Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) gene is driven by a promoter that cannot alone account for the temporal and tissue-specific regulation of the gene. This has led to the search for additional regulatory elements that cooperate with the basal promoter to achieve coordinated expression. We previously identified two alternative upstream exons of the gene that were mutually exclusive of the first exon, and one of which showed temporal regulation in the human and sheep lung. We now demonstrate that this alternative splice product generates a stable protein, which initiates translation at an ATG in exon 4, and thus lacks the N terminus of CFTR. The other splice variant inhibits translation of the protein. In a search for the promoter used by the upstream exons, we identified a novel element that contributes to the activity of the basal CFTR promoter in airway epithelial cells, but does not function independently. Finally, we demonstrate that, in primary airway cells, skin fibroblasts, and both airway and intestinal cell lines, the CFTR promoter is unmethylated, irrespective of CFTR expression status. Thus, methylation is not the main cause of inactivation of CFTR transcription.
Keywords: CFTR promoter, alternative exons, methylation
CLINICAL RELEVANCE.
This work investigates the mechanism of action of the cystic fibrosis transmembrane conductance regulator (CFTR) promoter in order to reveal aspects of developmental regulation of gene expression. It also identifies a novel element that may contribute to CFTR gene expression in airway cells. Finally, it addresses epigenetic mechanisms that influence the CFTR promoter in primary airway cells.
The cystic fibrosis transmembrane conductance regulator (CFTR) gene shows a complex pattern of expression that is not accounted for solely by regulatory elements that lie within the promoter region. However, this sequence is clearly required for gene expression, and thus warrants further investigation to determine its mechanism of action. Earlier studies dissected the function of sequences up to 4 kb 5′ to the translation start site, and demonstrated similarities with housekeeping gene promoters (1–3). For example, there is no TATA box, the sequence has a high GC content (65%), and contains three predicted sites for specificity protein 1 binding. Promoter deletion experiments identified both positive and negative cis-regulatory elements within the region (1, 2), and defined the minimal promoter as −226 to +98 with respect to the transcription start site (+1) at −132 bp 5′ to the first methionine codon. A negative element mapped between −345 and −277 bp 5′ to the start site (1). Several studies have demonstrated an important role in CFTR promoter activity of a variant cyclic adenosine monophosphate response element (3–5) that is highly conserved across species (6, 7), and an inverted CCAAT motif (Y box) (8, 9). In addition, a CArG-like motif that may bind serum response factor has been identified (10), and two binding sites for myeloid-specific zinc-finger protein 1 may contribute to gene repression (11).
Other features of the CFTR promoter that may contribute to both the temporal and spatial regulation of gene expression are the use of multiple transcription start sites (TSS) for the gene (2, 3) and the recruitment of alternative upstream exons. Distinct CFTR TSS were determined in adult lung and fetal lung (12), although the number of tissue samples evaluated was too small to exclude polymorphisms in TSS choice. However, a developmentally related switch in TSS might contribute to the down-regulation of CFTR expression that occurs during fetal lung maturation (13–15). The recruitment of alternative 5′ exons has also been implicated in the developmental down-regulation. These exons, which are usually mutually exclusive of exon 1, have been described in several species (2, 14, 16, 17). In human (exons −1a and 1a) and sheep (Ov1aS and Ov1aL) CFTR, the alternative 5′ exons are encoded close to the promoter (within 1 kb of the translational start site). They are spliced directly to exon 2 and exist in two forms: one that includes −1a spliced to 1a, and the other that includes only −1a (14). Inclusion of the −1a alternative transcript and the comparable Ov1aS in the sheep coincides with the start of developmental down-regulation of airway CFTR mRNA (13). Because the alternative upstream exons are mutually exclusive of exon 1, if translated, they would encode a protein with a different amino terminus from wild-type CFTR, probably with an ATG in exon 4 of the transcript.
Our aim was to elucidate the contribution of these alternative 5′ exons in human CFTR to the regulation of gene transcription, and to determine their effect on CFTR protein translation. Several mechanisms can be envisaged by which these exons could have a direct effect on CFTR expression, and these are not necessarily mutually exclusive. Here, we investigate the following hypotheses: (1) that inclusion of the alternative upstream exons, which are likely noncoding, and, in particular, the resultant loss of exon 1 from the CFTR transcript, generates a protein by using an initiation codon in exon 4, thus removing important cellular localization signals; (2) that mRNA secondary structure causes detachment of the 40S ribosomal subunit (18), which could contribute to loss of CFTR protein. We previously showed that Mfold evaluation of the alternative exons, Ov1aS and human −1a/1a, predicts Gibbs free energy values of −43.38 and −146.29, respectively, indicative of very stable secondary structure (14).
To identify the promoter element for the alternative upstream exons, we re-evaluated the CFTR promoter and its chromatin environment. By using more sensitive methods than were available for previous studies, we aimed to determine additional mechanisms of regulation of this complex element. Because actively transcribed genes are generally associated with promoter hypomethylation and inactive promoters with DNA hypermethylation, we evaluated the DNA methylation status of the CFTR promoter in relevant cell types. The CFTR gene promoter is associated with a strong CpG island (a High CpG promoter according to the classification of Weber and colleagues [19, 20]). In this study, we performed comprehensive DNA methylation analyses in a region encompassing approximately 1.7 kb of the CFTR promoter by sodium bisulfite sequencing. Our new data demonstrate important epigenetic features in the chromatin landscape of primary airway cells.
