Abstract
Sterol carrier protein-2 (SCP-2) is a non-specific intracellular lipid carrier protein. However, the molecular mechanism of ligand selectivity and the in vivo function of SCP-2 remain unclear. In this study, we used site directed mutagenesis to investigate ligand selectivity and in vivo function of the yellow fever mosquito sterol carrier protein-2 protein (AeSCP-2). Mutations to amino acids in AeSCP-2 known to interact with bound ligand also decreased NBD-cholesterol binding. Substitution of amino acids in the ligand cavity changed the ligand specificity of mutant AeSCP-2. Over-expressing AeSCP-2 wild-type in the Aedes aegypti cultured Aag-2 cells resulted in an increase in incorporation of [3H]cholesterol. However, over-expressing mutants that were deleterious to the binding of NBD-cholesterol in AeSCP-2 showed a loss in the ability to enhance uptake of [3H] cholesterol in cultured cells. Interestingly, when [3H]palmitic acid was used as the substrate for incorporation in vivo, there was no change in the levels of incorporation with over-expression of wild-type protein or mutated AeSCP-2s. The in vivo data suggest that AeSCP-2 is involved in sterol uptake, but not fatty acid uptake. This is the first report that the ability of cholesterol binding may directly correlate to AeSCP-2’s in vivo function in aiding the uptake of cholesterol.
Sterol carrier protein-2 (SCP-2) is a member of the SCP-2 gene family that includes genes encoding SCP-2, SCP-x, SCP-like-2, SCP-like-3, 17β-hydroxysteroid dehydrogenase type IV, and stomatin all of which possess the sterol-binding domain (1,2,3). SCP-2 is considered a nonspecific lipid carrier since the vertebrate version of SCP-2 binds to sterols, fatty acids, fatty acid acyl-CoA and phospholipids (4,5). However, the molecular mechanisms that underlie SCP-2’s ligand selectivity are unknown. The vertebrate SCP-2 has been characterized as a generalized lipid carrier protein thought to mediate cholesterol trafficking and metabolism (6) as well as fatty acid uptake and trafficking (7). The detailed molecular mechanisms of SCP-2’s function are not well understood such as whether SCP-2 plays a role in lipid metabolism or lipid uptake or both. Little data is available to date to show whether ligand-binding or ligand-selectivity in SCP-2 would impact the cellular function.
The yellow fever mosquito, Aedes aegypti, sterol carrier protein-2 protein (AeSCP-2) is highly expressed in the midgut, the main site of cholesterol absorption, but not in the head and hindgut (8). The recombinant AeSCP-2 binds radiolabelled cholesterol with a Kd of 10 nM (8). AeSCP-2 has been hypothesized to be involved in the uptake and delivery of cholesterol (an essential nutrient of insects) across the cellular barrier between the midgut and the hemocoel where it is then transported to sites of either storage or utilization (8, 9, 10). Binding of fatty acid in AeSCP-2 is shown in the protein crystal structure (9). AeSCP-2 was localized to the cytosol in the midgut epithelium and overexpression of the protein in Aedes aegypti cells resulted in increased incorporation of radiolabelled cholesterol (11). Knockdown of AeSCP-2 expression in the last instar larvae resulted in a reduction in cholesterol uptake, higher mortality and a reduction in fecundity (10). In the female adults, knockdown of AeSCP-2 expression leads to reduced uptake of cholesterol from the blood meal, but has little effect on palmitic acid uptake (2). In addition, several compounds were discovered from small molecular chemical libraries that inhibited binding of cholesterol to AeSCP-2 (12).
Mice with SCP-2 knockout show abnormal metabolism in branched fatty acid and bile acid formation (13,14), indicating that the mammalian SCP-2 is involved in lipid metabolism. However, the role of SCP-2 in cellular lipid uptake is not clear. Cholesterol can easily insert into the lipid bilayer and has a fast flip-flop rate (t1/2= 1–2 minutes) within the lipid bilayer (15). Diffusion of cholesterol into the lipid bilayer can happen readily, and an aqueous diffusion model has been proposed to account for some of the cholesterol uptake by cells (16). However, desorption of membrane-bound cholesterol is slow without the aid of a carrier protein (3,16). A lipid carrier protein, therefore, enhances desorption of membrane-bound cholesterol through collision with the membrane-bound cholesterol (17). Over-expression of the mammalian SCP-2 decreased the plasma membrane content cholesterol in L-cells (18), demonstrating the potential of lipid carrier proteins to enhance cellular lipid uptake. There is speculation that the ligand-binding and ligand transfer domains are distinct in SCP-2. The N-terminus of human SCP2 has been shown to be a membrane interaction domain (19). On the other hand, a single point mutation in Arabidopsis thaliana SCP-2 from Met100 to Leu100 abolished the sterol-binding (20).
Using the X-ray crystallographic structure of the AeSCP-2 and its palmitic acid-binding domain as a template (9), mutants of AeSCP-2 were created and studied for their ability to interact with ligands. In addition, cultured Aedes aegypti cells were transfected with over-expression vectors of AeSCP-2 wild type and mutants and tested for their abilities to enhance incorporation of either radiolabelled cholesterol or palmitic acid. In their natural environment, Aedes aegypti larvae feed on detritus which would include various sterols such as cholesterol, phytsterols, and fungal sterols. Little is known whether AeSCP-2 is involved in dietary sterol uptake or metabolism in mosquitoes. We report here for the first time that the binding of β-sitosterol, but not of 7-dehydrocholesterol, in AeSCP-2 occurs in vitro, and the selectivity of ligand can be altered via single point mutation that may affect the ligand cavity in the protein. Furthermore, the in vivo function of AeSCP-2 may be complex due to the selectivity in AeSCP-2-mediated cellular uptake of ligands. These results extend our knowledge of these critical interactions between AeSCP-2 and ligands as well as inhibitors.
Materials and Methods
Chemicals and Instrumentation
Chemicals and reagents were purchased from Sigma (St. Louis, MO), Fisher Scientific (Pittsburgh, PA) and ICN (Costa Mesa, CA) if their origins are not mentioned in the text. NBD-cholesterol was purchased from Molecular Probes (Eugene, OR). [1,2-3H (N)]-cholesterol (40 Ci/mMol) and [9,10-3H]-palmitic acid (60 Ci/mMol) were purchased from American Radiolabeled Chemicals, Inc. (St. Louis, MO, USA). AeSCP-2 inhibitors, SCPI-1 (N-4-{[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]amino}phenyl)aetamide hydrobromide) and SCPI-2 (8-chloro-2-(3-methoxyphenyl)-4,4-dimethyl-4,5-dihydroisothiazol[5,4-c]quinoline-1(2H)-thione), were purchased from ChemBridge Corporation (San Diego, CA, USA) with at least 90 % purity.
All fluorescent measurements were performed in a SpectraMax GeminiXS microplate fluorometer (Molecular Devices, Sunnyvale, CA) or a Synergy HT Multi-Detector Microplate Reader (BIO-TEK Instruments, Winooski, VT). Radiolabelled measurements were carried out in a 2500 TR Liquid Scintillation Analyzer (Canberra, Meriden, CT).
Cell Culture
The Aedes aegypti cell line (Aag-2) was maintained in Eagle’s medium (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (E5 complete medium) and grown at 28 °C in a 5% CO2 atmosphere (21,22). Cells were passed every 7 days in a 1:4 dilution of cells.
Plasmid Construction and Purification
Recombinant AeSCP-2 was expressed using a pGEX-4T-2 plasmid with the entire coding sequence of AeSCP-2 cloned in as previously described (9). For over-expression of AeSCP-2 in Aag-2 cells, the entire coding region of the AeSCP-2 was inserted into the pIE1hr expression vector (gift of Dr. Paul Frisson of the University of Wisconsin Madison) using HindIII and BglII sites in the multiple cloning region. The plasmids were purified using the Maxiprep kit (Qiagen, Valencia, CA). The quantity of DNA was determined by measurement of the UV spectrum at 260 nm.
Site-directed Mutagenesis
Mutations in the pGEX-4T-2/AeSCP-2 and pIE1hr/AeSCP-2 plasmids were introduced using the QuikChange II Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Mutant plasmids were sequenced to confirm the introduced point mutations using an automatic sequencer (ABI 377XL) and Big Dye labeling (American Pharmacia Biotech AB, Uppsala, Sweden).
Recombinant Protein Expression and Purification
The recombinant AeSCP-2 wild-type and mutants were expressed and purified as described (9) with the following changes. Purified samples of recombinant proteins were concentrated in either 50 mM Tris-HCl, pH 8.0 or 10 mM K2PO4, pH 7.5 in CENTRIPREP centrifugal filter devices (10K cutoff; Amicon, Billerica, MA) to a final volume of 1 mL after re-concentrating the sample from 10 mL of buffer 10 times. A sample (0.5 mg) of the AeSCP-2 was dialyzed against 4 × 4 liters of ddH2O, then lyophilized and subjected to mass spectroscopy in a Voyager DE-Pro MALDI-TOF Mass Spectrometer (Applied Biosystems, Foster City, CA) to demonstrate that the purified AeSCP-2 was in the apo-form. The protein concentration was determined using Coomassie Plus-The Better Bradford Assay Kit (Pierce, Rockford, IL) using the bovine serum albumin as a standard. The purification of recombinant AeSCP-2 wild type and mutants were monitored by Western blot analysis using an antibody raised against recombinant AeSCP-2 (8). All mutants of AeSCP-2 used in this study were recognized by the antibody and were similarly purified to homogeneity (data not shown).
Purified recombinant proteins were subjected to dynamic light scattering to determine whether the purified protein was in the monomeric state. AeSCP-2 wild type binds to ligand as a monomer (9); the soluble state of the protein was a concern since mutation in a protein may cause the instability of the protein structure that would result in aggregation of the protein. Dynamic Light Scattering (DLS) experiments were performed with a DynaPro™ (Protein Solutions, Santa Barbara, CA) in a 12 µL quartz cuvette at 23 oC. Protein samples buffered in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic, pH 7.4) were concentrated to approximately 1mM and passed through a 0.22 µM spin filter at 3000× g, for 3 minutes. Samples were stored on ice prior to the DLS measurements. The hydrodynamic radius, polydispersity, and presence of aggregates were analyzed for each mutant as well as wild-type recombinant AeSCP-2s. DLS has been used extensively to demonstrate a protein sample is absent of major aggregates prior to investing time in crystallization trials (23). The proteins analyzed in this study displayed a narrow unimodal distribution with polydispesity ranging from ≤30%, thereby demonstrating the protein samples were well folded and not in an aggregated state.
