Abstract
The major murine systemic lupus erythematosus (SLE) susceptibility locus Sle1 is syntenic to a chromosomal region linked with SLE susceptibility in multiple human studies. Congenic analyses have shown that Sle1 breaks tolerance to chromatin, a necessary step for full disease induction that can be suppressed by specific modifier loci. In the present study, our fine mapping analysis of the location of Sle1 has determined that three loci within this congenic interval, termed Sle1a, Sle1b, and Sle1c, can independently cause a loss of tolerance to chromatin. Each displays a distinctive profile of serological and cellular characteristics, with T and B cell functions being more affected by Sle1a and Sle1b, respectively. The epistatic interactions of Sle1 with other susceptibility loci to cause severe nephritis cannot be accounted, however, by these three loci alone, suggesting the existence of an additional locus, termed Sle1d. These findings indicate that the potent autoimmune phenotype caused by the Sle1 genomic interval reflects the combined impact of four, separate, susceptibility genes. This level of genetic complexity, combined with similar findings in other systems, supports the possibility that many complex trait loci reflect the impact of polymorphisms in linked clusters of genes with related functions.
Systemic lupus erythematosus (SLE) susceptibility is inherited as a multifactorial genetic disease (1). Thus far, linkage analyses in multiple murine models have detected 31 susceptibility loci distributed among 21 nonoverlapping genomic intervals, clearly illustrating the complexity of the genetic basis for susceptibility to systemic autoimmunity (2). In SLE patients, association and case-control studies have analyzed the contribution of numerous candidate genes, and several linkage analyses have been performed (3). Remarkably, one region of the genome, telomeric chromosome (chr) 1 in the mouse and its syntenic equivalent 1q21–44 in humans, has shown strong linkage in all human studies and in all genome scans conducted in the (NZB × NZW)F1 model and its derivative, the NZM2410 strain. In the mouse, three loci have been identified in that region: NZW-derived Sle1 in NZM2410 (4), and NZB-derived Lbw7 (5) and Nba2 (6) in (NZB × NZW)F1. In addition to linkage studies, association/case-control or gene disruption studies have shown that genes located in this region, such as those encoding for FcγRIIA, and FcγRIIIA (7, 8), ADPRP (9), FcRγ (10), and SAP (11), play a role in SLE susceptibility.
The characterization of the phenotypes of B6.Sle1, a congenic strain that carries the NZM susceptibility interval on a C57BL/6 (B6) background, has shown that Sle1 is associated with a selective loss of tolerance to chromatin and with a preferential targeting of H2A/H2B/DNA subnucleosomes (12, 13). Sle1 is expressed in B and T cells (ref. 13 and E. S. Sobel, unpublished data), and Sle1-expressing lymphocytes have a spontaneously activated phenotype, as indicated by an increased expression of B7–2 and CD69 on B and T cells, respectively (14). There is no evidence, however, of generalized B or T cell hyperactivity. Finally, B6.Sle1 mice exhibit a normal humoral response to antigenic challenge and normal rates of lymphocyte apoptosis. In many respects, this strain reflects what is seen in drug-induced lupus, which is also characterized by H2A/H2B/DNA autoantibodies in the absence of renal disease (15).
By combining the Sle1, Sle2, and Sle3 loci into a triple congenic strain, we have shown that these loci contain the minimal set of genes sufficient to reconstitute a fully penetrant SLE pathogenesis (16). Although a significant and unique role has been found for Sle2 (17) and Sle3 (18), the presence of Sle1 is necessary for the production of nephrophilic antibodies and clinical glomerulonephritis (GN) in the bicongenic combinations. Finally, we have identified a series of NZW-derived negative epistatic modifiers of Sle1. The most potent one, Sles1, specifically turns off all of the Sle1 immune phenotypes, leading to the suppression of the entire autoimmune pathological process triggered by Sle1 interactions with other Sle loci (19).
In summary, Sle1 is a potent SLE-susceptibility locus in both humans and in murine models. The primary defect associated with this locus is a break of tolerance to nucleosomes, which places Sle1 as a necessary step at the root of the SLE pathogenic cascade. We have identified specific loci that are able to shut down the entire autoimmune cascade and end organ pathogenesis by specifically suppressing Sle1. The combination of these genetic and functional characteristics identifies Sle1 as a key locus in the initiation of SLE pathogenesis and an attractive target for future therapeutic interventions.
Here, we present a fine-mapping analysis of the genetic location of Sle1 and demonstrate that Sle1 corresponds to a cluster of functionally related loci. Through congenic analysis, we show that three Sle1 loci share a common pathway leading to the loss of tolerance to chromatin but differ by various other serological and cellular phenotypes, suggesting a unique contribution to the pathogenic process. In addition, we show that the presence of these three loci could not account for the epistatic contribution of Sle1 to nephritis, suggesting the existence of a fourth locus.
Materials and Methods
Mice.
