Abstract
Smokers are more susceptible to respiratory viral infections, including influenza virus, but the mechanisms mediating this effect are unknown. To determine how epithelial cells contribute to the enhanced susceptibility seen in smokers, we established an in vitro model of differentiated nasal epithelial cells (NECs) from smokers, which showed enhanced mucin expression. The NECs from smokers responded to influenza infection with greater cytotoxicity, release of interleukin-6, and viral shedding than NECs from nonsmokers. Focusing on type I interferon (IFN) expression, we observed that influenza-infected NECs from smokers produced significantly less IFN-α than NECs from nonsmokers. Similarly, the expression of IRF7, a key transcription factor controlling the expression of IFN-α, was significantly decreased in influenza-infected and IFN-β–stimulated NECs from smokers. Furthermore, our data indicate that the DNA methylation of the IRF7 gene and expression of the DNA (cytosine-5-)-methyltransferase 1 was enhanced in NECs from smokers. To confirm these findings in vivo, we initiated a study in which smoking and nonsmoking healthy volunteers were inoculated nasally with the live-attenuated influenza virus (LAIV) vaccine, and nasal biopsies were obtained before and after the administration of LAIV. The LAIV-induced expression of IRF7 was lower in the nasal epithelium from smokers, supporting our in vitro observations. These data demonstrate that infection with influenza results in the reduced expression of transcription factor IRF7 in NECs from smokers, and that these effects may be mediated by an epigenetic modification of the IRF7 gene, thus providing a potential mechanism rendering smokers more susceptible to respiratory virus infections.
Keywords: influenza, IRF7, cigarette smoke, nasal epithelium
CLINICAL RELEVANCE.
This study describes potential cellular mechanisms for the suppressed antiviral defense responses seen in individuals chronically exposed (either actively or passively) to cigarette smoke. The data also suggest that epigenetic modifications at the level of the respiratory epithelium could contribute to modified immune responses in smokers.
Viral infections of the respiratory tract are a major cause of morbidity and mortality, especially in children and the elderly (1, 2). In the United States alone, more than 20,000 people die and more than 100,000 are hospitalized every year because of influenza virus-related diseases. Despite large-scale vaccination efforts and antiviral therapies, the morbidity and mortality associated with influenza have not significantly changed recently (3, 4). Epidemiologic studies show that smokers are more susceptible to influenza virus infections than nonsmokers (5–7), but the mechanisms mediating these effects are not known. Laboratory animal studies and human epidemiologic evidence indicate that adaptive immune responses, as marked by influenza-specific antibody production, are unaffected by chronic cigarette smoke exposure (6, 8, 9). Thus, the effects of cigarette smoke on susceptibility to influenza infection may involve antiviral defense responses of cells local to the airway, such as the host epithelium. Recent evidence suggests that antiviral defense responses, such as virus-induced apoptosis and type I interferon (IFN) signaling, are suppressed in cells exposed acutely to cigarette smoke (10, 11). However, whether and how chronic exposure to cigarette smoke affects antiviral defense responses remain unknown.
Airway epithelial cells are the primary site for influenza virus infection and replication. Virus-infected epithelial cells respond to influenza infection by synthesizing and releasing numerous cytokines, immunoregulatory molecules, and antiviral mediators. Among the mediators released by epithelial cells upon infection with influenza, regulated upon activation, normal T cell expressed and secreted, monocyte chemotactic protein-1, IL-8, IL-6, and eotaxin recruit and activate proinflammatory cells, whereas type I interferons (IFN-α and IFN-β) induce the synthesis and/or activity of mediators involved in turning off viral replication within the host cell. The expression of type I IFNs is controlled by interferon regulatory factors (IRFs) 3 and 7 (12). Specifically, viral infection triggers signaling cascades that culminate in the phosphorylation and activation of preexisting cytosolic IRF3 and IRF7, which stimulate predominantly the production of small amounts of IFN-β. Released IFN-β stimulates the type I IFN receptor (IFNAR) in an autocrine/paracrine fashion, leading to the activation of IFN-stimulated gene factor 3 and the de novo transcription of the IRF7 gene. Newly synthesized IRF7 further amplifies the type I IFN response by inducing the transcription of IFN-β and IFN-α, thus activating a larger “second wave” of type 1 IFN production and a positive feedback loop.
The differentiation of primary human epithelial cells under defined culture conditions in vitro results in a pseudostratified epithelium that emulates many characteristics found in human airway epithelium in vivo (13). We developed an in vitro model of nasal epithelial cells (NECs) obtained from current smokers, which when differentiated in vitro maintain the characteristics found in nasal epithelia in smokers in vivo and therefore provide an important tool for examining potential cellular mechanisms that mediate an enhanced susceptibility to influenza virus infections in smokers. Using this model as well as nasal epithelial biopsies obtained from subjects infected with the live-attenuated influenza virus (LAIV) vaccine, we performed experiments to test the hypothesis that chronic exposure to cigarette smoke modifies the ability of NECs to mount antiviral defense responses.
MATERIALS AND METHODS
Nasal Epithelial Cell Culture and Infection with Influenza
Primary human NECs were obtained from healthy smoking and nonsmoking adult volunteers by gently stroking the inferior surface of the turbinate several times with a Rhino-Probe curette (Arlington Scientific, Arlington, TX), which was inserted through a nasoscope. The selection criteria for subject recruitment were similar to those described previously (14). Briefly, subjects were aged 18 to 40 years, and identified themselves as generally healthy and without a diagnosis for any smoking-related disorder or history of asthma. Smoking status was assessed via questionnaire, and confirmed through urine cotinine analysis (14). All smokers recruited for the study were current smokers. This protocol was approved by the Institutional Review Board for Biomedical Research of the University of North Carolina School of Medicine.
Primary human NECs were expanded to passage 2 in bronchial epithelial growth medium (Cambrex Bioscience Walkersville, Inc., Walkersville, MD), plated on collagen-coated filter supports with a 0.4-μM pore size (Trans-CLR; Costar, Cambridge, MA), and cultured in a 1:1 mixture of bronchial epithelial cell basic medium and Dulbecco's minimal essential medium with SingleQuot supplements (Cambrex), bovine pituitary extracts (13 mg/ml), bovine serum albumin (1.5 μg/ml), and nystatin (20 units). Upon confluence, all-trans retinoic acid was added to the medium, and air–liquid interface (ALI) culture conditions (i.e., removal of the apical medium) were created to promote differentiation. Mucociliary differentiation was achieved 18 to 21 days after ALI.