Binding of NBD-cholesterol to AeSCP-2
The binding of 0.5 µM NBD-cholesterol in 50 mM Tris-HCl, pH 8.0, 25 oC to increasing concentrations of either AeSCP-2 wild-type or mutants was monitored by fluorescent spectroscopy. Sample volumes were 100 µL. Excitation was at 470 nm and emission was measured at 530 nm. Difference spectra were generated from the subtraction of buffer and ligand/inhibitor spectra without protein from the protein/ligand/inhibitor sample spectra. Kd values were obtained with data using a simple, single binding site, non-linear regression model in GraphPad PRISM® software version 4.0 (GraphPad Software Inc., San Diego, CA) using the equation:
Ligand Competition Assays
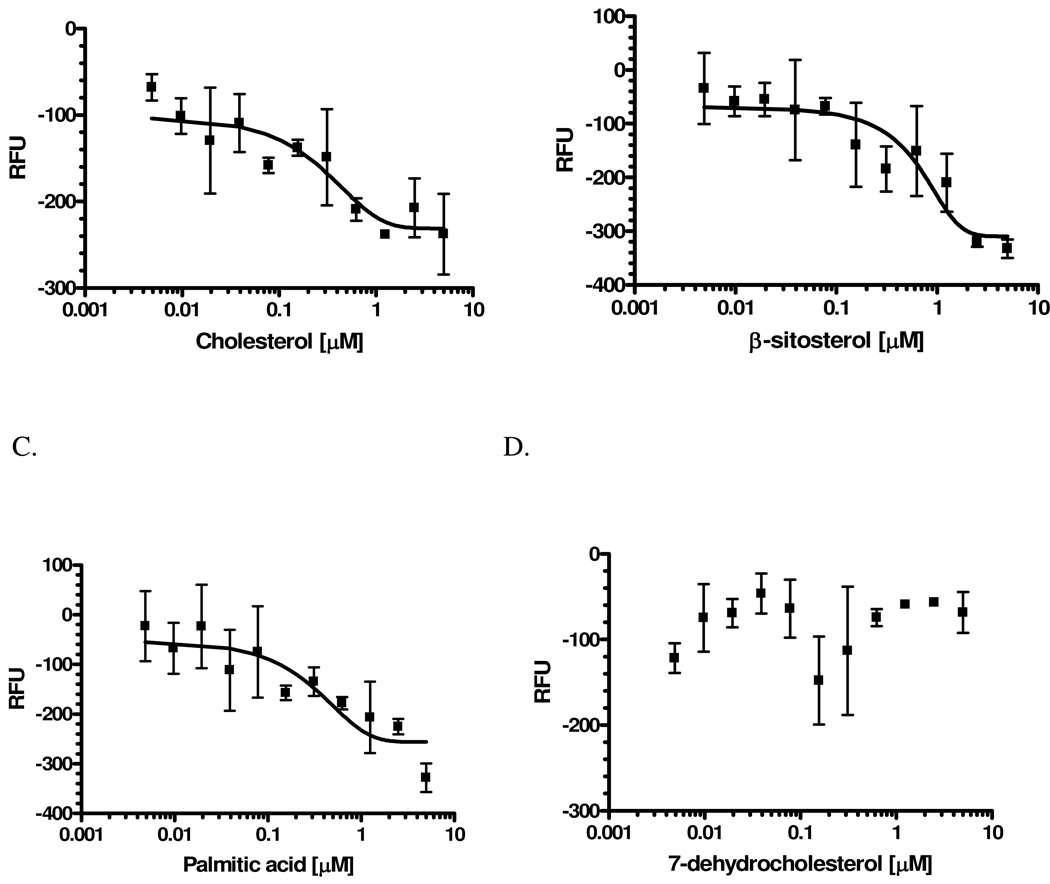
For initial ligand competition assays (Fig. 1), increasing concentrations of a competitor (cholesterol, 7-dehydrocholesterol, palmitic acid, or β-sitosterol) were added to 100 µL samples of 5 µM AeSCP-2 and 10 µM NBD-cholesterol in 10 mM KPO4, pH 7.5, 25 oC. Samples in 96-well plates were measured in the microplate fluorometer.
Figure 1.
Binding of NBD-cholesterol (1.25 µM) to AeSCP-2 under the conditions of a competitive ligand (0.005 to 5 µM). (A). Cholesterol. (B). β-sitosterol. (C). Palmatic acid. (D). 7-dehydrocholesterol. The background NBD-cholesterol fluorescence (NBD-cholesterol alone in the reaction buffer) was deducted from each assay. The trend line (the slope was shown by the trend line) was draw from the mean of three replicates and the bars represent standard deviation. R2 values of each non-linear regression curve were shown.
For binding assays, 0.1 µM or 0.5 uM NBD-cholesterol (Table 1 and Table 2, respectively) and increasing concentrations of wild type recombinant AeSCP-2 (0.02–20 µM) was mixed together in the presence of a competitor. Excitation was at 470 nm and emission was measured at 530 nm. Difference spectra were generated from the subtraction of buffer and ligand/inhibitor spectra without protein from the protein/ligand/inhibitor sample spectra. The results are the average of three experiments with standard deviations.
Table 1.
NBD-cholesterol binding to wild type AeSCP-2 in the presence of a competitive ligand
| Concentrations of the competitor (µM) |
0 | 0.1 | 2 | ||||
|---|---|---|---|---|---|---|---|
| Kd (µM)* | Kd (µM)* | mean diff.** vs. [0] |
p** | Kd (µM)* | mean diff. vs. [0] |
P | |
| cholesterol | 0.99 (0.21) | 3.77 (0.74) | 58.27 | <0.01 | 7.36 (2.07) | 50.07 | <0.01 |
| palmitic acid | 0.99 (0.21) | 11.32 (3.90) | 67.92 | <0.01 | 9.9 (3.49) | 51.15 | <0.01 |
| β-sitosterol | 0.99 (0.21) | 5.45 (2.11) | 48.79 | <0.01 | 5.51 (0.81) | 39.16 | >0.05 |
| 7- dehydrocholesterol |
0.99 (0.21) | 1.92 (0.81) | 30.22 | >0.05 | 1.35 (0.38) | 5.624 | >0.05 |
Mean Kd values of three replicates and the SE were shown.
The Kd values were compared using a repeated measures analysis of variance followed by Dunnett's post test and mean difference of the Dunnett's Multiple Comparison Test between the absence and the presence of a competitor are shown.
Data processed were net changes in NBD-cholesterol fluorescence in intensity (RFU = Relative fluorescence unit) of 0.1 µM NBD-cholesterol and increasing concentration of wild type recombinant AeSCP-2 (0.02–20 µM) in the presence of a competitor.
Table 2.
Difference between wild type AeSCP-2 and mutants in NBD-cholesterol binding in vitro
| wt | Y30A | F9W/W44 F |
F105 A |
F105 W |
M90L | W44E | F32W | F32A | |
|---|---|---|---|---|---|---|---|---|---|
| Kd (µM)* | 0.34 (0.16) |
0.25 (0.05) |
0.91 (0.62) | 1.15 (0.09) |
1.32 (0.45) |
2.75 (1.45) |
4.20 (1.67) |
4.94 (1.59) |
6.36 (2.11) |
| P**vs. wt | -- | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 |
| Fold of Kd change |
0.73 | 2.68 | 3.38 | 3.88 | 8.09 | 12.35 | 14.53 | 18.71 |
Mean Kd values (standard deviation) determined using the GraphPad PRISM4.0 (single binding site and non-linear regression model).
P values were given for Dunnett's post test between wild type (wt) and each mutant after a repeated measures analysis of variance.
Net changes in NBD-cholesterol fluorescence intensity (RFU = Relative fluorescence unit) in each concentration were processed (0.5 µM NBD-cholesterol with increasing protein concentrations (0.01 to 10 µM)).
For ligand competition assays with NBD-cholesterol in the presence of wild type AeSCP-2 or an AeSCP-2 mutant (Table 3), the 50% effective concentration (EC50) was obtained using a single site competition, non-linear regression model in the GraphPad PRISM software version 4.0 (GraphPad) using the equation:
Table 3.
