Abstract
The plasminogen activator inhibitor type–1 (PAI-1) effectively blocks the activities of free and receptor-bound urokinase-type plasminogen activator. Incubation of cultured human pleural mesothelial (Met5A) cells with TGF-β increased PAI-1 protein. TGF-β, phorbol myristate acetate, and the translation inhibitor cycloheximide induced PAI-1 mRNA and slowed its degradation, suggesting that PAI-1 mRNA could be regulated by interaction of a PAI-1 binding protein (PAI-1 mRNABp) with PAI-1 mRNA. We found that an approximately 60 kD cytoplasmic PAI-1 mRNABp is detectable in cytoplasmic extracts of MeT5A human pleural mesothelial and malignant mesothelioma cells. The PAI-1 mRNABp specifically binds to a 33-nt sequence in the 3′ untranslated region of PAI-1 mRNA. Insertion of this 33-nt sequence destabilizes otherwise stable β-globin mRNA, indicating that the binding sequence accelerates decay of endogenous PAI-1 mRNA. Competitive inhibition by overexpression of the 33-nt binding sequence in MeT5A cells reduced PAI-1 mRNA decay and increased PAI-1 protein and mRNA expression, indicating that the PAI-1 mRNABp destabilizes PAI-1 mRNA by its interaction with the endogenous 33-nt binding sequence. Incubation of Met5A cells with TGF-β attenuated the interaction of the PAI-1 mRNABp with the 33-nt sequence. By conventional and affinity purification, we isolated the PAI-1 mRNABp and confirmed its identity as 6-phospho-d-gluconate-NADP oxidoreductase, which specifically interacts with the full-length and the 33-nt sequence of the PAI-1 mRNA 3′ untranslated region. This newly recognized pathway could influence expression of PAI-1 by mesothelial or mesothelioma cells at the level of mRNA stability in the context of pleural inflammation or malignancy.
Keywords: PAI-1, mesothelial cells, post-transcriptional regulation
Serine proteases facilitate remodeling of the transitional stroma by breakdown of basement membranes and extracellular matrix proteins, including fibrin (1–3). Plasminogen activation can be facilitated by urokinase-type (uPA) and tissue-type (tPA) plasminogen activators. The uPA is mainly involved in extravascular proteolysis and is implicated in stromal remodeling as occurs in association with lung or pleural injury and neoplasia. Plasminogen activator inhibitor type–1 (PAI-1) binds and irreversibly inactivates uPA and tPA (4), thereby regulating expression of fibrinolytic activity and remodeling of intravascular or extravascular fibrin. The control of PAI-1 expression is therefore critical to these processes in a variety of organs, including the lung and pleural space.
Pathologic overexpression of PAI-1 has been linked to a broad range of inflammatory lung and pleural diseases (4, 5). For instance, a defect in uPA-related fibrinolytic activity, largely attributed to overexpression of PAI-1, has been associated with lung injuries, including acute respiratory distress syndrome and interstitial lung diseases (6, 7). PAI-1 is also increased in empyema, parapneumonic pleural effusions, and evolving pleurodesis and appears to be a critical determinant of intrapleural organization and loculation (8–10). In addition, PAI-1 is implicated in the maintenance of extravascular fibrin within the tumor stroma that characterizes human malignant mesothelioma (11). A variety of stimuli can influence expression of PAI-1 in a range of neoplastic epithelial or endothelial cells, including hormones, growth factors, hypoxia, endotoxin, glucocorticoids, and cytokines, acting at the transcriptional or post-transcriptional levels (12–17). Previous reports indicate that the presence of PAI-1 expression at the transcriptional level in response to TGF-β and other stimuli (18, 19). These observations, and our previous report that PAI-1 is alternatively regulated through post-transcriptional control at the level of mRNA stability in lung epithelial and lung carcinoma cells (19), suggest that pleural mesothelial cells might regulate PAI-1 expression in a similar manner. Pleural mesothelial cells are a rich source of PAI-1 (20, 21) that is inducible by proinflammatory cytokines, including TGF-β and TNF-α (22). Although limited PAI-1 mRNA degradation could contribute to this response, we are unaware of any previous studies that address this possibility or that define the responsible mechanism.
In this report, we show that stimuli implicated in the pathogenesis of pleural inflammation and malignancy stabilizes expression of PAI-1 mRNA in MeT5A human pleural mesothelial cells. We found that MeT5A and a range of human malignant mesothelioma cells express a 60-kD PAI-1 mRNA-binding protein (PAI-1 mRNABp). This binding protein acts as a trans-acting factor that regulates PAI-1 mRNA stability via interaction with a specific 33-nt cis-acting sequence of PAI-1 mRNA. Our results define a newly recognized pathway by which pleural mesothelial cells regulate PAI-1 expression and confirm that 6-phospho-d-gluconate-NADP oxidoreductase (6-PGD) is the PAI-1 mRNABp that interacts with and destabilizes PAI-1 mRNA.
MATERIALS AND METHODS
Media and Reagents
Culture media, penicillin, streptomycin, and FCS were purchased from Gibco BRL laboratories (Grand Island, NY); tissue culture plastics were from Becton Dickinson Lab ware (Linclon Park, NJ). BSA, ovalbumin, tris-base, aprotinin, dithiothreitol, PMSF, silver nitrate, ammonium persulfate, and phorbol myristate acetate (PMA) were from Sigma Chemical Co. (St. Louis, MO). Acrylamide, bisacrylamide, and nitrocellulose were from BioRad Laboratories (Richmond, CA). Anti–PAI-1 antibodies were obtained from American Diagnostics (Greenwich, CT). XAR X-ray film was purchased from Eastman Kodak (Rochester, NY).
Cell Cultures and Preparation of Cytosolic Extracts
Human pleural mesothelial (MeT5A) cells and human pleural malignant mesothelioma (MS-1, REN, M33K, and M9K) cells obtained from ATCC were maintained in RPMI 1640 medium containing 10% heat-inactivated FCS, 1% glutamine, 2% antibiotics, and 5 μg/ml plasmocin. Primary rabbit pleural mesothelial cells were harvested as we previously reported, characterized, and maintained under the same conditions (23). T75 flasks containing MeT5A and MS-1 cells were serum starved overnight and treated with or without TGF-β for 24 hours, washed with HBSS, and homogenized in extraction buffer (25 mM Tris-HCl [pH 7.9], 0.5 mM EDTA, and 0.1 mM phenylmethlsulfonyl fluoride) with several freeze-thaw cycles. The homogenates were then centrifuged at 12,000 × g for 15 minutes at 4°C, and the supernatants were collected. The protein content was measured with a Pierce BCA protein assay kit using various concentrations of serum albumin as standards.
Plasmid Construction
Human PAI-1 cDNA of the coding region, 3′UTR, and various deletion products were separately cloned into pCDNA3.1 vector (Invitrogen) by PCR amplification using full-length PAI-1 cDNA as a template. The orientation and sequence of the clones were confirmed by sequencing.
PAI-1 Deletion Products
Overlapping sections of the 3′UTR (∼800 bp) were systematically cloned into pcDNA3.1 vector. The 3′UTR was first divided into three overlapping regions (A, B, and C) of approximately 300 bp each. These regions were subdivided into three subsequent overlapping regions (A1, A2, A3, B1, B2, B3, C1, C2, and C3) each of approximately 100 bp. Region B3 (104 bp) was subdivided into regions B3a (33 bp), B3b (66 bp), and B3c (47 bp). As a control, 33 bp of random coding region were also cloned into the pcDNA3.1 vector.
In Vitro Transcription
The full-length 3′UTR and the deletion products of PAI-1 in pcDNA3.1 vector were linearized with XbaI, purified separately on agarose gels, extracted with phenol-chloroform, and used as a template for in vitro transcription with T7 polymerase. Sense mRNA was transcribed according to the supplier's (Ambion, Austin, TX) in vitro transcription protocol, except that 50 μCi (800 Ci/mmol) of [32P]UTP were substituted for unlabeled UTP in the reaction mixture. Passage through a NucAway (Ambion) column removed unincorporated radioactivity.
Mapping of the PAI-1 mRNABp–PAI-1 mRNA Interaction by UV Cross-Linking Assay
RNA-protein binding assays were performed using uniformly 32P-labeled transcripts corresponding to the PAI-1 3′UTR and various deletion regions. Transcripts (20,000 counts/min) were incubated with cytosolic extracts (100 μg) at 30°C in reaction buffer (15 mM KCl, 5 mM MgCl2, 0.25 mM EDTA, 0.25 mM dithiothreitol, 12 mM HEPES [pH 7.9], 10% glycerol, and Escherichia coli tRNA [200 ng/μl]) in a total volume of 40 μl for 30 minutes. The reaction mixtures were treated with 50 U of RNase T1 and incubated for 30 minutes at 37°C. Heparin (5 mg/ml) was added to decrease nonspecific protein binding, and the reactions were incubated at room temperature for 10 minutes. The reaction mixtures were transferred to a 96-well microtiter plate and irradiated on ice at 2,500 μJ for 30 minutes with an UV (UVC 500 Cross-linker) chamber apparatus (Amersham Biosciences). The samples were separated on an 8% SDS-PAGE under nonreducing conditions. The gels were dried and autoradiographed at −70°C.