MATERIALS AND METHODS
Plasmid Construction
Cosmid clones M2440 and F0424 (21) provided template DNA encompassing the 5′ proximal region of CFTR, and primers were designed by using the AC000111 sequence. Additional 5′ restriction sites (5′ primer Kpn I or Mlu I, 3′ primer Bgl II) were added to the primers to facilitate subsequent cloning (see Table E1 in the online supplement). PCR products were digested, gel purified, subcloned into the corresponding sites of the pGL3B Basic vector (Promega, Madison, WI), and verified by sequencing. The Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce the 944 A/G and 1,750 A/G mutations, and create pSP73 −1a1aATG/GTG, pGL3B −1a/1a CTG/GTG, and pSP73 −1a/1a2Val mutants. The cytomegalovirus (CMV) CFTR 936C plasmid (22) was used to generate pSP732–24 by exploiting a Pvu II restriction site at the exon 1/2 boundary, and exons 2–24 were cloned as a PvuII fragment into the PvuII site of pSP73. To create pSP731a/1a/2–24, −1a/1a RT-PCR product with flanking XhoI/PvuII sites was cloned into pSP732–24 cut with XhoI/PvuII. The XhoI site alone was used to create the pSP73−1a/2–24 construct. Mutants were generated as described previously here. To create pSP73Ov1aS/2–24, an Ov1aS (14) PCR product was generated with flanking XhoI and EcoRV sites and cloned into pSP732–24 cut with XhoI/PvuII. A similar approach was used to clone the Ov1aL PCR product with XhoI/PvuII sites into pSP732–24. All clones were verified by sequencing, and DNA prepared with the Promega MidiPrep Kit.
In Vitro Transcription and Translation
In vitro transcription reactions were performed from the relevant plasmid templates with T7 RNA polymerase and the Riboprobe In Vitro Transcription System (Promega). Transcripts were visualized by agarose gel electrophoresis. One step, in vitro transcription and translation, was performed with the TnT coupled reticulocyte lysate system (Promega) and 35S-methionine (Perkin Elmer, Waltham, MA). Radiolabeled products were examined by SDS-PAGE and analyzed by phosphorimager.
Cell Culture, Transient DNA Transfections, and Luciferase Assays
The 16HBE14o− (23) and Beas2B (24) (human bronchial epithelium) and Caco2 (25) (colon adenocarcinoma) cell lines were cultured in Dulbecco's modified Eagle medium supplemented with 10% FCS. Skin fibroblasts GM08333 (ATCC, Manassas, VA) were grown in Eagle's minimum essential medium, and primary tracheal epithelial cells were grown as described previously (26). Plasmid transfections into 16HBE14o−, Caco2, and Beas2B cells were performed with Lipofectin (Invitrogen, Carlsbad, CA), as specified by the manufacturer. Luciferase activities were measured with the Luciferase assay system (Promega). Transfection efficiencies were assessed by cotransfecting cells with CMV β-galactosidase DNA, measured by the β-galactosidase Enzyme Assay System (Promega). Transfections were done independently at least three times in triplicate with more than one plasmid preparation. Results were analyzed for statistical significance by one-way ANOVA, with Dunnett's multiple comparison, performed by using GraphPad Prism (GraphPad, San Diego, CA).
In Silico Analysis of the CFTR 5′ Region
The CFTR 5′ region (AC000111) was analyzed with a neural network promoter prediction program (http://www.fruitfly.org/seq_tools/promoter.html). The CFTR mRNA sequence was examined with Open Reading Frame (ORF) Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) to look for ORFs in the presence of alternative 5′ exons joined to exons 2–24.
DNA Methylation Analysis
Genomic DNA was extracted from the cells by cell lysis, followed by digestion with proteinase K and phenol/chloroform extraction. DNA was then subjected to sodium bisulfite treatment to modify unmethylated cytosine to uracil with the CpGenomeTM DNA Modification Kit (Chemicon International, Temećula, CA). Bisulfite-treated DNA was amplified by a nested-PCR protocol with primer sets designed to encompass the CFTR promoter (Table E3). The PCR reaction amplification control has been described previously (27). Amplified products were cloned, and a minimum of 12 clones were sequenced for each sample. The DNA sequence generated was subjected to bioinformatics analysis to summarize the CpG methylation status of these clones. Sequence reads were first trimmed of low-quality sequence by the phred program (28, 29), and vector sequence removed using the RepeatMasker program (http://www.repeatmasker.org). The sequences were correctly oriented with respect to the wild-type sequence with the use of the bl2 seq program (30), and then aligned by using the multalin program (31). Finally, perl scripts were written to identify the methylation status of all CpG sites in the sequences, and then to output a summary of all the methylation sites in text and graphic formats (the latter are illustrated in the relevant figures and supplementary figures). All clones displayed in the figures show 100% bisulfite conversion efficiency.
RESULTS
To further evaluate the mechanism of action and properties of the alternative 5′ exons of CFTR, a series of experiments was designed to test the hypotheses introduced above.
Inclusion of Human CFTR Alternative Exon −1a Alone into the CFTR Transcript Generates a Novel Protein, whereas Incorporation of Exons −1a/1a Does Not
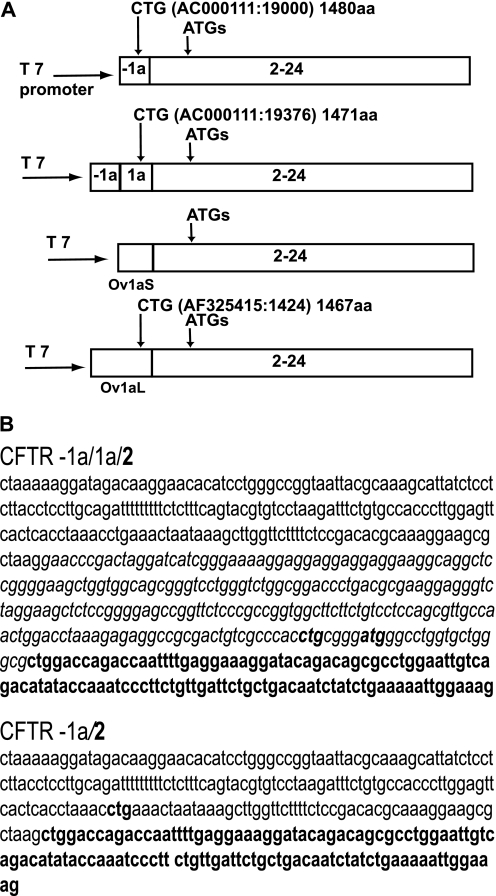
Alternative splicing of upstream exons of the CFTR gene suggested that a novel CFTR protein with a divergent function could be produced. However, there are no potential ATG initiation codons in exons −1a and 1a, suggesting that either an alternative CTG initiation codon may be used, or translation may start at an in-frame ATG in exons 3 or 4. Inspection of the sequence of the CFTR mRNAs with exon −1a either joined directly to exon 2 or to exon 1a, which is then spliced to exon 2, reveals a potential in-frame CTG in both exons −1a and 1a (Figures 1A and 1B). The alternative CTG in exon −1a (AC000111:19000) would generate a 1,480 amino acid (aa) protein, with the first 18 aa diverging from the normal N terminus of CFTR. However, when exon 1a is included in the transcript, this introduces a stop codon that disrupts the ORF. Use of the CTG in exon 1a (AC000111:19376) would result in a 1,471-aa protein, with the first 9 aa differing from the 1,480-aa wild-type CFTR protein. For the sheep CFTR transcript, exon Ov1aS contains no in-frame ATGs or CTGs, although exon Ov1aL includes a CTG (AF325415:1424), which is in frame and could generate a protein.
