Abstract
RNA transfection methods have not proven to be as popular as DNA methods due to the highly transient nature of the RNA inside the cell. However, there are many advantages in using RNA for gene over-expression, such as the rapidity of expression, the ability to express in all cell types without the need for cell-type specific promoters, and the ability to analyze the effects of gene over-expression in a transient manner. Therefore, we have developed a method (StabiLizingUtr: SLU) to stabilize the RNA for varying durations, using specific sequences from the 3′UTR of the Venezuelan equine encephalitis virus (VEEV). We have designed a plasmid for cloning genes upstream from repeated stabilizing sequences to generate mRNA with one or more VEEV stabilizing sequence motifs. We demonstrate this method in several cell and tissue types, including the mammalian cochlea, a tissue that has been difficult to transfect with other methods.
Introduction
For many years DNA transfection has been used successfully in a large number of different cell types and tissues (Mulligan and Berg, 1980; Mulligan and Berg, 1981; Itani et al., 1987; Keown et al., 1990; Gartner et al., 2006; Zeitelhofer et al., 2009). For cells that have been more difficult to transfect with DNA, viral vectors have been used for stable over-expression (Perri et al., 2000; Ehrengruber and Lundstrom, 2002; Ehrengruber and Lundstrom, 2007).
Fewer studies have used RNA for over-expressing genes (Malone et al., 1989; Dwarki et al., 1993; Mutzke et al., 2005), though studies in frog oocytes and early developmental studies have used RNA extensively, delivering it through direct microinjections. In addition, for short-term expression, RNA is being developed as an alternative to viral gene therapy (see Van Tendeloo et al., 2007 for review). RNA has several advantages over plasmid or viral approaches. Since gene expression from an RNA source does not require transcription, the protein product is produced rapidly after the transfection, and since the mRNA has to only gain access to the cytoplasm, rather than the nucleus, typical transfection methods produce results in an extremely high rate of transfection. Plasmid based approaches require that the promoter driving the expression of the gene of interest be active in the cells under study. While some of the more common promoters are active in a wide variety of cell lines, they are more limited in their expression in primary cells and tissues, particularly in postmitotic neurons. The transient nature of the expression could be useful when studying developmental processes that require only a brief “pulse” of a developmental signal or transcription factor (Sasagawa et al., 2002).
However, one of the main reasons that RNA transfection is less widely used than DNA is the limited stability of the RNA. Whereas viral over-expression persists, potentially for the life of the cell, and plasmid transfection can persist through multiple rounds of cell division, RNA is rapidly degraded. Because of the potential advantages of RNA based gene over-expression for certain applications, we sought to develop methods for producing mRNA with increased stability. In this report we describe a new vector that incorporates stabilizing sequences in the 3′UTR of the mRNA, which allows for a highly stable message, suitable for longer-term gene over-expression studies.
Methods
Synthesis of pSLU vector
The pSLU vector was made as follows. The sequences of Sindbis viral 3′ UTR and VEEV were obtained from the NCBI database (Sindbis virus; U90536.1, nucleotides 3793 to 4092: VEEV; NC_001449.1, nucleotides 11330 to 11427). Both sequences lack a 19-nucleotide conserved sequence element (3′ CSE) which is crucial for the duplication of the viral RNA. For construction of the pSLU vector, the DNA coding a T7 promoter, a multiple cloning site, 6x VEEV 3′ UTR and a T3 promoter were synthesized commercially by Integrated DNA Technologies (Coralville, IA), and then sub-cloned into pIDT SMERT v3. Each gene of interest was sub-cloned into the multiple cloning site of the pSLU using either BamHI/ XbaI (e.g. d2EGFP from Clonetech) or EcoRI/ XbaI (e.g. Mash1 and Math1).
mRNA production
To generate mRNA, we used the Ambion mMessage mMachine (AM1345; Ambion, Texas, USA) according to the manufacturer’s instructions. Linearized pSLU plasmid (1 μg of a stock at a concentration of 0.5-1 μg/μl) was added to a total reaction volume of 20 μl including 10 μl of ribonucleotides (2X NTP and ARCA:anti-reverse cap analog), 2 μl of 10X T7 reaction buffer, and 2 μl of T7 enzyme and RNase-free water. The reagents were mixed thoroughly, centrifuged briefly to collect the reactants together and incubated at 37°C for 2-2.5 hours. The RNA was then tailed using E-PAP according to the manufacturer’s directions, for 45 min, and then recovered by lithium chloride precipitation or phenol:chloroform extraction and isopropanol precipitation. 40-50 μg of RNA is a typical yield from a 20 μl reaction.