Effects of point mutation in AeSCP-2 on competitive binding of ligands (EC50 (µM))
| Competitor/protein | EC50 | EC50 90% CI | Dunnett's Test** |
Mean Diff. |
P value |
|---|---|---|---|---|---|
| cholesterol/wt | 0.211 | 0.01760 to 2.544 | wt | ||
| cholesterol/Y30A | 0.282 | 0.05466 to 1.452 | vs Y30A | −24.82 | >0.05 |
| cholesterol/F32W | 0.447 | 0.09762 to 2.049 | vs F32W | −68.86 | <0.01 |
| cholesterol/W44A | 0.072 | 0.02259 to 0.2292 | vs W44A | −6.014 | >0.05 |
| cholesterol/W44E | ND | vs W44E | 57.24 | <0.01 | |
| cholesterol/M90L | ND | 8.003e-016 to 3.009e+008 |
vs M90L | 35.35 | <0.05 |
| cholesterol/F105W | 0.474 | 0.009708 to 23.14 | vs F105W | 38.39 | <0.01 |
| β-sitosterol/wt | 0.142 | 0.002813 to 7.203 | wt | ||
| β-sitosterol/Y30A | 0.096 | 0.01598 to 0.5792 | vs Y30A | −53.42 | <0.01 |
| β-sitosterol/F32W | 0.061 | 0.002376 to 1.591 | vs F32W | −58.36 | <0.01 |
| β-sitosterol/W44A | 0.112 | 0.007834 to 1.607 | vs W44A | −6.309 | >0.05 |
| β-sitosterol/W44E | 0.042 | 0.003440 to 0.5018 | vs W44E | 130.1 | <0.01 |
| β-sitosterol/M90L | 0.046 | 0.005384 to 0.3945 | vs M90L | 128.7 | <0.01 |
| β-sitosterol/F105W | 0.083 | 0.008206 to 0.8347 | vs F105W | 74.13 | <0.01 |
| palmitic acid/wt | 0.348 | 0.06839 to 1.767 | wt | ||
| palmitic acid/Y30A | 0.100 | 0.02338 to 0.4259 | vs Y30A | −23.67 | >0.05 |
| palmitic acid/F32W | 0.398 | 0.1225 to 1.291 | vs F32W | −88.96 | <0.01 |
| palmitic acid/W44A | 0.127 | 0.03991 to 0.4065 | vs W44A | −23.87 | >0.05 |
| palmitic acid/W44E | 0.075 | 0.01329 to 0.4237 | vs W44E | 2.003 | >0.05 |
| palmitic acid/M90L | 0.127 | 0.03367 to 0.4777 | vs M90L | 66.14 | <0.01 |
| palmitic acid/F105W | 0.175 | 0.04578 to 0.6685 | vs F105W | 26.4 | >0.05 |
| 7-dehydrocholesterol/wt | ND | wt | |||
| 7- dehydrocholesterol/Y30A |
0.191 | 0.02403 to 1.513 | vs Y30A | −54.77 | <0.01 |
| 7- dehydrocholesterol/F32W |
0.260 | 0.005156 to 13.09 | vs F32W | −75.19 | <0.01 |
| 7- dehydrocholesterol/W44A |
ND | Vs W44A | −24.45 | >0.05 | |
| 7- dehydrocholesterol/W44E |
ND | 0.0005127 to 1364 | vs W44E | 52.63 | <0.01 |
| 7- dehydrocholesterol/M90L |
ND | 2.143e-006 to 6354 | vs M90L | 43.4 | <0.01 |
| 7- dehydrocholesterol/F105 W |
ND | vs F105W | 6.76 | >0.05 |
EC50 value (50% maximal effective concentration): mean (95% confidence intervals (95%CI)). ND: EC50 value cannot be determined due to “not converge” or extremely wide ranges of the EC50 95%CI (≥4 orders of magnitude). The average values of net changes in NBD-cholesterol fluorescence intensity (RFU = Relative fluorescence unit) of 4 independent samples were processed using the GraphPad PRISM4.0 (single competition site and non-linear regression model).
Mutant and wild-type (wt) EC50 values were compared using a repeated measures analysis of variance followed by Dunnett's post test.
In Vivo Incorporation of 3H-cholesterol by cells Over-expressing AeSCP-2 Wild Type and Mutants
The Aag-2 cells were grown to 40% confluence in a 65 mm tissue culture dish in E5 complete media. Cells were transfected with 12 µg of an over-expression plasmid/24 µL of Lipofectin/mL for 8 hours in the transfection media (E5 complete medium minus FBS and antibiotics). Cells were then transferred to E5 complete media. At 36 hours post transfection, the cells were transferred to steroid-free E5 complete media (25) for 12 hours. Media was replaced with 1 mL of steroid-free E5 complete media containing either 0.33 µCi [3H]-cholesterol/mL or [3H]palmitic acid/mL and incubated for 12 hours. The labeled cells are gently washed twice in 2 mLs of cold PBS in the dish. Lipids were extracted from the cells and the radioactivity in each sample was measured in a liquid scintillation counter as described (6). The protein pellets from each sample were re-dissolved in 200 µL of PBS and protein concentrations were determined using BCA kit (Pierce).
To quantify the over-expression of AeSCP-2 and mutants, an indirect enzyme-linked immunosorbent assay (ELISA) was performed in a 96-well microtiter plate using an antibody raised in a rabbit against purified recombinant AeSCP-2. Protein samples of 100 µL/well from Aag-2 cells were loaded into a 96-well plate (Immunoassay microplate, Thermo Scientific, Waltham, MA) and incubated for 1 hour at room temperature. The samples were removed and a 200 µL solution of blocking buffer (10 mg/mL bovine serum albumin, 1% goat serum) was added and incubated for 1 hour at room temperature. The primary antibody to AeSCP-2 (1:2000 in 100 µL PBS/well) was shaken in for 1 hour at room temperature. The wells were washed in a PBS/0.1% Tween-20 solution three times and the secondary antibody (goat anti-rabbit IGG) was added (1:2000 in 100 µL) and incubated for 1 hour at room temperature. The wells were washed in a PBS/0.1% Tween-20 solution three times and a 100 µL solution of 1.8 mM 2,2'-Azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) was added to each well. The plates were read in a VERSAmax microplate reader (Molecular Devices) at 405 nm. Purified recombinant AeSCP-2 in PBS was used in a concentration range of 0–1 µM to generate a standard curve. There were no significant differences in the levels of over-expressed AeSCP-2 and mutant in different samples of transfected Aag-2 cells (Suppl. Table 1). Therefore, the content of a [3H]-lipid is described as disintegrations/min/mg total cellular protein (DPM/mg protein).
Intrinsic Fluorescence Measurements of SCPIs Bound to AeSCP-2 Wild Type and Mutants
Fluorescence measurements from intrinsic sources are often useful for reporting conformational states of proteins (26). Fluorescence emission from tryptophan (Trp) residues in a protein is extremely sensitive to perturbations in the local environment, mostly due to different degrees of quenching. AeSCP-2 has only one tryptophan residue at position 44 that is mostly buried in the hydrophobic core of the protein (Suppl. Fig. 3; 9). Recombinant protein samples of wild type and mutants (20–50 µM) in a 96-well microplate were measured for their intrinsic fluorescence intensities in the absence and presence of either SCPI-1 or SCPI-2 (50–100 µM). SCPI-1 and SCPI-2 are two AeSCP-2 inhibitors identified via high throughput screening for AeSCP-2 chemical inhibitors (12). Sample volumes were 100 µL in 50 mM Tris-HCl, pH 8.0 at 25 °C. The samples were excited at 280 nm and emission spectra were measured from 320–390 nm. Difference spectra were generated by the subtraction of buffer and SCPI alone spectra from the spectra containing protein.
To ensure that the quenching of intrinsic tryptophan fluorescence was due to the interaction of inhibitor and the tryptophan in the native state of the protein, negative control spectra were collected after proteins were denatured in 6.5 M guandidine-HCl in 50 mM Tris-HCl, pH 7.4.
Statistic analysis
Binding or competitive binding assays presented in each table were always performed in parallel with 3–4 independent replicates. Therefore, Kd and EC50 values were compared using a repeated measures analysis of variance followed by Dunnett's post test using the GraphPad PRISM® software version 4.0 (GraphPad Software Inc., San Diego, CA) unless otherwise indicated. The data in this study were analyzed using a similar statistical method as described by others reporting similar studies on point mutation and ligand interactions (27).
Results
Ligand-binding and competition in AeSCP-2
NBD-cholesterol is a fluorescent analog of cholesterol, which has little fluorescence in hydrophilic solutions. After binding to a protein in the hydrophobic cavity of the protein, the fluorescent intensity of NBD-cholesterol increases. NBD-cholesterol has been used as the probe in studying sterol-binding proteins (27). Cholesterol has been shown to compete with NBD-cholesterol for binding to AeSCP-2 (12). Palmitic acid and cholesterol have been shown to bind AeSCP-2 (8, 9). Earlier reports show that Aedes aegypti larvae readily convert β-sitosterol to cholesterol (28,29), whereas conversion of cholesterol to 7-dehydrocholesterol is the first step in steroid hormone biosynthesis pathway (30). It is known that the vertebrate SCP-2 is a nonspecific intracellular lipid carrier (3). Therefore, potential competitors for binding in AeSCP-2 could be cholesterol, 7-dehydrocholesterol, palmitic acid, as well as β-sitosterol. Here we show that in addition to cholesterol, other ligands competed with the fluorescent cholesterol analog for binding to AeSCP-2. Cholesterol competed with the NBD-cholesterol for binding to AeSCP-2 in a dose-dependent fashion (Fig. 1A, cholesterol), consistent with what has been reported previously (12). In addition, palmitic acid and β-sitosterol also competed with NBD-cholesterol for binding to AeSCP-2 (Fig. 1B and C, β-sitosterol and palmitic acid, respectively). Interestingly, 7-dehydrocholesterol did not appear to compete with NBD-cholesterol for binding to AeSCP-2 (Fig. 1D, 7-d-cholesterol) suggesting that AeSCP-2 may not bind this metabolite of cholesterol. To further verify the potential ligands for AeSCP-2, NBD-cholesterol (0.1 µM) binding was measured with recombinant wild type AeSCP-2 under increasing protein concentrations (0.02–20 µM) in the presence of a competitor. At 5 µM AeSCP-2 and 0.1 µM NBD-cholesterol concentrations, the fluorescent intensity of bound NBD-cholesterol had reached maximal (Supplement Fig. 2). In the presence of a specific competitor under the same condition, binding of NBD-cholesterol would be reduced which would appear as an increase of Kd value for NBD-cholesterol compared to that of in the absence of a competitor. Binding of NBD-cholesterol to AeSCP-2 was reduced significantly in the presence of ≥ 0.1 µM for palmitic acid, and cholesterol, respectively (Table 1). The results suggest that cholesterol and palmitic acid might be natural ligands for AeSCP-2. On the other hand, significantly reduction of Kd values were only detected at 0.1 µM for β-sitosterol (Table 1, β-sitosterol), indicating β-sitosterol might be a natural ligand for AeSCP-2. However, the presence of 7-dehydrocholesterol did not significantly alter the Kd values under the same conditions (Table 1, 7-dehydrocholesterol), indicating 7-dehydrocholesterol might not be a specific competitive ligand for AeSCP-2. The results showed that cholesterol, palmitic acid, and β-sitosterol were likely ligands of AeSCP-2. This is the first report of β-sitosterol as a potential AeSCP-2 ligand.