For in vitro experiments, we used influenza A/Bangkok/1/79 (H3N2 serotype), which was propagated in 10-day-old embryonated hen's eggs. The virus was collected in the allantoic fluid and titered by a 50% tissue-culture infectious dose (TCID50) in Madin-Darby canine kidney cells and hemagglutination as described before (15). Stock virus was aliquoted and stored at −80°C until use. Unless otherwise indicated, for infection, approximately 5 × 105 cells were infected with approximately 128 hemagglutination units of influenza A/Bangkok/1/79, which resulted in approximately 10% of cells being infected with influenza 24 h after infection. Total RNA, basolateral supernatants, and apical washes were collected 24 h after infection.
RT-PCR
Total RNA was extracted using TRizol (Invitrogen, Carlsbad, CA) according to the supplier's instructions. First-strand cDNA synthesis and real-time RT-PCR were performed as described previously (15, 16). The mRNA analyses were performed using commercially available primer and probe sets (inventoried Taqman Gene Expression Assays) purchased from Applied Biosystems (Foster City, CA).
Cytokine and Lactate Dehydrogenase Analysis
Basolateral media were collected 24 hours after infection and analyzed for IFN-α or IL-6, using commercially available ELISA kits according to the supplier's instructions (Pierce, Rockford, IL). Cell viability was assessed by analyzing cell culture supernatants for lactate dehydrogenase (LDH) activity, using a commercially available kit according to the supplier's instructions (CytoTox 96; Promega, Madison, WI).
Influenza Virus Titer
Influenza virus titers in apical washes were determined by a TCID50 in Madin Darby canine kidney (MDCK) cells, and evaluated using the agglutination of red blood cells as an indicator, according to a modified protocol described previously (17). Briefly, MDCK cells grown in round-bottom 96-well plates were inoculated with virus-containing samples diluted in serum-free Eagle's minimum essential medium containing 20 μg/ml trypsin, using log10 dilutions. After 3 days of incubation, a suspension of human erythrocytes (0.5%) was added to each well. Wells exhibiting hemagglutination were considered positive, and virus titers were expressed as the TCID50.
Western Blotting
Whole cell lysates were prepared by lysing the cells in radioimmunoprecipitation assay buffer containing 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitors (Cocktail Set III; Calbiochem, San Diego, CA), as well as phosphatase inhibitors (0.5 mM NaVO4 and 1 mM β-glycerophosphate). Whole cell lysate (100 μg) was separated by SDS-PAGE followed by immunoblotting, using specific antibodies to IRF7, IRF3 (Santa Cruz Biotechnology, Santa Cruz, CA), or β-actin (1:2,000; US Biological, Swampscott, MA), which was used as a loading control. Antigen-antibody complexes were stained with anti-rabbit or anti-mouse, horseradish peroxidase (HRP)-conjugated antibody (1:4,000; Santa Cruz Biotechnology) and SuperSignal West Pico Chemiluminescent Substrate (Pierce). Chemiluminescent signals were acquired using a 16-bit CCD camera (GeneGnome System; Syngene, Frederick, MD), and visualized using GeneSnap software (Syngene). Densitometric quantification was performed using GeneTools analysis software (Syngene).
Analysis of DNA Methylation
Genomic DNA isolated from NECs of smokers and nonsmokers was analyzed for DNA methylation in the promoter site region of the human IRF7gene, using Methyl-Profiler DNA Methylation qPCR Assays according to the supplier's instruction (SABiosciences Corp., Frederick, MD). Briefly, Genomic DNA was isolated using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA), including the recommended removal of potential RNA contamination using RNase. The Methyl-Profiler DNA Methylation qPCR Assay is based on the digestion of unmethylated and methylated DNA, using methylation-sensitive and methylation-dependent restriction enzymes. The remaining DNA after digestion is quantified by real-time RT-PCR, using primers that specifically flank the region of interest, immediately upstream from the transcriptional start site of the IRF7 gene (18). For this analysis, the relative concentrations of differentially methylated DNA (specifically hypermethylated, unmethylated, and intermediary methylated DNA) are determined by comparing the amount of each digest with that of a mock digest. For each sample, data are expressed as the sum of the percent hypermediary and intermediary methylated DNA.
Immunohistochemistry
Cells were fixed with ice-cold acetone for 20 minutes, washed with Tris-buffered saline (TBS), and blocked with Powerblock (Biogenex, San Ramon, CA) for 1 hour at room temperature. Afterward, cells were incubated with primary antibody overnight at 4°C. The antibodies included anti-Muc5B (Millipore, Billerica, MA), anti-acetylated α-tubulin used at 1:800 (Invitrogen), which recognizes cilia, or anti-IRF7 (Santa Cruz Biotechnology). After incubation with the primary antibodies, samples were washed with TBS, followed by incubation with secondary antibodies at 1:200 for 1 hour at room temperature. For confocal microscopy, Alexa 568–conjugated goat anti-mouse and Alexa 488–conjugated goat anti-rabbit antibodies (both from Invitrogen) were used. After incubation with the secondary antibodies, samples were washed with TBS and visualized by confocal microscopy, using a Nikon C1Si, and the images were processed using EZ-C1 FreeViewer software (Nikon, Melville, NY). For the visualization of IRF7 levels, 5-μm sections were placed on Superfrost/plus slides (Fisher Scientific, Pittsburgh, PA) and stained for IRF7, using anti-IRF7 antibodies (Santa Cruz Biotechnology). After incubation with HRP-conjugated secondary antibodies, samples were washed with TBS and evaluated under light microscopy. As a control, sections were also incubated without the primary antibody, to detect nonspecific binding of the HRP-conjugated secondary antibody.