Competitive Inhibition by Sense mRNA, and Effects of SDS
Further experiments were designed to confirm the specificity of the PAI-1 mRNABp–mRNA interaction. Cytosolic extracts were pretreated with molar excess amounts of unlabeled full-length PAI-1 3′UTR or with a 33-nt deletion transcript of PAI-1 sense mRNA for 30 minutes before the addition of the 32P-labeled B3a transcript. In addition, cytosolic extracts were pretreated with SDS (0.1%) for 30 minutes at 30°C before the addition of 32P-labeled B3a mRNA transcript. We also pretreated 32P-labeled B3a transcript with RNase T1 before incubation with cytosolic extract to determine the specificity of the 60-kD PAI-1 mRNABp interaction. The PAI-1 mRNA–PAI-1 mRNABp interaction was determined by UV cross-linking assay as described previously.
Construction of β-Globin/33-bp PAI-1 Chimeric Message
Two 33-bp fragments, one corresponding to the PAI-1 mRNABp binding region and the other corresponding to a control nonbinding coding sequence, were prepared from PAI-1 cDNA, ligated to β-globin cDNA at the 3′ end (Age I sites added via PCR), and inserted into pcDNA3.1 vector (Invitrogen) at the HindIII/XbaI sites. The orientations and sequences of the chimeric clones were verified by sequencing. As a control, β-globin cDNA was inserted into the pcDNA3.1 vector. MeT5A cells were then transfected with the prepared chimeric plasmid constructs, β-globin in pcDNA3.1, and empty pcDNA3.1 vector using GeneJammer (Stratagene, La Jolla, CA), and stable transfectants were grown in culture flasks after antibiotic selection.
The stable transfectants were screened for the presence of the gene of interest by isolating total DNA (via TRI-Reagent) and using the DNA as a template for PCR. Positive clones were further screened to assess if mRNA was transcribed. Total RNA was isolated (via TRI-Reagent method), and cDNA was made using reverse transcriptase. Then PCR was performed using the forward and reverse primers of the transfected gene of interest, T7, BGH, or a combination of both. These stable cell transfectants were grown to confluence, and total RNA was isolated at various time points after the inhibition of transcription by 5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (DRB) (20 μg/ml). The decay of chimeric mRNA was determined by RT-PCR to determine if the 33-bp PAI-1 mRNABp-binding region of the PAI-1 mRNA 3′UTR region destabilizes the β-globin mRNA.
Total Protein Extraction and Western Blotting
Cells were grown to confluence and were serum starved overnight with RPMI-1640 media. The cells were treated with or without TGF-β or other agents for selected times in serum-free media. After these treatments, the condition media were collected, and cells were suspended in lysis buffer (10 mM Tris-HCl [pH 7.4] containing 150 mM NaCl, 1% Triton X-100, 15% glycerol, 1 mM Na3VO4, 1 mM NaF, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 3–10 μg aprotinin per 100 ml). The cell lysates were prepared using three freeze-thaw cycles. Proteins from conditioned media (CM) and cell lysates (CL) (50 μg) were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked with 1% BSA in wash buffer for 1 hour at room temperature followed by overnight hybridization with anti–PAI-1 monoclonal antibody in the same buffer at 4°C and washing, and PAI-1–immunoreactive proteins were detected by enhanced chemiluminescence. The membranes containing proteins from cell lysates were stripped with β-mercaptoethanol and subjected to Western blotting using β-actin monoclonal antibody as we described previously (24).
Random Priming of PAI-1 cDNA
The PAI-1 cDNA corresponding to the coding region (0.5 Kb) was subcloned to pGEMR-T vector (Promega, Madison, WI), and the sequence of the clones was confirmed by nucleotide sequencing. The PAI-1 insert was released by NcoI and PstI, purified on 1% agarose gels, extracted with phenol/chloroform, and labeled with [32P] deoxycytidine triphosphate (dCTP) using a Rediprime labeling kit (Promega). Passage through a Sephadex G-25 column removed unincorporated radioactivity. The specific activity of the product was 6 to 7 × 108 cpm/μg and used as a cDNA probe for Northern blotting.
PAI-1 mRNA Assessment by Northern Blotting
A Northern blotting assay was used to assess the level of PAI-1 mRNA. Confluent MeT5A cells were serum starved overnight in RPMI-BSA media. Total RNA was isolated using TRI reagent RNA (20 μg) separated on agarose/formaldehyde gels. After electrophoresis, the RNA was transferred to Hybond N+ according to the manufacturer's instructions. Prehybridization and hybridization were done at 65°C in NaCl (1 M)/SDS (1%) and 100 μg/ml salmon sperm DNA. Hybridization was performed with PAI-1 cDNA probes (1 ng/ml) labeled to approximately 6 to 7 × 108 cpm/μg of DNA overnight. After hybridization, the filters were washed twice for 15 minutes at 65°C, with, respectively, 2 × SSC, 1% SDS; 1 × SSC, 1% SDS; and 0.1% SSC, 1% SDS. The membranes were exposed to X-ray film at −70°C over night. The intensity of the bands was measured by densitometry and normalized against that of β-actin.
Purification of the PAI-1 mRNABp
Cytosolic extracts prepared as described previously were added to solid ammonium sulfate crystals at 40% saturation, and the precipitated proteins were discarded after checking the PAI-1 mRNA binding protein activity. Ammonium sulfate crystals were added to the 40% ammonium sulfate crystals to yield a final saturation of 60%. The precipitated proteins were collected, redissolved, and exhaustively dialyzed against extraction buffer containing 10% glycerol. The PAI-1 3′UTR binding activity was assessed by taking the aliquot (10 μg protein). The 40 to 60% ammonium sulfate fraction was passed through a blue-sepharose column (90 ml) in the same buffer containing 100 mM NaCl, and PAI-1 3′UTR mRNABp was eluted with a linear gradient (200 ml) of 0.1 to 1.0 M NaCl. Positive fractions were pooled and loaded onto a heparin-affigel column (90 ml) in the same buffer. After washing the unbound materials, PAI-1 mRNABp was eluted with a linear gradient (200 ml) of 0.0 to 0.5 M NaCl in extraction buffer. The PAI-1 mRNABp binding activity was eluted and pooled loaded onto a DEAE-sephacel column and eluted with linear NaCl gradient of 0 to 1 M NaCl in extraction buffer. Positive fractions were pooled, concentrated by ultrafiltration, and passed through mono-Q column fitted to a FPLC system. Unbound proteins were washed, and bound proteins were eluted by a linear gradient (40 ml) of 0 to 1 M NaCl. Fractions containing PAI-1 mRNABp activity were analyzed by UV cross-linking assays.
Positive fractions were pooled, dialyzed, and subjected to a final round of affinity purification using an RNA affinity column containing biotin-labeled 33-nt PAI-1 mRNABp binding sequences immobilized to streptavidin agarose, and the PAI-1 mRNA binding activity was assessed by UV cross-linking experiments. SDS-PAGE analyses of RNA affinity column eluate yielded two proteins with approximate molecular weight of 45 and 55 kD after silver nitrate staining. These two bands were excised and analyzed by mass spectroscopic analyses to determine the identity of the enriched proteins. Database analyses identified enriched proteins as citrate synthase (CS) and 6-PGD. To determine if CS or 6-PGD specifically binds to PAI-1 mRNA, we subjected purified yeast 6-PGD and porcine CS to PAI-1 mRNA binding by UV cross-linking assays. To further confirm the specificity of the 6-PGD interaction with the 33-nt PAI-1 mRNA 3′UTR sequence, we incubated purified yeast 6-PGD with the 33-nt PAI-1 mRNA binding sequence in the presence of a 200-fold molar excess of poly (A), (C), (G), and (U) ribonucleotides. Alternatively, we treated purified yeast 6-PGD with proteinase K or SDS before exposure to PAI-1 mRNA, or 6-PGD was exposed to 33-nt PAI-1 mRNA predigested with RNase A/T to ascertain the specificity of the 6-PGD binding interaction. Finally, 6-PGD was subcloned into eukaryotic expression vector pcDNA 3.1 and tranfected to H157 cells, and a stable cell line was prepared by antibiotic selection. Histidine-tagged 6-PGD containing a C-terminal V5 epitope was expressed in H157 cells and was affinity purified using a nickel column. Expression of the protein was confirmed by Western blotting of various fractions with anti-V5 and anti–6-PGD antibodies. The fractions containing recombinant 6-PGD were pooled, and the recombinant protein was tested for PAI-1 mRNA 3′UTR binding by a UV cross-linking assay as described previously (23). The specificity of 6-PGD–PAI-1 mRNA binding was confirmed by cold competition experiments.
