Abstract
Ecological and evolutionary studies of wild primates hold important keys to understanding both the shared characteristics of primate biology and the genetic and phenotypic differences that make specific lineages, including our own, unique. Although complementary genetic research on nonhuman primates has long been of interest, recent technological and methodological advances now enable functional and population genetic studies in an unprecedented manner. In the past several years, novel genetic data sets have revealed new information about the demographic history of primate populations and the genetics of adaptively important traits. In combination with the rich history of behavioral, ecological, and physiological work on natural primate populations, genetic approaches promise to provide a compelling picture of primate evolution in the past and in the present day.
Genetic studies of natural primate populations
Our closest living relatives, the nonhuman primates, are perennial subjects of public and scientific fascination because they occupy a unique place in evolutionary biology and ecology. The striking similarities we share with other primates make them important models for human physiology, behavior, and health [1–4]. At the same time, variation among primate populations and species provides a rich basis for comparative work (e.g. [5–7]) Such work is critical for understanding the common threads that tie primates together and the differences that make specific branches of the primate tree, including the human lineage, unique.
Within the larger primate literature, studies that focus on wild primates offer a unique perspective on how ecological and environmental factors influence evolutionarily important traits. Indeed, primates are well represented among systems for which extensive field data are available, much of which is extremely fine-grained and some of which is continuous over multiple decades (Table 1). As a result, for many species we now know a great deal about the relationship between ecological and environmental variation, social structure, demography, and physiology. Together these types of data contribute to a rich understanding of how primates evolve.
Table 1.
[JS1]Primate genomic resources are complemented by long-term field studies.
| Species | Representative long term field studies | Website or Representative Publication | Genomic Resources | |
|---|---|---|---|---|
| Apes | Common chimpanzee (Pan troglodytes) | Bossou, Nimba Mountains, Guinea | www.greenpassage.org | Genome sequenced |
| Budongo Forest, Uganda | culture.st-and.ac.uk/bcfs/index.html | |||
| Gombe Stream National Park, Tanzania | www.discoverchimpanzees.org | |||
| Mahale Mountains National Park, Tanzania | [96] | |||
| Kanyawara, Kibale National Park, Uganda | www.fas.harvard.edu/~kibale | |||
| Ngogo, Kibale National Park, Uganda | [97] | |||
| Taï National Park, Côte d’Ivoire | www.eva.mpg.de/primat/files/chimps.htm | |||
| Bonobo (Pan paniscus) | Lui Kotale, Salonga National Park, DRC | www.eva.mpg.de/primat/files/bonobo.htm | Genome sequence in progress | |
| Lomako Forest, DRC | www.uoregon.edu/~fwhite/Lomako Forest Bonobo Project.htm | |||
| Gorilla (Gorilla gorilla and Gorilla beringei) |
Bwindi Impenetrable National Park, Uganda | www-rcf.usc.edu/~stanford/bigape.html | Genome sequence in progress | |
| Karisoke, Virunga Mountains, Rwanda | www.gorillafund.org/conservation/karisoke_research_center.php | |||
| Mbeli Bai, Nouabalé-Ndoki National Park, Congo | [98] | |||
| Orangutan (Pongo sp.)c | Gunung Palung National Park, Indonesia | people.bu.edu/orang | Genome sequence in progress | |
| Ketambe, Indonesia; | [99] | |||
| Kutai Game Reserve, Indonesia; | [99] | |||
| Lower Kinabatangan, Indonesia; | [99] | |||
| Suaq Balimbing, Indonesia; | [99] | |||
| Tanjung Puting Reserve, Indonesia | [99] | |||
| White-cheeked gibbon (Nomascus leucogenys)d |
Genome sequence in progress | |||
| OW Mon keysa | Baboon (Papio sp.)e | Amboseli National Park, Kenya | www.princeton.edu/~baboon | Genome sequence in progress; genetic linkage map; ENCODE comparative sequence |
| Awash National Park, Ethiopia | [81] | |||
| Cape Peninsula, South Africa | www.baboonsonline.org/bru | |||
| De Hoop Nature Reserve, South Africa | [100] | |||
| Drakensberg Mountains, South Africa | [100] | |||
| Gashaka Gumti National Park, Nigeria | www.ucl.ac.uk/gashaka/home | |||
| Gombe Stream National Park, Tanzania | [101] | |||
| Kafue National Park, Zambia | [102] | |||
| Laikipia, Kenya | www.baboonsrus.com; www.rci.rutgers.edu/~palombit | |||
| Mikumi National Park, Tanzania | web.anglia.ac.uk/abru/info.htm | |||
| Moremi Game Reserve, Botswana | www.psych.upenn.edu/~seyfarth/Baboon research/baboon.htm | |||
| Vervet monkey (Cercopithecus aethiops) |
Amboseli National Park, Kenya | [15] | Genome sequence pending; genetic linkage map; ENCODE comparative sequence |
|
| Samburu National Park, Kenya | [103] | |||
| Long-tailed macaquef (Macaca fascicularis) |
Ketambe River, Sumatra, Indonesia | [104] | Genome sequence in progress | |
| Rhesus macaque (Macaca mulatta.) | Cayo Santiago, Puerto Ricog | cprc.rcm.upr.edu | Genome sequence complete; genetic linkage map |
|
| Eastern black and white colobus (Colobus guereza) |
Kakamega Forest National Reserve, Kenya | [105] | ENCODE comparative sequence | |
| Kibale National Park, Uganda | [106] | |||
| NW MonKEYSb | Owl monkey (Aotus nancymaae)h | ENCODE comparative sequence | ||
| Common marmoset (Callithrix jacchus) | Tapacura, Brazil | [107] | Genome sequence in progress; ENCODE comparative sequence |
|
| Squirrel monkey (Saimiri sp.) | Corcovado National Park, Costa Rica | [108] | Genome sequence pending; ENCODE comparative sequence |
|
| Manu National Park, Peru | [108] | |||
| Raleighvallen National Park, Suriname | [109] | |||
| Dusky titi monkey (Callicebus moloch) | Manu National Park, Peru | [110] | ENCODE comparative sequence | |
| Prosimians | Gray mouse lemur (Microcebus murinus) |
Kirindy Forest, Madagascar | [111] | Genome sequence in progress; ENCODE comparative sequence |
| Phillipine tarsier (Tarsius syrichta) | Corella, Bohol, Phillipines7 | www.tarsiusproject.org | Genome sequence in progress | |
| Greater bushbaby (Otolemur garnettii) | Gedi National Monument, Kenya | [112] | Genome sequence in progress; ENCODE comparative sequence |
|
Old World monkeys
New World monkeys
Field studies of both Bornean and Sumatran orangutans (variably considered species or subspecies of a single species, Pongo pygmaeus) are included here.
Other gibbon species are better studied in the wild (e.g., the agile gibbon, Hylobates agilis at Gunung Palung National Park, Indonesia [113]; the white-handed gibbon, Hylobates lar, and the siamang, Hylobates syndactylus, at Ketambe River, Indonesia [114]; and the white-handed gibbon at Khao Yai, Thailand [115])
Field studies of several allotaxa of Papio are listed here, including those focused on hybrids. Some taxonomies consider these allotaxa separate species and others, subspecies designations of a single species, Papio hamadryas. Genomic resource development has focused primarily on anubis baboons.