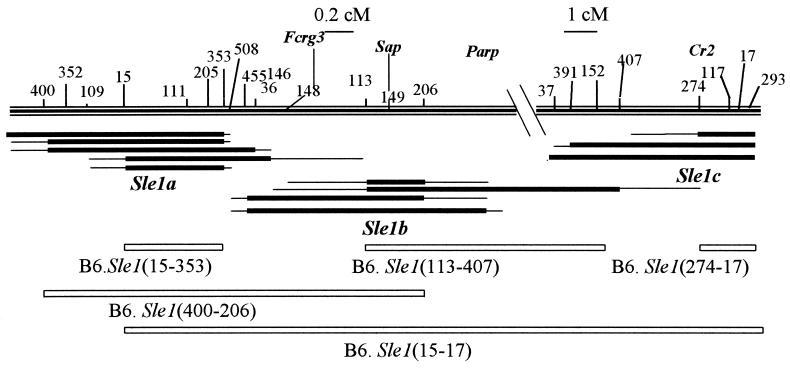
C57BL/6J (B6), NZW, and NZM2410 mice were originally obtained from The Jackson Laboratory and subsequently bred and maintained in conventional housing at the University of Florida. The production of the B6.Sle1 strain has been previously described (20). A high-resolution genetic map of telomeric chr 1 was produced with 46 microsatellite markers (21) that were polymorphic between NZM2410 and B6. The relative positions of these markers were mapped on a panel of 493 (NZM2410 × B6) meioses collected from two previous crosses (4, 22). Further resolution was achieved for some closely linked markers with recombinants generated in this study. (B6.Sle1 × B6) F1 × B6 progeny were PCR-screened at weaning for recombination within the Sle1 congenic interval with the 46-marker panel. By breeding to B6 mice, recombinants were subsequently expanded and used to generate further recombinants. A cohort of nonrecombinant mice that were either NZM/B6 (B6.Sle1het) or B6/B6 (B6 controls) across the whole Sle1 interval were kept as controls. Progeny from selected recombinants showing a positive phenotype (Fig. 1) were intercrossed to homozygosity and expanded for functional studies. The sublines generated (Table 1) were designated by the two termini markers of the corresponding subcongenic interval [such as B6.Sle1(15–353) contains the D1Mit15 to D1Mit353 NZM interval]. Although the large number of mice generated in this study has been staggered over 4 years, the functional studies were conducted concurrently. All mice were aged up to 12 months, and an approximately equal number of males and females was used.
Figure 1.
Genetic map of the B6. Sle1 recombinants that were positive for anti-chromatin Ab production, defining the three Sle1a, Sle1b, and Sle1c loci. The top of the figure shows chr 1 telomeric of D1Mit400, the microsatellite markers used to map the recombinants (such as 400 indicates the position of D1Mit400), and the position of relevant genes. Immediately under this map are shown the three clusters of recombinants that were anti-chromatin Ab positive. For each recombinant, the thick line indicates the NZM2410 interval, flanked by two thin lines showing the area of recombination between the NZM2410 and B6 genomes. Finally, the white rectangles show the homozygous intervals corresponding to the subcongenic strains produced for functional studies. Note the different scale for the Sle1a + b and Sle1c regions [0.2 centimorgan (cM) and 1 cM, respectively]. B6.Sle1(47–105), not represented on this figure, corresponds to the 7-cM interval located between D1Mit47 and D1Mit400.
Table 1.
Congenic lines and corresponding loci used in this study
| Congenic line | End markers* | Sle locus |
|---|---|---|
| B6.Sle1† | D1Mit47-D1Mit17 | Sle1 |
| B6.Sle1(47–105) | D1Mit47-D1Mit105 | None |
| B6.Sle1(15–353) | D1Mit15-D1Mit353 | Sle1a |
| B6.Sle1(113–407) | D1Mit113-D1Mit407 | Sle1b |
| B6.Sle1(113–206) | D1Mit113-D1Mit206 | Sle1b |
| B6.Sle1(274–17) | D1Mit274-D1Mit17 | Sle1c |
| B6.Sle1(400–206) | D1Mit400-D1Mit206 | Sle1a + Sle1b |
| B6.Sle1(15–17) | D1Mit15-D1Mit17 | Sle1a + Sle1b + Sle1c |
Serology.
Sera from B6.Sle1 recombinant mice were screened for anti-chromatin IgG antibodies (Ab) every other month from 6 to 12 months of age by ELISA as previously described (12). For interplate comparisons, a serial dilution of an NZM2410 serum was included on each plate to construct a standard curve. The OD value for a 1:100 dilution was assigned a value of 100 units. A mouse was deemed positive when at least two different serum samples scored greater than 25 units, the mean value +2 standard deviations of 50 12-month-old B6 controls. Subnucleosome specificity was assayed on 9-month-old sera as previously described (14), with each serum sample tested for the six different antigens on a same plate. Total serum IgM and IgG were also assayed on 9-month-old sera by capture ELISA as previously described, at a 1:3,000 and 1:5,000 serum dilution, respectively (12, 17). Ab response to ovalbumin (OVA) immunization was performed as previously described (17). Briefly, five 2-month-old females for each group were immunized i.p. with 100 μl of OVA (Sigma) in complete Freund's adjuvant and boosted 2 weeks later with the same dose of antigen in incomplete Freund's adjuvant. Sera were collected at 0, 2, and 6 weeks postimmunization and analyzed by ELISA for presence of anti-OVA IgM and IgG Ab at a 1:100 dilution.