To directly determine that 6-PGD regulates PAI-1 expression, stable MeT5A cells expressing 6-PGD or empty vector in pcDNA 3.1 were treated with or without TGF-β, and conditioned media were analyzed for PAI-1 expression. The cell lysates were analyzed for V5 epitope and β-actin by immunoblotting to confirm the expression of fusion protein. To confirm if PAI-1 expression is likewise regulated by 6-PGD in primary pleural mesothelial cells, we transduced primary rabbit mesothelial cells with adenovirus carrying 6-PGD or empty vector in the presence or absence of TGF-β and conditioned media, and cell lysates were analyzed for PAI-1 and 6-PGD protein expression by Western blotting.
To confirm that the 60-kD protein from Met5A cell lysates that specifically binds a 33-nt PAI-1 mRNA 3′UTR is 6-PGD, cell lysates from Met5A cells were incubated with a 32P-labeled PAI-1 mRNA 3′UTR probe. The reaction mixtures were UV cross-linked as described previously to immobilize the RNA–protein complex. After UV cross-linking, the reaction mixtures were immunoprecipitated with anti–6-PGD antibody or nonspecific mIgG overnight at 4°C. The immune complexes were precipitated using proteinA/G agarose, separated on the SDS-PAGE, dried, and autoradiographed.
RESULTS
Induction of PAI-1 in MeT5A and Mesothelioma Cells by TGF-β
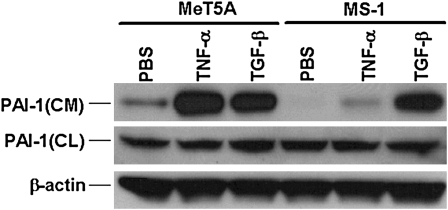
Nonmalignant Met5A and malignant pleural mesothelioma MS-1, REN, and M9K cells express different amounts of PAI-1 (25). We sought to confirm that TGF-β induces PAI-1 expression in MeT5A cells. MS-1 cells, which expressed relatively decreased amounts of PAI-1, were included for comparison in these analyses. The cells were cultured in 100-mm dishes and kept in serum-free media overnight. We later treated the cells with TGF-β (2 ng/ml) or with TNF-α (10 ng/ml), which have likewise been implicated in the pathogenesis of pleural injury and malignant mesothelioma, for 24 hours. Total proteins from the CM and CL were used for Western blotting using an anti–PAI-1 antibody. The data in Figure 1 demonstrate that TGF-β and TNF-α induce PAI-1 expression in MeT5A and MS-1 cells and that the increments are best appreciated in conditioned media because PAI-1 is a secreted protein. TGF-β and TNF-α induced more PAI-1 in MeT5A cells compared with that in MS-1 cells. MeT5A expressed relatively greater amounts of PAI-1 in the conditioned media than MS-1 cells under basal conditions (Figure 1).
Figure 1.
Expression of plasminogen activator inhibitor type–1 (PAI-1) by MeT5A human pleural mesothelial and MS-1 mesothelioma cells. Total proteins from conditioned media (CM) and cell lysates (CL) of MeT5A and MS-1 cells treated with PBS, TNF-α (10 ng/ml), or TGF-β (2 ng/ml) for 24 hours were subjected to Western blotting using an anti–PAI-1 antibody. The corresponding blots containing proteins from the CL were stripped and reprobed with β-actin monoclonal antibody for assessment of equal loading.
Induction of PAI-1 mRNA Expression by TGF-β in MeT5A cells
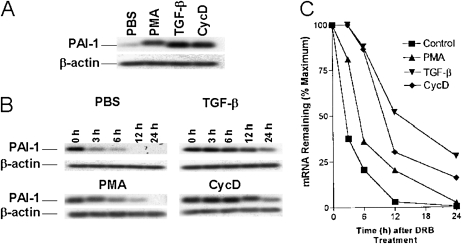
Having confirmed that TGF-β induces PAI-1 protein expression in MeT5A cells, we wanted to confirm whether increased expression of PAI-1 is due to the induction of PAI-1 mRNA. We assessed the steady-state levels of PAI-1 mRNA in TGF-β (2 ng/ml)-treated MeT5A cells by RT-PCR using 32P-labled dCTP in the reaction mixture. PMA (150 ng/ml) and cycloheximide (10 μg/ml) were used as additional treatments. TGF-β, cycloheximide, and PMA increase steady state levels of PAI-1 mRNA in MeT5A cells (Figure 2A).
Figure 2.
Effect of TGF-β, phorbol myristate acetate (PMA), and cycloheximide on PAI-1 mRNA expression in MeT5A cells. (A) MeT5A cells treated with PMA (150 ng/ml), TGF-β (2 ng/ml), or cycloheximide (10 μg/ml) for 6 hours. Total RNA was isolated, and PAI-1 mRNA was analyzed by RT-PCR using 32P-labeled deoxycytidine triphosphate in the reaction mixture. The 32P-labled PCR products were separated on urea/polyacrylamide gel electrophoresis, dried, and autoradiographed. (B) MeT5A cells were treated with PBS, PMA, TGF-β, or cycloheximide for 6 hours to induce maximum PAI-1 mRNA. The cells were later treated with 5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (DRB) to inhibit ongoing transcription, and the level of PAI-1 mRNA was determined by RT-PCR of total RNA collected at the selected time points. This data are representative of three repetitions. (C) Graphic depiction of the data illustrated in (B) showing the percentage of maximal (baseline) mRNA for each treatment condition plotted as a function of time.
Effect of TGF-β, PMA, and Cycloheximide on PAI-1 mRNA Stability
To determine if TGF-β, PMA, and cycloheximide increased steady-state expression of PAI-1 mRNA by increasing its stability, we treated these cells with PBS, TGF-β, PMA, or cycloheximide at the same concentrations described in the previous sections for 6 hours, at which point maximum amounts of PAI-1 mRNA were found to be induced in preliminary experiments. Previous studies, including our own (19, 23, 26), demonstrated that PMA and cycloheximide stabilize PAI-1 mRNA transcripts at the post-transcriptional level. Therefore, we used these two modulators as positive controls to evaluate PAI-1 mRNA stabilization by TGF-β. We then added DRB to the same media to inhibit ongoing transcription. Total RNA was isolated at subsequent selected time points over 0 to 24 hours, and PAI-1 mRNA was determined by RT-PCR. PAI-1 mRNA is highly unstable in MeT5A cells, with a half-life less than 3 hours (Figures 2B and 2C). However, PMA, TGF-β, and cycloheximide stabilized PAI-1 mRNA with a half-life approximating 8 to 12 hours. The response to the translational inhibitor cycloheximide suggests the possibility that a regulatory protein could contribute to the stabilization of PAI-1 mRNA in these cells.
PAI-1 mRNABp Binds the PAI-1 3′UTR mRNA, and the Interaction Is Attenuated by TGF-β
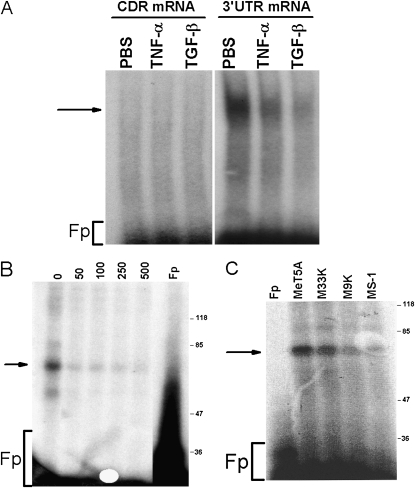
We next sought to elucidate the mechanism by which TNF-α and TGF-β increases the stability of PAI-1 mRNA and therefore sought to determine if the regulatory mechanism involves the interaction of a PAI-1 mRNA binding protein (PAI-1 mRNABp) with PAI-1 mRNA. We treated MeT5A cells with or without TNF-α (10 ng/ml) or TGF-β (2 ng/ml) for 24 hours, after which the cytoplasmic extracts were subjected to PAI-1 coding region (CDR) and 3′UTR mRNA binding analyses by gel mobility shift assays. There was no interaction of the PAI-1 mRNABp with the coding region of PAI-1 mRNA. We found that an approximately 60 kD PAI-1 mRNABp from the cytoplasmic extracts of MeT5A cells binds the 3′UTR of PAI-1 mRNA and that treatment of MeT5A cells with TGF-β– or TNF-α–inhibited PAI-1 mRNABp binding to the PAI-1 mRNA 3′UTR (Figure 3A). These data suggest the possibility that a cis-trans interaction between a binding element within the 3′UTR of PAI-1 mRNA and this 60-kD PAI-1 mRNABp regulates PAI-1 mRNA stability.
Figure 3.