Figure 1.
(A) Diagram of the constructs containing the alternative human exons −1a and 1a or sheep Ov1aS and Ov1aL exons joined to cystic fibrosis transmembrane conductance regulator (CFTR) exons 2–24. Vertical arrows denote putative alternative initiation codons (together with their location on AC000111 or AF325415, and the predicted amino acid [aa] length resulting from their usage). The ATGs in exons 3 and 4 are also shown. (B) Sequence of exons −1/1a joined to exon 2 and exon −1a joined to exon 2. Sequence of alternative exons is presented as following: exon −1a, Roman font; exon 1a, italics; exon 2, bold. The potential CTG and ATG initiation codons are marked in bold.
The CFTR −1a/2–24 transcript uses an ATG in exon 4 to generate a novel protein.
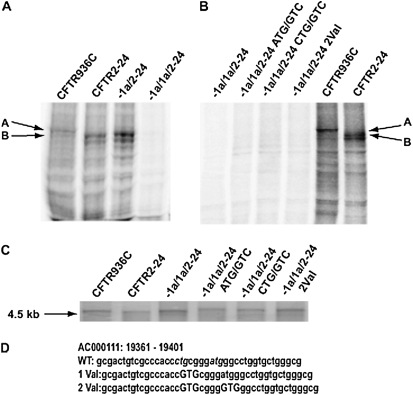
To determine whether the −1a/2 splice variant generated novel translation products, we constructed the pSP73−1a/2–24 plasmid (Figure 1A). This construct provided a template for in vitro transcription/translation reactions by using the T7 promoter in pSP73. Proteins were labeled with [35S] methionine. Control plasmids containing the whole CFTR transcript (exons 1–24, pCMVCFTR936C) or exons 2–24 alone (pSP73CFTR2–24) were also evaluated. In vitro transcription and translation of pCMVCFTR936C yielded a protein migrating at approximately 140 kD, as expected for unglycosylated CFTR. In contrast, both pSP73CFTR2–24 and pSP73−1a/2–24 produced two major species that migrated at a faster mobility (Figure 2A.). We demonstrated by mutagenesis that the proteins produced from pSP73CFTR2–24 arise from the use of ATGs in exon 4 (at aas M150, M152 or M156) (Figure E1) (32). Hence, based on their migration on SDS/PAGE, inclusion of exon −1a spliced to exon 2 apparently generates proteins that also initiate translation at these methionines in exon 4.
Figure 2.
In vitro transcription and translation of constructs containing alternative upstream exons joined to CFTR exons 2–24. (A and B) In vitro transcription and translation. 35S-labeled proteins separated by SDS-PAGE and visualized by autoradiography. The products from full-length CFTR (936C) and exons 2–24 are marked by arrows (A and B, respectively). (A) Inclusion of exon −1a in the transcript generates the same products as exons 2–24 alone, whereas inclusion of exons −1a/1a together inhibits translation. (B) Mutation of the potential CTG and ATG initiation codons in exon 1a does not restore translation from more distal ATGs in exon 4. (C) In vitro transcription. RNA transcripts were generated with T7 polymerase, denatured after RNase-free DNase treatment, and separated on a formaldehyde agarose gel, poststained by ethidium bromide. Stable RNA was generated from all constructs. (D) Partial sequence of exon 1a and location of potential initiation sites. The CTG and ATG potential initiation sites are in italics, and the mutations in capitals.
Incorporation of exons −1a and 1a together into the mRNA inhibits translation in vitro.
The longer human (exon −1a/1a/2) and sheep (Ov1aL) alternative 5′ exons are predicted to exhibit secondary structure that might interfere with efficient translation (14). To explore the role of these CFTR alternative 5′ exons, we next inserted exons −1a/1a, Ov1aS, and Ov1aL into the pSP73CFTR2–24 vector (Figure 1A). In the same in vitro transcription and translation system described previously here, no protein was generated from the pSP73−1a/1a/2–24 construct (Figure 2A), whereas the controls produced proteins of the expected size. Very low levels of protein were generated from the pSP73Ov1aS/2–24 and pSP73Ov1aL/2–24 constructs, thus it was not possible to conclusively determine the product (Figure E2).
Data from other genes suggest alternative 5′ AUGs associated with ORFs may exert negative translational control on the major protein (33). As such, we next examined this possibility for the CFTR 5′ CTG and ATG. The putative CTG initiation codon in exon 1a was destroyed by site-directed mutagenesis, converting the CTG to GTG (Leu/Val) (pSP73–1/1a/2–24 CTG/GTG [Figures 2B and 2D]). In addition, the ATG, located 4 bp 3′ to this CTG was mutated on both the wild-type and mutant CTG plasmid (pSP73−1/1a/2–24 ATG/GTG and pSP73−1/1a/2–24 2Val). However, none of these alterations generated novel proteins from the constructs incorporating the alternative 5′ exons (Figure 2B). This suggests that no individual upstream ORF is responsible for absence of translational efficiency observed with these constructs, and negative translational control caused by usage of 5′ translation start sites is unlikely to be important in regulating CFTR expression.