Flow Cytometry.
Splenocytes were depleted of red blood cells with 0.83% NH4Cl, and single-cell suspensions were prepared. FACS analysis was performed as previously described (23). All primary Abs [CD4 (RM4–5), CD8a (53–6.7), CD45R/B220 (RA3–6B2), CD69 (H1.2F3), and CD86/B7.2 (GL1), CD5 (53–7.3), CD44 (IM7), and CD62L (MEL-14)] were purchased from PharMingen and used at pretitrated dilutions. Cells were first blocked on ice with staining medium (PBS/5% horse serum/0.05% sodium azide) containing 10% rabbit serum. Cells were then stained with optimal amounts of conjugated primary Abs diluted in staining medium for 30 min. After two washes, biotin-conjugated Abs were revealed using streptavidin-Quantum red (Sigma). Cell staining was analyzed using a FACScan (Becton Dickinson Immunocytometry Systems). Dead cells were excluded on the basis of scatter characteristics, and 10,000 events were acquired per sample. Positive staining for each given primary Ab was determined relative to the isotype controls, all purchased from PharMingen. Analyses were conducted on 9- to 12-month-old mice, with at least one age-matched B6 mouse included in each assay.
Histology.
Kidneys were fixed and stained with hematoxylin and eosin and periodic acid-Schiff. Mice were scored as GN positive when ≥25% of their glomeruli showed a qualitative (segmental and/or global mesangial, hyaline, or proliferative) lesion on multiple sections (score 3–4). We have previously shown that only proliferative lesions are significantly correlated with proteinuria and mortality (16).
Results
The Sle1 Congenic Interval Contains Three Nonoverlapping Regions Mediating Anti-Chromatin Ab Production.
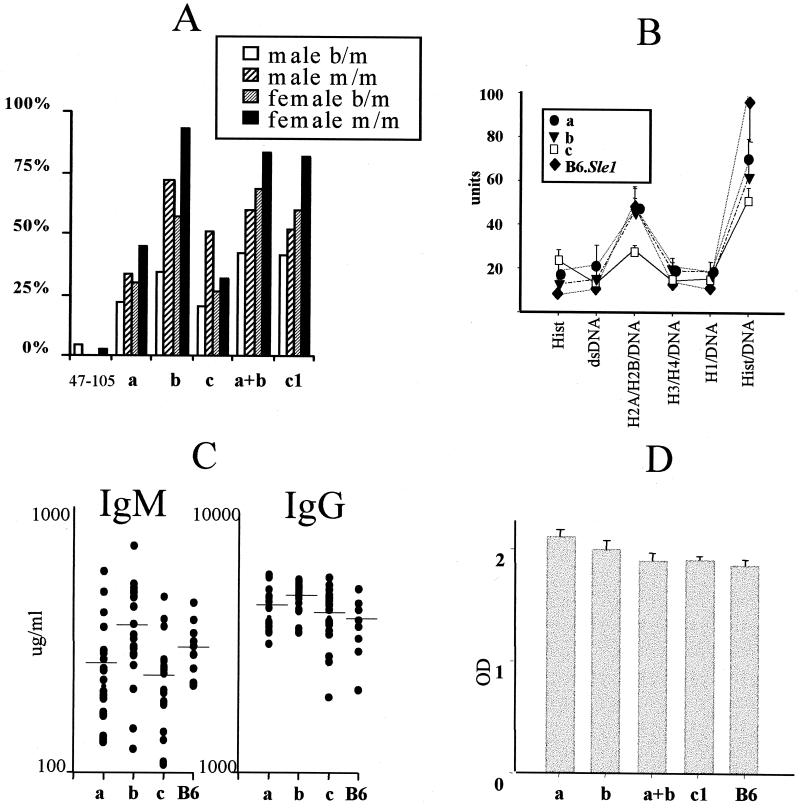
Sle1-mediated anti-chromatin IgG production is expressed with an allele dose inheritance, the difference between the presence of one or two copies of the NZM2410 allele resulting in a greater penetrance and earlier age of onset (12). Consequently, Sle1 was mapped in heterozygous B6.Sle1 recombinants, with one copy of NZM2410 recombinant intervals on a B6 background. We identified and analyzed a collection of 290 overlapping heterozygous congenic recombinants from a cohort of >2,000 testcross progeny produced from (B6.Sle1 × B6)F1 × B6 crosses. These congenic recombinant mice were bred with B6 to rescue their recombinant chromosomes and produce multiple progeny for phenotypic analysis. More than 800 mice were assessed for anti-chromatin IgG production during the analysis of these B6.Sle1 congenic recombinant sublines. Three nonoverlapping regions, named a, b, and c, that resulted in anti-chromatin Ab production were identified (Fig. 1). Sle1a maps to an approximately 1-cM interval between D1Mit15 and D1Mit353, Sle1b is located in an interval of about the same size between D1Mit113 and D1Mit206, and Sle1c maps to the very telomeric end of chr 1 to an approximate 2-cM interval telomeric of D1Mit274. The highest penetrance was associated with Sle1b (57% in females, 34% in males), whereas the penetrance in Sle1a and Sle1c reached around 25% (Fig. 2A). Interestingly, the whole heterozygous Sle1 interval penetrance (60% in females, 41% in males) was reconstituted in intervals combining Sle1a + b (68% in females, 42% in males). The penetrance of anti-chromatin Ab in intervals centromeric to Sle1a was very low (2%), corresponding to background variations. Finally, although a higher penetrance was observed in females for each locus, only Sle1b showed a significant gender disparity (P = 0.03) similar to that of the whole Sle1 interval (12).