Synonymous with crab-eating macaque or cynomolgus monkey (more common in the medical literature).
The Cayo Santiago rhesus macaque population is free-ranging, but not natural (rhesus macaques are native to Asia, not the Americas).
Other Aotus species are better studied in the wild (e.g., the night monkey, Aotus trivirgatus, in Manu National Park, Peru [110]; Azara’s night monkey, Aotus azarae, in Estancia Guaycolec, Argentina: see www.sas.upenn.edu/~eduardof/EstanciaGuaycolec.html)
Unlike most of the other projects listed here, this is a shorter-term study focused primarily on data collection via radio telemetry (tarsiers are nocturnal
In contrast, we know relatively less about the evolutionary genetics of wild primate populations. More so than for behavioral and ecological studies, research in this field has been constrained by the available technology. Thus, whereas observational methods for collecting behavioral data have remained relatively consistent over the past several decades, the possibilities for genetic analysis have only recently expanded from allozyme analyses of one or a few protein-coding loci, to analyses of modest microsatellite data sets, to the current ability to produce the kinds of large data sets amenable for highly powered population and functional genetic studies [8, 9].
Primate studies are well positioned to take advantage of these new approaches. In particular, the increasing ease of genetic data collection (Figure 1) addresses one of the historical challenges of working on nonhuman primates. Already, the number of full genome sequences available for primates supersedes that for most other groups of animals [10], a testament to the importance of primate studies to the scientific community and to research relevant to human evolution and health. Meanwhile, the collection of phenotypic and environmental data remains a core strength of primate field studies. Such data act as an important scaffold for genetic studies, providing crucial ecological and behavioral insight into the causes and consequences of genetic patterns.
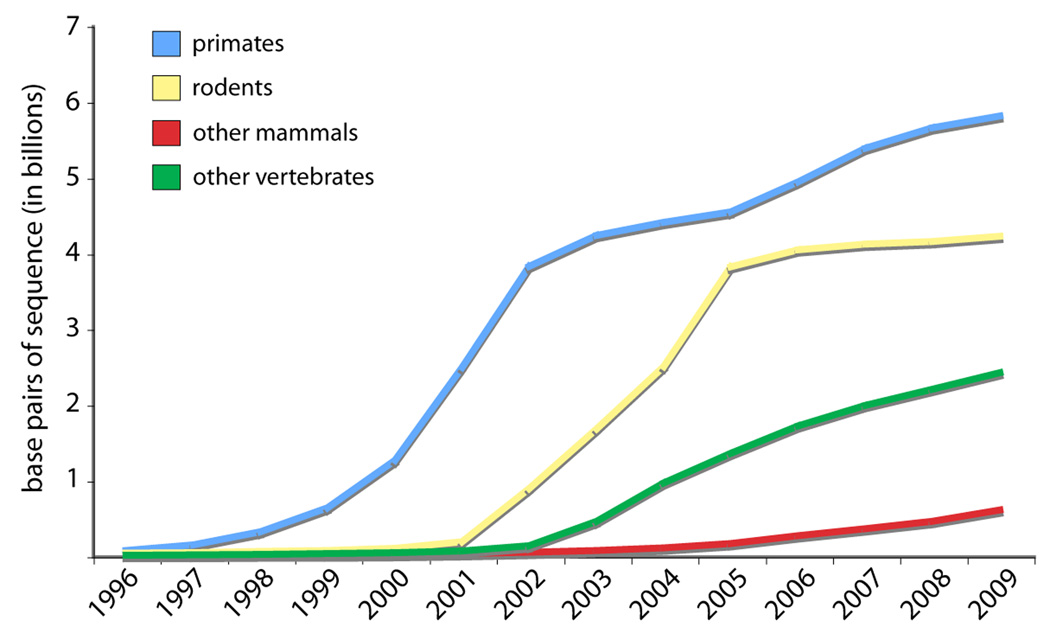
Figure 1.
The amount of sequence data for primates has rapidly expanded. The availability of sequence-based resources sets the stage for work that integrates genetic perspectives with ecological, behavioral, and demographic data on the same species. Other groups (one other mammalian order, rodents; all non-primate, non-rodent mammals; and all non-mammalian vertebrates) are shown for comparison. Data reflect the amount of sequence data available at the end of each calendar year (in the December 15 yearly Genbank release), and were downloaded from NCBI (ftp.ncbi.nih.gov/genbank/release.notes/) on March 1, 2010.
Here, we consider the possibilities for integrating genetic data and analysis with the extensive field-based data sets on natural primate populations, drawing from recent examples in the literature (we focus on conceptual approaches for this integration; for a detailed review of methods for obtaining samples and molecular techniques useful in primate research, see [8]). This direction offers the opportunity to combine genetic, phenotypic, and environmental perspectives on the same individuals, an approach that has already proven instrumental for testing long-standing hypotheses in primate behavioral ecology (e.g., [11–13]). Although some of the most exciting findings are undoubtedly yet to come, results from completed work already illustrate the potential of such work for understanding the population and functional genetics of primate populations.
Population structure and gene flow in a behavioral and ecological context
A central project of evolutionary geneticists focuses on understanding population history and demography, which are mediated by individual dispersal patterns and differential reproductive success—parameters that have also been of great interest to primate field researchers. In particular, research on natural primate populations has focused intensively on the behavioral and ecological factors that contribute to variation in these characteristics (e.g., [14–16]). Placing genetic data and inference in the context of this work therefore presents the opportunity to link changes in the genetic composition of a population with its proximate causes and resulting phenotypic effects.
In many primates, natural populations are subdivided into stable social groups. The size and composition of such groups vary within and between species, as do the patterns of dispersal that produce change in group membership over time [17]. In many cases, sex-biased dispersal (in which one sex typically disperses while the other remains in the natal group; this pattern can leave distinctive marks on different parts of the genome [18]) results in outbred social groups in which many individuals are nonetheless closely related and form tight social bonds [19]. This pattern closely matches the “breeding group” model developed to account for the population genetic properties of species organized into stable social groups that also exhibit sex-biased dispersal and reproductive skew [20, 21]. Because these properties (for example, co-residency of close, but not inbred, relatives within a social group) are likely to be important for the evolution of kin-biased behavior, they have also been of great interest to behavioral field biologists.
The results of this work have provided deeper resolution into the kinds of behavioral variation that influence population structure. Indeed, studies in primates (in addition to other taxa; see for example [22–24]) have highlighted how behavioral patterns can directly predict population genetic structure, thus providing a mechanistic explanation for the patterns embedded in genetic data. For instance, genetic structure in wild baboons (Papio cynocephalus) closely matches predictions arising from observational data on mate guarding and dominance rank, emphasizing the importance of male reproductive skew in shaping population structure in this species [25]. Similarly, the values of classical F statistics (which evaluate the proportion of genetic variance in a population explained by subgroups within the population) in Venezuelan red howler monkeys (Alouatta seniculus) can be directly interpreted in the light of levels of reproductive skew, female philopatry, and male dispersal known from years of intensive field observations [26].