Effect of TGF-β on the PAI-1 mRNABp–PAI-1 mRNA binding interaction. (A) MeT5A cells were treated with PBS or TNF-α (10 ng/ml) or TGF-β (2 ng/ml) for 24 hours at 37°C in serum-free RPMI media. The cytosolic extracts were prepared and separately incubated with 32P-labeled PAI-1 mRNA CDR or 3′UTR in the presence of tRNA. The reaction mixtures were later digested with RNase T1 and heparin as described in Materials and Methods to avoid nonspecific interaction. After heparin digestion, the reaction mixtures were separated on native polyacrylamide gel (4%), dried, and autoradiographed. Fp = free probe. (B) Specificity of the PAI-1 mRNABp–PAI-1 mRNA 3′UTR interaction. Cytosolic extracts of MeT5A cells were incubated with varying amounts of unlabeled transcript before exposure to 32P-labeled transcript. The reaction mixtures were later digested with RNase T1 and heparin and UV irradiated at 4°C. The immobilized RNA–protein complexes were separated on a SDS-PAGE, dried, and autoradiographed. (C) Cytosolic extracts from MeT5A, M33K, M9K, and MS-1 cells were subjected to PAI-1 mRNA 3′UTR binding analyses as described in (A) in the presence of tRNA. After RNase T1 and heparin digestion, the reaction mixtures were subjected to UV cross-linking and autoradiography. The panels are representative of two independent repetitions.
To determine the specificity of the binding interaction, we incubated MeT5A cell lysates with a 200-fold molar excess of unlabeled cold sense transcript and analyzed the PAI-1 mRNABp–PAI-1 3′UTR mRNA interaction by UV cross-linking. Pretreatment of MeT5A cell lysates with a molar excess of unlabeled PAI-1 3′UTR sense transcript abolished PAI-1 mRNABp binding with the 32P-labeled PAI-1 mRNA 3′UTR in a dose-dependent manner (Figure 3B). Treatment of MeT5A lysates with SDS or digestion of 32P-labled PAI-1 mRNA 3′UTR transcript with RNaseT1 abolished PAI-1 mRNABp binding to PAI-1 mRNA, demonstrating the specificity of the interaction (data not shown). UV cross-linking experiments indicated that PAI-1 mRNABp from the cytosolic extracts of nonmalignant MeT5A cells bound to PAI-1 mRNA to a greater extent than to the M33K, M9K, and MS-1 mesothelioma cell lines (Figure 3C). The apparent inconsistency in PAI-1 expression and PAI-1 mRNABp interaction between nonmalignant MeT5A and malignant MS-1 cells may be attributable to variable associated changes in PAI-1 mRNA transcription or its translation (4).
Mapping of the PAI-1 mRNABp Binding Sequence of the 3′UTR of PAI-1 mRNA and its Function in mRNA Degradation and PAI-1 Expression
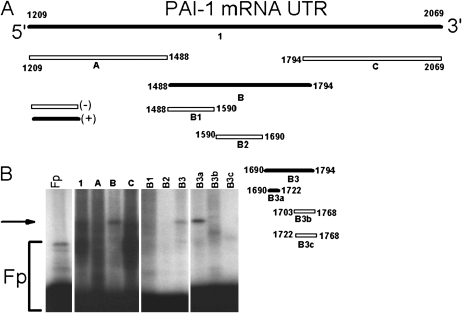
To identify the minimal PAI-1 mRNABp binding sequences of the PAI-1 mRNA 3′UTR, we made a series of 32P-labeled deletion mRNAs by in vitro transcription (Figure 4A). These transcripts were individually tested for their ability to bind the 60-kD PAI-1 mRNABp by UV cross-linking experiments. The PAI-1 mRNABp specifically bound to a 33-nt (nt 1690–1722, B3a) sequence of the PAI-1 mRNA 3′UTR (Figure 4B). In separate experiments, a 200-fold excess of unlabeled PAI-1 3′UTR, B, and B3a sense mRNA abolished the binding interaction of the 60-kD PAI-1 mRNABp with the 32P-labeled B3 transcript. Excess amounts of all the other unlabeled sense mRNAs failed to abolish the binding interaction between the 60-kD PAI-1 mRNABp and the 32P-labeled B3a transcript (data not shown), supporting the specificity of the interaction.
Figure 4.
Identification of the PAI-1 mRNABp binding sequences of the PAI-1 3′UTR mRNA. (A) Deletion map indicating the PAI-1 mRNABp binding site represented in the PAI-1 mRNA 3′UTR. Positive interactions between the PAI-1 mRNABp and the specific deletion transcript are indicated as (+) or open bars; negative (−) interactions are indicated as solid bars. (B) Met5A cell lysates were incubated with 32P-labeled full-length 3′UTR or 3′UTR deletion transcripts. Transcript–protein complex formation was analyzed by UV cross-linking assay. The arrow indicates the transcript–protein complex. Fp = 32P-labeled PAI-1 mRNA 3′UTR probe incubated with buffer alone.
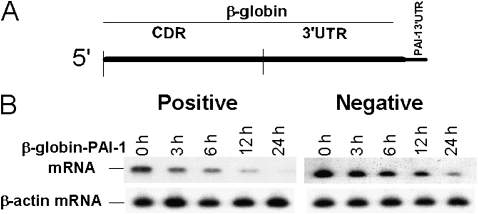
To determine if the binding of the PAI-1 mRNABp to the 33-nt PAI-1 mRNA 3′UTR sequence (nt 1690–1722) contains information for message stability, we inserted this 33-nt base pair sequence into β-globin cDNA (Figure 5A). We chose this strategy because we have found that β-globin is a stable gene amenable to the creation of chimeras that can be assessed for altered stability. The chimeric β-globin–PAI-1 3′UTR cDNA was next subcloned into the eukaryotic expression vector pcDNA3.1. We also prepared a chimeric β-globin–PAI-1 cDNA containing a control 33-nt non–PAI-1 mRNABp-binding CDR sequence. MeT5A cells were then transfected with the chimeric β-globin–PAI-1 cDNA constructs and with β-globin cDNA alone. The decay of chimeric β-globin–PAI-1 mRNA was determined by RT PCR after inhibiting ongoing transcription. Insertion of the PAI-1 mRNABp-binding 33-nt PAI-1 3′UTR sequence (Positive) destabilized otherwise stable β-globin mRNA (Figure 5B). However, insertion of a control non–PAI-1 mRNABp binding CDR sequence of similar size (Negative) failed to alter the stability of β-globin mRNA, and β-globin mRNA was likewise stable (data not shown), indicating that the PAI-1–binding 33-nt PAI-1 3′UTR sequence contains regulatory information for mRNA degradation. Based on these observations, we inferred that the interaction of this destabilization determinant with the PAI-1 mRNABp renders endogenous PAI-1 mRNA unstable in MeT5A cells.
Figure 5.
Determination of the destabilization function of the PAI-1 mRNABp binding sequence of the 3′UTR of PAI-1 mRNA. (A) Structure of β-globin–PAI-1 3′UTR chimeric mRNA. The 33-nt (1,690–1,722) PAI-1 3′UTR mRNABp binding sequence and a control non–PAI-1 mRNABp binding sequence of similar size from the coding region were inserted into the 3′UTR of β-globin cDNA, after which the chimeric cDNA was subcloned into pcDNA 3.1. (B) Met5A cells transfected with the chimeric β-globin–PAI-1 3′UTR gene containing the 33 nt PAI-1 mRNABp binding (positive) or nonbinding control (negative) sequence of PAI-1 in pcDNA 3.1. Total RNA was isolated at different time intervals after treatment with DRB as described in the legend to Figure 2, and the level of chimeric mRNA was analyzed by RT-PCR using β-globin forward and PAI-1 reverse primers.
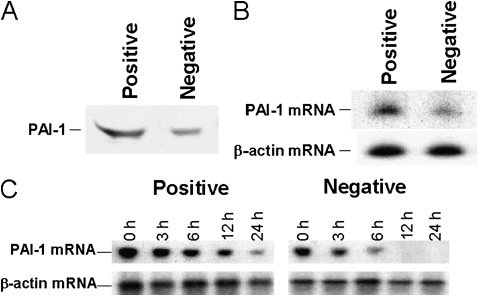
We next directly tested the effect of the PAI-1 mRNABp binding to the 33-nt PAI-1 mRNA 3′UTR sequence on cellular PAI-1 expression. Stable MeT5A cells expressing chimeric β-globin–PAI-1 mRNA containing the 33-nt PAI-1 mRNABp-binding (positive) or a control 33-nt non–PAI-1 mRNABp-binding (negative) sequence were switched to serum-free media for 12 hours to competitively inhibit PAI-1 mRNABp binding to endogenous PAI-1 mRNA. The CM was analyzed for PAI-1 expression. Overexpression of MeT5A cells with chimeric β-globin–PAI-1 mRNA containing the 33-nt PAI-1 mRNABp-binding but not the 33-nt control nonbinding sequence induced PAI-1 expression (Figure 6A). Similar results were obtained in the analysis of PAI-1 mRNA in MeT5A cells overexpressing the PAI-1 mRNABp binding or control sequence (Figure 6B). These results indicate that the interaction of the 60-kD PAI-1 mRNABp with the 33-nt PAI-1 mRNA 3′UTR binding sequence down-regulates cellular PAI-1 expression and that the interaction is subject to competition. This response is the anticipated effect of competition of the expressed binding sequence with the endogenous binding sequence, resulting in increased PAI-1 mRNA stability and increased PAI-1 expression.