Figure 2.
Serological characteristics of the three Sle1 loci. (A) Penetrance of anti-chromatin IgG Ab production in heterozygous (b/m) and homozygous (m/m) congenic recombinants. Heterozygous congenic intervals have been distributed into three nonoverlapping clusters defining the three loci, a, b, and c, as shown on Fig. 1. () groups intervals centromeric to D1Mit105, which do not contain Sle1a, Sle1b, or Sle1c. Within each heterozygous group, a number of overlapping intervals are represented, each of them in multiple mice. Number of mice for each group: (), 62; Sle1a, 54; Sle1b, 18; Sle1c, 65; Sle1a + b, 122; B6.Sle1het, 15. For homozygous mice, each locus is represented by a specific interval (Table 1): B6.Sle1(47–105), 70 mice; B6.Sle1(15–353), 39; B6.Sle1(113–274), 32; B6.Sle1(274–17), 26; B6.Sle1(400–206), 42; B6.Sle1, 60. (B) Subnucleosomal specificities of IgG Ab. Mean +SE on 12 sera for each substrain tested simultaneously for the six chromatin antigens by ELISA as described in ref. 13. (C) Total serum IgM and IgG levels compared with B6 controls. Horizontal lines indicate mean values for each group. (D) IgG response to OVA immunization 6 weeks postimmunization. Mean +SE on five mice for each substrain immunized simultaneously.
Functional Characterization of Sle1a, Sle1b, and Sle1c.
To functionally characterize the three Sle1 loci, we produced and analyzed the subcongenic B6.Sle1 strains described in Fig. 1 and Table 1. The penetrance of anti-chromatin IgG Ab associated with each homozygous Sle1 locus (Fig. 2A) paralleled the results obtained with the corresponding heterozygous loci, showing that the Sle1 allele dose inheritance (12) applies to its individual loci. The contribution Sle1b was the strongest (93% of females, 72% males), its penetrance being even greater than that of the whole Sle1 interval. In addition, the age of onset was earliest in Sle1b mice, with most mice seropositive by 7 months of age and exhibiting maximal penetrance around 9 months of age. This phenotype was at least as robust as that for the intact Sle1 interval (12) and stronger than that for Sle1a or Sle1c (data not shown). Anti-chromatin Ab production was not significantly different between B6.Sle1b(113–274) and B6.Sle1b(113–206), indicating that loci located between D1Mit206 and D1Mit407 did not contribute to Sle1b anti-chromatin Ab phenotype. The combinations of either Sle1a + b or Sle1a + b + c loci were equivalent to the whole Sle1 interval, whereas the homozygous B6.Sle1(47–105) only generated background levels of anti-chromatin Ab.
Although the three Sle1 loci were associated with a loss of tolerance to chromatin, we observed serological differences between them. Both Sle1a and Sle1b showed the same enhanced response to the H2A/H2B/DNA subnucleosomal particle (Fig. 2B), which we have described as a characteristic of the whole Sle1 interval (14). Sle1c, however, did not show a marked specificity for any specific chromatin component. In addition, only Sle1b was associated with a significant increase in total serum IgM and IgG over B6 controls (Fig. 2C, P = 0.04 and P = 0.01, respectively). As with the intact Sle1 interval (12), this increase mostly corresponded to antinuclear Abs, with the spontaneous Ab production against other auto or foreign antigens being similar between B6.Sle1b and B6 (data not shown). Finally, Sle1a was associated with an increased IgG response to OVA immunization as compared with B6 controls (Fig. 2D, P < 0.01) without any significant difference in the IgM response (data not shown). Interestingly, Sle1a mediated a response that was higher than that of the intact Sle1 interval or the Sle1a + b combination (P < 0.01 and P = 0.05, respectively). There was a trend showing a higher Sle1b IgM response to OVA immunization (P = 0.06), and Sle1c did not shown any difference from either IgM or IgG B6 response (data not shown).