Inclusion of human or sheep CFTR alternative 5′ exons does not inhibit CFTR transcription.
The pSP73CFTR2–24 construct produced two major products resulting from initiation at in-frame downstream AUGs in exon 4 (M150, M152, and M156 [Figure E1] and Ref. 32), and the inclusion of the upstream alternative −1a/1a exons abolished the production of these proteins in vitro. However, this might have been due to failure of translation alone or of transcription, causing lack of production of a stable mRNA. To evaluate these two possibilities, we performed in vitro transcription reactions on relevant constructs. T7 RNA polymerase was used to generate RNA transcripts from two controls, pCMV936C and pSP73CFTR2–24, and the following constructs: pSP73−1a/1a2–24; pSP73Ov1aS2–24; pSP73Ov1aL2–24; pSP73−1a/1a2–24ATG/GTG; pSP73−1a/1a2–24CTG/GTG; and pSP73−1a/1a2–24.2Val. Transcripts were visualized on agarose gels, and all the constructs containing alternative 5′ exons produced an mRNA that was indistinguishable from the control products (Figure 2C). Thus, incorporation of 5′ alternative exons into the CFTR exon 2–24 cDNA did not inhibit transcription.
Re-evaluation of the CFTR Promoter
In an attempt to identify the cis-acting element that was used for the transcription of exons −1a and 1a, we evaluated 5 kb of sequence upstream of the major CFTR translational start site.
In silico analysis of the CFTR promoter sequence.
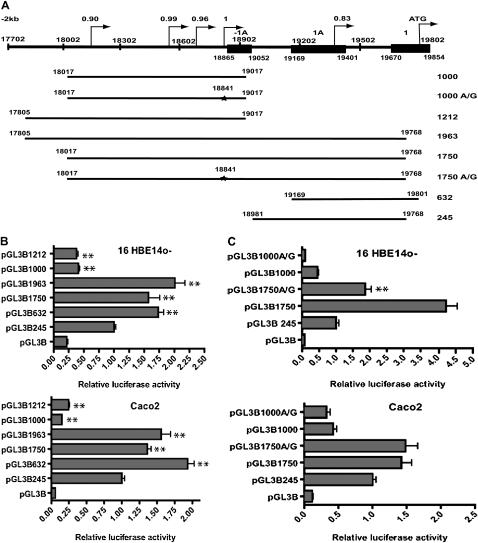
We used a time-delay neural network promoter prediction program (34) (http://www.fruitfly.org/seq_tools/promoter.html) to inspect AC000111:1–20,000, focusing on sequence 5 kb 5′ to the ATG in CFTR exon 1. With this program, the optimal promoter is given a score of 1 on a scale from 0 to 1. Sequences of particular interest were identified approximately 2 kb 5′ to the ATG, and also upstream of exons −1a, where four alternative promoters with scores of 0.90 or higher were predicted (Figure 3A). The most distal of these (AC000111:18,126–18,176) has a score of 0.90, the next at AC000111:18,553–18,603 has a score of 0.99; another, with a score of 0.96, is located at AC000111:18,769–18,814, and the most proximal, with a score of 1.0, lies just in front of CFTR exon −1a, at AC000111:18,830–18,880. This program also predicts a promoter with a score 0.83 within the minimal CFTR promoter with a TSS 421 bp 5′ to the translation initiation codon AC000111:19,802 (Figure 3A). This may correspond to the basal promoter of the gene.
Figure 3.
CFTR promoter analysis. (A) Diagram of the 5′ region of the human CFTR gene (to scale). The alternative exons, −1a and 1a, and exon 1 are shown (filled boxes). Putative promoters, with their predicted scores, are shown by arrows. Numbering refers to AC000111. At the bottom are shown each of the constructs used in reporter gene assays, with the name denoting the length of promoter fragment. The star denotes the A/G mutation that disrupts the putative promoter with a score of 1. (B and C) Luciferase reporter gene assays with CFTR promoter fragment constructs. Each bar chart shows the luciferase activities for each construct relative to pGL3B245 (CFTR basal promoter construct = 1) in B, 16HBE14o− and Caco2 cells. (B and C) Luciferase activities were normalized for transfection efficiency by cotransfection with pCMV/β-galactosidase. Each bar is the average of at least three transfection experiments, with each sample assayed in triplicate. Error bars represent SD. (B) Stars indicate statistical significance of a comparison between pGL3B245 values and those of the other construct (**P < 0.01). (C) The effect of the 18,841 A/G mutation in the 1,750 construct on promoter activity in 16HBE14o− and Caco2 cells. Stars indicate statistical significance of the pGL3B1750 to pGL3B1750 A/G comparison in 16HBE14o− cells (P < 0.01).
Transcriptional activity of the CFTR gene promoter proximal region in human bronchial epithelial cells lines and the Caco2 colon carcinoma cell line.
Earlier studies on CFTR defined a GC-rich promoter that lacked a TATA box and had many features characteristic of housekeeping gene promoters (1–3). However, the new in silico analysis described here, in combination with our previous data on the tissue-specific and temporal regulation of the exon −1a–containing splice form (14), encouraged us to re-evaluate CFTR promoter activity. We generated luciferase reporter gene constructs (pGL3B) encompassing up to 2,000 nucleotides upstream of the ATG in CFTR exon 1 (Figure 3A). These constructs were transiently transfected into the human bronchial epithelial cell lines 16HBE14o− and Beas2B, and Caco2 intestinal cells. 16HBE14o− and Caco2 express high levels of CFTR mRNA, whereas the transcript is barely detectable and often lost from Beas2B.