Social behavior and the structure of social groups can in turn be influenced by the spatial distribution of resources and local ecological and environmental patterns, a relationship that forms the core of the socioecological model for primate behavioral ecology [27, 28]. Ecological and behavioral data were key, for example, in understanding patterns of population structure in the eastern mountain gorilla (Gorilla beringei beringei) population of the Bwindi Impenetrable Forest in Uganda [29]. Among the Bwindi gorillas, population structure emerges primarily from the distribution of breeding females; males exhibit almost no discernible population structure. This sex difference occurs despite the fact that both male and female gorillas can disperse from their natal groups and the fact that female gorillas travel substantial distances in daily life. Ecological data on spatial variation in plant community composition revealed that females, but not males, express a behavioral preference for remaining in areas where the plant community is familiar [29]. Thus, local ecology exerts an important effect on the distribution of genetic variation in this population via sex differences in foraging behavior. This effect might also help explain genetic data that indicate male-mediated historical gene flow between eastern gorillas and western gorillas [30, 31]. If the patterns in Bwindi also held for gorilla populations in the past, primarily male-mediated gene flow between these species might have resulted in part from female preferences to maintain closer ties to a known resource base. Hence, ecological and behavioral studies of modern primate populations can aid in interpreting past population history, especially for species for which extensive population genetic analyses are also available [31–35] (Box 1).
Box 1: Genetic data and demographic history in Pan
Genetic data carry the signature of past demographic events for many generations. Thus, they can make an important contribution to understanding historical population structure, growth, and change, which in turn provides valuable context for genetic studies of field populations in the present. The power of genetic analysis in reconstructing the past is illustrated by the extensive genetic investigations of population history in chimpanzees and bonobos [32–34, 76–78].
These studies demonstrate the nature of the insights that can be drawn from both modestly sized multilocus studies and from more recent genome-scale studies. Encouragingly, inferences from these studies have been qualitatively consistent even across different data sets. For example, a recent large resequencing data set dated the divergence time between chimpanzees and bonobos at 1.29 million years [33], consistent with previous estimates (∼800 – 1.8 million years: [77, 78]) but with a much smaller window of uncertainty. Smaller data sets have also suggested hypotheses that can be tested further. For example, a survey of nine unlinked intergenic regions revealed an excess of rare alleles relative to neutral expectations in central chimpanzees, a pattern suggestive of recent population expansion [77]. Indeed, a more recent analysis indicates that the effective population size of central chimpanzees has increased by at least four-fold since the split between central and western chimpanzees, whereas the western chimpanzee population has contracted during the same time [33].
Genetic data have also produced a better understanding of gene flow in chimpanzees. Both PCA and STRUCTURE analyses support the genetic distinctness of bonobos from chimpanzees and of chimpanzee subspecies from one another [32]. However, several studies also indicate past gene flow between western chimpanzees and central chimpanzees [33, 76, 79]. Thus, an isolation and migration model might better describe the demographic history of these groups than a simple split. One interesting possibility is that this link might have been enabled by the proposed fourth subspecies of chimpanzee, P. t. vellerosus, which could be genetically more similar to western chimpanzees but is geographically closer to central chimpanzees [79].
Taken together, work on the demographic history of chimpanzees provides a template for expanding genetic inference of demographic history in other primates. Additionally, recent work illustrates how powerful inferences can be drawn from relatively small numbers of individuals, given large amounts of sequence data [33]. Data sets of these dimensions are readily generated through new sequencing technologies, and will be increasingly available in the near future.
Finally, field data can highlight the sensitivity of population structure to behavioral and demographic change. For instance, a long-term study of red deer (Cervus elaphus) showed that decreasing levels of observed population structure among females likely resulted from a decrease in reproductive skew (a behavioral pattern) caused by rapid recent population growth (a demographic effect) [24]. Similar kinds of data are available for many natural primate populations, making it possible to connect estimates of population genetic change with the underlying behavioral processes that generate it [36, 37]. Such studies can illustrate how behavioral variation among individuals and behavioral plasticity within individuals—hallmarks of many primates—can influence the maintenance and distribution of genetic variation within populations (and also between populations and species in the more extreme case of hybridization: Box 2).
Box 2: Genetic and phenotypic analyses of hybridization in primates
Genetic analyses and field observations can be merged to study an extreme case of gene flow, naturally occurring hybridization between primate species. Hybridization is a common phenomenon among primates, and has been proposed to play an important role in the evolution of the primate lineage [80]. Studying hybridization in wild primates is therefore of great interest for investigating the emergence of genetic and phenotypic differences between divergent groups.
Phenotypic data and genetic estimates of admixture can be combined, for instance, to investigate how hybridization influences fitness-related traits. In baboons (genus Papio), naturally occurring hybridization occurs at the geographic boundaries between all five species. Long-term observations in a hybrid zone in Ethiopia between anubis baboons (P. anubis) and hamadryas baboons (P. hamadryas) [81] suggest that behavioral prezygotic isolation does not play a strong role in checking this process, despite markedly different social structures that characterize these two species. Although hybrid males suffer a disadvantage in gaining mates in some groups, they do equally as well as other males in groups that include many hybrids [82]. In Kenya, where yellow baboons (P. cynocephalus) and anubis baboons sometimes hybridize, genetic and phenotypic data suggest that hybrids might in fact enjoy a fitness advantage [36, 37]. More anubis-like individuals mature earlier, especially males [36]; anubis-like males also appear to be more successful in competing for mates (Tung, Alberts, and J. Altmann, unpublished data). These data contrast sharply with work on hybridization in New World howler monkeys. Although mantled howler monkeys (Alouatta palliata) and black howler monkeys (A. pigra) naturally hybridize where their ranges overlap in Mexico, observational and genetic evidence suggest that only female hybrids are viable and fertile [83]. This case represents perhaps the best evidence for the accumulation of intrinsic postzygotic isolation in naturally hybridizing primates, suggesting fertile ground for comparison between cases of hybridization across different primate taxa.
Future work on these systems will be able to both investigate how genetic background correlates with interesting traits, and attempt to identify the loci responsible for phenotypic differences between hybridizing species. Admixture mapping approaches, which investigate how ancestry-informative genetic markers and trait variation cosegregate among admixed individuals, will be particularly appropriate for this line of work [84]. Additionally, increasing amounts of genetic data on hybridizing populations will enable investigators to complement data on hybridization in the present with estimates of the timing and rate of gene flow between species in the past.
Testing the selective import of functional variation
The action of natural selection on functional genetic variants (i.e., polymorphisms that have a causal relationship with trait variation) is responsible for much of the phenotypic diversity in behavior, physiology, and morphology exhibited by primates, including traits that differentiate humans from other primates. Identifying loci that have been targets of natural selection via sequence analysis is therefore a vital area of research in primate genetics. However, even when a locus is strongly marked by natural selection, understanding the reasons underlying its selective history can be greatly enhanced by studying its role within a natural ecological context (particularly in cases in which selection is thought to be strong). Combining sequence-based evidence for selection with phenotypic observations in the field therefore represents a natural integration of genetics, ecology, and behavior in wild populations.
This approach is nicely illustrated by work on the adaptive significance of trichromacy (i.e., full color vision). Despite the long history of research on color vision in primates, the selective advantage(s) of trichromacy continue to be the subject of debate [38–40]. “Allelic trichromacy,” in which individuals within a species can be either dichromatic (red-green colorblind) or trichromatic depending on genotype, presents the opportunity to study the role of different visual systems in a natural ecological setting. Indeed, sequence-based analyses already support a role for natural selection in maintaining the polymorphisms responsible for variation in visual system [41, 42], and the functional effects of the variants themselves are well characterized. The signature of selection in sequence data alone, however, is compatible with several different selective mechanisms, including heterozygote advantage, frequency-dependent selection, and niche differentiation by genotype.