Figure 6.
Effect of PAI-1 mRNABp binding sequence overexpression on PAI-1. (A) Stable MeT5A cells overexpressing chimeric β-globin–PAI-1 mRNA containing the 33-nt PAI-1 mRNABp binding PAI-1 3′UTR sequence or nonbinding 33-nt CDR sequence were grown to confluence. The cells were switched to serum-free media overnight, and the CM was analyzed for PAI-1 expression by Western blotting using anti–PAI-1 antibody. (B) Total RNA isolated from MeT5A cells overexpressing the PAI-1 mRNABp binding or control sequence was analyzed for PAI-1 mRNA expression by RT-PCR using 32P-labeled deoxycytidine triphosphate (dCTP) in the reaction mixture as described in Figure 2. RNA from the same samples was later analyzed for β-actin mRNA. (C) The MeT5A cells expressing the PAI-1 mRNABp binding or nonbinding control PAI-1 mRNA sequences were treated with DRB to inhibit ongoing transcription and the level of endogenous PAI-1 mRNA was determined by RT-PCR of total RNA collected at different time points. PAI−1 mRNA levels were analyzed by RT-PCR using 32P-labeled dCTP in the reaction mixture. The same sample was analyzed for β-actin mRNA.
Based on these observations, we sought to confirm the role of the PAI-1 mRNABp in the post-transcriptional regulation of PAI-1 mRNA stability. MeT5A cells overexpressing chimeric β-globin–PAI-1 mRNA containing the 33-nt PAI-1 mRNABp binding sequence or control non–PAI-1 mRNABp binding sequence were treated with DRB to inhibit ongoing transcription, and the decay of endogenous PAI-1 mRNA was determined by RT-PCR or Northern blotting. Overexpression of the 33-nt PAI-1 mRNABp binding sequence in MeT5A cells slowed the decay of endogenous PAI-1 mRNA (Figure 6C). Overexpression of the nonbinding control sequence in MeT5A cells failed to slow PAI-1 mRNA degradation. These results indicate that competitive inhibition of the binding of PAI-1 mRNABp to the PAI-1 mRNA 3′UTR promotes PAI-1 protein expression through the induction of PAI-1 mRNA stability.
Purification of the PAI-1 mRNABp and Determination of Its Ability to Bind to the 33-nt PAI-1 3′UTR Sequence
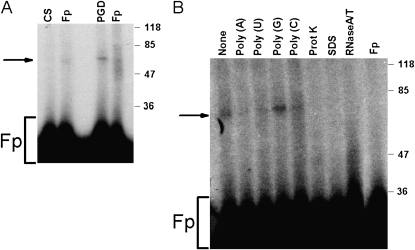
Isolation of the PAI-1 mRNABp was next accomplished using combined conventional chromatography and a PAI-1 mRNA affinity column, as described in Materials and Methods. This purification strategy yielded an enriched preparation that contained two specific bands with approximate molecular weights of 45 and 55 kD. Mass spectrophotometric analysis of isolated polypeptides and database analyses indicated high homology of these bands to CS and 6-PGD, respectively. To determine whether CS or 6-PGD specifically interacts with the 33-nt PAI-1 mRNA 3′UTR, we initially tested proteins expressed in E. coli for PAI-1 mRNA binding activity by UV cross-linking assay. CS and 6-PGD expressed in a prokaryotic system failed to interact with PAI-1 mRNA. We suspected that post-translational modification of proteins is required for PAI-1 mRNA 3′UTR binding. Therefore, we tested the ability of commercially available porcine CS and yeast 6-PGD to bind the PAI-1 3′UTR by UV cross-linking assay using full-length PAI-1 3′UTR and the 33-nt PAI-1 mRNABp binding sequence. UV cross-linking analyses indicated that only 6-PGD binds to the full-length and the 33-nt PAI-1 mRNA 3′UTR (Figure 7A). The specificity of 6-PGD binding to the full-length PAI-1 3′UTR was confirmed by incubation of 6-PGD in the presence of a 200-fold molar excess of cold sense transcript corresponding to the full-length 3′UTR or the 33-nt PAI-1 mRNABp binding sequence. Both transcripts inhibited binding to 32P-labeled PAI-1 3′UTR mRNA. The presence of a molar excess of unlabeled poly (A), poly (C), and poly (U) but not poly (G) inhibited the interaction of 6-PGD with the 32P-labeled 33-nt PAI-1 mRNA 3′UTR. Similarly, pretreatment of 6-PGD with SDS or predigestion of the 32P-labeled 33-nt PAI-1 mRNA transcript with RNase T1 abolished its binding (Figure 7B), confirming the specificity of the interaction.
Figure 7.
Binding of 6-phosphogluconate dehydrogenase (6-PGD) to the 3′UTR PAI-1 mRNA. (A) Porcine citrate (CS) and yeast-derived 6-PGD (10 μg) were incubated with the 32P-labeled PAI-1 mRNA 3′UTR in the presence of tRNA. The reaction mixtures were later digested with RNase T1 and heparin as described in Materials and Methods to avoid nonspecific interaction. After heparin digestion, the reaction mixtures were UV irradiated at 4°C. The immobilized RNA–protein complexes were separated by SDS-PAGE, dried, and autoradiographed. Fp = free probe. (B) Specificity of the yeast 6-PGD–PAI-1 mRNA 3′UTR interaction. Yeast-derived 6-PGD was incubated with a molar excess of unlabeled transcript or homopoly (A), poly (U), poly (G), poly (C) ribonucleotides, or proteinase K or SDS before exposure to 32P-labeled transcript. The reaction mixtures were later digested with RNase T1 and heparin and subjected to UV cross-linking assay as described in (A).
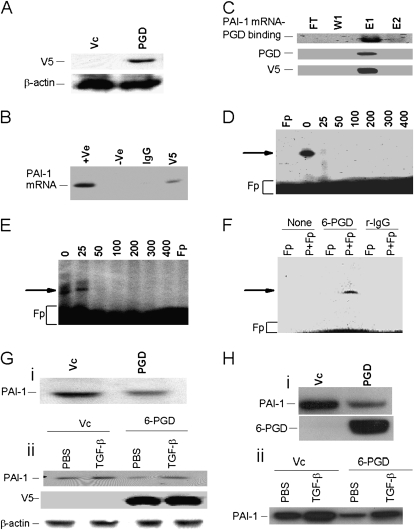
To independently confirm that 6-PGD binds to PAI-1 mRNA 3′UTR, we subcloned 6-PGD cDNA in pcDNA3.1 and transfected lung carcinoma H157 cells by lipofection. We chose to use H157 cells in these analyses because they likewise express PAI-1 mRNA and the 60-kD PAI-1 mRNABp (19), as do MeT5A and selected mesothelioma cells. H157 proliferate considerably faster than Met5A cells and express recombinant protein more efficiently, representing critical advantages for their use in these specific analyses. 6-PGD cDNA was therefore transfected in these cells to express recombinant 6-PGD protein and to determine its ability to bind PAI-1 mRNA. The stable cell lines were selected by antibiotic selection. Cell lysates from 6-PGD and vector cDNA-transfected stable cell lines were initially tested for expression of recombinant fusion protein by Western blotting using anti-V5 antibody. Only cells transfected with 6-PGD cDNA expressed V5 antigen, indicating the expression of recombinant 6-PGD (Figure 8A). To confirm that recombinant 6-PGD interacts with PAI-1 mRNA, we sequentially immunoprecipitated lysates from H157 cells transfected with 6-PGD cDNA with nonspecific mouse IgG and anti-V5 monoclonal antibody. Total RNA isolated from mIgG and anti–V5-immune complexes were analyzed for PAI-1 mRNA by RT-PCR. PAI-1 mRNA coprecipitated with immunoprecipitated anti–V5-immune complex (Figure 8B). No PAI-1 mRNA transcript was detected in the mIgG immune complex. We next purified the histidine-tagged recombinant 6-PGD from the lysates of 6-PGD–transfected H157 cells using a nickel affinity column. Recombinant 6-PGD (r6-PGD) bound to the nickel column and eluted in a single fraction (Figure 8C). Further analysis of the nickel column eluate confirmed PAI-1 mRNA binding activity by UV–cross-linking assay. The same fraction was positive for anti–6-PGD and V5 antibodies by immunoblotting. To further confirm the specificity of r6-PGD binding to the PAI-1 mRNA 3′UTR, we incubated 32P-labeled PAI-1 mRNA 33-nt binding sequence with r6-PGD in the presence of a molar excess of unlabeled 33-nt PAI-1 mRNABp binding sequence and the PAI-1 full-length 3′UTR. The full-length PAI-1 mRNA 3′UTR sequence abolished r6-PGD binding to the PAI-1 mRNA 33-nt 6-PGD 3′UTR binding sequence (Figure 8D). The unlabeled 33-nt PAI-1 mRNABp binding sequence also competitively abolished the binding interaction (Figure 8E). These results demonstrate the specificity of the interaction of 6-PGD with the PAI-1 mRNA 3′UTR.