Explant Electroporation
The mRNA was centrifuged for 15,000 x g for 6-8 minutes, and the supernatant removed. The pelleted mRNA was washed once in 80% RNA grade ethanol and resuspended in 21 μl RNase free water and incubated at 68-72° C for 2-3 minutes to dissolve the pellet. We used 1 μl to check the concentration, which should be between 2 and 3 μg/μl. The RNA was kept on ice until ready for use. When co-transfection with multiple genes was required, two or more mRNAs were mixed together. For example, for co-electroporation of 2 mRNAs, 0.4 μg of each mRNA was mixed with PBS-glycerol stock to make a final solution of 0.8μg total mRNA in a 10% glycerol-PBS buffer containing 1μl of RNAse inhibitor.
For cochlear, cortical or retinal explants, we dissected the tissue from the animal after sacrifice. For the embryonic dissections, the embryos were removed from the dam and placed in PBS on ice. To dissect the retinas, the eyes were removed from the animals and the lens was removed; next the scleral tissue and attached pigmented epithelium were removed leaving the neural retina (Roberts et al., 2006). The cerebral cortex was dissected by removing the skull, to expose the cerebral cortex, and then by dissecting the cortex from the subcortical telencephalon. The cochleas were dissected as follows: first, the hindbrain was removed and the otic capsule visualized. The capsule was dissected from the surrounding temporal bone, and then the cochlear duct isolated from the capsule. The roof of the duct was removed to allow penetration of the mRNA, and the explants were placed on collagen coated Millicel tissue culture inserts (Hayashi et al., 2008a; Hayashi et al., 2008b). Immediately after dissection (retinas and cortex) or after overnight culture (cochlea), the explants were placed in an electroporation chamber filled with HBSS; the positive electrode was placed below the explant and the negative electrode above. The RNA solution was carefully added over the explant, such that it and rested on top of the explant. Voltage was then applied; three 55 msec pulses of 10 Volts for cochlea and 30 V for retina were optimal for our tissues, but other tissues may require different settings that need to be empirically derived. (We used a BTX Electro Square Porator T-820.) The explants were then returned to the tissue culture medium for up to 36 hrs. For all experiments in which we used primary mouse tissues, the mice were housed in the University of Washington Department of Comparative Medicine and the experimental methods and all procedures were approved by the Institutional Animal Care and Use Committee.
Cell Line Transfection
For transfection of cell lines, 24-well tissue culture plates were coated with poly-D-Lysine for 30 minutes and then rinsed well with water three to five times. HEK-293 and rat Muller cells were plated in DMEM-F12 media with 10% fetal bovine serum and penicillin/streptomycin. Two hours prior to transfection, media was changed to Opti-mem media without antibiotics. One of three different lipid-based transfection methods (Trans-IT (Mirus)/Lipofectamine (InVitrogen)/Genejammer(Stratagene)) were used as follows: the transfection reagent was mixed with the RNA in a 3:1 ratio (3μl/1μg); after 20 minutes at room temperature the mix was added to the wells. The plates were then returned to the incubator and GFP expression was analyzed 12-24 hours later.
The H-1 (WA-01) human embryonic stem cell line was obtained from WiCell Research Institute. The cells were cultured and passaged on a feeder layer made of irradiated mouse embryonic fibroblasts. Retinal induction was performed as previously described (Lamba et al., 2006). Briefly, embryoid bodies (EBs) were formed by treating undifferentiated hES cell colonies with type IV collagenase (Invitrogen) and resuspending approximately 150-100-cell clumps per ml in a 6 well ultra-low attachment plate (VWR). These EBs were cultured for 3 days in the presence of mouse noggin (R&D Systems), human recombinant Dkk-1 (R&D Systems) and human recombinant insulin-like growth factor-1 (IGF-1) (R&D Systems). On the fourth day, embryoid bodies were plated onto poly-D-lysine-Matrigel (Collaborative Research, Inc) coated plates and cultured in the presence of DMEM:F12, B-27 supplement, N-2 Supplement (Invitrogen), mouse noggin, human recombinant Dkk-1, human recombinant IGF-1 and human recombinant basic fibroblast growth factor (bFGF) (R&D Systems). The media was changed every 2-3 days for up to 3 weeks. Thereafter, the cells were cultured in the above media without any growth factors prior to transfection with mRNA.