Significant differences were also observed between the splenic lymphocyte distribution and activation status associated with each of the three Sle1 loci (Table 2). The combinations of the Sle1a + b loci or all three loci resulted in similar lymphocyte characteristics as observed for the intact Sle1 interval, with a significant increase of B7–2 expression on B cells and the early activation marker CD69 on CD4+ T cells (data not shown). The Sle1b interval resulted in values that were not significantly different from the whole Sle1 interval or the Sle1a + b or Sle1a + b + c combinations. These results suggest that, as for the serological phenotypes, Sle1b is responsible for the strongest contribution Sle1 phenotypes. In contrast, high B7–2 or CD69 expression was not observed with either Sle1a or Sle1c, which were instead associated with lymphocytic characteristics that were masked by the expression of the whole Sle1 interval. Sle1a was associated with a significant reduction in T cell numbers, which affected both CD4+ and CD8+ subsets. This reduction was compensated by a moderate increase in B220+ cells (data not shown). Although reduced in numbers, CD4+ T cells from B6.Sle1a displayed a higher level of late activation status, with reduced CD62L and increased CD44 expression, and showed an increased proportion of CD62Llo CD44hi memory cells. Finally, Sle1c was associated with an increased percentage of CD4+ CD62Llo CD44hi memory cells.
Table 2.
Characteristics of splenocytes associated with each of the three Sle1 loci
| %
total
splenocytes
|
||||||
|---|---|---|---|---|---|---|
| Na | Sle1a | Sle1b | Sle1c | Sle1 | B6 | |
| B220+ B7-2hi | 22–48 | 49.34 ± 2.34 | 55.77 ± 2.56*** | 48.03 ± 1.55 | 53.02 ± 1.07*** | 49.35 ± 0.82 |
| T cells | 18–55 | 17.15 ± 0.82*** | 23.98 ± 0.83 | 23.85 ± 1.21 | 24.31 ± 1.32 | 25.15 ± 0.84 |
| CD4+ CD69+ | 18–45 | 33.28 ± 1.64 | 38.56 ± 2.22*** | 27.58 ± 1.38 | 34.47 ± 1.98** | 25.51 ± 0.86 |
| CD4+ CD62L+ | 7–22 | 19.40 ± 1.83*** | 23.18 ± 1.05 | 25.67 ± 3.03 | 22.57 ± 1.57 | 27.53 ± 3.29 |
| CD4+ CD44+ | 7–19 | 65.80 ± 1.96*** | 55.01 ± 3.67 | 54.21 ± 4.98 | 57.06 ± 2.39 | 51.17 ± 4.03 |
| CD4+ CD62Llo CD44hi | 7–16 | 24.37 ± 1.78* | 22.55 ± 1.73 | 25.50 ± 1.62* | 21.81 ± 2.27 | 18.00 ± 2.74 |
Results are mean ± standard error. Significance level in a one-tailed t test comparisons with B6 values: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Number of 9- to 12-month-old mice used in each group.
Contribution of the Sle1 Loci to Lupus Nephritis.
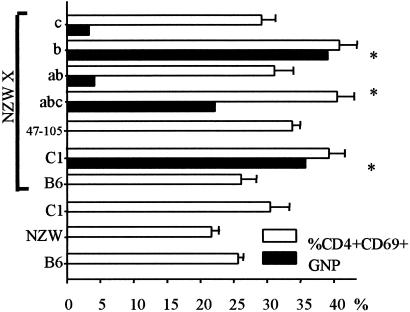
We previously discovered that homozygosity for the Sle1 interval is required for the development of fatal GN via epistatic interactions in the B6/NZW hybrid system (19). We investigated which, if any, of the Sle1a, Sle1b, or Sle1c loci were responsible for this nephrogenic phenotype by crossing Sle1 subintervals to NZW, resulting in NZW homozygosity at the corresponding Sle1 locus and NZW/B6 heterozygosity at the rest of the genome. We compared the kidney histology and lymphocyte activation status of NZW crossed to B6.Sle1(15–17) for Sle1a + b + c, B6.Sle1(400–206) for Sle1a + b, B6.Sle1(113–407) for Sle1b (mapping of Sle1b to the smaller 113–206 interval was achieved after the completion of this experiment), B6.Sle1(274–17) for Sle1c, and B6.Sle1(47–105) to a cohort of (NZW × B6.Sle1)F1 mice and a number of appropriate controls, all aged concurrently to up to 12 months of age (Fig. 3). Surprisingly, the combination of Sle1a + b, which resulted in identical serological and cellular phenotypes as the whole Sle1 interval (see above), resulted in a significantly lower (P < 0.01) penetrance of proliferative GN in [NZW × B6.Sle1(400–206)]F1 mice than in (NZW × B6.Sle1)F1. The [NZW × B6.Sle1(113–407)]F1 mice, however, showed a similar penetrance of proliferative GN to that of (NZW × B6.Sle1)F1 and [NZW × B6.Sle1(15–17)]F1 (χ2 = 1.22 and 1.49, respectively, P > 0.05). Since the Sle1b locus is contained in both (400) and () intervals, this result suggests the existence of an additional locus, either a nephrogenic locus located between D1Mit206 and D1Mit407 or a negative modifier located between Sle1a and Sle1b. Whether or not this fourth locus, Sle1d, acts independently or requires Sle1c for full expression of its phenotype remains to be determined.