Inoculation of Subjects with LAIV
Healthy smoking and nonsmoking volunteers between ages 18 and 35 years were recruited for this study, and assessed for their smoking status by questionnaire and urine cotinine analysis. Specific exclusion criteria included a history of asthma, chronic obstructive pulmonary disease (COPD), cardiac disease, or any chronic cardiorespiratory condition; any type of immunodeficiency; a previous known illness diagnosed specifically as influenza; current pregnancy; or egg allergy. The screening protocol included specific testing to rule out HIV infection and pregnancy. After an initial screening visit (∼3–6 wk previously), volunteers were inoculated nasally with the LAIV vaccine (FluMist; MedImmune, Inc., Gaithersburg, MD), according to the manufacturer's recommendations. Throughout the study, FluMist 2006–2007 and 2007–2008 were used, which are based on the strains A/New Caledonia/20/99 (H1N1) for 2006–2007 and A/Solomon Islands/3/2006 (H1N1) for 2007–2008, as well as A/Wisconsin/67/2005 (H3N2) and B/Malaysia/2506/2004 for both years. Nasal biopsies from both nostrils were obtained 3–6 wk before (screening visit) and 4 days after inoculation with LAIV, as described previously. Tissue from one nostril was immediately fixed in 4% paraformaldehyde for subsequent analysis using immunohistochemistry, whereas the sample from the other nostril was immediately processed for isolation of RNA. Although some overlap existed, the LAIV inoculation study was separate from the study obtaining NECs for in vitro culture described previously, and in consequence, the subject pools were generally different. This protocol was approved by the Institutional Review Board for Biomedical Research of the University of North Carolina School of Medicine.
Statistical Analysis
All data are expressed as mean ± SEM. Data were analyzed using either the Student t test or a one-way ANOVA and Newman-Keuls post hoc analysis. A value of P < 0.05 was considered significant.
RESULTS
In Vitro Model of NECs from Smokers
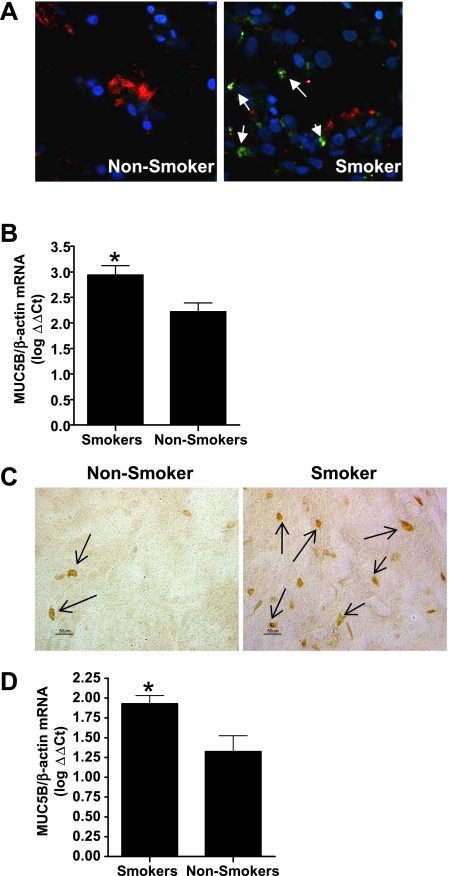
Biopsies obtained from smokers and nonsmokers were analyzed for the expression of mucin. One of the major mucins produced by differentiated airway epithelial cells (19, 20), Muc5B, was enhanced in nasal biopsies from smokers, as shown by immunohistochemistry (Figure 1A) and Muc5B mRNA concentrations (Figure 1B). Differentiated NECs from smokers and nonsmokers were analyzed for the production of mucosubstances and the expression of specific mucins. Differences in the expression of Muc5B were maintained after culturing NECs in vitro, as shown by the increased number of cells staining positively for Muc5B (Figure 1C) and the increased Muc5B mRNA concentrations seen in NECs from smokers compared with nonsmokers (Figure 1D). These data confirm the notion that Muc5B expression is enhanced in smokers (21), and that this change is maintained in differentiated NECs in long-term in vitro cell culture.
Figure 1.
Increased Muc5B expression in nasal epithelial cells from smokers. (A) Nasal biopsies were immunohistochemically labeled using anti-acetylated α-tubulin (cilia, red), Muc5B (green), and nuclei (4′,6-diamidino-2-phenylindole; blue), and visualized using confocal microscopy (arrows indicate Muc5B-positive cells). (B) Total RNA isolated from nasal biopsies was analyzed for levels of Muc5B mRNA, and normalized for expression of β-actin (n = 20 smokers; n = 13 nonsmokers). (C) In vitro differentiated NECs were immunohistochemically labeled using anti-Muc5B and HRP-conjugated secondary antibodies. Sections were visualized en face by light microscopy (arrows indicate Muc5B-positive cells). (D) Total RNA isolated from differentiated NECs from smokers and nonsmokers was analyzed for concentrations of Muc5B mRNA and normalized for the expression of β-actin (n = 12 smokers; n = 16 nonsmokers). *Significantly different from nonsmoker NECs (P < 0.05).
Effects of Influenza Virus on NECs from Smokers and Nonsmokers
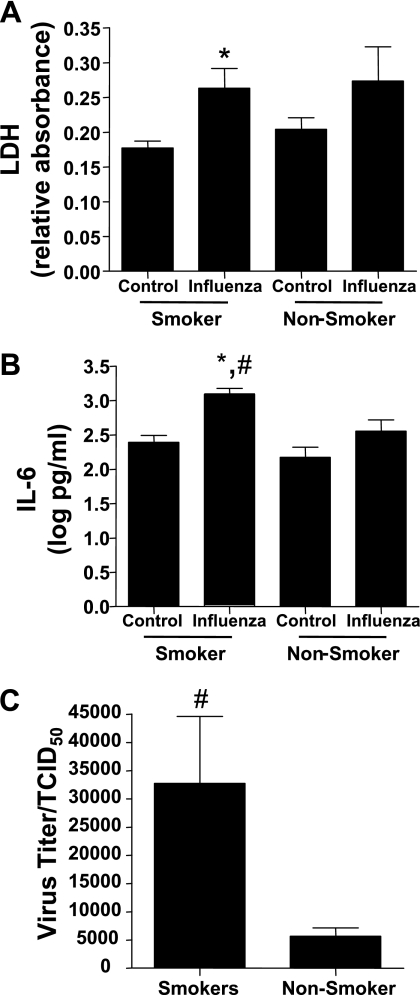
We next determined whether NECs from smokers exhibited modified influenza-induced cell responses. Using a similar infection protocol as described elsewhere (15, 16), we measured concentrations of LDH released into the basolateral supernatants as a marker of influenza-induced cytotoxicity, concentrations of IL-6 as a marker of influenza-induced inflammatory mediator release, and viral replication in NECs. Influenza-induced cytotoxicity was significantly enhanced in NECs from smokers, but not nonsmokers, compared with their noninfected control subjects (Figure 2A). Similarly, the influenza-induced release of IL-6 into basolateral supernatants was significantly greater in NECs from smokers than in nonsmokers (Figure 2B). Moreover, Figure 2C shows that influenza virus replication was significantly greater in NECs from smokers.