Immunohistochemistry
The explants were fixed with 4% paraformaldehyde, and either cryoembedded and sectioned on a Leica CM1850 cryostat at 10-16 microns or processed as whole mounts. The whole mounts and tissue sections were immunolabeled with antibodies to tissue-specific cell types. For the cochlea, we used rabbit anti-Myosin 6 (Proteus Biosciences) at 1:1000; guinea pig anti-Gfi1 (gift from Hugo Bellen, Baylor College of Medicine, Texas) at 1:1000; mouse anti-Cre (Chemicon) at 1:250. For the retina, we used Ascl1(Mash1) antibody (Abcam), and for the cortex, TuJ1 (Covance). We incubated the sections or whole mounts in primary antibody for a minimum of two hours, and typically 24 hrs. We then rinsed off the unbound primary antibody for up to two hours prior to incubation in appropriate secondary antibody, conjugated with AlexaFluor (InVitrogen). After the incubation period in secondary antibody, the tissue was washed again twice in PBS for 10-20 minutes and then mounted with Fluoromount-G (Southern Biotech).
Results
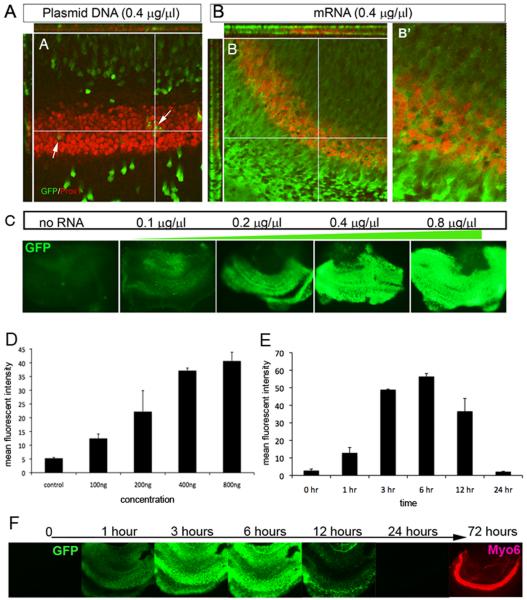
Figure 1 demonstrates some of the differences between mRNA and plasmid cDNA transfection in the developing mouse organ of Corti, a tissue that has proven difficult to transfect with existing methods. A typical example of a plasmid DNA transfection by electroporation in the embryonic organ of Corti is shown in Figure 1 A. Many cells express the GFP in regions adjacent to the sensory region, but few cells express GFP in the sensory region, which is labeled with Prox1 (red); arrows point out a few examples of double-labeled support cells in the sensory region. By contrast, when mRNA is transfected using the same electroporation method in a sister explant, instead of a few scattered cells, nearly every cell in the explant expresses some level of GFP (Figure 1 B). It is difficult to make out the individual cells in these explants since the GFP expression is so widespread, however, the confocal sectioning Z-axis reconstructions show that the cells with the highest levels of expression in the Prox1+ region are likely support cells, though hair cells may also express the GFP. A higher magnification image of this region shown in Figure 1B’ also confirms that most Prox1+ cells express GFP. The “dose-response” of the expression of GFP from the mRNA is shown in a series of micrographs taken of other transfected cochlear explants (Figure 1 C,D). The level of GFP expression directly correlates with the amount of mRNA used in the electroporation (Figure 1D). However, when we analyzed the time course of expression of the GFP, using a destabilized eGFP so that persistence of the GFP does not affect our interpretation of message stability, we find that expression is rapid: protein is already visible at 1 hour and the level peaks around 6 hours (Figure 1E,F); however the expression is noticeably reduced by 12 hours and no longer detectable by 24 hours (Figure 1 E,F). These data show that mRNA transfection is an extremely efficient approach for gene over-expression, even in tissue that has proven difficult to transfect with plasmid based approaches, but that the over-expression is highly transient.
Figure 1.