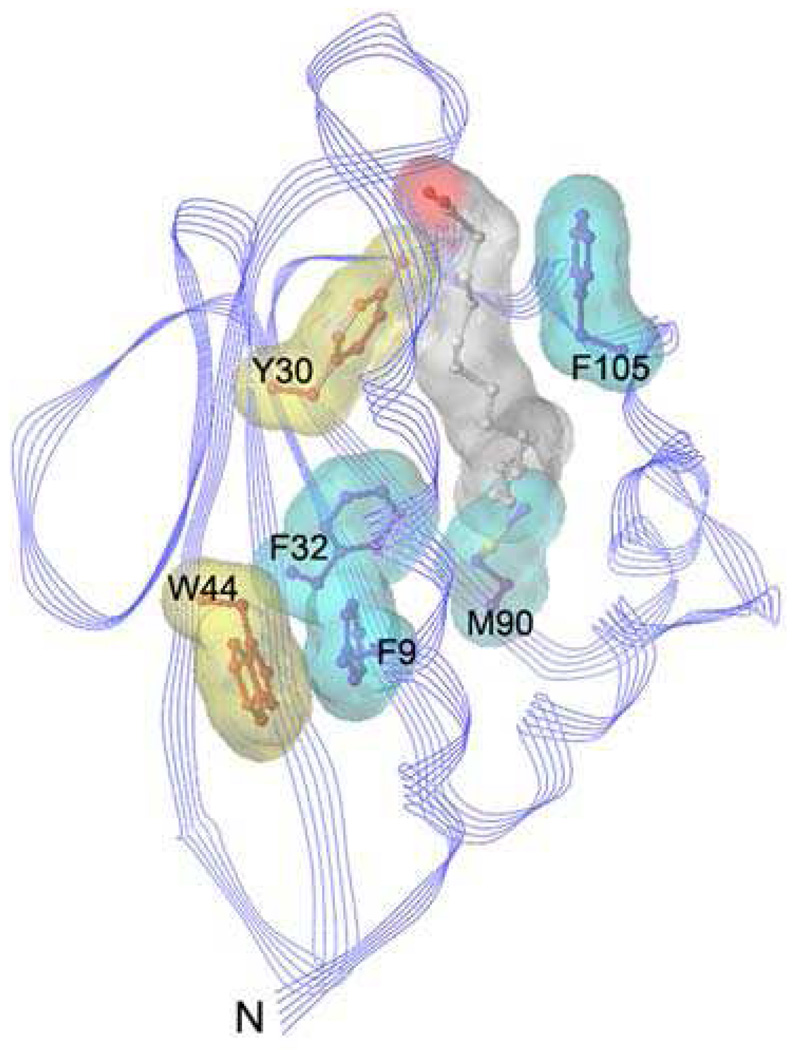
Ligand-binding in AeSCP-2 mutants
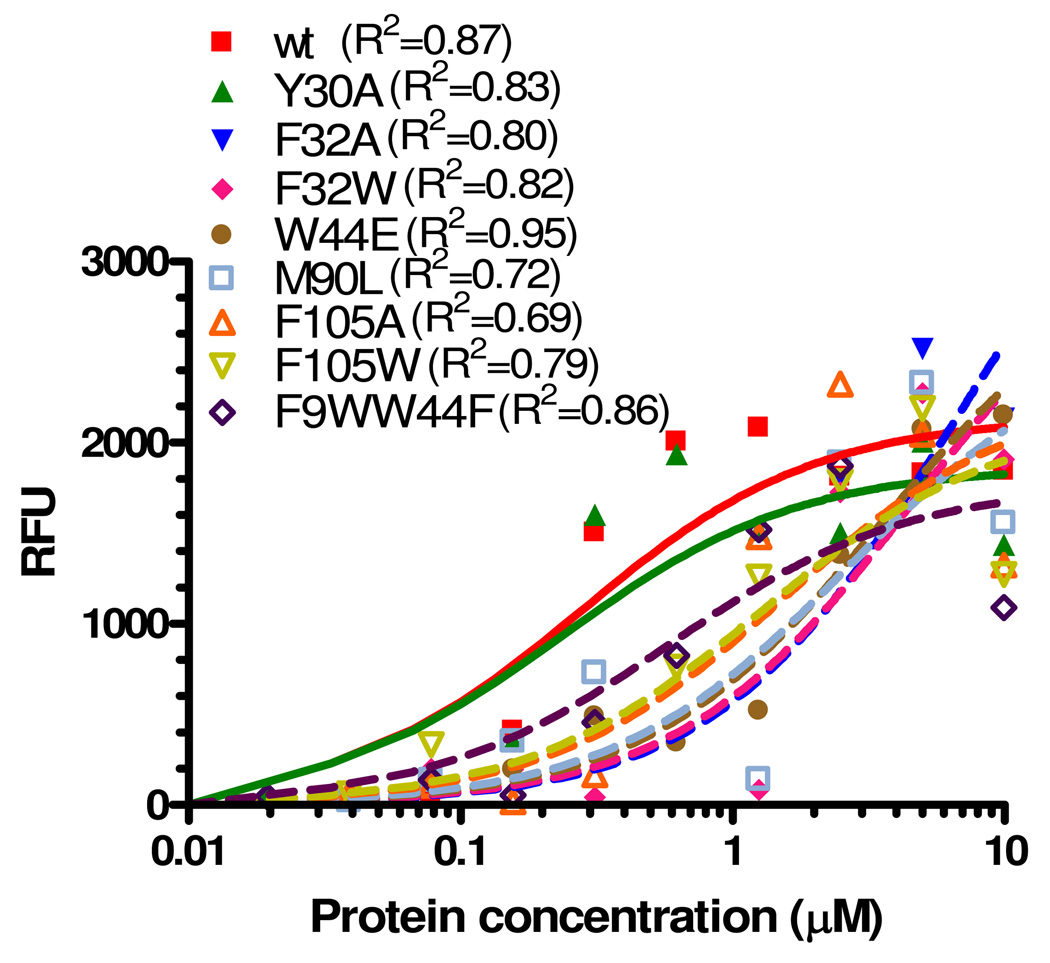
The 3-dimensional protein structure of AeSCP-2 shows that hydrophobic amino acid residues line the interior ligand-binding cavity (9). Based on the protein structure of AeSCP-2, we selected several amino acid residues (Figure 2, F9, Y30, F32, W44, M90 and F105) in AeSCP-2 for point mutation experiments to shed light on the mechanism of ligand-binding in AeSCP-2. The Y30 residue does not directly interact with bound ligand, although Y30 resides in the hydrophobic core of the protein (9), whereas the only tryptophan residue at position 44 seems to form π-π stocking with F9 that may stabilize the ligand cavity (1). All of the point mutations except Y30A in AeSCP-2 caused reductions in binding of NBD-cholesterol (Fig. 3). The wild type AeSCP-2 and Y30A mutant have a Kd value of 0.34 and 0.25 µM for NBD-cholesterol, respectively (Table 2). Mutations of F9W/W44F, F105A, F105W F32A, F32W, W44E, and M90L in AeSCP-2 reduced the ability of the mutants to bind NBD-cholesterol compared to the wild type (Fig. 3 and Table 2). However, F32W was only mutant that caused significant change in Kd values compared to the wild type (Table 2). Mutant F32W reduced NBD-cholesterol binding affinity by more than 10-fold (Table 2), which also significantly decreased the sensitivity to cholesterol competitive binding (Table 3). Interestingly, mutants that only resulted in moderate reductions in NBD-cholesterol binding (Table 2) had severely affected the sensitivity to cholesterol binding (Table 3). Both W44E and M90L lost the ability for cholesterol competitive binding, whereas retained palmitic acid binding capacity (Table 3). These results further established that AeSCP-2 binds NBD-cholesterol and that critical amino acids in the protein structure, when changed, appeared to be responsible for reduced binding affinity to the ligand. It is interesting to note that all mutant AeSCP-2s studied bound NBD-cholesterol at higher concentrations of the protein (Fig. 3, ≥5 µM proteins), suggesting that none of the individual amino acid mutations were severe enough to abolish NBD-cholesterol binding to the protein.
Figure 2.
Ribbon diagrams (Sybyl) of the AeSCP-2 (pdb entry 1PZ4) at 1.35 Å resolution and selected amino acid residues for point mutations that directly interact with bound ligand (blue colored sticks, F9, F32, M90, and F105) and that are not in direct contact with the ligand (orange colored stick, Y30 and W44). The C16 fatty acid is highlighted as a ball and stick model with carbons (white) and oxygen (red).
Figure 3.
Binding of NBD-cholesterol to recombinant wild type and mutant AeSCP-2s. The background NBD-cholesterol fluorescence (NBD-cholesterol alone in the reaction buffer) was deducted from each assay. Shown are net changes in NBD-cholesterol fluorescence in intensity (RFU = Relative fluorescence unit) at 0.5 µM concentration in the presence of increasing concentrations of each protein (0.01 to 10 µM). The data represent mean values from three replicates and processed using GraphPad PRISM4.0 (single binding site and non-linear regression model). R2 values of each non-linear regression curve were shown.
To investigate whether point mutations in AeSCP-2 also affect ligand-binding specificity, we used the mutant AeSCP-2s to perform competitive binding assays. We performed competitive binding assays with 5 µM purified protein and 1.25 µM NBD-cholesterol under conditions of increasing concentrations of a competitor. In the wild type AeSCP-2, the only ligand that did not show competitive binding was 7-dehydrocholesterol (Table 3), which is consistent with previous results (Table 1 and Fig. 1). There was little change in competitive binding of palmitic acid in tested mutants (Table 3, palmitic acid). However, in mutants M90L and W44E, cholesterol lost its ability for completive binding to the proteins, respectively (Table 3). On the other hand, β-sitosterol was able to compete with NBD-cholesterol for binding to M90L and W44E (Table 3). Interestingly, Y30A and F32W mutations enabled 7-dehydrocholesterol to compete with NBD-cholesterol for binding to the proteins (Table 3), whereas the wild type AeSCP-2 did not (Fig. 1, Table 1, and Table 3). The results indicate that cholesterol and palmitic acid, two ligands that are known to bind to AeSCP-2 (8,9), may be accommodated via different amino acid residues in AeSCP-2.