Figure 3.
Nephrogenicity associated with the Sle1 loci on an NZW hemizygous background. Black bars indicate the penetrance of proliferative GN (GNP) associated with each strain combination. White bars indicate the corresponding percentage of CD4+ CD69+ lymphocytes at sacrifice (mean +SE, 20–25 mice per group). The asterisks indicate the strain combinations that showed a significant amount of GNP, which are also those with the highest CD69 expression in CD4+ cells.
The increased renal pathology in [NZW × B6.Sle1(113–407)]F1 mice was not associated with an increased autoantibody production, either in amounts of Ab produced or in the spectrum of targeted antigens (data not shown). The increased renal pathology correlated, however, with an increased number of CD4+CD69+ T cells in both B6.Sle1(113–407) and [NZW × B6.Sle1(113–407)]F1 mice (Table 2 and Fig. 3), suggesting that early activation of T cells plays a critical role in the contribution of the Sle1 loci to lupus nephritis.
Discussion
A high-resolution genetic map of the Sle1 locus has been constructed as a first step to identify the corresponding susceptibility gene. Congenic recombinants were generated from B6.NZMc1 and screened for anti-chromatin Ab production. In this process, it became clear that Sle1 corresponded to several genes, in that three groups of nonoverlapping recombinant intervals were able to confer antinuclear Ab production. These loci have been termed Sle1a, Sle1b, and Sle1c starting from the most centromeric locus. Interestingly, the position Sle1a corresponds to the GN peak linkage in the initial backcross analysis (4) and to anti-dsDNA Ab in the (NZM2410 × B6)F2 progeny (22). The Sle1b position corresponds to the peak linkage for a variety of autoantibodies and splenomegaly in the (NZM2410 × B6)F2 progeny (22) and to the position of Nba2 (6).
A functional relationship among these loci is supported by the observation that each can independently mediate a loss of tolerance to nuclear antigens. However, the immunological phenotypes mediated by these genes are clearly discrete as each expresses a unique combination of the various component phenotypes associated with the Sle1 interval. The phenotypes of Sle1a and Sle1b are especially similar in this respect, with Sle1c being more divergent. Sle1b serological and cellular characteristics are the closest to that of the whole Sle1 interval. In addition of its high penetrance of anti-chromatin Ab, higher levels of serum IgM, IgG, and B7–2 expression on B cells suggest that Sle1b mostly affects B cell function. Sle1a was associated with a much lower anti-chromatin Ab penetrance and with phenotypes that mostly affected T cells that, although in significantly lower numbers, expressed higher levels of late activation markers. In addition, a higher IgG response to antigenic challenge with no difference in T cell-independent humoral response also suggested that Sle1a affected T cell functions. Interestingly, the phenotypes linked to Sle1a and Sle1b positions in a (NZM2410 × B6)F2 analysis (22) already suggested a differential effect of each of these two loci on T cells and B cells, respectively. Mixed chimeras have already shown that Sle1 is expressed in both T and B cells (ref. 14 and E. S. Sobel, unpublished data). Our results now suggest that two different Sle1 loci might be expressed in those two lymphocyte compartments, a hypothesis that will have to be directly addressed by additional mixed chimera experiments. The phenotypes associated with Sle1c are more difficult to interpret, and the fact that the H2A/H2B/DNA subnucleosome is not the main antigenic determinant suggests a more distant functional relationship with the other two Sle1 loci. The expression of the Cr2 (complement receptor 2) gene, which is located in the very telomeric chr 1, is significantly decreased in human SLE patients and in the MRLlpr mouse (24), making Cr2 an interesting candidate for Sle1c.