RNA can be efficiently electroporated into tissues, but expression lasts only a short time. Either plasmid DNA (A) or mRNA (B) coding for destabilized eGFP was transfected into E15 cochlear explants, and the explants fixed in paraformaldehyde either 24 hours (A) or six hours (B) later to show the difference in expression of the plasmid cDNA and mRNA transfections. The explants were immunolabeled for Prox1 (red) to show the sensory epithelium of the organ of Corti. The plasmid shows many GFP-expressing cells in the region adjacent to the sensory region, the greater epithelial ridge (GER) with relatively few cells within the sensory region (arrows). By contrast, the mRNA (B) shows that nearly every cell in the explant expresses GFP in both the GER and the Prox1+ sensory region. Confocal sectioning shows that many of the GFP+ cells are Prox1+ support cells. C. D. The expression of GFP in the explants correlates well with the amount of mRNA used for the transfection, from 0.1 μg/μl to 0.8 μg/μl, where it begins to plateau. E.F. The transient nature of the mRNA-induced expression is seen when the same explant is imaged live at intervals of 1, 3, 6, 12, 24 and 72 hours after the transfection. Expression of GFP can already be observed as early as 1 hour post-transfection (F, E), and this peaks between 3 hrs and 6 hrs. There is a noticeable decline in the level of GFP at 12 hours and by 24 hours has declined to pre-transfection levels. Myosin6 (Myo6, red) was used to label the explant after fixing at 72 hours to show the sensory epithelium is maintained throughout this period.
Because of the potential advantages of RNA based gene over-expression for certain applications, we sought to develop methods for producing mRNA with increased stability by slowing its degradation. Polyadenylated mRNAs undergo multiple rounds of translation, but with each round, deadenylases shorten the polyA tail. Once the tail is removed, the mRNA is degraded from the 3′ end via the exosome. RNA viruses have evolved many different mechanisms to elude the cell’s RNA degradation machinery. Sindbis virus and Venezuelan equine encephalitis virus (VEEV) are RNA viruses that have genomes that closely resemble eukaryotic mRNA, with a 5′ cap and a polyadenylate tail and importantly they have evolved stabilizing elements in the 3′ UTR (Garneau et al., 2008).
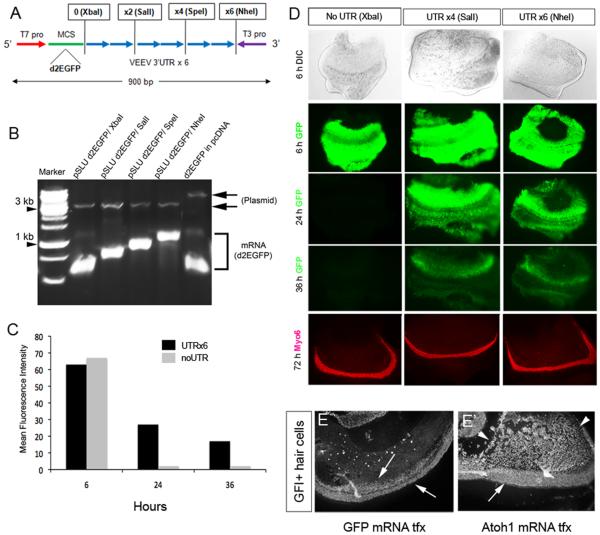
We tested whether incorporating these sequences from the 3′ UTR of the Venezuelan equine encephalitis virus (VEEV) or the Sinbis virus could be useful for stabilizing the mRNA. We first tested a single repeat and found this was not effective when taken from either the Sinbis virus (300 bp) or the VEEV virus (100 bp). However, since the VEEV sequence was only 100 bp long, we generated multimers of the sequence and tested whether multimers of this sequence would be more effective than the monomers (Figure 2A). We found that 2 or more repeats of the sequence at the 3′ end of the transcript would stabilize the message and allow expression for up to 24 hours (Figure 2 C,D). When 4 or 6 repeats of the sequence were used, the destabilized eGFP expression was observed as long as 36 hours (Figure 2 C,D). We call this technique the StabiLized UTR approach, or SLU. We designed a plasmid for cloning genes with varying numbers of repeats of the stabilizing sequences, called pSLU (Figure 2 A).
Figure 2.