Figure 2.
The NECs from smokers are more susceptible to influenza virus than are the cells from nonsmokers. The NECs from smokers and nonsmokers were infected with influenza A/Bangkok/2/79. Basolateral supernatants collected 24 hours after infection were assessed for (A) LDH and (B) IL-6 (n = 13 smokers; n = 15 nonsmokers). (C) Apical supernatants collected 24 hours after infection were analyzed for viral replication (n = 21 smokers; n = 13 nonsmokers). *Significantly different from noninfected cells (P < 0.05). #Significantly different from nonsmokers (P < 0.05).
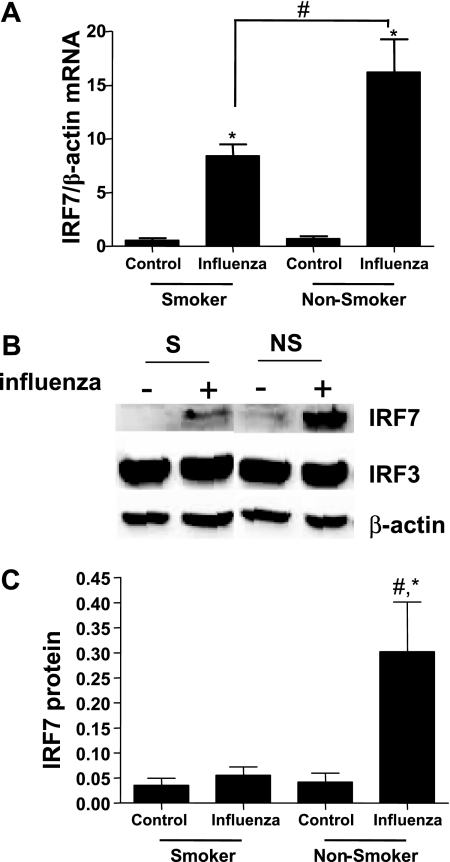
To determine potential mechanisms mediating the enhanced effects of influenza virus infections seen in NECs from smokers, we assessed whether the influenza-induced expression of antiviral defense mediators was modified. Specifically, we analyzed the influenza-induced expression of IFN-α in NECs from smokers and nonsmokers. Whereas infection with influenza significantly increased the release of IFN-α over noninfected cells in NECs from nonsmokers, the influenza-induced release of IFN-α was significantly reduced in differentiated NECs from smokers compared with nonsmokers (Figure 3A). Furthermore, the expression of IFN-β was significantly enhanced in NECs from both smokers and nonsmokers, with similar influenza-induced IFN-β mRNA concentrations in both groups (Figure 3B). Because the influenza-induced IFN-β expression did not appear to be different in NECs from smokers and nonsmokers, and because of its key role in regulating the expression of IFN-α, we measured influenza-induced IRF7 expression in NECs from smokers and nonsmokers. Influenza-induced IRF7 mRNA (Figure 4A) concentrations were greater in NECs from nonsmokers than smokers. Similarly, protein levels of IRF7 increased in response to influenza infection in NECs from nonsmokers, whereas no such increase was observed in smokers (Figure 4B). The expression of IRF3 was the same in both infected and noninfected NECs from smokers and nonsmokers. A densitometric analysis demonstrated that influenza-induced IRF7 expression was significantly greater in NECs from nonsmokers than smokers (Figure 4C). These data indicate that the ability to enhance IRF7 expression in response to influenza virus is significantly reduced in NECs from smokers.
Figure 3.
Influenza-induced IFN-α and IFN-β expression by NECs from smokers and nonsmokers. The NECs were infected with influenza A Bangkok/2/79 or left uninfected, and analyzed 24 hours after infection. (A) Basolateral supernatants were analyzed for IFN-α concentrations (n = 7 smokers; n = 12 nonsmokers). (B) Total RNA was analyzed for IFN-β mRNA concentrations (n = 6 smokers; n = 9 nonsmokers). *Significantly different from noninfected cells (P < 0.05). #Significantly different from smokers (P < 0.05).
Figure 4.
Expression of IRF7 in NECs from smokers and nonsmokers. The NECs were infected with influenza A Bangkok/2/79 or left uninfected, and analyzed for IRF7 expression 24 hours after infection. (A) Total RNA was analyzed for IRF7 mRNA and normalized to β-actin mRNA concentrations (n = 16 smokers; n = 14 nonsmokers). (B) Whole cell lysates were analyzed for IRF7 and IRF3 protein levels by Western blotting. Membranes were stripped and analyzed for β-actin to assure equal loading. Representative immunoblots are shown. (C) Densitometric analysis of IRF7 protein levels (n = 6 smokers; n = 6 nonsmokers). *Significantly different from noninfected cells (P < 0.05). #Significantly different from smokers (P < 0.05).