Our previous analyses, either with the NZM2410 or the (NZW × B6.Sle1)F1 models, have highlighted that homozygosity for NZW alleles was necessary for Sle1 contribution to GN, whereas anti-chromatin Ab production was allele dose-dependent (4, 12, 19, 22). Consistent with these results, the anti-chromatin Ab mediated by Sle1a, Sle1b, or Sle1c showed allele dose inheritance. Although the Sle1a + b combination reconstituted the serological and cellular phenotypes of the entire Sle1 interval, these loci were not sufficient for GN induction in the (NZW × B6.Sle1)F1 model. In this model, however, a level of GN similar to that (NZW × B6.Sle1)F1 was obtained with NZW crossed to B6.Sle1(113–407), a congenic strain containing Sle1b plus an NZM2410-derived region extending about 5 cM telomeric (Figs. 1 and 3). These results suggest the presence of a fourth locus, Sle1d, with a critical function in the pathway leading to nephritis. Sle1d could be a locus located between D1Mit206 and D1Mit407, with, as our results suggest, no effect on autoantibody production and lymphocyte activation. This would place Sle1d in the third functional pathway that we have previously defined for SLE pathogenesis (2, 25), which contains loci affecting the end-organ susceptibility to autoimmune damage. The D1Mit206 to D1Mit407 interval contains ADPRP, which has been strongly associated with human SLE (9), and this region is large enough to contain many other potential candidates. Further characterization of additional congenic recombinants and their F1 hybrids with NZW will be necessary to determine the location and function of Sle1d.
A final intriguing result supporting the possible functional relationship of all four loci in the Sle1 interval is the suppressive effects of Sles1 on all of the phenotypes expressed by Sle1. We previously demonstrated that the introgression of Sles1 (located on chromosome 17 in an interval including H2) onto the B6.Sle1 genome resulted in a complete suppression of autoantibody production and cellular activation on a B6 background and fatal GN when crossed with NZW (19). This finding, coupled with the fine mapping analysis presented here, suggests that Sles1 is capable of suppressing Sle1a, Sle1b, Sle1c, and Sle1d. The ability of Sles1 to suppress each of these loci separately is now being tested with the individual truncated congenic intervals. In addition, we are in the process of assessing the cell lineages involved in this suppressive effect and in fine mapping the location of Sles1.
Clustering of Susceptibility Loci Is Not a Special Feature of Sle1.
We have shown by linkage analysis that the Sle3 congenic interval contains another susceptibility locus, Sle5 (22), but these two loci may belong to two different functional pathways (26). The Sle2 congenic interval contains the negative modifier locus Sles2 (19), and initial genetic mapping with congenic recombinants also showed several loci contributing to the Sle2 phenotypes (L.M., unpublished data). Congenic strains in other systems have also shown that some of the strongest loci clearly correspond to clusters of loci. The pristane-induced plasmacytoma resistance locus on chr 4 corresponds to at least two loci, Pctr1 and Pctr2 (27). Several of the QTL insulin-dependent diabetes loci in the NOD mouse correspond to multiple loci, such as the initial Idd3 on chr 3 now includes Idd10, Idd17, and Idd18 (28), or even H2-linked Idd1 (29). A similar level complexity probably exists in humans, as recent linkage analyses of SLE susceptibility suggested up to five closely linked loci in the 1q31–42 genomic region (3).
The significance of this locus clustering is unclear. It is possible that a large number of susceptibility loci contribute to polygenic diseases, and it is only when some of these loci are linked and interact to produce a strong phenotypic effect that the corresponding genomic region is detected by linkage analysis. Alternatively, clustering of susceptibility loci may result from functional and evolutionary pressures, and consequently, weak isolated loci would be rare. The fact that clustered loci tend to belong to the same functional pathway, such as loss of tolerance to nuclear antigens for Sle1 (this study) or B cell hyperactivation for Sle2 (L.M., unpublished results), supports the latter hypothesis. In addition to ongoing efforts toward the molecular identification of the corresponding genes, the upcoming challenge will be to define how they interact within a cluster and with genes from other clusters to produce the complex SLE pathogenesis.
Acknowledgments
We thank Chandra Mohan and Eric S. Sobel for critical reading of the manuscript. This work was supported by National Institutes of Health Grant PO1 AI39824 (to E.K.W.) and an Arthritis Foundation Investigator Award (to L.M.).