The pSLU plasmid incorporates sequences from the VEEV 3′UTR to stabilize mRNA for longer periods in cells. (A) The pSLU plasmid was designed to allow different numbers of UTR repeats, depending on the restriction digestion for preparing the template. (B) mRNA synthesized from the templates prepared with one of four different restriction enzymes shows that the message has the appropriate length for multimers of the VEEV sequences. (C,D) Cochlear explants were transfected with mRNA with either no VEEV sequences (Xba1), 4 repeats (Sal1) or 6 repeats (Nhe1) and the level of GFP expression analyzed at 6, 12, 24, and 36 hours after transfection. GFP expression is robust in all at 6 hours, but with no VEEV sequences expression is absent at 12 hours. With either 4 or 6 repeats, expression is strong (approximately 50% of the 6 hr level) at 24 hours and still apparent at 36 hours. Note in all experiments, destabilized eGFP is used to prevent persistence of the GFP protein after the message has been degraded. (E,E’) The mRNA is functional in an assay of hair cell production in the cochlea. GFI, a marker of hair cells is expressed in the sensory epithelium of the cultures (arrows) similar to that of myo6 in earlier panels. When Atoh1 mRNA is transfected into the explants, there is a large increase in the number of GFI+ hair cells (arrowheads) throughout the GER (E’).
In addition to GFP, we tested several other constructs to determine whether transient expression of proteins could reach functional levels within this time-frame. Previous experiments have shown that over-expression of Atoh1, a gene that is necessary and sufficient for cochlear hair cell development, and can direct cells to become hair cells in the GER (greater epithelial ridge), a region, which would not normally give rise to hair cells (Bermingham et al., 1999; Zheng and Gao, 2000; Gubbels et al., 2008). We carried out a similar experiment with stabilized mRNA. We found that transfection of the stabilized mRNA coding for Atoh1 into the developing GER induces a large number of hair cells in the GER (Figure 2E,E’), many more than had previously been shown using plasmid transfection. Figure 2 E and E’ show this effect; nearly every cell in the GER expresses the hair cell marker (Gfi) after Atoh1 mRNA electroporation. Thus, the method allows for a high level of rapid expression of genes that can act as developmental switches, like transcription factors. In this case, a relatively short pulse of Atoh1 was sufficient to direct the GER cells to the stable hair cell fate (see supplemental Figure 1). We have used Gfi1 as a hair cell marker because it is a gene that is expressed earlier in hair cell development than Myosin 6 (Wallis et al., 2003; Hertzano et al., 2004); however, we have also shown that all Gfi-immunoreactive cells also express Myosin 6 (Supplemental Figure 1).
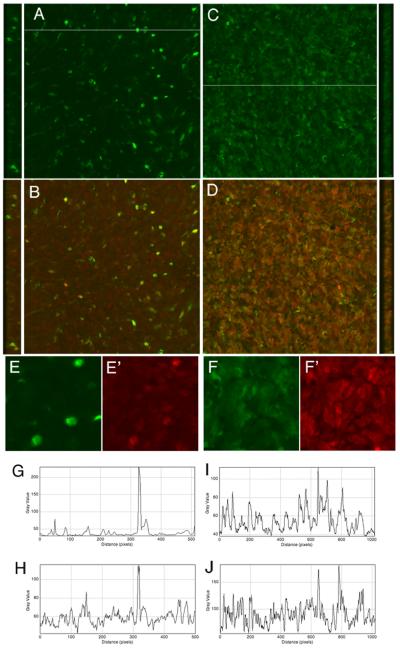
In addition to the cochlea, we tested this method on a variety of other tissues and cells. We transfected mRNA coding for the transcription factor Mash1/Ascl1 into the embryonic retina and assayed its expression (Figure 3). Panels A, B and E/E’ show the results of transfection of retina with a plasmid encoding Mash1/Ascl1 after 24 hours, when labeled with an antibody against Mash1/Ascl1. Scattered retinal cells co-express GFP and a higher level of the Mash1/Ascl1 than the background endogenous level. By contrast, when a sister retinal explant is co-transfected with mRNA for Mash1/Ascl1 and GFP (Figure 3 C,D, F/F’), there is a much broader domain of over-expression, and as was observed in the cochlea, nearly every cell expresses GFP. Nearly all cells in the field also express a higher level of Mash1/Ascl1 than the endogenous level (compare the levels in F’ with the un-transfected cells in E’). The results are quantified in Figure 3 G,I (GFP) and H,J (Mash1/Ascl1), where it can be appreciated that the overall level of Mash1/Ascl1 expression from the mRNA (average 100 in J) is nearly as high as the few cells that over-express Mash1/Ascl1 from the plasmid (H). These results show that the mRNA derived from the pSLU vector can be used in developing retina as well as the cochlea.