Field studies provide a way to test these alternative hypotheses. In these studies, genotypic information on functional opsin variation is integrated with phenotypic information on individual behavior collected under natural conditions. For example, one prediction of a heteroyzygote advantage hypothesis is that heterozygous individuals exhibit an overall foraging advantage relative to homozygotes (all trichromatic individuals are heterozygotes in these species; homozygotes are dichromatic). However, when tested in the field, no general trichromat advantage has been identified for either wild spider monkeys (Ateles geoffroyi) [43] or wild white-faced capuchins (Cebus capucinus) [44]. Alternatively, a niche differentiation hypothesis predicts that dichromats and trichromats will excel at different kinds of foraging tasks. Indeed, in capuchins, dichromats detect camouflaged insects better in low light, whereas trichromats enjoy an advantage in obtaining embedded, noncryptic insects [45], although they do not spend different amounts of time feeding on specific resources [46]. These studies have thus made early inroads into understanding the selective import of different visual systems in natural populations. Comparative work across other species that exhibit allelic trichromacy (most New World monkeys and several lemurs), but that experience different ecological circumstances, should shed further light on these questions.
Thus, field data can help fill in even a well-studied evolutionary picture by providing a testing ground for investigating how genetically different individuals differ phenotypically in the wild. In cases where similar functional genetic variants segregate in both humans and nonhuman primates, they can also shed light on our own evolutionary past. For example, both humans and chimpanzees (Pan troglodytes) harbor genetic variation at the TAS2R38 gene which alters individual sensitivity to bitter-tasting compounds [47], and which appears to have evolved under selection in humans [48]. Studying the proximate impetus for natural selection on human populations is difficult, given the dramatic changes in the human diet from ancient to modern times. The parallel relationship in chimpanzees, however, presents the opportunity to study how similar variants influence dietary choices in a natural context in our closest living relatives (although the selective regime on TAS2R38 in chimpanzees probably differed from that in humans: [47]).
Functional genetics and the genotype-phenotype relationship in wild primates
One of the most exciting possibilities for genetic research in natural primate populations lies in the prospect of identifying functional genetic variation that influences ecologically and adaptively relevant traits (Box 3). This area of research is in the early phase of development. Indeed, the genome-scale approaches that will likely move this area forward have, until recently, been impracticable in wild primates. Thus, functional studies in primates have largely relied on candidate gene approaches motivated by existing information on humans, and genotype-phenotype mapping studies have largely been confined to captive animals (Box 4). However, even at this modest scale, several of the unique contributions that arise from combining genetic approaches with behaviorally and ecologically well-studied populations are already apparent.
Box 3: Why conduct functional genetic research in wild primates?
To date, most functional genetic studies in primates have been conducted in a captive or laboratory setting, where individual subjects can be manipulated and in vitro tools (such as luciferase reporter assays in cell culture) can be exploited. However, although these approaches are of great value, they are also limited. In vitro effects do not always recapitulate in vivo biology [85], and a functional effect identified in vitro might not be relevant to animals in their natural environment. Gene-environment interactions, in which functional genetic effects change in direction or magnitude across environmental conditions, can also confound attempts to extrapolate from the laboratory to natural populations. Additionally, captivity itself can produce artifacts, particularly in stress and immune-related pathways [86].
These factors strongly argue for inclusion of individuals sampled under natural conditions in functional genetic studies, and the most obvious choice for such work are systems for which observational field data are readily available. Genetic work on these populations comes with some natural advantages. In particular, they bring to the table a wealth of existing knowledge about the distribution of phenotypic variation, the distribution of ecological and environmental variation that individuals within the sample experience, and the relationship between these two factors. Given that phenotypic variation is classically modeled as the result of a combination of environmental factors, genetic factors, and the dependent interactions between them [87], this means that several major parts of the overall picture are already in place. Indeed, recent trait mapping work in human genetics suggests that incorporating environmental effects in genotype-phenotype modeling can improve the ability to detect genetic associations [88]. Finally, these data also position natural primate populations as good models for studying the evolutionary relevance of genotype-environment interactions.
Furthermore, natural primate populations provide a unique window into human biology and evolution. Unlike studies in modern human populations, behavioral and demographic observations of nonhuman primates occur in real-time and can be extremely fine-grained (in some cases, occurring on a near-daily basis). Nonhuman primates are therefore particularly well suited for certain kinds of studies, including longitudinal studies of maturation and aging [5], and studies that investigate the relationship between genetic effects and sociosexual behaviors. Because the ecological circumstances of nonhuman primates have not changed over time as dramatically as they have for modern humans, they also allow researchers to interrogate the selective effects of genetic variation in an environment more similar to that in which it evolved.
Box 4: Captive primate populations
Although the goals of research on captive primates often differ from the goals of work on natural populations, the availability of large research communities that work in both milieus creates the opportunity for collaboration and interplay between research done in captive settings and in the wild. This can take at least three forms:
Resource development
Because of the strong biomedical focus of many captive primate facilities, development of genetic resources has been a priority for species that are medical models for human disease. These priorities have influenced both major institutional priorities, such as the choice of genome sequencing targets, and also the generation of additional resources, especially genome-wide linkage maps [89–91]. These markers often translate well between captive and natural populations. As marker development becomes a greater priority for field populations as well, this exchange of resources is likely to accelerate and become increasingly bidirectional.
Hypothesis generation and an alternative testing arena
Studies in captivity can be important for providing the first tests of ecological and evolutionary hypotheses. For example, foraging tasks set for captive animals have been key to understanding the selective maintenance of allelic trichromacy (e.g., [92, 93]). In other cases, work on captive animals has focused on phenotypes or environments that are less obviously relevant to evolution in natural populations. Captive studies in rhesus macaques, for instance, have delved into the genetic and environmental basis for alcoholism and for aggressive behavior in animals reared by peers [1]. Although these tests are not obviously mirrored by naturally occurring situations, they have identified genetic variants and broad environmental categories that motivate research on other traits with a potentially shared mechanistic basis (e.g., [62]).
Replication
In nonhuman primates, sample sizes for genetic studies will generally be relatively small. Where possible, the existence of both captive and wild populations for a given species can be leveraged to test replication of apparent genetic effects (although some caveats attach to such comparisons: see Box 3). For example, hybridization between yellow baboons and anubis baboons is known to influence morphological traits in the wild [94]. A QTL mapping study in captive baboons (a colony that also includes anubis-yellow hybrids) has identified candidate regions of the genome that influence morphological variation [95], providing insight into the possible basis for this effect. As the genetic basis for such traits becomes more clear, checking for consistency between multiple populations should therefore be of considerable interest.