In an attempt to define the promoter for the −1a-containing transcript, we cloned two fragments that encompass the region 5′ to this exon into pGL3B, pGL3B1000 (AC000111:18,017–19,017), and pGL3B1212 (AC000111:17,805–19,017) (Figure 3A). Both fragments contain all four promoters predicted in silico with a score of greater than 0.9. Transient transfection data demonstrate that, although luciferase activity driven by these two regions is slightly higher than a promoterless control vector alone (pGL3B), the difference is not statistically significant (P > 0.05), and activity is significantly lower than that of the CFTR basal promoter in all three cells lines (P < 0.01; Figure 3B). Hence, the predicted promoters in this region do not appear to have independent activity in these cells lines, and, furthermore, initiation of transcription of exon −1a probably does not occur within the region (AC000111:17,702–18,902).
A novel cis-acting promoter element.
A series of constructs (Figure 3A) were designed to re-evaluate CFTR promoter activity in several cell types that had not been investigated in earlier studies (1, 2). Moreover, the constructs were tailored to incorporate newer data on the CFTR promoter region. The results of transient transfection assays with the basal CFTR promoter, pGL3B245 (AC000111:18,981–19,768) (35, 36), and additional constructs pGL3B632 (19,169–19,801), pGL3B1750 (18,017–19,768), and pGL3B1963 (17,805–19,768), are shown in Figure 3B and Figure E3. In all cell types, the pGL3B632 fragment produced about twice the luciferase levels of the basal promoter (245). In contrast, the pGL3B1750 and pGL3B1963 constructs produced the highest levels of luciferase in comparison with the basal promoter, this effect being most evident in the airway lines 16HBE14o− and Beas2B (2- and 2.25-fold respectively; P < 0.01). The activity of these two constructs was lower in Caco2 cells (1.35-fold for pGL3B1750 and 1.60-fold for pGL3B1963; P < 0.01), but both data sets implicated cis-acting elements in this region of the 5′ untranslated region that augment basal promoter activity.
To further investigate these elements, we evaluated the 5′ regions that encompass them in pGL3B constructs lacking the basal promoter region (pGL3B1000 and pGL3B1212) (Figure 3A). These constructs have no promoter activity alone (see section Transcriptional activity of the CFTR gene promoter proximal region in human bronchial epithelial cells lines and the Caco2 colon carcinoma cell line); however, their ability in cis to augment CFTR basal promoter activity suggests that they contain functional elements. The first candidate element was the putative promoter with a score of 1.0 at AC000111:18,841, predicted by in silico analysis (above); hence, this was destroyed by site-directed mutagenesis (A to G) in the pGL3B1750 and pGL3B1000 constructs. This mutagenesis reduced the score of putative promoter from 1.0 to 0.44. The two mutant constructs, pGL3B1750A/G and pGL3B1000A/G, were transfected into Caco2 and 16HBE14o− cells alongside the wild-type constructs, and luciferase activity measured (Figure 3C). Although, in Caco2 cells, the A/G mutation had no significant effect on luciferase activity (P > 0.05), in 16HBE14o− cells it resulted in a 2.5-fold decrease in activity (P < 0.01) (Figure 3C). This effect was only observed in the longer pGL3B1750 construct, but not in the pGL3B1000A/G (P > 0.05).
These results provide further evidence that this cis-acting element is particularly important in airway expression of the CFTR gene.
DNA Methylation of the CFTR Promoter Region
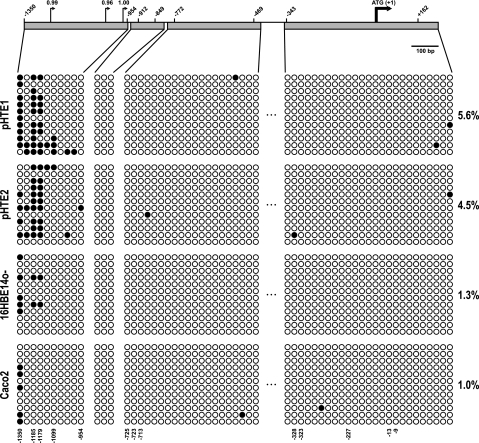
To evaluate whether methylation plays a role in CFTR promoter activity, a 5′ upstream region, including the CFTR promoter and −1a/1a exons, was evaluated by bisulfite sequencing. DNA methylation at each CpG in a segment upstream of the ATG translation initiation site of CFTR (AC000111:18,421–20,000) was evaluated in the following cell types: primary human tracheal epithelial cells (pHTE); 16HBE14o−, Beas2B, and Calu3 airway cell lines; Caco2 colon carcinoma cells; MCF7 breast cancer epithelial cells; and skin fibroblasts. All cell types, except MCF7 and fibroblasts, express the CFTR gene. Specific bisulfite DNA sequencing assays were designed to investigate the methylation of four fragments in the promoter region (for primers, see Table E1). Bisulfite-modified DNA was PCR amplified, cloned, and sequenced, and a minimum of 12 clones was analyzed for each region. A total of 58 CpGs was evaluated in a region of approximately 1.7 kb. A diagram representing the location of each of the CGs is shown in Figure 4, with individual CGs being numbered with respect to their location in base pairs upstream (−) or downstream (+) of the CFTR translational start site (1). With the exception of CG −1,512, there are no CGs in more than 500 bp 5′ to the region analyzed. Figure 4 first shows data from two independent cultures of pHTE cells, which express low levels of CFTR and show a distinct pattern of methylation at the CGs between −1,099 and −1,350, but no methylation of the more proximal promoter. No methylation is evident in 16HBE14o− and Caco2 cells that both have transcriptionally active CFTR genes, although there is evidence for partial methylation of CG −1,350 in both lines (Figure 4).
Figure 4.