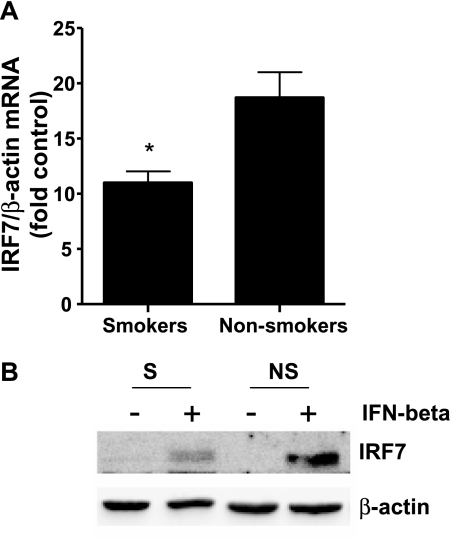
To test further whether the reduced release of IFN-α seen in Figure 3A was mediated by impaired type I IFN signaling and the ability to enhance IRF7 expression, we stimulated NECs from smokers and nonsmokers with IFN-β, which induces the production of IFN-α via an IRF7-dependent positive feedback loop (22). Our previous studies demonstrated that the receptor for IFN-β is predominantly localized on the basolateral side of the epithelium, which is why we only added IFN-β to the basolateral compartment to stimulate IFN-dependent gene expression (23). Figure 5 shows that the IFN-β–induced expression of IRF7 was significantly lower in NECs from smokers than in those from nonsmokers, at both the mRNA (Figure 5A) and protein (Figure 5B) levels.
Figure 5.
Interferon-induced IRF7 expression in NECs from smokers and nonsmokers. The NECs were stimulated with 1 ng/ml IFN-β from the basolateral side, and analyzed for IRF7 expression 24 hours after infection (n = 7 smokers; n = 7 nonsmokers). (A) Total RNA was analyzed for IRF7 mRNA, normalized to β-actin mRNA levels, and expressed as fold induction over the respective control. (B) Whole cell lysates were analyzed for IRF7 protein levels by Western blotting. Membranes were stripped and analyzed for β-actin to assure equal loading. Representative immunoblots are shown. *Significantly different from nonsmokers (P < 0.05).
Potential Role of Epigenetic Mechanisms Mediating Suppressed IRF7 Expression
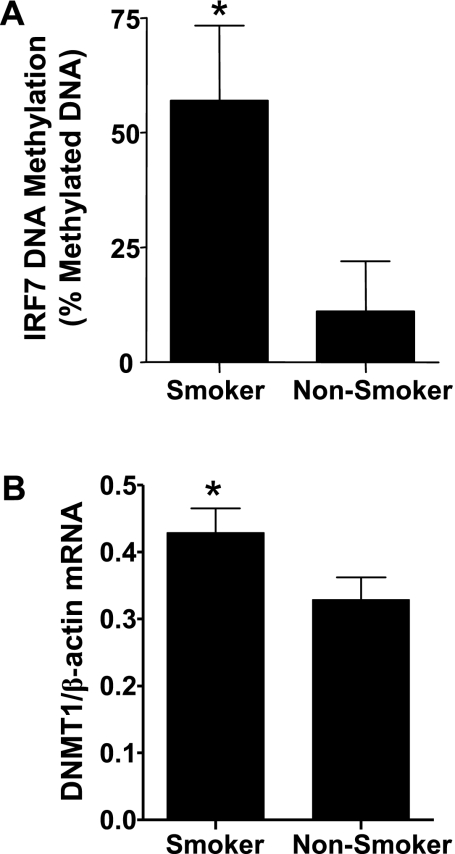
Figure 1 and previous reports indicate that airway epithelial cells obtained from defined diseased populations can maintain specific functional characteristics after differentiation in vitro (24, 25). To determine the potential mechanisms mediating the long-term maintenance of phenotypic and functional characteristics ex vivo, which are also reflected by a decreased ability to produce antiviral mediators, we assessed epigenetic modifications of the IRF7 gene. Previous studies demonstrated that methylation of the IRF7 gene results in gene silencing and a decreased ability of type I IFN to induce gene expression (26). Using a quantitative PCR-based DNA methylation analysis, we found that the IRF7 gene was significantly more methylated in NECs from smokers compared with nonsmokers (Figure 6A). In addition, we examined the expression of DNA (cytosine-5-)-methyltransferase 1 (DNMT1), a DNA methyltransferase that copies DNA methylation patterns within the DNA to the daughter strands during DNA replication. Figure 6B shows that DNMT1 mRNA levels were higher in cultured NECs from smokers compared with nonsmokers. These data are consistent with the hypothesis that chronic smoke exposure reduces type I IFN responses to influenza virus, and that this may be associated with epigenetic modifications of the IRF7 gene.
Figure 6.
IRF7 gene DNA methylation and expression of DNMT1 in NECs. (A) Genomic DNA isolated from NECs from smokers and nonsmokers was analyzed for DNA methylation of the IRF7 gene, using a quantitative PCR-based DNA methylation assay. The data are presented as percent methylated DNA (n = 6 smokers; n = 6 nonsmokers). (B) Total RNA isolated from NECs of smokers and nonsmokers was analyzed for DNMT1 mRNA concentrations, and normalized for the expression of β-actin mRNA levels (n = 11 smokers; n = 13 nonsmokers). *Significantly different from nonsmokers (P < 0.05).
In Vivo Confirmation of Suppressed Influenza-Induced IRF7 Expression in NECs from Smokers
To test whether similar effects occur in smokers in vivo, we obtained nasal epithelial biopsies at baseline and 4 d after the inoculation of volunteers with LAIV. The expression of IRF7 was examined in nasal biopsies, using real-time RT-PCR and immunohistochemistry. The LAIV-induced IRF7 mRNA concentrations were greater in nasal biopsies from nonsmokers compared with smokers, although not at a statistically significant level (P = 0.05) (Figure 7A). Immunohistochemical staining demonstrated that IRF7 was minimally expressed at baseline, and was robustly expressed in nonsmokers 4 d after infection with LAIV (Figure 7B, upper panels), but not in smokers (Figure 7B, lower panel). These results appeared consistent with our observations made in vitro regarding the suppression of influenza-induced IRF7 expression in smokers.
Figure 7.
Influenza-induced IRF7 expression in vivo. Nasal biopsies were obtained from smokers and nonsmokers 2–4 weeks before (baseline) and 4 days after administration of LAIV, and were examined for the expression of IRF7. (A) Total RNA was analyzed for IRF7 mRNA, normalized to β-actin mRNA levels, and expressed as fold induction over the subject-specific baseline level (n = 12 smokers; n = 13 nonsmokers). (B) Nasal biopsies were fixed, and paraffin-embedded sections were immunohistochemically stained using antibodies against IRF7 and visualized by light microscopy. C, cilia.