First, field studies enable research into traits that might not be relevant or variable in captive primates, but are important in natural populations and are relevant to human health and disease. Wild primates are subject to naturally occurring infection by a wide variety of parasites and pathogens that do not occur in captive colonies. Simian immunodeficiency virus (SIV) is both common and highly pathogenic in the wild chimpanzee population of Gombe National Park, for example, but had previously been thought to be rare and nonpathogenic based on studies in captive chimps [49]. In such cases, field data are critical to understanding genetic variation that influences trait variation. For instance, like humans, nonhuman primates act as hosts for the group of parasites that cause malaria in humans. Indeed, in several cases, these parasites have been shown to cross-infect across multiple primate species (Plasmodium falciparum, for instance, has been shown to infect wild chimpanzees as well as humans: [50]) or have undergone evolutionary transitions to move from nonhuman primates to humans [51]. However, little is known about how genetic variation in wild primates influences risk of infection. Baboon populations in east Africa exhibit high rates of natural infection by Hepatocystis kochi, a parasite nested within the primate Plasmodium clade, offering an opportunity to pursue such work [52]. Indeed, recent evidence supports a link between Hepatocystis susceptibility in wild baboons and genetic variation at the baboon Duffy antigen receptor for chemokines (DARC; also abbreviated FY), which also influences gene expression of the baboon DARC gene in vivo [53]. This association parallels the known link in humans between DARC genetic variation and infection by the malarial parasite Plasmodium vivax, an effect that is mediated by variation in DARC gene expression [54, 55]. Other disease-related traits might be influenced by similar parallelisms. For example, copy number variation at the chemokine (C-C motif) ligand 3-like (CCL3L) locus has been linked in some studies to HIV or SIV progression in humans [56] and captive rhesus macaques (Macaca mulatta) [57]. Segregating copy number variation at CCL3L is also found in chimpanzees [56, 57], and represents a potential target for such work.
Second, field data are necessary to understand the role of functional genetic variation in natural populations, even when the genetic variants are well studied in captive animals. For example, the serotonin transporter gene, SLC6A4, encodes a protein that plays an important part in controlling circulating serotonin concentration. Like humans, rhesus macaques harbor segregating genetic variation in both the promoter and 3’ untranslated region of this gene that influences levels of SLC6A4 expression [58–60] and cerebrospinal fluid serotonin metabolites [58]. Because levels of these metabolites, especially 5-hydroxyindoleacetic acid (5-HIAA), have been linked with natal dispersal timing in free-ranging male rhesus macaques [61], genetic variation in SLC6A4 was hypothesized to also associate with differences in male dispersal timing. Indeed, a functional promoter variant at this locus significantly associates with timing of male natal dispersal (the low expressing allele is linked to delayed dispersal: [62]) and, perhaps because of the dispersal effect, with timing of male reproduction [63]. This example highlights the type of trait that will be of particular interest for primate field studies: even though dispersal timing is impossible to study in the laboratory, it is an important fitness-related trait in many primates, and one that has been extensively studied in natural populations [19].
What lies ahead for the genetics of natural primate populations?
Field-based ecological, behavioral, and demographic data already play an important role in genetic studies of nonhuman primates, and will be increasingly important in the future as the scope and scale of genetic data on wild primates grow. Recent technological and methodological advances, especially high throughput sequencing approaches, represent a major advance for the field. Indeed, the extensive data collection these tools enable mean that, for the first time, the quality and quantity of genetic data on wild primates will complement the rich behavioral and ecological data sets already in place for many of these species. This expansion of the available data will undoubtedly have dramatic consequences for studies of primate genetics, and we outline some of the possibilities below.
Novel data sets for resource development and genomic exploration
As a direct consequence of the falling cost of genomic technologies, population-based data collection is becoming increasingly feasible to conduct, even for non-model organisms. As a result, individual investigators are now able to collect large genetic data sets tailored to their specific species and/or study populations, at a much more rapid rate than in the past. This development will not only lead to a vast increase in the availability of genetic markers, but will also allow primate evolutionary geneticists to explore aspects of genome function that go beyond variation at the sequence level (including quantitative measurements of gene expression, epigenetic patterns, protein-DNA binding, and copy number variation: [64]), particularly for tissues that are often feasible to sample in the field, such as skin and blood (see also “Challenges”). Conducting such assays on a genome-wide level will allow investigators to ask how change at the genome sequence level translates into change in genome function, including at adaptively relevant traits. This research question is already being addressed in interspecific comparisons between humans, chimpanzees, and rhesus macaques (the primates that currently boast the highest quality genome sequence) [65, 66]. However, we still know very little about intraspecific variation in genome structure and function in primates, including how variation is partitioned among populations that are exposed to different ecological regimes (a major area of research in human genetics, but for which no comparative data in other primates currently exist). We anticipate that the next several years will witness a vast expansion in the data sets needed to address these questions.
Population history and demography
Previous studies have provided important insights into demographic processes such as admixture and gene flow [18]. New waves of genetic data are positioned to build on this work by providing much more fine-grained estimates of these processes. These results will eventually allow us to investigate, for example, how and why the evolutionary histories of species that experienced similar patterns of historic environmental change might have differed. Genetic data can also be combined with high-resolution geographic information system (GIS) data to understand how features of the environment affect the distribution of genetic variation in modern populations (part of the emerging field of landscape genetics: [67]). Such analyses can be leveraged both to understand the behavioral and ecological determinants of population structure in the present, and to help interpret the possible role of ecological and environmental factors that influenced genetic exchange in the more distant past. Studies with relatively limited genetic data sets have already revealed clues to how anthropogenic environmental change has influenced the distribution of several endangered primates [68, 69]. With the greater power provided by larger data sets, these analyses will soon become generalizable to a larger number of systems and accompanied by improved inference into relationship between geographic variation and population history.
Functional genetics and the genotype-phenotype relationship
Trait mapping studies will also be able to take advantage of larger-scale genotyping and resequencing data sets. For candidate gene studies, these approaches will provide much improved ability to correct for potential confounds, such as cryptic population structure and relatedness. Even more importantly, they will allow the field to expand its perspective beyond candidate genes to investigate previously uncharacterized loci. These studies will help address, for instance, the degree to which parallel genotype-phenotype relationships between species (broadly defined here as functional genetic variants at homologous loci, which influence the same or similar phenotypes: [47, 53, 57, 62, 70]) relate to shared ancestral selection pressures. Finally, genetic studies will be able to better exploit the historic strengths of primate field research: individual-centered environmental and phenotypic data collection over the life course. For example, early life effects play an important role in influencing phenotypic variation in humans and in nonhuman primates [71–74]. The ability to scan the genome for environmentally induced epigenetic modifications presents a new opportunity to understand the possible mechanisms connecting early life with trait variation later in life.
Challenges
A central theme of this review is to emphasize how new technological and methodological developments are making it increasingly possible to conduct genetic studies in natural primate populations, where genetic inferences can be combined with behavioral, ecological, and other sources of data. Although these developments are unquestionably expanding the possibilities for research in primate genetics, some important challenges remain.
First, and especially for collection of genomic data sets other than sequence or genotyping data, obtaining tissue samples from wild primates will often be very difficult (especially for some tissue types of great interest, such as brain or liver). Thus, the first waves of analysis will likely focus on sample types that are easier to obtain, such as blood, skin, or samples suitable for microbiome analysis (fecal samples or vaginal, buccal, or nasal swabs); field studies have already proven adept at gathering such samples from a variety of primates (e.g., [25, 75]). Opportunistic sampling from natural deaths can also serve as a strategy for building up sample sets over time (akin to strategies for recovering tissues from zoo primates), although such opportunities will be rare. Although these approaches will not give us a comprehensive look at all aspects of genetic and genomic variation, research in humans has demonstrated that we can learn a great deal from blood cells alone (all the HapMap cell lines, for example, are lymphocytes). Such samples are also highly relevant for a major focus of primate research, the evolution of the immune system and disease resistance. With optimization, the new generation of genomic tools might also become applicable to noninvasively collected samples such as shed hair and feces, greatly expanding the possibilities for population-based research.