Figure 8.
Binding of recombinant 6-PGD (r6-PGD) to the 3′UTR PAI-1 mRNA. (A) r6-PGD was expressed in H157 cells. Total lysates from H157 cells transfected with empty pcDNA 3.1 vector (Vc) or 6-PGD cDNA were analyzed for expression of V5 fusion epitope by Western blotting using anti-V5 antibody. (B) Immunoprecipitation of PAI-1 mRNA with r6-PGD. Cell lysates from stable H157 cells transfected with 6-PGD cDNA were sequentially immunoprecipitated with nonspecific mouse IgG and anti-V5 antibodies. Total RNA was isolated from the immune complex, and PAI-1 mRNA was analyzed by RT-PCR in the presence of 32P-labeled dCTP. The PCR reaction containing a PAI-1 cDNA template was used as a positive (+Ve) control, and the reaction lacking template was used as negative (−Ve) control. (C) Purification of r6-PGD. Total lysates from stable H157 cells were passed through a nickel column, after which flow through (FT), wash (W1), and eluates (E1 and E2) were (5 μg) subjected to UV cross-linking (PAI-1 mRNA PGD binding) as described previously to assess the PAI-1 mRNA binding activities of the purified fractions. The same fractions were subjected immunoblotting using anti–6-PGD and anti-V5 antibodies respectively. Specificity of the r6-PGD–PAI-1 mRNA 3′UTR interaction. r6-PGD was incubated with 0- to 400-fold molar excess of unlabeled transcript corresponding to the unlabeled full-length PAI-1 mRNA 3′UTR (D) or 33-nt 6-PGD binding sequence (E) before exposure to 32P-labeled transcript corresponding to the 33-nt PAI-1 mRNABp binding sequence. The reaction mixtures were later digested with RNase T1 and heparin and subjected to UV cross-linking assay as described in Figure 7A. (F) Demonstration of the 6-PGD interaction with PAI-1 mRNA in Met5A cells. Cytosolic extracts (100 μg) were incubated with the 32P-labeled 33-nt 6-PGD binding sequence of PAI-1 mRNA 3′UTR in the presence of tRNA. The reaction mixtures were later digested with RNase T1 and heparin to avoid nonspecific interaction. After UV irradiation of the reaction mixtures at 4°C, the immobilized RNA–protein complexes were immunoprecipitated with protein AG agarose alone (none) or with protein AG agarose conjugated with anti–6-PGD polyclonal antibody (6-PGD) or rabbit IgG (r-IgG). Agarose beads were washed three times with lysis buffer and separated by SDS-PAGE, dried, and autoradiographed. Fp = free probe. (Gi) Stable Met5A cells expressing empty vector (Vc) or 6-PGD cDNA in pcDNA3.1 were switched to serum-free RPMI media overnight. The conditioned media was then analyzed for PAI-1 expression by Western blotting. (Gii) Stable Met5A cells expressing vector or 6-PGD cDNA as described in Figure 8Gi were treated with PBS or TGF-β for 24 hours. The conditioned media was analyzed for PAI-1, and the cell lysates were tested for V5-epitope and β-actin by Western blotting. (Hi) Primary rabbit mesothelial cells cultured in 60-mm dishes were transduced with empty pacAd5CMV vector (Vc) or the same vector carrying 6-PGD cDNA. After 48 hours, the conditioned media were analyzed for PAI-1 expression by Western blotting. (Hii) Primary rabbit mesothelial cells transduced with adenovirus carrying empty vector or 6-PGD cDNA as described in (Hi) were treated with PBS or TGF-β and the conditioned media was analyzed for PAI-1 by Western blotting.
To confirm that the protein from the Met5A lysates that binds to PAI-1 mRNA is 6-PGD, we incubated Met5A lysates with the 32P-labeled 33-nt PAI-1 3′UTR binding sequence after sequential RNase T1, heparin digestion, and UV cross-linking. We subjected the reaction mixtures to immunoprecipitation using anti–6-PGD polyclonal antibody or rabbit IgG. The immune complexes were resolved on SDS-PAGE and autoradiography. Immunoprecipitation of 6-PGD antibody but not rabbit IgG showed the presence of radioactive protein and PAI-1 mRNA complex, indicating that 6-PGD binds to the 33-nt PAI-1 mRNA 3′UTR (Figure 8F).
We next expressed 6-PGD in Met5A cells and analyzed the effects on PAI-1 expression by Western blotting. Stable cells expressing 6-PGD but not vector cDNA reduced PAI-1 expression (Figure 8Gi). To determine how 6-PGD regulates TGF-β–induced PAI-1 expression, we treated Met5A cells transfected with 6-PGD cDNA with PBS or TGF-β for 24 hours. Conditioned media were analyzed for PAI-1 expression. TGF-β induced PAI-1 expression in vector and 6-PGD cDNA transfected cells; however, PAI-1 expression in Met5A cells overexpressing 6-PGD was at least 50% less than that of vector cDNA transfected cell (Figure 8Gii). To confirm that 6-PGD regulates PAI-1 expression in primary mesothelial cells, we transduced primary rabbit mesothelial cells with adenovirus expressing 6-PGD and used adenovirus expressing empty vector (Ad5CMV) as a control. The conditioned media were analyzed for PAI-1 expression, and lysates were tested for 6-PGD expression. The cells expressing 6-PGD but not empty vector reduced PAI-1 expression by greater than 50% (Figure 8Hi). Consistent with the findings in Met5A cells, 6-PGD overexpression in rabbit mesothelial cells blunted TGF-β–induced PAI-1 expression.
DISCUSSION
In this report, we extend previous studies in which we examined how components of the fibrinolytic system are regulated at the post-transcriptional level of mRNA stability. We previously showed that the urokinase receptor uPAR is regulated at this level in pleural mesothelial, malignant mesothelioma, and lung epithelial cells (24). We also showed that the regulation of uPA and uPAR at the post-transcriptional level involves cis–trans interactions of novel mRNA binding proteins with specific sequences within the respective mRNAs. Binding proteins were identified and characterized, which either increased or decreased the stability of the target mRNA (26, 27). These studies provide proof of the principle that the fibrinolytic system is subject to post-transcriptional regulation and raised the possibility that PAI-1 expression is regulated at this level in pleural mesothelial cells. We used MeT5A cells as a primary model system to address this possibility and elucidate the regulatory mechanism.
In this study, we found that TGF-β induces expression of PAI-1 in MeT5A cells. Because PAI-1 is a secreted protein, we found the majority of PAI-1 protein in the conditioned media where changes in PAI-1 expression were best appreciated. Unlike conditioned media, cell lysates contain PAI-1 bound to uPAR and uPA in a trimeric complex that can undergo internalization and degradation. In addition, TGF-β and TNF-α may differentially induce uPA, uPAR, and PAI-1 transcription and expression in different cell lines (18, 28, 29), which likely accounts for the lack of detectable changes in PAI-1 expression in Met5A or MS-1 cell lysates as assessed by Western blotting. We also found that PAI-1 is regulated at the level of mRNA stability in these cells and that human malignant pleural mesothelioma cells likewise express a PAI-1 mRNABp that could regulate PAI-1 expression in a similar fashion. TGF-β and PAI-1 are elaborated by human malignant mesothelioma cells, and both are implicated in the pathogenesis of this neoplasm (30, 31). This newly recognized pathway provides a versatile regulatory system through which TGF-β could regulate PAI-1 mRNA and protein expression, and we speculate that this pathway could operate in the context of pleural inflammation or neoplasia. Supporting this inference, we recently reported that uPAR expression is altered in part through post-transcriptional regulation via the uPAR mRNABps PGK and hnRNPC in acute lung injury induced by LPS in mice (32). This report raises the possibility that the post-transcriptional control of PAI-1 we describe herein could be operative in vivo. We plan to pursue this possibility in the context of pleural inflammation in a follow-up to this report.
In studies reported by other groups, PAI-1 was previously reported to be regulated at the post-transcriptional level in rat hepatoma and endothelial cells (33) by PMA, insulin, IGF, and cyclic nucleotide analogs (33–36). We previously found that post-transcriptional regulation of PAI-1 occurs in lung cancer-derived cell lines, including the H157 line, and in nonmalignant lung epithelial cells (19). In this study, we found that PAI-1 expression in lung cancer cells was inversely correlated with the association of a similar 60-kD PAI-1 mRNABp with the PAI-1 3′UTR. Our present data suggest that this same protein is expressed by pleural mesothelial and malignant mesothelioma cells and provide a strong justification to examine the regulatory pathway in the MeT5A and H157 model systems. Although the regulation of PAI-1 in mesothelial or other cells can be regulated at the transcriptional level by acidosis, hypoxia, fibrin derivatives, or TGF-β (18, 28, 29, 37), our findings suggest that the effects could involve post-transcriptional control and demonstrate that the underlying mechanism regulating PAI-1 expression at the level of mRNA stability involves the interaction of a newly described PAI-1 mRNABp with a specific sequence we localized to the PAI-1 mRNA 3′UTR. Because proinflammatory cytokines such as TNF-α and TGF-β induce PAI-1 mRNA synthesis, the observed differences between PAI-1 expression and the PAI-1 mRNA–PAI-1 mRNABp interaction in the different cell types or those stimulated by the cytokines we used could be attributable to alternate regulation at the transcriptional level (4).