In vivo function of AeSCP-2 mutants
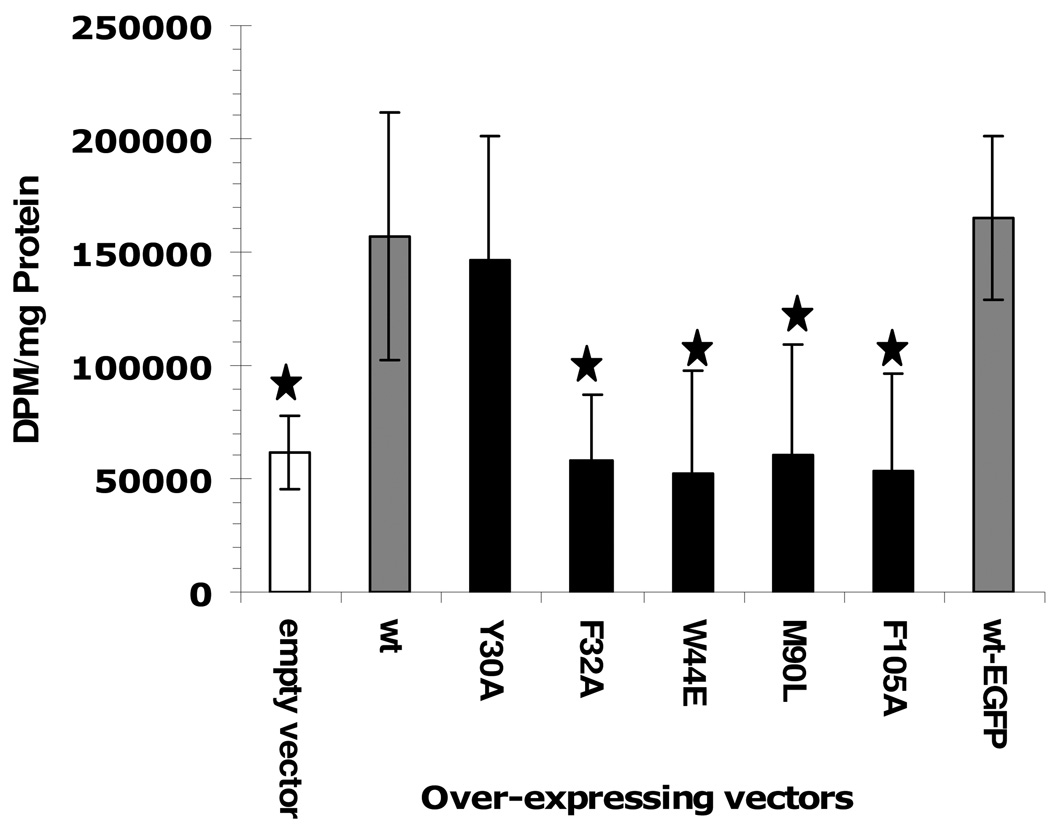
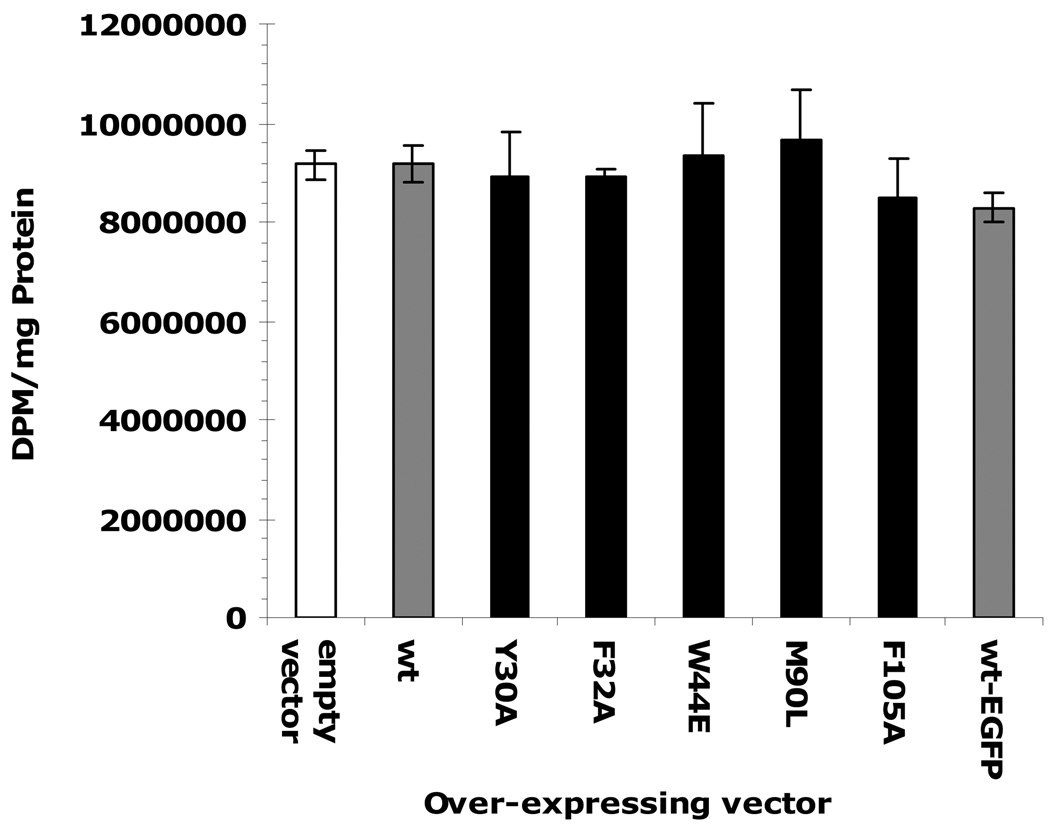
Studies of the incorporation of radio labeled cholesterol and palmitic acid into cells were conducted using the Aedes aegypti cell line Aag-2 that is embryonic origin (31). The mosquito cells were transfected with over-expression vectors of either wild type or mutant AeSCP-2s, and the AeSCP-2 over-expressing cells were then incubated with labeled ligand. Previously, incorporation of [3H]cholesterol was shown to increase in cells over-expressing AeSCP-2 (11). To determine if the mutations in AeSCP-2 that showed reduced affinity for cholesterol in vitro (Fig. 3 and Table 2) or impaired competitiveness for cholesterol binding (Table 3) would have any effect on AeSCP-2’s function in vivo, expression vectors were constructed to over-express AeSCP-2 mutants in Aag-2 cells. Incorporation of [3H]cholesterol increased significantly compared to empty vector when wild type AeSCP-2 was over expressed in the Aag-2 cells (Fig. 4, wt vs. empty vector), which was consistent with previously reported studies (11). Over-expressing the wild type AeSCP-2/EGFP fusion protein had similar levels of [3H] cholesterol incorporation compared to the wild type AeSCP-2 (Fig. 4, wt-EGFP vs. empty vector), indicating that the EGFP-tag at the C-terminus of the wild type AeSCP-2 did not affect the in vivo function of AeSCP-2. However, incorporation of the [3H]cholesterol did not increase in cells over-expressing AeSCP-2 mutants F32A, W44E, M90L, and F105A when compared to wild type (Fig. 4). On the other hand, over-expression of Y30A mutant had similar increased levels of [3H] cholesterol incorporation as that of the wild type AeSCP-2 (Fig. 4). The Y30A mutation had little effect on cholesterol-binding to the protein in vitro or sensitivity for cholesterol competitive binding (Fig. 3, Table 2, and Table 3); whereas, F32A, W44E, M90L and F105A showed moderately decreased affinity to NBD-cholesterol (Fig. 3, Table 2, and Table 3) and W44E and M90L lost sensitivity for cholesterol competitive binding. The results indicate that cholesterol binding in AeSCP-2 directly correlated to the in vivo function of AeSCP-2 in enhancing cholesterol uptake.
Figure 4.
The effect of point mutation in AeSCP-2 on [3H]cholesterol uptake in Aedes aegypti Aag-2 cells. Aag-2 cells were transfected with over-expression vectors (see Materials and Methods) and the empty vector was used as negative controls. Over-expression of each protein was verified via western blotting analysis (Suppl. Fig. 2). The mean and standard deviation are shown (N=3). Stars mark significant differences (p<0.05) between wild type AeSCP-2 and mutant AeSCP-2s and the negative control.
In contrast, when [3H]palmitic acid was used as the ligand, there was no increase in [3H]palmitic acid incorporation when the cells were transfected with either empty or AeSCP-2 wild type expression vectors (Fig. 5). The results are consistent with a previous report that shows over-expression of wild type AeSCP-2 did not enhance [3H]palmitic acid uptake in cultured cells (31). Over-expression of AeSCP-2 mutants in Aag-2 cells did not affect [3H]palmitic acid incorporation (Fig. 5). The results suggest that AeSCP-2 does not play a significant role in palmitic acid incorporation in the Aag-2 cells, despite the fact that palmatic acid bound specifically to AeSCP-2 and the mutants in vitro (Fig. 1, Table 1, and Table 3).
Figure 5.
The effect of point mutation in AeSCP-2 on [3H]palmitic acid uptake in Aedes aegypti Aag-2 cells. Aag-2 cells were transfected with over-expression vectors (see Materials and Methods) and the empty vector was used as negative controls. Over-expression of each protein was verified via western blotting analysis (Suppl. Fig. 2). The mean and standard deviation are shown (N=3).