DNA methylation analyses by sodium bisulfite sequencing of the CFTR 5′ region. The figure depicts representative examples of four regions sequenced after sodium bisulfite treatment for two primary tracheal epithelial cells (pHTE) samples, 16HBE14o−, and Caco2. (Top) Scaled map of the 5′ region of the CFTR gene. The bold arrow represents the ATG, and numbers in each sequenced region represent the first and last CpG dinucleotide. Small arrows represent scores for putative promoters analyzed in silico. (Bottom) Each circle represents a CpG dinucleotide. A total of 12 clones or alleles were sequenced for each region. A total of 58 CpGs in all regions were analyzed by this method. Unmethylated, open circles; methylated, filled circles. Percentages of DNA methylation are indicated on the right of the panel.
Additional cell lines analyzed are shown in Figure E4. MCF7 is almost completely methylated at all CGs of the regions analyzed in 1,700 bp flanking the ATG. In contrast, no other cell type demonstrates extensive methylation of the promoter region irrespective of CFTR expression level. Nonexpressing fibroblasts show only inconsistent methylation of a few CGs in the basal promoter region, and partial methylation of CGs at −772, −767, −756, and −954, but complete methylation of CG −1350. The Beas2B line (in which CFTR expression is barely detectable and sometimes absent) showed partial methylation across the whole promoter region, and complete methylation of CGs 1350, −1185, and −1179. In contrast, high CFTR expression levels were associated with complete absence of CG methylation in Calu3 cells.
DISCUSSION
Studies from many laboratories have demonstrated that the promoter region of the CFTR gene lacks elements that drive its tissue-specific expression. However, this region is clearly essential for driving gene expression, and we have demonstrated that it interacts directly with intronic enhancers to coordinate transcription (37). Moreover, several groups have determined that the inclusion of alternative 5′ exons may contribute to the CFTR regulatory mechanisms, although the precise role of these exons has not been elucidated.
We previously demonstrated that the recruitment of one alternative upstream exon (−1a), which was mutually exclusive with exon 1 and spliced directly to exon 2 of CFTR, showed temporal regulation in lung development. The alternative form of CFTR mRNA appeared just before the start of down-regulation of the full-length CFTR transcript, which is a normal feature of lung development, and disappeared before birth. We speculated that this splice form might contribute to the down-regulation of the gene during development.
We now demonstrate that this splice form can generate a truncated CFTR protein that initiates translation at an ATG within exon 4. This protein lacks essential membrane localization signals, which would likely cause the majority of the protein to remain elsewhere in the cell. These data are consistent with in vivo analysis of a novel mutation in CFTR, which removes the first methionine from the protein (38). It is of interest that another splice form of CFTR (−1a/1a2–24), which does not show temporal regulation of expression, is not translated in vitro, although a stable mRNA is detected. This might suggest that the predicted RNA secondary structure that we observed in silico for the −1a/1a exons (14) may indeed be responsible for detachment of the 40S ribosomal subunit from the mRNA transcript during translation.
The probable importance of exon −1a in temporal regulation of CFTR expression led us to search for a cis element that might provide a promoter for this transcript, because exon −1a lies 5′ to the region previously identified as the basal promoter for CFTR. We first used in silico analysis of the genomic sequence 5′ of exon −1a to identify predicted promoters, and found four elements with high scores. To evaluate these for promoter activity, we performed a series of transient transfections of luciferase constructs, driven from different promoter fragments, in both airway and intestinal epithelial cells. For all previous experiments, we used the “245” fragment for the basal CFTR promoter in reporter gene assays, which encompasses 787 bp of the promoter region (AC000111:18,981–19,767) (35, 36). It was of interest that, in the current experiments, a shorter promoter fragment, 632, that lacked 188 bp at the 5′ end of 245, had significantly greater promoter activity (P < 0.01) in both airway and intestinal epithelial cells. It may be relevant that this fragment lacks the sequence that encodes parts of exon −1a, which is present in “245.” These transient assays also revealed a cis-acting element that augmented promoter activity in the 1 kb 5′ to the “245” fragment, because the 1,750 and 1,963 fragments showed approximately two-fold greater activity than “245” alone in the airway cell lines, 16HBE14o− and Beas2B, but a smaller increase in Caco2 cells. This 1-kb region alone had no independent promoter activity, but apparently contributed to the overall strength of the CFTR promoter fragment in driving luciferase expression. To further evaluate this region, the putative promoter at AC000111:18,841 was mutated (A > G) to reduce its score from 1 to 0.44. The 1,750A/G construct generated luciferase values that were significantly reduced (P < 0.01) in comparison with the wild-type 1,750 construct, but only in 16HBE14o− cells, and not in Caco2 cells in which the mutation was silent. These data suggest that the additional promoter activity seen in the region 5′ to exon −1a may show some degree of cell-type specificity, being fully functional only in certain airway epithelial cells, and not in intestinal cell lines.