Figure 3.
mRNA coding for the transcription factor Ascl1 was transfected along with mRNA for GFP into embryonic retina, either as a plasmid (A,B,E,E’) or as stabilized mRNA made from the six repeat form of pSLU (C,D,F,F’). The retinal explant was allowed to survive in vitro for 24 hr and then the level of Ascl1 expression was assessed with an antibody against Ascl1. The plasmid transfection results in scattered cells throughout the retina expressing both GFP (A,E, green) and Ascl1 (B,E’, red), the latter clearly expressed above the dim red labeling of the endogenous Ascl1 (E). However, mRNA transfection results in nearly every cell expressing high levels of both Ascl1 (D,F’ red) and GFP (C, F, green). The level of expression was quantified for the plasmid transfection in G for GFP and H for Ascl1, by taking a profile of a single line across the field shown in A. Note the correspondence between the peak for GFP and Ascl1 in G and H. However, panels I and J show the much broader and higher efficiency of transfection, as well as co-expression achieved with mRNA (note the scale change panel J from H).
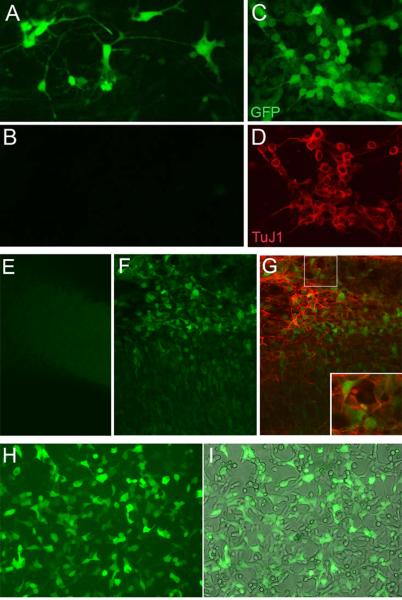
To determine whether this method would be applicable to experiments in other developing tissues and cells, we transfected mRNA coding for GFP produced from the pSLU plasmid with six repeats of the VEEV sequence into several other cell types. We found that mRNA generated from the pSLU plasmid drove GFP expression in neurons derived from human ES cells (Figure 4 A-D). Figure 4 A and B compare transfected (A) or untransfected (B) neurons, and panels C and D show a field of TuJ1+ neurons expressing GFP from the mRNA. Mouse cerebral cortex slices were also transfected efficiently with the mRNA and again, many TuJ1+ neurons were found to express the GFP within 24 hours (Figure 4 E-G). The same GFP mRNA works well with HEK cells transfected with lipid-based transfection methods (Figure 4 H,I), and so should have general applicability. In another example of an application that might be of use to many investigators we electroporated retinas from an mT/mG with mRNA for Cre recombinase and showed that we get robust Cre protein expressed and Cre excision activity (Supplemental Figure 2). The mT/mG double fluorescent Cre reporter mouse line which expresses membrane-targeted tandem dimer Tomato (mTomato) prior to Cre-mediated excision and membrane-targeted green fluorescent protein (mGFP) after excision (Muzumdar et al., 2007).
Figure 4.
mRNA transfection with pSLU synthesized message shows efficient expression in a variety of cell types. (A-D) Human embryonic stem cells were used to generate retinal neurons and then transfected by electroporation with stabilized GFP mRNA containing six repeats using pSLU (A, C, D) and photographed 48 hours later. Panel B shows a field of untransfected cells from a sister culture as panel A, also cultured for 48 hours. Panels C and D show the same field, showing that many of the transfected cells expressing GFP are Tuj1 + retinal neurons. (E-G) mRNA for GFP was transfected into embryonic mouse cerebral cortex in vitro (F,G), while the panel in E shows an un-transfected cortical slice from the same mouse. Expression of the transfected GFP mRNA was present both in the migrating neuroblasts (NB) and the neurons (TuJ1+); the inset in panel G shows a higher magnification view of neurons expressing GFP. (H,I) HEK cells were transfected with stabilized GFP mRNA made from the 6 repeat form of pSLU using lipofectamine (H and I show the fluorescent and phase view of the same field).