Effects of SCPIs on intrinsic Trp fluorescence
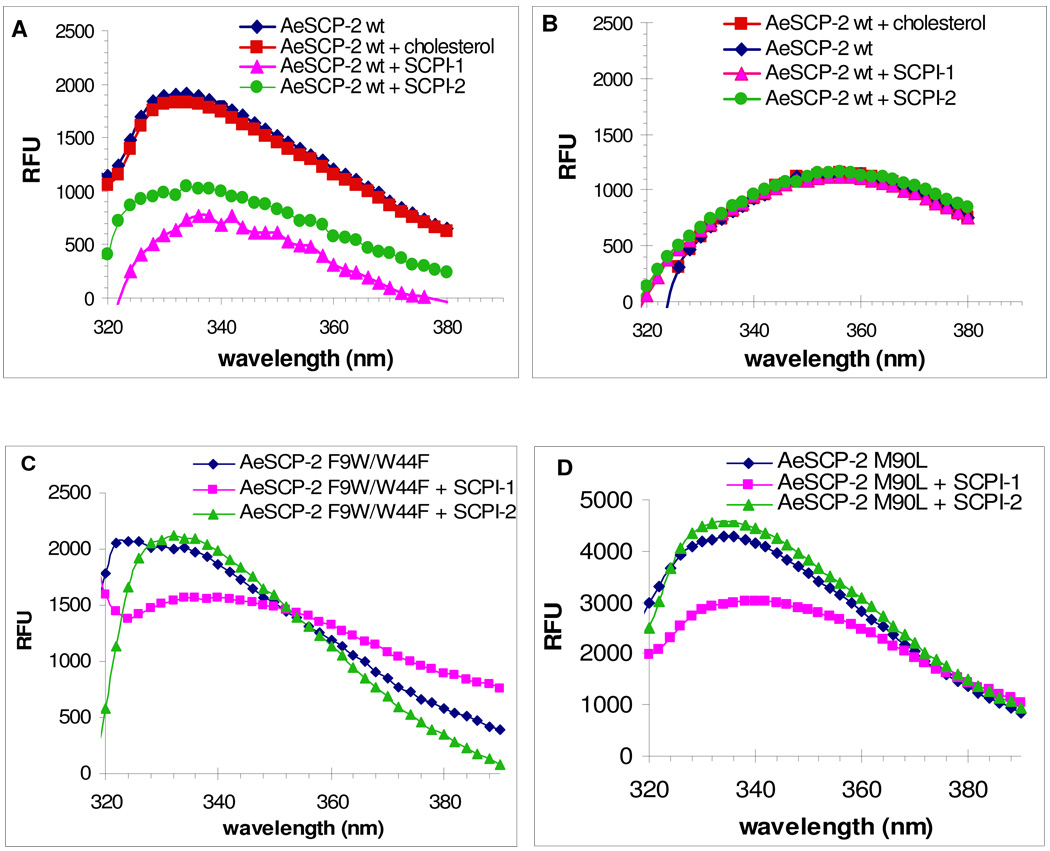
Both SCPI-1 and SCPI-2 suppress NBD-cholesterol binding in AeSCP-2 at IC50 less than 0.4 µM concentrations (12). However, molecular interactions between AeSCP-2 and SCPIs are unclear. Whether SCPIs bind to AeSCP-2 similarly as that of the natural ligand is unknown. The intrinsic fluorescent emission of the Trp residue in proteins is sensitive to perturbations in the local environment, due mostly to different degrees of quenching. A Trp residue buried in a hydrophobic domain of a native protein has maximal fluorescent intensity usually at 330–333 nm, whereas in a denatured protein the Trp residue’s fluorescent emission shows a red shift to 350–353 nm (32). AeSCP-2 has only one tryptophan residue that is situated within the hydrophobic ligand binding cavity, though it does not directly interact with the ligand (9). Wild type recombinant AeSCP-2 showed a tryptophan fluorescent emission peak at about 332 nm (Fig. 6A, AeSCP-2 wt), indicating that the W44 residue is most likely buried in a hydrophobic domain. In the presence of either 50 µM SCPI-1 or 50 µM SCPI-2, there is a significant decrease in the tryptophan intrinsic fluorescence signal of wild type AeSCP-2 at 10 µM (Fig. 6A, AeSCP-2 wt + SCPI-1 or + SCPI-2). The quenching effects of SCPI-1 on intrinsic fluorescence of Trp in wild type AeSCP-2 were consistent with that of an earlier report (34). However, this is the first report that SCPI-2 quenched Trp intrinsic fluorescence in the wild type AeSCP-2 in a similar fashion as that of SCPI-1. In contrast, cholesterol does not produce a quenching of the intrinsic Trp fluorescent signal in AeSCP-2 (Fig. 6A, AeSCP-2 wt + cholesterol). For negative controls, the protein was denatured in 6.5 M guanidine hydrochloride and had a tryptophan intrinsic fluorescent emission peak at 352 nm that is typical of Trp fluorescence in denatured protein (32), showing no significant quenching of the signal in the presence of SCPI-1 or SCPI-2 (Fig. 6B). These data suggested a strong link of inhibitor binding in a region close to an intrinsically fluorescing amino acid. The most likely candidate for this interaction is the single tryptophan at position 44 (W44) in the polypeptide chain. Interestingly, SCPI-1 still quenched the tryptophan intrinsic fluorescence in mutants F9W/W44F and M90L (Fig. 6C and D, SCPI-1), whereas SCPI-2 did not affect the intrinsic fluorescence of W44 in the same mutants (Fig. 6C and D, SCPI-2).
Figure 6.
Intrinsic tryptophan fluorescent emission in wild type and mutants. A. Wild type AeSCP-2 recombinant proteins in the presence or absence of ligand or inhibitors. B. Wild type AeSCP-2 recombinant protein denatured in 50 mM Tris-HCl, 6.5 M GuHCl, pH 7.4 in the presence and absence of ligand or inhibitors. C. F9W/W44F mutant AeSCP-2 in the presence and absence of inhibitors. D. M90L mutant AeSCP-2 in the presence and absence of inhibitors.
Discussion
Ligand specificity of AeSCP-2
In their natural environment, the dietary sterols that mosquito larvae feed on are mostly various phytosterols in the degrading leaf debris (27,28). Cholesterol is an essential nutrient to insects due to the lack of de novo cholesterol biosynthesis (28). The ability of AeSCP-2 to bind β-sitosterol is of interest because Aedes aegypti larvae effectively convert β-sitosterol to cholesterol (27), whereas conversion of phytosterols to cholesterol most likely takes place in the insect midgut (35,36,37,38). Furthermore, AeSCP-2 expression is high in the midgut during the larval feeding stage (8). The results suggest that AeSCP-2 may be involved in trafficking of phytosterols in the midgut for conversion to cholesterol. Further experiments are required to verify the involvement of AeSCP-2 in phytosterol conversions in the midgut.
The vertebrate SCP-2 has been shown to stimulate androgen production in adrenal glands (39,40). In insects, cholesterol is the precursor for the synthesis of ecdysteroids, the insect molting hormones. In the ecdysteroid hormone biosynthesis pathway, cholesterol is first metabolized to 7-dehydrocholesterol in the cytoplasm (29). It was speculated that sterol carrier proteins such as SCP-2 may be in involved in transfer of 7-dehydrocholesterol into mitochondria for ecdysteroid biosynthesis (28). We did not detect binding of 7-dehydrocholesterol to wild type AeSCP-2 in vitro (Fig. 1 and Table 1). In addition, there is a lack of evidence of AeSCP-2 localizing in the mitochondria (11), where steroid biosynthesis takes place. The results suggest that AeSCP-2 may not be involved in transport of 7-dehydrocholesterol to mitochondria, the early steps in the ecdysteroid biosynthesis (29). Surprisingly, single point mutations in AeSCP-2 effectively altered ligand specificity (Table 3). Three sterols examined in this study responded to point mutations in the ligand cavity differently. F32W introduced a bulkier side chain residue into the ligand cavity (Fig. 2), yet, the mutation enabled 7-dehydrocholesterol to compete with NBD-cholesterol for binding to the protein (Table 3). On the other hand, F32W significantly reduced affinity of the protein to NBD-cholesterol (Figure 3 and Table 2) and decreased the cholesterol binding (Table 3). However, F32W significantly enhanced β-sitosterol competitive binding to the protein (Table 3). Binding of sterols in AeSCP-2 has not been resolved at the molecular structural level; therefore, the role of F32 in sterol-binding is unknown. It is likely that F32 plays a role in stabilization of the bound sterol ligand. Y30A mutation is not a conservative substitution (41); therefore, it might alter the hydrophobic ligand cavity allowing 7-dehydrocholesterol-binding. The structural difference between cholesterol and the 7-dehydrocholesterol is the formation of a double-bond in the steroid ring between carbon 7 and 8. It is possible that 7-dehydrocholesterol may adapt to a different conformation from that of cholesterol regarding the trans/cis status of the sterol ring (42), therefore binding of 7-dehydrocholesterol may require a slightly different cavity space that Y30 in AeSCP-2 could not provide. It is noticed that amino acid residues at the equivalent positions in AeSCP-2-like proteins are not conserved in all members of the family (1), suggesting that other members of the AeSCP-2 like proteins in the protein family may bind to 7-dehydrocholesterol.
It is possible that W44E and M90L mutations resulted in altered affinity only to cholesterol (Fig. 3 and Table 3). This is a likely scenario since palmitic acid and β-sitosterol did not show the same sensitivity to compete with NBD-cholesterol at very low concentrations as that of cholesterol in W44E and M90L mutants (Table 3). Interestingly, AeSCP-2 is the only member of the mosquito SCP-2 protein family that has methionine at the equivalent positions of M90 (1), and AeSCP-2 was the only protein among AeSCP-2 and AeSCP-2like proteins to affect cholesterol uptake in vivo (Fig. 4; 2,41). However, sensitivity to palmitic acid competition with NBD-cholesterol in AeSCP-2 mutants was not impaired in all tested mutants (Table 3). A single point mutation in SCP-2 that alters the sensitivity and may affect the selectivity of sterol ligand has been reported. In the A. thaliana SCP-2, an M to L mutation at the equivalent position of M90 in AeSCP-2 results in an abolished sterol sensitivity in BODIPY-phosphatidylcholine transfer assays (20). The results from single point mutations in AeSCP-2 suggest that sterol ligand selectivity may be strongly influenced by the specific residues in the hydrophobic core, whereas a fatty acid ligand could be accommodated in the ligand cavity with diverse non-polar amino acid residues.
In the 3-dimensional AeSCP-2 protein structure, W44 is only partially buried in the hydrophobic core of AeSCP-2 (Suppl. Fig. 3). In addition, the tryptophan is coordinated by π-π stacking with phenylalanine (F9) which interacts with ligand (Fig. 2 and Suppl. Fig. 3). Changes in this π-π-stacking arrangement may lead to disruption of this region of the ligand cavity and the weakening of cholesterol binding. To test the overall sensitivity of this π-π-stacking structure to changes in ligand-binding, double mutants were created that mutated the tryptophan around the aromatic triad (Fig. 2, F9, F32, and W44). The recombinant F32W/W44F protein was unstable when purified which was determined via dynamic light scattering (23) as having >40% polydispersity. Therefore, a conservative double mutation at those positions in AeSCP-2 may reduce the biological activity. The recombinant F9W/W44F protein had only a moderate effect on in binding to NBD-cholesterol (Fig. 3 and Table 2), whereas W44A did not affect NBD-cholesterol/cholesterol competition compared to the wild type (Table 3). Therefore, W44 and F9 π-π stacking may not play a critical role in ligand binding, at least for cholesterol binding to the protein. It must be noted that all mutants, when presented at high concentrations, bound the NBD-cholesterol (Fig. 3), suggesting that none of the mutations were severe enough so as to cause complete disruption of the ligand binding.