Our observations implicating limited cell-type–specific mechanisms in promoter function encouraged us to re-evaluate the DNA methylation status of the CFTR promoter in a number of cell types to determine whether this showed any correlation with CFTR expression levels. DNA methylation is one of the epigenetic mechanisms responsible for gene regulation in normal cells, and it is already known that disruption of this mechanism is associated with several diseases (39). Our results are in contrast to previous reports on long-term cell lines, which revealed that expression of CFTR was associated with an unmethylated promoter, and that methylation was generally associated with an inactive CFTR promoter (2). However, a difference between CFTR promoter methylation in tissues and cell lines has been demonstrated (40): whereas CpG sites were not methylated in high and low CFTR–expressing cell lines, in very low or nonexpressing lines, the CpG sites were partially or completely methylated. In contrast, none of these sites were methylated in tissues, irrespective of CFTR expression levels. Our data are completely consistent with the CFTR promoter being associated with a strong CpG island (high CpG promoter [19, 20]), as these are generally unmethylated, even when the promoter is inactive. The only cell line in which we observe extensive methylation (93.4%) of the promoter is the MCF7 breast carcinoma cell line, which does not express CFTR. This observation is consistent with our previous work (41). In contrast, human skin fibroblasts, which also lack CFTR expression, show only very low levels (7.8%) of methylation, suggesting that this modification is not required to inactivate the CFTR promoter. Methylation of specific CGs in the basal promoter may accompany transcriptional repression of CFTR subsequent to the unfolded protein response (42). Of particular interest is the methylation of the CFTR promoter in pHTE cells, which are the only primary cell type that we have evaluated, and which express only low levels of CFTR mRNA. Here, the promoter is only 5.6% methylated, but the methylated CGs, rather than being located across the whole promoter, are clustered in the region CG −1350 to −1099. Four CGs (−1350, −1185, −1,179, and −1099) are partially methylated, wheareas additional CGs in this region remain unmethylated. Caco2 and 16HBE14o− cells, which both express abundant CFTR mRNA, have very low levels of promoter methylation (∼1%), with an increase in methylation frequency only at CG −1,350. These data are consistent with a model in which DNA methylation starts 5′ to a gene promoter and progressively migrates toward the TSS of the gene (43). In this model, DNA methylation might be a late event in gene inactivation. First, a combination of different factors, such as loss of transcription factors and proteins that bind to that promoter, such as histones, can turn the gene off. This can lead to the spreading of DNA methylation in the promoter region. In this case, DNA methylation is a consequence rather than the cause of the inactivation of the gene, as has already been observed in the promoters of other genes (44, 45). This hypothesis could explain the fact that, in our study, primary fibroblasts, which do not express CFTR, are almost completely unmethylated. On the other hand, all regions analyzed here are highly methylated in MCF7, which also does not express CFTR. It is of interest that CGs −987 and −1,018 that are immediately upstream of the in silico–predicted promoter with the highest score (AC000111:18,830–188,841) and just 5′ to exon −1a remain unmethylated, both in pHTE and Beas2B cells.
In the context of CG methylation, it is relevant to consider other modifications of chromatin structure that are associated with active or inactive promoters. We previously evaluated the chromatin landscape at the CFTR promoter in a number of airway, intestinal, and fibroblast cell lines through chromatin immunoprecipitation with antibodies specific for modifications that are associated with active chromatin (acetylated H3, acetylated H4, and dimethylated H3, K4) (46). Enrichment of acetylated H3 and dimethylated H3, K4 was limited to active CFTR promoters, whereas H4 acetylation was not markedly enriched, irrespective of CFTR promoter activity. Thus, a combination of epigenetic modifications clearly contributes to the multiple mechanisms regulating the promoter of the CFTR gene.
Supplementary Material
Acknowledgments
The authors thank Drs. Calvin Cotton, Shih-Hsing Leir, and Christopher J. Ott for their contributions, and Kelly Ardnt, Christina Smith, Selva Musa, and Melissa Arvide for excellent technical assistance.
This work was supported in part by Vaincre La Mucoviscidose (France), the Cystic Fibrosis Foundation (USA), National Institutes of Health grant R01 HL094585, the Children's Memorial Research Center, and by the Maeve McNicholas Memorial Foundation (F.F.C.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2009-0149OC on October 23, 2009
Author Disclosure: M.B.S. received a patent for Technology for Normalization of cDNA Libraries, licensed by Columbia University, and receives royalties from it for $1001–$5000. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Chou JL, Rozmahel R, Tsui LC. Characterization of the promoter region of the cystic fibrosis transmembrane conductance regulator gene. J Biol Chem 1991;266:24471–24476. [PubMed] [Google Scholar]
- 2.Koh J, Sferra TJ, Collins FS. Characterization of the cystic fibrosis transmembrane conductance regulator promoter region: chromatin context and tissue-specificity. J Biol Chem 1993;268:15912–15921. [PubMed] [Google Scholar]
- 3.Yoshimura K, Nakamura H, Trapnell BC, Dalemans W, Pavirani A, Lecocq JP, Crystal RG. The cystic fibrosis gene has a “housekeeping”-type promoter and is expressed at low levels in cells of epithelial origin. J Biol Chem 1991;266:9140–9144. [PubMed] [Google Scholar]
- 4.McDonald RA, Matthews RP, Idzerda RL, McKnight GS. Basal expression of the cystic fibrosis transmembrane conductance regulator gene is dependent on protein kinase A activity. Proc Natl Acad Sci USA 1995;92:7560–7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews RP, McKnight GS. Characterization of the cAMP response element of the cystic fibrosis transmembrane conductance regulator gene promoter. J Biol Chem 1996;271:31869–31877. [DOI] [PubMed] [Google Scholar]
- 6.Vuillaumier S, Dixmeras I, Messai H, Lapoumeroulie C, Lallemand D, Gekas J, Chehab FF, Perret C, Elion J, Denamur E. Cross-species characterization of the promoter region of the cystic fibrosis transmembrane conductance regulator gene reveals multiple levels of regulation. Biochem J 1997;327:651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams S, Mouchel N, Harris A. A comparative genomic analysis of the cow, pig and human CFTR genes identifies potential intronic regulatory elements. Genomics 2003;81:628–639. [DOI] [PubMed] [Google Scholar]