DISCUSSION
Epidemiologic evidence and experimental data suggest that exposure to cigarette smoke increases the susceptibility to and severity of respiratory virus infections (5, 6, 8, 9). We developed an in vitro model of NECs from smokers that maintains the phenotypic changes observed in the nasal epithelium in this group in vivo. Using this experimental model, our data indicate that NEC from smokers may be more susceptible to influenza virus infections, and that this susceptibility is associated with a suppressed ability to upregulate IFN-α expression after infection. In addition, we found that the ability of virus to induce expression of the key IFN pathway transcription factor IRF7 was also suppressed in cells from smokers, and that this suppression was associated with increased DNA methylation of the IRF7 gene. Using responses induced by the temperature-sensitive LAIV vaccine as a model for transient, self-limited infection, we also found that the influenza-induced expression of IRF7 was suppressed in the nasal epithelium in smokers in vivo. Thus, our data suggest that chronic exposure to cigarette smoke modifies the antiviral defense responses of the respiratory epithelium, and that epigenetic modifications of a key antiviral defense gene could mediate these effects.
Our study focused on early events in the course of viral infection, that is, the results of the initial interaction between virus and host epithelial cells. The respiratory epithelium is the main target of respiratory viruses, is the main site for viral replication, and plays an important role during initial antiviral defense responses. Viral infection normally induces the production of type I IFNs, which in turn activate the synthesis of interferon-stimulated genes, and in consequence, limit viral replication (29). Previous studies demonstrated that virus-induced interferon production is suppressed in epithelial cells from diseased populations. Specifically, bronchial epithelial cells from patients with asthma exhibited blunted IFN-β and IFN-λ expression after infection with rhinovirus (27, 28). Thus, in bronchial epithelial cells from patients with asthma, virus-induced interferon production is significantly reduced. However, the present data indicate that influenza infection resulted in a similar upregulation of IFN-β expression, although the ability of influenza to enhance IFN-α expression was significantly reduced in NECs from smokers. As indicated previously, a positive feedback loop that amplifies type I IFN production after the initial virus-dependent production of IFN-β is dependent on IRF7 (12). In addition to infection with influenza, stimulation with IFN-β also resulted in the suppressed expression of IRF7 in NECs from smokers. Thus, in NECs from smokers, the action, rather than the production, of IFN-β appears to be blunted. Other factors being equal, we would expect this to result in a poorer ability to clear virus from the nasal passages. Consequently, the suppression of elements of type I IFN signaling could play a role in the increased susceptibility to infection observed in epidemiologic studies of smokers and those exposed to second-hand smoke.
An interesting finding in our study involved the retention of histologic and functional differences between NECs from smokers and nonsmokers, despite the long-term culture of cells in vitro. Several groups demonstrated that epithelial cells obtained from defined disease populations, when differentiated in vitro, maintain many of their phenotypic and functional characteristics found in vivo (24, 25). For example, bronchial epithelial cells obtained from patients with asthma maintain increased basal cytokine production in vitro (24). Cultured bronchial epithelial cells from patients with COPD demonstrate a greater ability to induce Muc5AC expression than cells obtained from normal subjects, which corresponds to similar observations made in vivo (30). In our model, the expression of Muc5B, the major mucin found in the sputum of patients with COPD (21), was enhanced both in nasal biopsies from smokers in vivo and in differentiated NECs from smokers in vitro. In addition, recent data from our group also indicate that ciliary phenotypes, such as percent ciliated cells and ciliary beat frequency, are different in the nasal epithelium of smokers, which is a phenotype also maintained in differentiated NECs in vitro (17) (J. L. Carson, personal communication). Thus, differentiated NECs obtained from smokers maintain characteristics found in the nasal epithelium in these individuals in vivo, and therefore provide a viable model for studying cellular mechanisms by which exposure to cigarette smoke modifies antiviral defense responses at the level of the epithelium.
One mechanism for producing lasting changes in cellular function and gene expression is gene silencing through the DNA methylation of discrete CpG islands. Patterns of DNA methylation are faithfully propagated through cell division by copying existing methylation patterns during DNA replication, using the parental strand as a template. Previous studies demonstrated that hypermethylation of the IRF7 gene results in a decreased ability of type I IFNs to induce gene expression (18, 26). The present data indicate that the IRF7 gene is methylated, and that the expression of DNMT1 is increased in NECs from smokers. The region of the IRF7 gene that was analyzed here is in close proximity to the transcriptional start site, is within the promoter region of the IRF7 gene, and was previously shown to be important for silencing the IRF7 gene in response to stimulation with IFN-α (18). In addition to IRF7, a number of other host defense genes, such as IFNγ, STAT1, COX2, and ICSBP/IRF8, which also play important roles in host defense responses against viruses, were shown to be silenced by DNA methylation under various circumstances, including exposure to cigarette smoke (31). Therefore, cigarette smoke-induced epigenetic modification is likely to exert an impact on additional genes beyond IRF7 that are important in the host response to viral infection.
Our data demonstrate that the expression of DNMT1 is enhanced in NECs from smokers. Whereas DNMT3a and DNMT3b are capable of a de novo methylation of symmetrically unmethylated CpGs, DNMT1 faithfully propagates DNA methylation patterns through cell divisions, and the enhanced expression of DNMT1 is associated with hypermethylation, in bronchial epithelial cells (32). A number of compounds in cigarette smoke are capable of promoting DNA methylation (33, 34), and it has become increasingly apparent that this is one of the major mechanisms of gene silencing of tumor-suppressor genes (35, 36). Nicotine-derived nitrosamine ketone, a component of cigarette smoke, increased the expression and activity of DNMT1 in respiratory epithelial cells of mouse lungs (37), and DNMT1 is highly expressed in lung tumors from smokers, in association with increased DNA methylation and the gene silencing of tumor-suppressor genes such as p16(INK4a) (38). In addition, the decreased expression of DNMT1 resulted in a reversal of both hypermethylation and gene silencing in bronchial epithelial cells (32), further supporting the significance of this enzyme in the context of epigenetic changes.