Discussion
In this report we describe a new technique for stabilizing mRNA for use in over-expression experiments. The use of mRNA for testing sufficiency of genes in developmental processes has many advantages over plasmid or viral based approaches; transfection of mRNA allows very rapid, widespread expression without the need for cell-type specific promoters (see Van Tendeloo et al, 2007 for review). Moreover, tissues that have been difficult to transfect with other methods are robustly transfected with mRNA. Another recent study has also demonstrated the increased efficiency of transfection using RNA, as compared with DNA for both cell lines and mixed neuronal cultures using lipid based transfection (Zou et al., 2010). However, the transient nature of mRNA has limited its use for many experiments. We have found that mRNA stabilized with the SLU method persists for up to 36 hours, sufficient time for many developmental processes. The ability to stabilize mRNA provided by the SLU technique should therefore allow investigators to combine the advantages of mRNA with those of other over-expression methods. In addition, the SLU technique will allow studies of the effects of transient over-expression to be carried out more efficiently than existing systems, since the length of time for over-expression can be determined by the number of VEEV sequences incorporated into the gene of interest.
Using this technique in the developing cochlea, we have found that only a transient expression of Atoh1 is sufficient to drive the GER cells to a hair cell fate; this is the type of study for which this approach is ideal. Frequently in development, transcription factors act as stable “on” switches in the logic of developmental circuits. Atoh1 is an example of this type of sustained “on” switch, since after a relatively brief period of development it is no longer required. The use of SLU for controllable transient over-expression will be very useful in identifying which transcriptional regulators act in this manner and to better define the specific windows of developmental time over which these factors act. The SLU technique may also have advantages over other approaches for gene over-expression where it is important not to permanently modify the genome. For example, when creating iPS cells, methods that allow efficient expression of pluripotency factors to reprogram cells that do not use viruses are desirable, and the SLU method may find applicability in this area.
In summary, we have developed a way to stabilize mRNA for up to 36 hours in cells, sufficient time for analysis of effects of gene over-expression in many developmental experiments. This will allow investigators to better take advantage of the benefits of mRNA over other gene over-expression approaches, including the rapid, highly efficient, and widespread over-expression that is not limited to specific cell types.
Supplementary Material
Supplemental Figure 1 mRNA encoding mAtoh1 was electroporated into E 15 cochlear explants and maintained in culture for 4 days. A large number of ectopic hair cells were produced outside of the sensory epithelium in the GER (Panels A and B), these ectopically produced hair cells co-express two markers Gfi1 (Red) and Myosin6 (green, panel B). The normal region of sensory cells can been seen in panel A (indicated with bracket) where the p75 (green) labels the rows of pillar cells between the inner and outer hair cells (myosin6, red) and also labels the Claudius cells adjacent to the organ of Corti. The scale bar is 200μm on the left panel and 100μm on the right panel.
Supplemental Figure 2 P3 explanted retinas from mT/mG mice were electroporated with mRNA for Cre recombinase and cultured for two days. The explants were fixed with 4% paraformaldehyde, cryoembedded and sectioned. The tissue sections were immunolabeled with an antibody to Cre (purple) and imaged. The green cells (mGFP) indicate where the mT/mG locus has been recombined and Dapi (blue) reveals all nuclei.
Acknowledgements
We acknowledge the members of the Reh and Bermingham-McDonogh labs for their helpful criticisms during the development of this project. We thank Dr. Hugo Bellen for the gift of anti-Gfi1 antisera. We also thank Dr. J. Brzezinski for helpful comments on the manuscript and we are much obliged to Dr. Byron Hartman for his confocal artistry. This work was supported by grants from the NIH to T.A.R. (R01 EY013475, P01GM081619) and to O.B-McD. (R01 DC005359) and from an unrestricted gift to T.A.R. from an anonymous donor.