- 8.Pittman N, Shue G, LeLeiko NS, Walsh MJ. Transcription of cystic fibrosis transmembrane conductance regulator requires a CCAAT-like element for both basal and cAMP-mediated regulation. J Biol Chem 1995;270:28848–28857. [DOI] [PubMed] [Google Scholar]
- 9.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, LeLeiko NS, Walsh MJ. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem 1999;274:7803–7815. [DOI] [PubMed] [Google Scholar]
- 10.Rene C, Taulan M, Iral F, Doudement J, L'Honore A, Gerbon C, Demaille J, Claustres M, Romey MC. Binding of serum response factor to cystic fibrosis transmembrane conductance regulator CArG-like elements, as a new potential CFTR transcriptional regulation pathway. Nucleic Acids Res 2005;33:5271–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulatowski LM, Whitmore KL, Romigh T, VanderWyden AS, Satinover SM, Drumm ML. Strain-specific variants of the mouse Cftr promoter region reveal transcriptional regulatory elements. Hum Mol Genet 2004;13:1933–1941. [DOI] [PubMed] [Google Scholar]
- 12.White NL, Higgins CF, Trezise AE. Tissue-specific in vivo transcription start sites of the human and murine cystic fibrosis genes. Hum Mol Genet 1998;7:363–369. [DOI] [PubMed] [Google Scholar]
- 13.Broackes-Carter FC, Mouchel N, Gill D, Hyde S, Bassett J, Harris A. Temporal regulation of CFTR expression during ovine lung development: implications for CF gene therapy. Hum Mol Genet 2002;11:125–131. [DOI] [PubMed] [Google Scholar]
- 14.Mouchel, N., Broackes-Carter, F. and A., H. Alternative 5′ exons of the CFTR gene show developmental regulation. Hum Mol Genet 2003;12:759–769. [DOI] [PubMed] [Google Scholar]
- 15.Trezise AE, Chambers JA, Wardle CJ, Gould S, Harris A. Expression of the cystic fibrosis gene in human foetal tissues. Hum Mol Genet 1993;2:213–218. [DOI] [PubMed] [Google Scholar]
- 16.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 1989;245:1059–1065. [DOI] [PubMed] [Google Scholar]
- 17.Davies WL, Vandenberg JI, Sayeed RA, Trezise AE. Cardiac expression of the cystic fibrosis transmembrane conductance regulator involves novel exon 1 usage to produce a unique amino-terminal protein. J Biol Chem 2004;279:15877–15887. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 1991;266:19867–19870. [PubMed] [Google Scholar]
- 19.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 2005;37:853–862. [DOI] [PubMed] [Google Scholar]
- 20.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007;39:457–466. [DOI] [PubMed] [Google Scholar]
- 21.Smith DJ, Nuthall HN, Majetti ME, Harris A. Multiple potential intragenic regulatory elements in the CFTR gene. Genomics 2000;64:90–96. [DOI] [PubMed] [Google Scholar]
- 22.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 1990;63:827–834. [DOI] [PubMed] [Google Scholar]
- 23.Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol 1994;10:38–47. [DOI] [PubMed] [Google Scholar]
- 24.Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, Brash DE, Park JB, Rhim JS, Harris CC. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res 1988;48:1904–1909. [PubMed] [Google Scholar]
- 25.Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst 1977;58:209–214. [DOI] [PubMed] [Google Scholar]
- 26.Davis, P.B., Silski, C.L. and Perez, A. cAMP does not regulate [Ca2+]i in human tracheal epithelial cells in primary culture. J Cell Sci 1994;107:2899–2907. [DOI] [PubMed] [Google Scholar]
- 27.Costa FF, Paixão VA, Cavalher FP, Ribeiro KB, Cunha IW, Rinck JA Jr, O'Hare M, Mackay A, Soares FA, Brentani RR, et al. SATR-1 hypomethylation is a common and early event in breast cancer. Cancer Genet Cytogenet 2006;165:135–143. [DOI] [PubMed] [Google Scholar]
- 28.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998;8:186–194. [PubMed] [Google Scholar]
- 29.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998;8:175–185. [DOI] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988;16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramalho AS, Lewandowska MA, Farinha CM, Mendes F, Gonçalves J, Barreto C, Harris A, Amaral MD. Deletion of CFTR translation start site reveals functional isoforms of the protein in CF patients. Cell Physiol Biochem 2009;24:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Park EH, Couture G, Harvey I, Garneau P, Pelletier J. An upstream open reading frame impedes translation of the huntingtin gene. Nucleic Acids Res 2002;30:5110–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 2001;26:51–56. [DOI] [PubMed] [Google Scholar]
- 35.Smith AN, Barth ML, McDowell TL, Moulin DS, Nuthall HN, Hollingsworth MA, Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J Biol Chem 1996;271:9947–9954. [DOI] [PubMed] [Google Scholar]
- 36.Phylactides M, Rowntree R, Nuthall H, Ussery D, Wheeler A, Harris A. Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. Eur J Biochem 2002;269:553–559. [DOI] [PubMed] [Google Scholar]
- 37.Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med 2009;13:680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramalho AS, Lewandowska MA, Farinha CM, Mendes F, Gonçalves J, Barreto C, Harris A, Amaral MD. Lack of N-terminus causes major impairment on CFTR processing but does not abolish function. Pediatr Pulmonol 2005;S28:207. [Google Scholar]
- 39.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457–463. [DOI] [PubMed] [Google Scholar]
- 40.Denamur E, Chehab FF. Methylation status of CpG sites in the mouse and human CFTR promoters. DNA Cell Biol 1995;14:811–815. [DOI] [PubMed] [Google Scholar]
- 41.Smith AN, Wardle CJ, Harris A. Characterization of DNASE I hypersensitive sites in the 120kb 5′ to the CFTR gene. Biochem Biophys Res Commun 1995;211:274–281. [DOI] [PubMed] [Google Scholar]
- 42.Bartoszewski R, Rab A, Twitty G, Stevenson L, Fortenberry J, Piotrowski A, Dumanski JP, Bebok Z. The mechanism of cystic fibrosis transmembrane conductance regulator transcriptional repression during the unfolded protein response. J Biol Chem 2008;283:12154–12165. [DOI] [PubMed] [Google Scholar]
- 43.Turker M. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene 2002;21:5388–5393. [DOI] [PubMed] [Google Scholar]
- 44.Costa FF, Verbisck NV, Salim AC, Ierardi DF, Pires LC, Sasahara RM, Sogayar MC, Zanata SM, Mackay A, O'Hare M, et al. Epigenetic silencing of the adhesion molecule ADAM23 is highly frequent in breast tumors. Oncogene 2004;23:1481–1488. [DOI] [PubMed] [Google Scholar]
- 45.Oyer JA, Chu A, Brar S, Turker MS. Aberrant epigenetic silencing is triggered by a transient reduction in gene expression. PLoS One 2009;4:e4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem J 2007;408:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.