Interestingly, our observations appear to be in contrast to those in a previous study demonstrating that mice exposed to cigarette smoke for 2 weeks and subsequently stimulated with polyinosinic:polycytidylic acid had increased levels of type I IFNs in bronchoalveolar lavage fluid (39). Similarly, airway epithelial cells exposed to cigarette smoke condensate and infected with respiratory syncytial virus showed a greater expression of IRF7 (40). However, mice exposed to cigarette smoke for 3 to 5 months and infected with a low dose of influenza virus showed decreased inflammatory responses, whereas mice infected with high doses of influenza after smoke exposure showed increased inflammatory responses (8). Thus, the effects of cigarette smoke on viral infection and subsequent inflammatory or antiviral responses may depend on the specific viral pathogen, level of infection, and chronicity of smoke exposure.
The overall response of the respiratory tract to viral infection involves not only direct virus–epithelium interactions, but also a large array of innate and adaptive immune responses that determine the outcome of infection. Thus, studies of systemic immune responses to virus in smokers are relevant to our study. In this regard, influenza-specific antibody levels were reported not to be different in smokers compared with nonsmokers (6). In addition, influenza-specific memory responses upon rechallenge were not altered by chronic exposure to cigarette smoke in mice (8). Further research is required to define the impact of cigarette smoke-associated suppression of epithelial antiviral responses on the complex overall response to influenza in the human respiratory tract.
In conclusion, our data strongly suggest that chronic exposure to cigarette smoke suppresses epithelial antiviral (type 1 IFN) pathways in human nasal epithelium. This likely results in an increased susceptibility to viral infection, and these effects may be mediated by the gene silencing of key antiviral defense pathway mediators, including IRF7. Viral infections remain a major public health concern and a likely cause of exacerbations of smoking-related diseases, such as COPD (41). Understanding the mechanisms by which exposure to cigarette smoke enhances susceptibility to viral infections could lead to new strategies for minimizing influenza-induced morbidity and even mortality. For example, potential therapeutic strategies could include interventions or reversals of epigenetic modifications induced by cigarette smoke, which were already shown to improve glucocorticoid sensitivity in patients with COPD (42), and to reverse the markers of cigarette smoke–induced cancer using in vitro and animal in vivo models (43).
Acknowledgments
The authors thank Sally Ivins and Margret Herbst for assistance in developing an institutional review board-approved protocol and recruiting subjects for the present study. The contents of this study are solely the responsibility of the authors, and do not necessarily represent the official views of the National Heart, Lung, and Blood Institute at the National Institutes of Health. Although the research described in this article was funded wholly or in part by the United States Environmental Protection Agency through cooperative agreement CR829522 with the Center for Environmental Medicine, Asthma, and Lung Biology (University of North Carolina at Chapel Hill, Chapel Hill, NC), it has not been subjected to that agency's required peer and policy review, and therefore does not necessarily reflect the views of the Agency, and no official endorsement should be inferred. The mention of trade names or commercial products does not constitute endorsement or recommendation for use.
This work was supported by the United States Environmental Protection Agency through cooperative agreement CR829522 with the Center for Environmental Medicine, Asthma, and Lung Biology (University of North Carolina at Chapel Hill, Chapel Hill, NC), by grant HL095163 from the National Heart, Lung, and Blood Institute of the National Institutes for Health (I.J. and T.L.N.), and by the Flight Attendants Medical Research Institute (I.J. and T.L.N.).
Originally Published in Press as DOI: 10.1165/rcmb.2009-0254OC on October 30, 2009
Author Disclosure: I.J. has received industry-sponsored grants from Entegrion ($10,001–$50,000), and sponsored grants from the National Institutes of Health (NIH) ($100,000) and the Flight Attendants Medical Research Institute ($50,001–$100,000). J.L.C. has received sponsored grants from the Flight Attendants Medical Research Institute ($50,001–$100,000). T.L.N. has received sponsored grants from the NIH ($100,001 or more), the Cystic Fibrosis Foundation ($100,001 or more), and the Flight Attendants Medical Research Institute ($100,001 or more). None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Monto AS. Occurrence of respiratory virus: time, place and person. Pediatr Infect Dis J 2004;23(Suppl 1)S58–S64. [DOI] [PubMed] [Google Scholar]
- 2.Klimov A, Simonsen L, Fukuda K, Cox N. Surveillance and impact of influenza in the United States. Vaccine 1999;17(Suppl):S42–S46. [DOI] [PubMed] [Google Scholar]
- 3.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003;289:179–186. [DOI] [PubMed] [Google Scholar]
- 4.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA 2004;292:1333–1340. [DOI] [PubMed] [Google Scholar]
- 5.Kark JD, Lebiush M. Smoking and epidemic influenza-like illness in female military recruits: a brief survey. Am J Public Health 1981;71:530–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kark JD, Lebiush M, Rannon L. Cigarette smoking as a risk factor for epidemic A (H1N1) influenza in young men. N Engl J Med 1982;307:1042–1046. [DOI] [PubMed] [Google Scholar]
- 7.Arcavi L, Benowitz NL. Cigarette smoking and infection. Arch Intern Med 2004;164:2206–2216. [DOI] [PubMed] [Google Scholar]
- 8.Robbins CS, Bauer CM, Vujicic N, Gaschler GJ, Lichty BD, Brown EG, Stampfli MR. Cigarette smoke impacts immune inflammatory responses to influenza in mice. Am J Respir Crit Care Med 2006;174:1342–1351. [DOI] [PubMed] [Google Scholar]
- 9.Lebiush M, Rannon L, Kark JD. An outbreak of A/USSR/90/77 (H1N1) influenza in army recruits: clinical and laboratory observations. Mil Med 1982;147:43–48. [PubMed] [Google Scholar]
- 10.Groskreutz DJ, Monick MM, Babor EC, Nyunoya T, Varga SM, Look DC, Hunninghake GW. Cigarette smoke alters respiratory syncytial virus-induced apoptosis and replication. Am J Respir Cell Mol Biol 2009;41:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Fu WC, Liu J, Harty RN, Fuchs SY. Cigarette smoking products suppress anti-viral effects of type I interferon via phosphorylation-dependent downregulation of its receptor. FEBS Lett 2008;582:3206–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes B, Lubyova B, Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res 2002;22:59–71. [DOI] [PubMed] [Google Scholar]