References
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zohbi HY. Math1:an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Dwarki VJ, Malone RW, Verma IM. Cationic liposome-mediated RNA transfection. Methods Enzymol. 1993;217:644–654. doi: 10.1016/0076-6879(93)17093-k. [DOI] [PubMed] [Google Scholar]
- Ehrengruber MU, Lundstrom K. Semliki Forest virus and Sindbis virus vectors. Curr Protoc Hum Genet. 2002 doi: 10.1002/0471142905.hg1202s33. Chapter 12:Unit 12 12. [DOI] [PubMed] [Google Scholar]
- Ehrengruber MU, Lundstrom K. Alphaviruses: Semliki Forest virus and Sindbis virus vectors for gene transfer into neurons. Curr Protoc Neurosci. 2007 doi: 10.1002/0471142301.ns0422s41. Chapter 4:Unit 22. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J. The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and Mammalian cells. J Virol. 2008;82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Collin L, Lalli G. Nucleofection of primary neurons. Methods Enzymol. 2006;406:374–388. doi: 10.1016/S0076-6879(06)06027-7. [DOI] [PubMed] [Google Scholar]
- Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, Brigande JV. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455:537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Kokubo H, Hartman BH, Ray CA, Reh TA, Bermingham-McDonogh O. Hesr1 and Hesr2 may act as early effectors of Notch signaling in the developing cochlea. Dev Biol. 2008a;316:87–99. doi: 10.1016/j.ydbio.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci. 2008b;28:5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yucel R, Frankel WN, Rechavi G, Moroy T, Friedman TB, Kelley MW, Avraham KB. Transcription profiling of inner ears from Pou4f3(dd1/dd1) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- Itani T, Ariga H, Yamauchi N, Tadakuma T, Yasuda T. A simple and efficient liposome method for transfection of DNA into mammalian cells grown in suspension. Gene. 1987;56:267–276. doi: 10.1016/0378-1119(87)90143-0. [DOI] [PubMed] [Google Scholar]
- Keown WA, Campbell CR, Kucherlapati RS. Methods for introducing DNA into mammalian cells. Methods Enzymol. 1990;185:527–537. doi: 10.1016/0076-6879(90)85043-n. [DOI] [PubMed] [Google Scholar]
- Lamba DA, Karl MO, Ware CB, Reh TA. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci U S A. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan RC, Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980;209:1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- Mulligan RC, Berg P. Factors governing the expression of a bacterial gene in mammalian cells. Mol Cell Biol. 1981;1:449–459. doi: 10.1128/mcb.1.5.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutzke T, Schubkegel G, Teufel R, Ketterer T, Probst J, Scheel B, Carralot JP, Pascolo S, Ghoreschi K, Weigert C. Co-transfection of messenger RNA and siRNA as a method to study the efficiency of siRNA. Nucleosides Nucleotides Nucleic Acids. 2005;24:147–152. doi: 10.1081/NCN-51908. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Perri S, Driver DA, Gardner JP, Sherrill S, Belli BA, Dubensky TW, Jr., Polo JM. Replicon vectors derived from Sindbis virus and Semliki forest virus that establish persistent replication in host cells. J Virol. 2000;74:9802–9807. doi: 10.1128/jvi.74.20.9802-9807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci U S A. 2006;103:6218–6223. doi: 10.1073/pnas.0509981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa S, Takabatake T, Takabatake Y, Muramatsu T, Takeshima K. Improved mRNA electroporation method for Xenopus neurula embryos. Genesis. 2002;33:81–85. doi: 10.1002/gene.10094. [DOI] [PubMed] [Google Scholar]
- Van Tendeloo VF, Ponsaerts P, Berneman ZN. mRNA-based gene transfer as a tool for gene and cell therapy. Curr Opin Mol Ther. 2007;9:423–431. [PubMed] [Google Scholar]
- Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, Zoghbi HY, Orkin SH, Bellen HJ. The sinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M, Vessey JP, Thomas S, Kiebler M, Dahm R. Transfection of cultured primary neurons via nucleofection. Curr Protoc Neurosci. 2009 doi: 10.1002/0471142301.ns0432s47. Chapter 4:Unit4 32. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- Zou S, Scarfo K, Nantz MH, Hecker JG. Lipid-mediated delivery of RNA is more efficient than delivery of DNA in non-dividing cells. Int J Pharm. 389:232–243. doi: 10.1016/j.ijpharm.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 mRNA encoding mAtoh1 was electroporated into E 15 cochlear explants and maintained in culture for 4 days. A large number of ectopic hair cells were produced outside of the sensory epithelium in the GER (Panels A and B), these ectopically produced hair cells co-express two markers Gfi1 (Red) and Myosin6 (green, panel B). The normal region of sensory cells can been seen in panel A (indicated with bracket) where the p75 (green) labels the rows of pillar cells between the inner and outer hair cells (myosin6, red) and also labels the Claudius cells adjacent to the organ of Corti. The scale bar is 200μm on the left panel and 100μm on the right panel.
Supplemental Figure 2 P3 explanted retinas from mT/mG mice were electroporated with mRNA for Cre recombinase and cultured for two days. The explants were fixed with 4% paraformaldehyde, cryoembedded and sectioned. The tissue sections were immunolabeled with an antibody to Cre (purple) and imaged. The green cells (mGFP) indicate where the mT/mG locus has been recombined and Dapi (blue) reveals all nuclei.