Second, dissecting genotype-phenotype relationships remains a major challenge across all organisms, including in humans and in model systems. Even in the most extensively studied nonhuman primates, long life histories and relatively low densities mean that sample sizes are will likely remain modest. This area invites collaboration between researchers working on different populations, including those focused on captive populations (Box 4). Investigators interested in pursuing trait associations will also often need to include alternative sources of information—particularly about the mechanisms through which genetic effects act—into genotype-phenotype mapping studies. Indeed, in the examples of genotype-phenotype studies published thus far in primates, in vitro functional tests and in vivo measurements of molecular intermediaries such as gene expression have bolstered the overall case for a genotype-phenotype relationship.
Finally, much of this work will rely extensively on computational and statistical modeling skills, in addition to expertise in field research, behavior, and ecology. Developing skills in statistics and programming will therefore be an important training priority. Taking account of population and species-level characteristics in analyses of large-scale data sets will therefore require substantial investment in developing models appropriate for each individual system.
Conclusions
Genetic studies in primates present the exciting possibility that genetic inferences can be placed in the context of complementary behavioral and ecological data about the same systems, gathered under natural conditions. This opportunity has been made possible by generations of primate field research, which have produced a well-developed framework for understanding the causes and consequences of genetic evolution. As genomic resources for these species proliferate, natural primate populations will become increasingly good subjects for evolutionary genetics research. This area of research therefore has the potential to spur remarkable new collaborations that bridge lab-based molecular genetics, computational modeling and data analysis, and field-based data collection on natural populations.
Additional Materials
Acknowledgments
We thank G.H. Perry for directing us to some relevant pieces of literature and C.C. Babbitt and four anonymous reviewers for helpful comments on the manuscript. J.T. is supported by a Duke University Katherine Goodman Stern Dissertation Fellowship; S.C.A. is supported by NSF grants DEB-0846286 and IOS-0919200 and NIH grants AG034513-01 and AG031719-01A1; G.A.W. is supported by NSF grants DEB-0846286 and BCS-0827552 and NIH grant 5P50-GM-081883-02.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barr CS, et al. The utility of the non-human primate model for studying gene by environment interactions in behavioral research. Genes Brain Behav. 2003;2:336–340. doi: 10.1046/j.1601-1848.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 2.Bellino FL, Wise PM. Nonhuman Primate Models of Menopause Workshop. Biol Reprod. 2003;68:10–18. doi: 10.1095/biolreprod.102.005215. [DOI] [PubMed] [Google Scholar]
- 3.Jolly CJ. A proper study for mankind: analogies from the papionin monkeys and their implications for human evolution. Yrbk Phys Anthropol. 2001;116:177–204. doi: 10.1002/ajpa.10021. [DOI] [PubMed] [Google Scholar]
- 4.Rogers J, Hixson JE. Baboons as an animal model for genetic studies of common human disease. Am J Hum Genet. 1997;61:489–493. doi: 10.1086/515527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronikowski AM, et al. The aging baboon: Comparative demography in a non-human primate. P Natl Acad Sci USA. 2002;99:9591–9595. doi: 10.1073/pnas.142675599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunn CL, et al. Promiscuity and the primate immune system. Science. 2000;290:1168–1170. doi: 10.1126/science.290.5494.1168. [DOI] [PubMed] [Google Scholar]
- 7.Van Schaik CP, Kappeler PM. Infanticide risk and the evolution of male-female association in primates. P Roy Soc Lond B Bio. 1997;264:1687–1694. doi: 10.1098/rspb.1997.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Fiore A, Gagneux P. Molecular Primatology. In: Campbell CJ, et al., editors. Primates in Perspective. Oxford University Press; 2007. pp. 369–393. [Google Scholar]
- 9.Goodman M, et al. Moving primate genomics beyond the chimpanzee genome. Trends Genet. 2005;21:511–517. doi: 10.1016/j.tig.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Siepel A. Phylogenomics of primates and their ancestral populations. Genome Res. 2009;19:1929–1941. doi: 10.1101/gr.084228.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchan JC, et al. True paternal care in a multi-male primate society. Nature. 2003;425:179–181. doi: 10.1038/nature01866. [DOI] [PubMed] [Google Scholar]
- 12.Perry S, et al. Kin-biased social behaviour in wild adult female white-faced capuchins, Cebus capucinus. Anim Behav. 2008;76:187–199. [Google Scholar]
- 13.Vigilant L, et al. Paternity and relatedness in wild chimpanzee communities. P Natl Acad Sci USA. 2001;98:12890–12895. doi: 10.1073/pnas.231320498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberts SC, Altmann J. Balancing costs and opportunities: dispersal in male baboons. Amer Natur. 1995;145:279–306. [Google Scholar]
- 15.Cheney DL, et al. Reproductive success in vervet monkeys. In: Clutton-Brock TH, editor. Reproductive Success. University of Chicago Press; 1988. [Google Scholar]
- 16.Lawler RR. Fitness and extra-group reproduction in male Verreaux's sifaka: An analysis of reproductive success from 1989–1999. Am J Phys Anthropol. 2007;132:267–277. doi: 10.1002/ajpa.20507. [DOI] [PubMed] [Google Scholar]
- 17.Smuts BB, et al., editors. Primate Societies. Chicago University Press; 1987. [Google Scholar]
- 18.Vigilant L. Elucidating population histories using genomic DNA sequences. Curr Anthropol. 2009;50:201–212. doi: 10.1086/592025. [DOI] [PubMed] [Google Scholar]
- 19.Pusey AE. Sex-biased dispersal and inbreeding avoidance in birds and mammals. Trends Ecol Evol. 1987;2:295–299. doi: 10.1016/0169-5347(87)90081-4. [DOI] [PubMed] [Google Scholar]
- 20.Chesser RK. Influence of gene flow and breeding tactics on gene diversity within populations. Genetics. 1991;129:573–583. doi: 10.1093/genetics/129.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugg DW, et al. Population genetics meets behavioral ecology. Trends Ecol Evol. 1996;11:338–342. doi: 10.1016/0169-5347(96)20050-3. [DOI] [PubMed] [Google Scholar]
- 22.Archie EA, et al. Fine-scale population genetic structure in a fission-fusion society. Mol Ecol. 2008;17:2666–2679. doi: 10.1111/j.1365-294X.2008.03797.x. [DOI] [PubMed] [Google Scholar]
- 23.Foerster K, et al. A spatial genetic structure and effects of relatedness on mate choice in a wild bird population. Mol Ecol. 2006;15:4555–4567. doi: 10.1111/j.1365-294X.2006.03091.x. [DOI] [PubMed] [Google Scholar]
- 24.Nussey DH, et al. Rapidly declining fine-scale spatial genetic structure in female red deer. Mol Ecol. 2005;14:3395–3405. doi: 10.1111/j.1365-294X.2005.02692.x. [DOI] [PubMed] [Google Scholar]
