters in intact plants. J Exp Bot. 2001;52:301–8. [PubMed] [Google Scholar]
- 24.Christmann A, Hoffmann T, Teplova I, Grill E, Muller A. Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol. 2005;137:209–19. doi: 10.1104/pp.104.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, et al. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat Chem Biol. 2007;3:716–21. doi: 10.1038/nchembio.2007.32. [DOI] [PubMed] [Google Scholar]
- 26.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–4. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mertens R, Deus-Neumann B, Weiler EW. Monoclonal antibodies for the detection and quantitation of the endogenous plant growth regulator, abscisic acid. FEBS Lett. 1983;160:269–272. [Google Scholar]
- 28.Huang D, Wu W, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot. 2008;59:2991–3007. doi: 10.1093/jxb/ern155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–34. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleywegt GJ, Jones TA. Efficient rebuilding of protein structures. Acta Crystallogr D Biol Crystallogr. 1996;52:829–32. doi: 10.1107/S0907444996001783. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–55. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.