In vivo function of AeSCP-2
Multiple steps are involved in the cellular lipids uptake. Lipid carriers such as low density lipoproteins (LDL in the vertebrate and LDLp in the insects) donate lipids to the extracellular layer of the cytoplasmic membrane where the free lipids flip-flop across the cytoplasm membrane bilayer; cytosolic lipids carriers such as cytosolic fatty acid binding protein and SCP-2 desorbs the free lipids from the cytoplasm membrane and deliver the lipids to the site of storage or metabolism (43,44). It is speculated that the ability to bind cholesterol would be critical for AeSCP-2’s in vivo function in aiding cholesterol uptake. The correlation between affinity to cholesterol and the ability in transport the cholesterol ligand suggests an important role for AeSCP-2 in cholesterol uptake in vivo (Figure 3 and Fig. 4). The mechanisms underlining AeSCP-2-mediated cellular cholesterol uptake is unclear. AeSCP-2 is not a membrane bound protein (11), therefore AeSCP-2-mediated lipid absorption is unlike the membrane transporters such as Niemann-Pick C1-like 1 (NPC1-L1) (45). AeSCP-2 may enhance desorption of membrane-bound ligand through collision with the membrane-bound cholesterol (15), which in turn may lead to an enhanced cellular uptake of cholesterol. The SCP-2/membrane collision model is the proposed mode of action for fatty acid transport by the SCP-2 of Yarrowia lipolytica (46). However, the SCP-2/membrane collision model cannot explain AeSCP-2’s ligand selectivity in aiding cholesterol but not palmitic acid uptake in cultured mosquito cells (Fig. 4 and Fig. 5 wt vs. empty vector). It has been shown that the vertebrate SCP-2 increases sterol transfer from the cholesterol-rich membrane domain (47). Therefore, the interaction of apo-AeSCP-2 and the cholesterol-rich domains in the cytoplasm membrane may be the key in AeSCP-2-mediated selective desorption of membrane-bound cholesterol, which results in protein-mediated diffusion of cholesterol into the cell. This process may be specifically important to insects since receptor or lipoprotein mediated endocytosis and/or exocytosis is not involved in cellular lipid transfer (48,49). Further studies are needed to shed light on the mechanism of AeSCP-2-mediated selective uptake of cholesterol in mosquito cells.
Understanding the role of ligand specificity is complex in regard to the biological function of members of the SCP-2 family. In the mouse L-cells, over-expression of SCP-2 did not affect palmitic acid uptake but reduces uptake of phytanic acid (50), whereas it has been shown that SCP-2 over-expression enhances uptake of NBD-stearate, analog of the C18 fatty acid (51). However, over expression of wild type AeSCP-2 or mutants did not change the levels of radiolabeled palmitic acid incorporation in mosquito cultured cells (Fig. 5), consistent with the report that knockdown expression of AeSCP-2 did not affect [3H]palmitic acid uptake from the bloodmeal in female adults (2). The results suggest that AeSCP-2 may not be involved in cellular palmitic acid uptake in mosquitoes or AeSCP-2 may only play a minor role in cellular palmitic acid uptake. Therefore, binding of a ligand in AeSCP-2 in vitro does not predict the in vivo role of AeSCP-2 in ligand transport. Similar observation has been reported for another mammalian sterol carrier protein (27). Aedes aegypti has the cytosolic fatty acid binding protein (AAEL005997-RA) that may play a major role in protein-mediated desorption of membrane-bound free fatty acid in mosquito cells. Since AeSCP-2 does bind to palmitic acid in vitro (9; Fig. 1 and Table 1), AeSCP-2 may be involved in other aspects of cellular fatty acid metabolism. Further work is needed to address the biological role of AeSCP-2 in fatty acid metabolism.
Experiments were carried out to explore the interaction of inhibitors of cholesterol binding on AeSCP-2 wild type and mutants. A significant quenching of the absorption spectra was observed when either SCPI-1 or SCPI-2 was added to the protein. The differences in quenching tryptophan fluorescence emission between SCP-2 inhibitors −1 and −2 suggested that the inhibitors bind similarly but not identically to the protein. However, this region of the molecular interaction between SCPIs and AeSCP-2 is not affected by bound cholesterol (Fig. 5B). Further work is needed to address the how SCPIs suppress binding of cholesterol to AeSCP-2 at the molecular level.
In summary, results from the in vitro and in vivo studies of wild type and mutant AeSCP-2s presented here further affirm the hypothesis that SCP-2 in Aedes aegypti plays a crucial role in the cellular uptake of cholesterol, an essential nutrient for the insect. Amino acid residues have been identified that appear to play critical roles in the proper binding of cholesterol in AeSCP-2, both in the ligand cavity and in overall uptake of cholesterol in live cells.
Supplementary Material
Acknowledgments
This work was supported by grant W9113M-05-1-0006 from the Deployed War Fighter Protection Research Program (DWFP) administered by the U.S. Armed Forces Pest Management Board (AFPMB), and by the National Institutes of Health Research Grant #5R01AI067422 to Q.L
Abbreviations used are
- AeSCP-2
(Aedes aegypti sterol carrier protein-2)
- SCPI
(Aedes ageypti sterol carrier protein-2 inhibitor)
- DTT
(dithiothreitol)
- EDTA
(ethylene diamine tetra acetic acid)
- GST
(glutathione S-transferase)
- SCP-2
(sterol carrier protein-2)
- NBD
cholesterol: 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-o.
Footnotes
Supporting Information Available
Online at ACS site: http://pubs.acs.org
Supplementary Table 1: Quantitative analysis of AeSCP-2s over-expression in Aag-2 cells.
Supplementary Figure 1: Western Blot analysis of AeSCP-2 over-expression in Aag-2 cells.
Supplementary Figure 3: Partial surface diagrams (Pymol) of the AeSCP-2 (pdb entry 1PZ4) at 1.35 Å resolution and showing π-π stacking of F9 and W44.
References
- 1.Dyer DH, Vyazunova I, Lorch JM, Forest KT, Lan Q. Characterization of the yellow fever mosquito sterol carrier protein-2 like 3 gene and ligand-bound protein structure. Mol Cell Biochem. 2009;326:67–77. doi: 10.1007/s11010-008-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyer DH, Wessely V, Forest KT, Lan Q. Three-dimensional structure/function analysis of SCP-2-like2 reveals differences among SCP-2 family members. J. Lipid Res. 2008;49:644–653. doi: 10.1194/jlr.M700460-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Gallegos AM, Atshaves BP, Storey SM, Starodub O, Petrescu AD, Huang H, McIntosh AL, Martin GG, Chao H, Kier AB, Schroeder F. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog. Lipid Res. 2001;40:498–563. doi: 10.1016/s0163-7827(01)00015-7. [DOI] [PubMed] [Google Scholar]
- 4.Frolov A, Cho TH, Billheimer JT, Schroeder F. Sterol carrier protein-2, a new fatty acyl coenzyme A-binding protein. J. Biol. Chem. 1996;271:31878–31884. doi: 10.1074/jbc.271.50.31878. [DOI] [PubMed] [Google Scholar]
- 5.Colles SM, Woodford JK, Moncecchi D, Myers-Payne SC, McLean LR, Billheimer JT, Schroeder F. Cholesterol interaction with recombinant human sterol carrier protein-2. Lipids. 1995;30:795–803. doi: 10.1007/BF02533954. [DOI] [PubMed] [Google Scholar]
- 6.Moncecchi D, Murphy EJ, Prows DR, Schroeder F. Biochim. Biophys. Acta. 1996;1302:110–116. doi: 10.1016/0005-2760(96)00044-6. [DOI] [PubMed] [Google Scholar]
- 7.Murphy EJ. Sterol carrier protein-2: Not just for cholesterol any more. Mol. Cell Biochem. 2002;239:87–93. [PubMed] [Google Scholar]
- 8.Krebs KC, Lan Q. Isolation and expression of a sterol carrier protein-2 gene from yellow fever mosquito. Aedes aegypti. Insect Mol. Biol. 2003;12:51–60. doi: 10.1046/j.1365-2583.2003.00386.x. [DOI] [PubMed] [Google Scholar]
- 9.Dyer DH, Lovell S, Thoden JB, Holden HM, Rayment I, Lan Q. The Structural Determination of an Insect Sterol Carrier Protein-2 with a Ligand-bound C16 Fatty Acid at 1.35-Å Resolution. J. Biol. Chem. 2003;278:39085–39091. doi: 10.1074/jbc.M306214200. [DOI] [PubMed] [Google Scholar]
- 10.Blitzer EJ, Vyazunova I, Lan Q. Functional analysis of AeSCP-2 using gene expression knockdown in the yellow fever mosquito. Aedes aegypti. Insect Mol. Biol. 2005;14:301–307. doi: 10.1111/j.1365-2583.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 11.Lan Q, Massey RJ. Subcellular localization of the mosquito sterol carrier protein-2 and sterol carrier protein-x. J. Lipid Res. 2004;45:1468–1474. doi: 10.1194/jlr.M400003-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Kim M, Wessely V, Lan Q. Identification of mosquito sterol carrier protein-2 inhibitors. J. Lipid Res. 2005;46:650–657. doi: 10.1194/jlr.M400389-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Seedorf U, Raabe M, Ellinghaus P, Kannenberg F, Fobker M, Engel T, Denis S, Wouters F, Wirtz KWA, Wanders RJA, Maeda N, Assmann G. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-x gene function. Genes. Dev. 1998;12:1189–1201. doi: 10.1101/gad.12.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs M, Hafer A, Münch C, Kannenberg F, Teichmann S, Scheibner J, Stange EF, Seedorf U. Disruption of the Sterol Carrier Protein 2 Gene in Mice Impairs Biliary Lipid and Hepatic Cholesterol Metabolism. J. Biol. Chem. 2001;276:48058–48065. doi: 10.1074/jbc.M106732200. [DOI] [PubMed] [Google Scholar]