- 25.Altmann J, et al. Behavior predicts genetic structure in a wild primate group. P Natl Acad Sci USA. 1996;93:5797–5801. doi: 10.1073/pnas.93.12.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pope TR. The influence of dispersal patterns and mating system on genetic differentiation within and between populations of the red howler monkey (Alouatta seniculus) Evolution. 1992;46:1112–1128. doi: 10.1111/j.1558-5646.1992.tb00623.x. [DOI] [PubMed] [Google Scholar]
- 27.Van Schaik CP. The ecology of social relationships amongst female primates. In: Standen V, Foley RA, editors. Comparative Socioecology: The Behavioral Ecology of Humans and Other Mammals. Blackwell Press; 1989. pp. 195–218. [Google Scholar]
- 28.Wrangham RW. An ecological model of female-bonded primate groups. Behaviour. 1980;75:262–300. [Google Scholar]
- 29.Guschanski K, et al. Females shape the genetic structure of a gorilla population. Curr Biol. 2008;18:1809–1814. doi: 10.1016/j.cub.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Ackermann RR, Bishop JM. Morphological and molecular evidence reveals recent hybridization between gorilla taxa. Evolution. 2010;64:271–290. doi: 10.1111/j.1558-5646.2009.00858.x. [DOI] [PubMed] [Google Scholar]
- 31.Thalmann OH, et al. The complex evolutionary history of gorillas: Insights from genomic data. Mol Biol Evol. 2007;24:146–158. doi: 10.1093/molbev/msl160. [DOI] [PubMed] [Google Scholar]
- 32.Becquet C, et al. Genetic structure of chimpanzee populations Plos Genet 20073,p- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caswell JL, et al. Analysis of chimpanzee history based on genome sequence alignments. Plos Genet. 2008;4 doi: 10.1371/journal.pgen.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer A, et al. Demographic history and genetic differentiation in apes. Curr Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez RD, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- 36.Charpentier MJE, et al. Age at maturity in wild baboons: genetic, environmental and demographic influences. Mol Ecol. 2008;17:2026–2040. doi: 10.1111/j.1365-294X.2008.03724.x. [DOI] [PubMed] [Google Scholar]
- 37.Tung J, et al. Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Mol Ecol. 2008;17:1998–2011. doi: 10.1111/j.1365-294X.2008.03723.x. [DOI] [PubMed] [Google Scholar]
- 38.Dominy NJ, Lucas PW. Ecological importance of trichromatic vision to primates. Nature. 2001;410:363–366. doi: 10.1038/35066567. [DOI] [PubMed] [Google Scholar]
- 39.Mollon JD. Tho She Kneeld in That Place Where They Grew … The Uses and Origins of Primate Color-Vision. J Exp Biol. 1989;146 doi: 10.1242/jeb.146.1.21. 21-&. [DOI] [PubMed] [Google Scholar]
- 40.Surridge AK, et al. Evolution and selection of trichromatic vision in primates. Trends Ecol Evol. 2003;18:198–205. [Google Scholar]
- 41.Hiwatashi T, et al. An Explicit Signature of Balancing Selection for Color-Vision Variation in New World Monkeys. Mol Biol Evol. 2010;27:453–464. doi: 10.1093/molbev/msp262. [DOI] [PubMed] [Google Scholar]
- 42.Surridge AK, Mundy NI. Trans-specific evolution of opsin alleles and the maintenance of trichromatic colour vision in Callitrichine primates. Mol Ecol. 2002;11:2157–2169. doi: 10.1046/j.1365-294x.2002.01597.x. [DOI] [PubMed] [Google Scholar]
- 43.Hiramatsu C, et al. Importance of achromatic contrast in short-range fruit foraging of primates. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel ER, et al. Effect of color vision phenotype on the foraging of wild white-faced capuchins, Cebus capucinus. Behav Ecol. 2007;18:292–297. [Google Scholar]
- 45.Melin AD, et al. Effects of colour vision phenotype on insect capture by a free-ranging population of white-faced capuchins, Cebus capucinus. Anim Behav. 2007;73:205–214. [Google Scholar]
- 46.Melin AD, et al. Polymorphic color vision in white-faced capuchins (Cebus capucinus): Is there foraging niche divergence among phenotypes? Behav Ecol Sociobiol. 2008;62:659–670. [Google Scholar]
- 47.Wooding S, et al. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- 48.Wooding S, et al. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. P Natl Acad Sci USA. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escalante AA, et al. A monkey's tale: the origin of Plasmodium vivax as a human malaria parasite. P Natl Acad Sci USA. 2005;102:1980–1985. doi: 10.1073/pnas.0409652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips-Conroy JE, et al. Hepatocystis in populations of baboons (Papio hamadryas s.l.) of Tanzania and Ethiopia. J Med Primatol. 1988;17:145–152. [PubMed] [Google Scholar]
- 53.Tung J, et al. Evolution of a malaria resistance gene in wild primates. Nature. 2009;460:388–392. doi: 10.1038/nature08149. [DOI] [PubMed] [Google Scholar]
- 54.Miller LH, et al. Resistance factor to Plasmodium vivax in blacks - Duffy Blood Group genotype, Fyfy. New Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 55.Tournamille C, et al. Disruption of a GATA motif in the Duffy gene promoter abolishes erthyroid gene-expression in Duffy negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 56.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 57.Degenhardt JD, et al. Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in rhesus macaques (Macaca mulatta) Plos Genet. 2009;5 doi: 10.1371/journal.pgen.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bennett AJ, et al. Early experience and serotonin transporter gene variation interact to influence primate CNS function. Mol Psychiatr. 2002;7:118–122. doi: 10.1038/sj.mp.4000949. [DOI] [PubMed] [Google Scholar]
- 59.Lesch KP, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 60.Vallender EJ, et al. Functional variation in the 3 ' untranslated region of the serotonin transporter in human and rhesus macaque. Genes Brain Behav. 2008;7:690–697. doi: 10.1111/j.1601-183X.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- 61.Kaplan JR, et al. Delayed dispersal and elevated monoaminergic activity in free-ranging rhesus monkeys. Am J Primatol. 1995;35:229–234. doi: 10.1002/ajp.1350350305. [DOI] [PubMed] [Google Scholar]
- 62.Trefilov A, et al. Natal dispersal in rhesus macaques is related to serotonin transporter gene promoter variation. Behav Genet. 2000;30:295–301. doi: 10.1023/a:1026597300525. [DOI] [PubMed] [Google Scholar]
- 63.Krawczak M, et al. Male reproductive timing in rhesus macaques is influenced by the 5HTTLPR promoter polymorphism of the serotonin transporter gene. Biol Reprod. 2005;72:1109–1113. doi: 10.1095/biolreprod.104.038059. [DOI] [PubMed] [Google Scholar]
- 64.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 65.Babbitt CC, et al. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol Evol. 2010;2010:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blekhman R, et al. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manel S, et al. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 2003;18:189–197. [Google Scholar]