- 13.Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol 1996;14:104–112. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Wang X, Brighton L, Hazucha M, Jaspers I, Carson JL. Increased nasal epithelial ciliary beat frequency associated with lifestyle tobacco smoke exposure. Inhal Toxicol 2009;21:875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaspers I, Zhang W, Brighton LE, Carson JL, Styblo M, Beck MA. Selenium deficiency alters epithelial cell morphology and responses to influenza. Free Radic Biol Med 2007;42:1826–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaspers I, Ciencewicki JM, Zhang W, Brighton LE, Carson JL, Beck MA, Madden MC. Diesel exhaust enhances influenza virus infections in respiratory epithelial cells. Toxicol Sci 2005;85:990–1002. [DOI] [PubMed] [Google Scholar]
- 17.Farag-Mahmod FI, Wyde PR, Rosborough JP, Six HR. Immunogenicity and efficacy of orally administered inactivated influenza virus vaccine in mice. Vaccine 1988;6:262–268. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Tang L, Roberts PC, Kraniak JM, Fridman AL, Kulaeva OI, Tehrani OS, Tainsky MA. Interferon regulatory factors IRF5 and IRF7 inhibit growth and induce senescence in immortal Li-Fraumeni fibroblasts. Mol Cancer Res 2008;6:770–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon JH, Moon HJ, Seong JK, Kim CH, Lee JJ, Choi JY, Song MS, Kim SH. Mucociliary differentiation according to time in human nasal epithelial cell culture. Differentiation 2002;70:77–83. [DOI] [PubMed] [Google Scholar]
- 20.Bernacki SH, Nelson AL, Abdullah L, Sheehan JK, Harris A, Davis CW, Randell SH. Mucin gene expression during differentiation of human airway epithelia in vitro: muc4 and muc5b are strongly induced. Am J Respir Cell Mol Biol 1999;20:595–604. [DOI] [PubMed] [Google Scholar]
- 21.Kirkham S, Kolsum U, Rousseau K, Singh D, Vestbo J, Thornton DJ. MUC5B is the major mucin in the gel phase of sputum in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008;178:1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy DE, Marie I, Smith E, Prakash A. Enhancement and diversification of IFN induction by IRF-7-mediated positive feedback. J Interferon Cytokine Res 2002;22:87–93. [DOI] [PubMed] [Google Scholar]
- 23.Ciencewicki JM, Brighton LE, Jaspers I. Localization of type I interferon receptor limits interferon-induced TLR3 in epithelial cells. J Interferon Cytokine Res 2009;29:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayram H, Devalia JL, Khair OA, Abdelaziz MM, Sapsford RJ, Sagai M, Davies RJ. Comparison of ciliary activity and inflammatory mediator release from bronchial epithelial cells of nonatopic nonasthmatic subjects and atopic asthmatic patients and the effect of diesel exhaust particles in vitro. J Allergy Clin Immunol 1998;102:771–782. [DOI] [PubMed] [Google Scholar]
- 25.Pierrou S, Broberg P, O'Donnell RA, Pawlowski K, Virtala R, Lindqvist E, Richter A, Wilson SJ, Angco G, Moller S, et al. Expression of genes involved in oxidative stress responses in airway epithelial cells of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007;175:577–586. [DOI] [PubMed] [Google Scholar]
- 26.Lu R, Au WC, Yeow WS, Hageman N, Pitha PM. Regulation of the promoter activity of interferon regulatory factor-7 gene: activation by interferon snd silencing by hypermethylation. J Biol Chem 2000;275:31805–31812. [DOI] [PubMed] [Google Scholar]
- 27.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, Holgate ST, Davies DE. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 2005;201:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, Kebadze T, Mallia P, Stanciu LA, Parker HL, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med 2006;12:1023–1026. [DOI] [PubMed] [Google Scholar]
- 29.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev 2001;14:778–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, Morton JD, Battaile JT, Patel AC, Patterson GA, Castro M, et al. Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J Clin Invest 2006;116:309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med 2007;7:85–102. [DOI] [PubMed] [Google Scholar]
- 32.Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res 2008;68:9005–9014. [DOI] [PubMed] [Google Scholar]
- 33.Kerr KM, Galler JS, Hagen JA, Laird PW, Laird-Offringa IA. The role of DNA methylation in the development and progression of lung adenocarcinoma. Dis Markers 2007;23:5–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res 1999;424:127–142. [DOI] [PubMed] [Google Scholar]
- 35.Schrump DS, Nguyen DM. Targeting the epigenome for the treatment of thoracic malignancies. Thorac Surg Clin 2006;16:367–377. [DOI] [PubMed] [Google Scholar]
- 36.Hutt JA, Vuillemenot BR, Barr EB, Grimes MJ, Hahn FF, Hobbs CH, March TH, Gigliotti AP, Seilkop SK, Finch GL, et al. Life-span inhalation exposure to mainstream cigarette smoke induces lung cancer in B6C3F1 mice through genetic and epigenetic pathways. Carcinogenesis 2005;26:1999–2009. [DOI] [PubMed] [Google Scholar]
- 37.Belinsky SA, Nikula KJ, Baylin SB, Issa JP. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA 1996;93:4045–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer 2007;55:205–213. [DOI] [PubMed] [Google Scholar]
- 39.Kang MJ, Lee CG, Lee JY, Dela Cruz CS, Chen ZJ, Enelow R, Elias JA. Cigarette smoke selectively enhances viral PAMP- and virus-induced pulmonary innate immune and remodeling responses in mice. J Clin Invest 2008;118:2771–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castro SM, Kolli D, Guerrero-Plata A, Garofalo RP, Casola A. Cigarette smoke condensate enhances respiratory syncytial virus-induced chemokine release by modulating NF-kappa B and interferon regulatory factor activation. Toxicol Sci 2008;106:509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caramori G, Ito K, Contoli M, Di Stefano A, Johnston SL, Adcock IM, Papi A. Molecular mechanisms of respiratory virus-induced asthma and COPD exacerbations and pneumonia. Curr Med Chem 2006;13:2267–2290. [DOI] [PubMed] [Google Scholar]
- 42.Cosio BG, Tsaprouni L, Ito K, Jazrawi E, Adcock IM, Barnes PJ. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. J Exp Med 2004;200:689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, Baylin SB. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res 2003;63:7089–7093. [PubMed] [Google Scholar]