We identified a 60-kD regulatory PAI-1 mRNA binding protein in MeT5A cells based on its ability to form a specific mRNA–protein complex with a 33-nt PAI-1 mRNA cis element that is located in the 3′UTR of PAI-1 mRNA. N-terminal amino acid sequence data identified the candidate binding protein as 6-GPD oxidoreductase (E.C.1.1.1.44). We expressed 6-PGD and analyzed its binding properties. Our results demonstrate that the 6-PGD specifically binds to a 33-nt PAI-1 mRNA 3′UTR. Another candidate binding moiety, CS, was not found to bind the 33-nt sequence, and rigorous analyses confirmed that the 60-kD PAI-1 mRNABp is 6-PGD. The analyses were conducted using recombinant preparations of 6-PGD expressed in prokaryotic and eukaryotic expression systems, adding to the power of the approach. The destabilizing effect of the interaction of 6-PGD with the target sequence was confirmed by independent analyses in three model systems, in which MeT5A, primary rabbit mesothelial cells, or H157 cells were used and different confirmatory strategies were applied to strengthen the conclusions. We found that r6-PGD specifically binds to a 33-nt sequence that contains A (7nt), U (10 nt), C (6 nt), and G (10 nt). These findings suggest that regulation of PAI-1 at the level of mRNA in mesothelial and malignant mesothelioma or lung epithelial cells exhibits specificity because a number of PAI-1 mRNA binding proteins have previously been described in rat hepatocytes (35, 38, 39). Our data extend the finding of that study and show that 6-PGD, the dominant PAI-1 mRNABp we found by gel shift and UV–cross-linking analyses in MeT5A, human mesothelioma, or lung epithelial cells, interacts with a specific 33-nt PAI-1 mRNA binding sequence and destabilizes PAI-1 mRNA and that the machinery is found in H157 as well as MeT5A and primary pleural mesothelial cells.
We used chimeric gene analyses to confirm that the interaction of 6-PGD with the 33-nt PAI-1 mRNA 3′UTR binding sequence regulates PAI-1 mRNA stability. Along these lines, we found that insertion of 33-nt 6-PGD binding sequences into β-globin mRNA destabilized the chimeric message in MeT5A cells, indicating that the 6-PGD binding sequence contains information for message instability. Inhibition of 6-PGD binding to the 33-nt PAI-1 mRNA 3′UTR sequence by TGF-β treatment of MeT5A cells and induction of PAI-1 expression by competitive inhibition of 6-PGD binding to the 33-nt PAI-1 mRNA 3′UTR but not a nonbinding coding region control sequence clearly indicates that 6-PGD negatively regulates PAI-1 expression. Overexpression of 6-PGD in Met5A cells and primary mesothelial cells suppressed basal and TGF-β–induced PAI-1 expression. In addition, induction of PAI-1 mRNA by stabilization of endogenous transcript clearly indicates the existence of a novel mode of regulation of PAI-1 expression by 6-PGD.
6-PGD is an important enzyme of the pentose phosphate metabolic pathway. The enzyme catalyzes the oxidative decarboxylation of 6-phosphogluconate to ribulose-5-phosphate and CO2 with a concomitant reduction of NADP+ to NADPH. In the case of NADPH deficiency, the concentration of reduced glutathione in living systems declines, resulting in cell death. For this reason, 6-PGD is defined as an antioxidant enzyme (40, 41). Like other dehydrogenases, this molecule possesses a nucleotide binding domain, which may account for its ability to bind a specific RNA sequence. Our data demonstrate that 6-PGD binds PAI-1 mRNA and thereby regulates expression of PAI-1 at the level of mRNA stability. This pleiotropism has a precedent that we recently reported in the context of the regulation of the urokinase receptor or uPAR. We reported that uPAR expression is regulated in a number of different cell types, including mesothelial cells, by destabilization of uPAR mRNA by phosphoglycerate kinase (PGK) (26). Like 6-PGD, PGK plays a key alternative role in cell metabolism. PGK is a key glycolytic enzyme that catalyzes the reversible conversion of 1, 3-diphosphoglycerate to 3-phosphoglycerate. Our findings extend our understanding about the regulation of PAI-1 expression at the post-transcriptional level and support the concept that the regulation of the fibrinolytic system involves the newly recognized contribution of mRNA binding proteins that otherwise may play key roles in the regulation of cellular metabolism.
In summary, we demonstrate that the 6-PGD 33-nt PAI-1 mRNA 3′UTR interaction regulates PAI-1 mRNA stability. TGF-β stimulates expression of PAI-1 by MeT5A cells in culture by stabilization of PAI-1 mRNA. The TGF-β–induced stabilization of PAI-1 mRNA involves altered 6-PGD interaction with PAI-1 mRNA 3′UTR. If operative in vivo, this pathway could contribute to the relative local overexpression of PAI-1 and the paucity of pleural fibrinolytic capacity associated with inflammatory pleural diseases (8, 9). This newly identified pathway is, to our knowledge, the first description of the ability of 6-PGD to regulate the expression of PAI-1 in any cell type.
Acknowledgments
The authors thank Andrea Hannos for technical support.
This work was supported by National Heart, Lung, and Blood Institute Grants P01HL-076406 and R01 HL071147.
Originally Published in Press as DOI: 10.1165/rcmb.2009-0046OC on October 23, 2009
Author Disclosure: S.I. received lecture fees from Brahms for $5,001 to $10,000 along with an industry-sponsored grant from Attenuon, LLC for $10,001 to $50,000. He has received consultancy fees from the National Institutes of Health for less than $1,000 and a sponsored grant for more than $100,001. He has received a sponsored grant from FAMRI for more than $100,001, and book fees for less than $1,000 from Hodder Arnold Publishers and Wolters Kluwer Publishers and $1,001 to $5,000 from Williams and Wilkens Publishers. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Dano K, Andreasen PA, Grondahl-Hansen J, Kristensen P, Nielsen LS, Skriver L. Plasminogen activators, tissue degradation, and cancer. Adv Cancer Res 1985;44:139–266. [DOI] [PubMed] [Google Scholar]
- 2.Liotta LA, Stetler-Stevenson WG, Steeg PS. Cancer invasion and metastasis: positive and negative regulatory elements. Cancer Invest 1991;9:543–551. [DOI] [PubMed] [Google Scholar]
- 3.Mignatti P, Rifkin DB. Biology and biochemistry of proteinases in tumor invasion. Physiol Rev 1993;73:161–195. [DOI] [PubMed] [Google Scholar]
- 4.Sprengers ED, Kluft C. Plasminogen activator inhibitors. Blood 1987;69:381–387. [PubMed] [Google Scholar]
- 5.Mazar AP, Henkin J, Goldfarb RH. The urokinase plasminogen activator system in cancer: implications for tumor angiogenesis and metastasis. Angiogenesis 2000;3:15–32. [DOI] [PubMed] [Google Scholar]
- 6.Idell S, James KK, Levin EG, Schwartz BS, Manchanda N, Maunder RJ, Martin TR, McLarty J, Fair DS. Local abnormalities in coagulation and fibrinolytic pathways predispose to alveolar fibrin deposition in the adult respiratory distress syndrome. J Clin Invest 1989;84:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertozzi P, Astedt B, Zenzius L, Lynch K, LeMaire F, Zapol W, Chapman HA Jr. Depressed bronchoalveolar urokinase activity in patients with adult respiratory distress syndrome. N Engl J Med 1990;322:890–897. (see comments). [DOI] [PubMed] [Google Scholar]
- 8.Idell S, Girard W, Koenig KB, McLarty J, Fair DS. Abnormalities of pathways of fibrin turnover in the human pleural space. Am Rev Respir Dis 1991;144:187–194. [DOI] [PubMed] [Google Scholar]
- 9.Idell S, Allen T, Chen S, Koenig KB, Mazar A, Azghani A. Intrapleural activation, processing, efficacy, and duration of protection of single-chain urokinase in evolving tetracycline-induced pleural injury in rabbits. Am J Physiol Lung Cell Mol Physiol 2007;292:L25–L32. [DOI] [PubMed] [Google Scholar]