Abbreviations
- SLE
systemic lupus erythematosus
- chr
chromosome
- B6
C57BL/6J
- GN
glomerulonephritis
- OVA
ovalbumin
- cM
centimorgan
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031336098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031336098
References
- 1.Winchester R. In: Systemic Lupus Erythematosus. Lahita R G, editor. New York: Churchill Livingstone; 1992. pp. 65–85. [Google Scholar]
- 2.Wakeland E K, Wandstrat A E, Liu K, Morel L. Curr Opin Immunol. 1999;11:701–707. doi: 10.1016/s0952-7915(99)00039-4. [DOI] [PubMed] [Google Scholar]
- 3.Harley J B, Moser K L, Gaffney P M, Behrens T W. Curr Opin Immunol. 1998;10:690–696. doi: 10.1016/s0952-7915(98)80090-3. [DOI] [PubMed] [Google Scholar]
- 4.Morel L, Rudofsky U H, Longmate J A, Schiffenbauer J, Wakeland E K. Immunity. 1994;1:219–229. [PubMed] [Google Scholar]
- 5.Kono D H, Burlingame R W, Owens D G, Kuramochi A, Balderas R S, Balomenos D, Theofilopoulos A N. Proc Natl Acad Sci USA. 1994;91:10168–10172. doi: 10.1073/pnas.91.21.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake C G, Rozzo S J, Hirschfeld H F, Smarnworawong N P, Palmer E, Kotzin B L. J Immunol. 1995;154:2441–2447. [PubMed] [Google Scholar]
- 7.Salmon J E, Millard S, Schachter L A, Arnett F C, Ginzler E M, Gourley M F, Ramsey-Goldman R, Peterson M G E, Kimberly R P. J Clin Invest. 1996;97:1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J, Edberg J C, Redecha P B, Bansal V, Guyre P M, Coleman K, Salmon J E, Kimberly R P. J Clin Invest. 1997;100:1059–1070. doi: 10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsao B P, Cantor R M, Grossman J M, Shen N, Teophilov N T, Wallace D J, Arnett F C, Hartung K, Goldstein R, Kalunian K C, et al. J Clin Invest. 1999;103:1135–1140. doi: 10.1172/JCI5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clynes R, Dumitru C, Ravetch J V. Science. 1998;279:1052–1054. doi: 10.1126/science.279.5353.1052. [DOI] [PubMed] [Google Scholar]
- 11.Bickerstaff M C, Botto M, Hutchinson W L, Herbert J, Tennent G A, Bybee A, Mitchell D A, Cook H T, Butler P J, Walport M J, et al. Nat Med. 1999;5:694–697. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]
- 12.Morel L, Mohan C, Croker B P, Tian X-H, Wakeland E K. J Immunol. 1997;158:6019–6028. [PubMed] [Google Scholar]
- 13.Mohan C, Alas E, Morel L, Yang P, Wakeland E K. J Clin Invest. 1998;101:1362–1372. doi: 10.1172/JCI728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobel E S, Mohan C, Morel L, Schiffenbauer J, Wakeland E K. J Immunol. 1999;162:2415–2421. [PubMed] [Google Scholar]
- 15.Rubin R L, Bell S A, Burlingame R W. J Clin Invest. 1992;90:165–173. doi: 10.1172/JCI115832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morel L, Croker B P, Blenman K R, Mohan C, Huang G, Gilkeson G, Wakeland E K. Proc Natl Acad Sci USA. 2000;97:6670–6675. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan C, Morel L, Yang P, Wakeland E K. J Immunol. 1997;159:454–465. [PubMed] [Google Scholar]
- 18.Mohan C, Yu Y, Morel L, Yang P, Wakeland E K. J Immunol. 1999;162:6492–6502. [PubMed] [Google Scholar]
- 19.Morel L, Tian X-H, Croker B P, Wakeland E K. Immunity. 1999;11:131–139. doi: 10.1016/s1074-7613(00)80088-6. [DOI] [PubMed] [Google Scholar]
- 20.Morel L, Yu Y, Blenman K R, Caldwell R A, Wakeland E K. Mamm Genome. 1996;7:335–339. doi: 10.1007/s003359900098. [DOI] [PubMed] [Google Scholar]
- 21.Dietrich W F, Miller J, Steen R, Merchant M A, Damron Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O'Connor T J, et al. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 22.Morel L, Mohan C, Yu Y, Schiffenbauer J, Rudofsky U H, Tian X-H, Longmate J, Wakeland E K. Mamm Genome. 1999;10:176–181. doi: 10.1007/s003359900964. [DOI] [PubMed] [Google Scholar]
- 23.Mohan C, Morel L, Yang P, Watanabe H, Croker B, Gilkeson G, Wakeland E K. J Clin Invest. 1999;103:1685–1695. doi: 10.1172/JCI5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K, Kozono Y, Waldschmidt T J, Berthiaume D, Quigg R J, Baron A, Holers V M. J Immunol. 1997;159:1557–1569. [PubMed] [Google Scholar]
- 25.Wakeland E K, Morel L, Mohan C, Yui M. J Clin Immunol. 1997;17:272–281. doi: 10.1023/a:1027370514198. [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, Morel L, Mohan C, Croker B P, Wakeland E K. FASEB J. 1998;12:A609. [Google Scholar]
- 27.Potter M, Mushinski E B, Wax J S, Hartley J, Mock B A. Cancer Res. 1994;54:969–975. [PubMed] [Google Scholar]
- 28.Lyons P A, Wicker L S. In: in Genes and Genetics of Autoimmunity. Theofilopoulos A N, editor. Basel: Karger; 1998. pp. 208–225. [Google Scholar]
- 29.Hattori M, Yamato E, Itoh N, Senpuku H, Fujisawa T, Yoshino M, Fukuda M, Matsumoto E, Toyonaga T, Nakagawa I, et al. J Immunol. 1999;163:1721–1724. [PubMed] [Google Scholar]