- 15.Leventis R, Silvius JR. Use of Cyclodextrins to Monitor Transbilayer Movement and Differential Lipid Affinities of Cholesterol. Biophys. J. 2001;81:2257–2267. doi: 10.1016/S0006-3495(01)75873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips MC, Johnson WJ, Rothblat GH. Mechanisms and consequences of cellular cholesterol exchange and transfer. Biochimica et Biophysica Acta. 1987;906:223–276. doi: 10.1016/0304-4157(87)90013-x. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigueza WV, Phillips MC, Williams KJ. Structural and metabolic consequences of liposome-lipoprotein interactions. Advanced Drug Delivery Reviews. 1998;32:31–43. doi: 10.1016/s0169-409x(97)00130-0. [DOI] [PubMed] [Google Scholar]
- 18.Atshaves BP, Gallegos AM, McIntosh AL, Kier AB, Schroeder F. Sterol Carrier Protein-2 Selectively Alters Lipid Composition and Cholesterol Dynamics of Caveolae/Lipid Raft vs Nonraft Domains in L-Cell Fibroblast Plasma Membranes. Biochemistry. 2003;42:14583–14598. doi: 10.1021/bi034966+. [DOI] [PubMed] [Google Scholar]
- 19.Huang H, Gallegos AM, Zhou M, Ball JM, Schroeder F. Role of the Sterol Carrier Protein-2 N-Terminal Membrane Binding Domain in Sterol Transfer. Biochemistry. 2002;41:12149–12162. doi: 10.1021/bi0260536. [DOI] [PubMed] [Google Scholar]
- 20.Viitanen L, Nylund M, Eklund DM, Alm C, Eriksson A-K, Tuuf1 J, Salminen TA, Mattjus P, Edqvist J. Characterization of SCP-2 from Euphorbia lagascae reveals that a single Leu/Met exchange enhances sterol transfer activity. FEBS Journal. 2006;273:5641–5655. doi: 10.1111/j.1742-4658.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 21.Peleg J. Growth of Arboviruses In Primary Tissue Culture of Aedes Aegypti Embryos. Am. J. Trop. Med. & Hyg. 1968;17:219–223. doi: 10.4269/ajtmh.1968.17.219. [DOI] [PubMed] [Google Scholar]
- 22.Lan Q, Fallon M. Small heat shock proteins distinguish between two mosquito species and confirm identity of their cell lines. Am. J. Trop. Med. Hyg. 1990;43:669–676. doi: 10.4269/ajtmh.1990.43.669. [DOI] [PubMed] [Google Scholar]
- 23.Zulauf M, D'Arcy A. Light scattering of proteins as a criterion for crystallization. J. Cryst. Growth. 1992;122:102–106. [Google Scholar]
- 24.Avdulov NA, Chochina SV, Igbavboa U, Warden CS, Vassiliev AV, Wood WG. Lipid binding to amyloid beta-peptide aggregates: preferential binding of cholesterol as compared with phosphatidylcholine and fatty acids. J Neurochem. 1997;69:1746–1752. doi: 10.1046/j.1471-4159.1997.69041746.x. [DOI] [PubMed] [Google Scholar]
- 25.Lan Q, Gerenday A, Fallon AM. Cultured Aedes albopictus mosquito cells synthesize hormone-inducible proteins. In Vitro Cell Dev Biol. 1993;29A:813–818. doi: 10.1007/BF02634349. [DOI] [PubMed] [Google Scholar]
- 26.Lakowicz JR. Fluorescence spectroscopic investigations of the dynamic properties of proteins, membranes and nucleic acids. J Biochem Biophys Methods. 1980;2:91–119. doi: 10.1016/0165-022x(80)90077-9. [DOI] [PubMed] [Google Scholar]
- 27.Andersen J, Olsen L, Hansen KB, Taboureau O, Jørgensen FS, Jørgensen AM, Bang-Andersen B, Egebjerg J, Strømgaard K, Kristensen AS. Mutational mapping and modeling of the binding site for (S)-citalopram in the human serotonin transporter. J Biol Chem. 2010;285:2051–2063. doi: 10.1074/jbc.M109.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker BY, Epand RF, Epand RM, Miller WL. Cholesterol Binding Does Not Predict Activity of the Steroidogenic Acute Regulatory Protein, StAR. J. Biol. Chem. 2007;282:10223–10232. doi: 10.1074/jbc.M611221200. [DOI] [PubMed] [Google Scholar]
- 29.Svoboda JA, Thompson MJ, Herbert EW, Jr, Shortino TJ, Szczepanik-Vanleeuwen PA. Utilization and Metabolism of Dietary Sterols in the Honey Bee and the Yellow Fever Mosquito. Lipids. 1982;17:220–225. doi: 10.1007/BF02535107. [DOI] [PubMed] [Google Scholar]
- 30.Warner SA, Sovocool GW, Domnas AJ, Jaronski ST. The Composition of Sterols in Mosquito Larvae Is Optimal for Zoosporogenesis in Lagenidium giganteum. J. of Invertebrate Pathology. 1984;43:293–296. doi: 10.1016/0022-2011(84)90154-x. [DOI] [PubMed] [Google Scholar]
- 31.Grieneisen ML, Warren JT, Gilbert LI. Early Steps in Ecdysteroid Biosynthesis: Evidence for the Involvement of Cytochrome P-450 Enzymes. Insect Biochem. Molec. Biol. 1993;23:13–23. doi: 10.1016/0965-1748(93)90077-6. [DOI] [PubMed] [Google Scholar]
- 32.Vyazunova I, Lan Q. Insect sterol carrier protein-2 gene family: structures and functions. Chapter 11. In: Liu Nannan., editor. Recent Advances in Insect Physiology, Toxicology and Molecular Biology. Research Signpost, Kerala, India.: 2008. pp. 173–198. [Google Scholar]
- 33.Reshetnyak YK, Burstein EA. Decomposition of Protein Tryptophan Fluorescence Spectra into Log-Normal Components. II. The Statistical Proof of Discreteness of Tryptophan Classes in Proteins. Biophysical J. 2001;81:1710–1734. doi: 10.1016/S0006-3495(01)75824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larson RT, Wessely V, Jiang Z, Lan Q. Larvicidal activity of sterol carrier protein-2 inhibitor in four species of mosquitoes. J. Medical Entomol. 2008;45:439–444. doi: 10.1603/0022-2585(2008)45[439:laoscp]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dwivedy AK. Dietary cholesterol utilization by the housefly, Musca domestica. Insect Biochem Mol Biol. 1985;15:137–140. [Google Scholar]
- 36.Kuthiala A, Ritter KS. Esterification of cholesterol and cholestanol in the whole body, tissues, and frass of Heliothis zea. Arch Insect Biochem Physiol. 1988;7:237–248. [Google Scholar]
- 38.Komnick H, Giesa U. Intestinal absorption of cholesterol, transport in the haemolymph, and incorporation into the fat body and Malpighian tubules of the larval dragonfly Aeshna cyanea. Comp Biochem Physiol Comp Physiol. 1994;107:553–557. doi: 10.1016/0300-9629(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 39.McNamara BC, Jefcoate CR. The role of sterol carrier protein 2 in stimulation of steroidogenesis in rat adrenal mitochondria by adrenal cytosol. Arch Biochem Biophys. 1989;75:53–62. doi: 10.1016/0003-9861(89)90349-4. [DOI] [PubMed] [Google Scholar]
- 40.Mendis-Handagama SM, Aten RF, Watkins PA, Scallen TJ, Berhman HR. Peroxisomes and sterol carrier protein-2 in luteal cell steroidogenesis: a possible role in cholesterol transport from lipid droplets to mitochondria. Tissue Cell. 1995;27 doi: 10.1016/s0040-8166(05)80056-4. 483-90.41. [DOI] [PubMed] [Google Scholar]
- 41.Hentikoff S, Hentikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tantuco Deretey E, Csizmadia IG. Stabilities for the eight isomeric forms of the steroid skeleton (perhydrocyclopentanophenanthrene) J. Mol. Structure (Theochem) 2000;503 97–111.43. [Google Scholar]
- 43.Stremmel W, Pohl L, Ring A, Herrmann T. A new concept of cellular uptake and intracellular trafficking of long-chain fatty acids. Lipids. 2001;36:981–989. doi: 10.1007/s11745-001-0809-2. [DOI] [PubMed] [Google Scholar]
- 44.Maxfield FR, Menon AK. Intracellular sterol transport and distribution. Curr Opin Cell Biol. 2006;18:379–385. doi: 10.1016/j.ceb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Chu BB, Ge L, Li BL, Yan Y, Song BL. Membrane topology of human NPC1L1, a key protein in enterohepatic cholesterol absorption. J Lipid Res. 2009;50:1653–1662. doi: 10.1194/jlr.M800669-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falomir-Lockhart LJ, Burgardt NI, Ferreyra RG, Ceolin M, Ermácora MR, Córsico B. Fatty acid transfer from Yarrowia lipolytica sterol carrier protein 2 to phospholipid membranes. Biophys J. 2009;97:248–256. doi: 10.1016/j.bpj.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallegos AM, McIntosh AL, Atshaves BP, Schroeder F. Structure and cholesterol domain dynamics of an enriched caveolae/raft isolate. Biochem J. 2004;382:451–461. doi: 10.1042/BJ20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jouni ZE, McGill B, Wells MA. Beta-cyclodextrin facilitates cholesterol efflux from larval Manduca sexta fat body and midgut in vitro. Comp Biochem Physiol B Biochem Mol Biol. 2002;132:699–709. doi: 10.1016/s1096-4959(02)00084-2. [DOI] [PubMed] [Google Scholar]
- 49.Atshaves BP, Storey SM, Petrescu A, Greenberg CC, Lyuksyutova OI, Smith R, 3rd, Schroeder F. Expression of fatty acid binding proteins inhibits lipid accumulation and alters toxicity in L cell fibroblasts. Am J Physiol Cell Physiol. 2002;283:C688–C703. doi: 10.1152/ajpcell.00586.2001. [DOI] [PubMed] [Google Scholar]
- 50.Atshaves BP, Storey SM, Schroeder F. Sterol carrier protein-2/sterol carrier protein-x expression differentially alters fatty acid metabolism in L cell fibroblasts. J. of Lipid Research. 2003;44:1751–1762. doi: 10.1194/jlr.M300141-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Murphy EJ. Sterol carrier protein-2 expression increases NBD-stearate uptake and cytoplasmic diffusion in L cells. Am J Physiol Gastrointest Liver Physiol. 1998;275:237–243. doi: 10.1152/ajpgi.1998.275.2.G237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.