- 10.Philip-Joet C, Alessi M-C, Philip-Joet C, Aillaud M, Barriere J-R, Arnaud A, Juhan-Vague I. Fibrinolytic and inflammatory processes in pleural effusions. Eur Respir J 1995;8:1352–1356. [DOI] [PubMed] [Google Scholar]
- 11.Idell S, Pueblitz S, Emri S, Gungen Y, Gray L, Kumar A, Holiday D, Koenig KB, Johnson AR. Regulation of fibrin deposition by malignant mesothelioma. Am J Pathol 1995;147:1318–1329. [PMC free article] [PubMed] [Google Scholar]
- 12.Damjanovich T, Turzó C, Adány R. Factors involved in the plasminogen activation system in human breast tumours. Thromb Haemost 1994;71:684–691. [PubMed] [Google Scholar]
- 13.Emeis JJ, Kooistra T. Interleukin 1 and lipopolysaccharide induce an inhibitor of tissue-type plasminogen activator in vivo and in cultured endothelial cells. J Exp Med 1986;163:1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawdey M, Podor TJ, Luskutoff DJ. Regulation of type 1 plasminogen activator inhibitor gene expression in cultured bovine aortic endothelial cells: induction by transforming growth factor-beta, lipopolysaccharide, and tumor necrosis factor-alpha. J Biol Chem 1989;264:10396–10401. [PubMed] [Google Scholar]
- 15.Andreasen PA, Pyke C, Riccio A, Kristensen P, Nielsen LS, Lund LR, Blasi F, Dano K. Plasminogen activator inhibitor type 1 biosynthesis and mRNA level are increased by dexamethasone in human fibrosarcoma cells. Mol Cell Biol 1987;7:3021–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kooistra T, Bosma PJ, Tons HAM, Van den Berg AP, Meyer P, Princen HMG. Plasminogen activator inhibitor 1: biosynthesis and mRNA level are increased by insulin in cultured human hepatocytes. Thromb Haemost 1989;62:723–728. [PubMed] [Google Scholar]
- 17.Mayer M, Lund LR, Riccio A, Skouv J, Nielsen LS, Stacey SN, Dano K, Andreasen PA. Plasminogen activator inhibitor type-1 protein, mRNA and gene transcription are increased by phorbol esters in human rhabdomyosarcoma cells. J Biol Chem 1988;263:15688–15693. [PubMed] [Google Scholar]
- 18.Bergstrom M, Falk P, Holmdahl L. Effect of acidosis on expression of mesothelial cell plasminogen activator inhibitor type-1. Surg Endosc 2006;20:1448–1452. [DOI] [PubMed] [Google Scholar]
- 19.Shetty S, Idell S. Posttranscriptional regulation of plasminogen activator inhibitor-1 in human lung carcinoma cells in vitro. Am J Physiol Lung Cell Mol Physiol 2000;278:L148–L156. [DOI] [PubMed] [Google Scholar]
- 20.Rougier JP, Guia S, Hagege J, Nguyen G, Ronco PM. PAI-1 secretion and matrix deposition in human peritoneal mesothelial cell cultures: transcriptional regulation by TGF-beta 1. Kidney Int 1998;54:87–98. [DOI] [PubMed] [Google Scholar]
- 21.Idell S, Zwieb C, Kumar A, Koenig KB, Johnson AR. Pathways of fibrin turnover of human pleural mesothelial cells in vitro. Am J Respir Cell Mol Biol 1992;7:414–426. [DOI] [PubMed] [Google Scholar]
- 22.Holmdahl L, Ivarsson ML. The role of cytokines, coagulation, and fibrinolysis in peritoneal tissue repair. Eur J Surg 2002;165:1012–1019. [DOI] [PubMed] [Google Scholar]
- 23.Shetty S, Idell S. A urokinase receptor mRNA binding protein from rabbit lung fibroblasts and mesothelial cells. Am J Physiol 1998;274:L871–L882. [DOI] [PubMed] [Google Scholar]
- 24.Idell S. Endothelium and disordered fibrin turnover in the injured lung: newly recognized pathways. Crit Care Med 2002;30:S274–S280. [DOI] [PubMed] [Google Scholar]
- 25.Tucker TA, Dean C, Komissarov A, Koenig K, Mazar A, Pendurthi U, Allen TC, Idell S. The urokinase receptor supports tumorigenesis of human malignant pleural mesothelioma cells. Am J Respir Cell Mol Biol (In press) [DOI] [PMC free article] [PubMed]
- 26.Shetty S, Idell S. Urokinase induces expression of its own receptor in Beas2B lung epithelial cells. J Biol Chem 2001;276:24549–24556. [DOI] [PubMed] [Google Scholar]
- 27.Shetty S, Muniyappa H, Halady PKS, Idell S. Regulation of urokinase receptor expression by phosphglycerate kinase. Am J Respir Cell Mol Biol 2004;31:100–106. [DOI] [PubMed] [Google Scholar]
- 28.Olman MA, Hagood JS, Simmons WL, Fuller GM, Vinson C, White KE. Fibrin fragment induction of plasminogen activator inhibitor transcription is mediated by activator protein-1 through a highly conserved element. Blood 1999;94:2029–2038. [PubMed] [Google Scholar]
- 29.Kutz SM, Hordines J, McKeown-Longo PJ, Higgins PJ. TGF-beta1-induced PAI-1 gene expression requires MEK activity and cell-to-substrate adhesion. J Cell Sci 2001;114:3905–3914. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki E, Kim S, Cheung HK, Corbley MJ, Zhang X, Sun L, Shan F, Singh J, Lee W-C, Albelda SM, et al. A novel small-molecule inhibitor of transforming growth factor beta type I receptor kinase (SM16) inhibits murine mesothelioma tumor growth in vivo and prevents tumor recurrence after surgical resection. Cancer Res 2007;67:2351–2359. [DOI] [PubMed] [Google Scholar]
- 31.Kumar-Singh S, Weyler J, Martin MJ, Vermeulen PB, Van Marck E. Angiogenic cytokines in mesothelioma: a study of VEGF, FGF-1 and -2, and TGF beta expression. J Pathol 1999;189:72–78. [DOI] [PubMed] [Google Scholar]
- 32.Bhandary YP, Velusamy T, Shetty PK, Shetty RS, Idell S, Cines DB, Jain D, Bdeir K, Abraham E, Tsuruta Y, et al. Posttranscriptional regulation of uPAR expression in lipopolysaccharide-induced acute lung injury. Am J Respir Crit Care Med (In press) [DOI] [PMC free article] [PubMed]
- 33.Fattal PG, Schneider DJ, Sobel BE, Billadello JJ. Post-transcriptional regulation of expression of plasminogen activator inhibitor type 1 mRNA by insulin and insulin-like growth factor 1. J Biol Chem 1992;267:12412–12415. [PubMed] [Google Scholar]
- 34.Bosma PJ, Kooistra T. Different induction of two plasminogen activator inhibitor 1 mRNA species by phorbol ester in human hepatoma cells. J Biol Chem 1991;266:17845–17849. [PubMed] [Google Scholar]
- 35.Heaton JH, Tilmann-Bogush M, Leff NS, Gelehrter TD. Cyclic nucleotide regulation of type-1 plasminogen activator-inhibitor mRNA stability in rat hepatoma cells: identification of cis-acting sequences. J Biol Chem 1998;273:14261–14268. [DOI] [PubMed] [Google Scholar]
- 36.Tilmann-Bogush M, Heaton JH, Gelehrter TD. Cyclic nucleotide regulation of PAI-1 mRNA stability: identification of cytosolic proteins that interact with an a-rich sequence. J Biol Chem 1999;274:1172–1179. [DOI] [PubMed] [Google Scholar]
- 37.Kietzmann T, Samoylenko A, Roth U, Jungermann K. Hypoxia-inducible factor-1 and hypoxia response elements mediate the induction of plasminogen activator inhibitor-1 gene expression by insulin in primary rat hepatocytes. Blood 2003;101:907–914. [DOI] [PubMed] [Google Scholar]
- 38.Heaton JH, Dlakic WM, Dlakic M, Gelehrter TD. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the type-1 plasminogen activator inhibitor mRNA. J Biol Chem 2001;276:3341–3347. [DOI] [PubMed] [Google Scholar]
- 39.Hirashima Y, Kobayashi H, Suzuki M, Tanaka Y, Kanayama N, Terao T. Transforming growth factor-{beta}1 produced by ovarian cancer cell line HRA stimulates attachment and invasion through an up-regulation of plasminogen activator inhibitor type-1 in human peritoneal mesothelial cells. J Biol Chem 2003;278:26793–26802. [DOI] [PubMed] [Google Scholar]
- 40.Kozar RA, Weibel CJ, Cipolla J, Klein AJP, Haber MM, Abedin MZ, Trooskin S. Antioxidant enzymes are induced during recovery from acute lung injury. Crit Care Med 2000;28:2486–2491. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava SK, Beutler E. Gluthatione metabolism of erythrocyte: the enzymic cleavage of glutathione-haemoglobin preperations by glutathione reductase. Biochem J 1989;119:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]