- 68.Liu ZJ, et al. The effect of landscape features on population genetic structure in Yunnan snub-nosed monkeys (Rhinopithecus bieti) implies an anthropogenic genetic discontinuity. Mol Ecol. 2009;18:3831–3846. doi: 10.1111/j.1365-294X.2009.04330.x. [DOI] [PubMed] [Google Scholar]
- 69.Quemere E, et al. Landscape genetics of an endangered lemur (Propithecus tattersalli) within its entire fragmented range. Mol Ecol. 2010;19:1606–1621. doi: 10.1111/j.1365-294X.2010.04581.x. [DOI] [PubMed] [Google Scholar]
- 70.Loisel DA, et al. Ancient polymorphism and functional variation in the primate MHC-DQA1 5 ' cis-regulatory region. P Natl Acad Sci USA. 2006;103:16331–16336. doi: 10.1073/pnas.0607662103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barker DJP, et al. Fetal origins of adult disease: strength of effects and biological basis. Intl J Epidemiol. 2002;31:1235–1239. doi: 10.1093/ije/31.6.1235. [DOI] [PubMed] [Google Scholar]
- 72.Coplan JD, et al. Persistent elevations of cerebrospinal fluid concentrations of corticotropin-releasing factor in adult nonhuman primates exposed to early-life stressors: Implications for the pathophysiology of mood and anxiety disorders. P Natl Acad Sci USA. 1996;93:1619–1623. doi: 10.1073/pnas.93.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Onyango PO, et al. Persistence of maternal effects in baboons: Mother's dominance rank at son's conception predicts stress hormone levels in subadult males. Horm Behav. 2008;54:319–324. doi: 10.1016/j.yhbeh.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roseboom TJ, et al. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Mol Cellul Endocrinol. 2001;185:93–98. doi: 10.1016/s0303-7207(01)00721-3. [DOI] [PubMed] [Google Scholar]
- 75.Glander KE, et al. Field methods for capture and measurement of 3 monkey species in Costa Rica. Folia Primatol. 1991;57:70–82. doi: 10.1159/000156567. [DOI] [PubMed] [Google Scholar]
- 76.Becquet C, Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17:1505–1519. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fischer A, et al. Evidence for a complex demographic history of chimpanzees. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- 78.Yu N, et al. Low nucleotide diversity in chimpanzees and bonobos. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- 80.Arnold ML, Meyer A. Natural hybridization in primates: One evolutionary mechanism. Zoology. 2006;109:261–276. doi: 10.1016/j.zool.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 81.Phillips-Conroy JE, Jolly CJ. Changes in the structure of the baboon hybrid zone in the Awash National Park, Ethiopia. Am J Phys Anthropol. 1986;71:337–350. [Google Scholar]
- 82.Bergman TJ, et al. Behavioral variation and reproductive success of male baboons (Papio anubis x Papio hamadryas) in a hybrid social group. Am J Primatol. 2008;70:136–147. doi: 10.1002/ajp.20467. [DOI] [PubMed] [Google Scholar]
- 83.Cortes-Ortiz L, et al. Hybridization in large-bodied new world primates. Genetics. 2007;176:2421–2425. doi: 10.1534/genetics.107.074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buerkle CA, Lexer C. Admixture as the basis for genetic mapping. Trends Ecol Evol. 2008;23:686–694. doi: 10.1016/j.tree.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 85.Cirulli ET, Goldstein DB. In vitro assays fall to predict in vivo effects of regulatory polymorphisms. Hum Mol Genet. 2007;16:1931–1939. doi: 10.1093/hmg/ddm140. [DOI] [PubMed] [Google Scholar]
- 86.Kennerly E, et al. A gene expression signature of confinement in peripheral blood of red wolves (Canis rufus) Mol Ecol. 2008;17:2782–2791. doi: 10.1111/j.1365-294X.2008.03775.x. [DOI] [PubMed] [Google Scholar]
- 87.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Longman Group; 1996. [Google Scholar]
- 88.Igl W, et al. Modeling of environmental effects in genome-wide association studies identifies SLC2A2 and HP as novel loci influencing serum cholesterol levels. Plos Genet. 2010;6:e1000798. doi: 10.1371/journal.pgen.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jasinska AJ, et al. A genetic linkage map of the vervet monkey (Chlorocebus aethiops sabaeus) Mamm Genome. 2007;18:347–360. doi: 10.1007/s00335-007-9026-4. [DOI] [PubMed] [Google Scholar]
- 90.Rogers J, et al. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 2006;87:30–38. doi: 10.1016/j.ygeno.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 91.Rogers J, et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- 92.Caine NG, et al. Dichromatic and trichromatic Callithrix geoffroyi differ in relative foraging ability for red-green color-camouflaged and non-camouflaged food. Int J Primatol. 2003;24:1163–1175. [Google Scholar]
- 93.Leonhardt SD, et al. Seeing red: behavioral evidence of trichromatic color vision in strepsirrhine primates. Behav Ecol. 2009;20:1–12. [Google Scholar]
- 94.Alberts SC, Altmann J. Immigration and hybridization patterns of yellow and anubis baboons in and around Amboseli, Kenya. Am J Primatol. 2001;53:139–154. doi: 10.1002/ajp.1. [DOI] [PubMed] [Google Scholar]
- 95.Sherwood RJ, et al. A genomewide linkage scan for quantitative trait loci influencing the craniofacial complex in baboons (Papio hamadryas spp.) Genetics. 2008;180:619–628. doi: 10.1534/genetics.108.090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nishida T, editor. The Chimpanzees of the Mahale Mountains: Sexual and Life History Strategies. University of Tokyo Press; 1990. [Google Scholar]
- 97.Mitani JC, Watts DP. Why do chimpanzees hunt and share meat? Anim Behav. 2001;61:915–924. [Google Scholar]
- 98.Stokes EJ, et al. Female dispersal and reproductive success in wild western lowland gorillas (Gorilla gorilla gorilla) Behav Ecol Sociobiol. 2003;54:329–339. [Google Scholar]
- 99.van Schaik CP, et al. Orangutan cultures and the evolution of material culture. Science. 2003;299:102–105. doi: 10.1126/science.1078004. [DOI] [PubMed] [Google Scholar]
- 100.Barrett L, et al. Market forces predict grooming reciprocity in female baboons. P Roy Soc Lond B Bio. 1999;266:665–670. [Google Scholar]
- 101.Packer C, et al. Problems with primate sex ratios. Philos T Roy Soc B. 2000;355:1627–1635. doi: 10.1098/rstb.2000.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weyher A, et al. Molecular identification of Schistosoma mattheei from feces of kinda (Papio cynocephalus kindae) and grayfoot baboons (Papio cynocephalus griseipes) in Zambia. J Parasitol. 2009:1. doi: 10.1645/GE-2186.1. [DOI] [PubMed] [Google Scholar]
- 103.Whitten PL. Effects of patch quality and feeding subgroup size on feeding success in vervet monkeys (Cercopithecus aethiops) Behaviour. 1988;105:35–52. [Google Scholar]
- 104.Van Noordwijk MA, Van Schaik CP. The effects of dominance rank and group size on female lifetime reproductive success in wild long-tailed macaques, Macaca fascicularis. Primates. 1999;40:105–130. doi: 10.1007/BF02557705. [DOI] [PubMed] [Google Scholar]
- 105.Fashing PJ, et al. Influence of plant and soil chemistry on food selection, ranging patterns, and biomass of Colobus guereza in Kakamega Forest, Kenya. Int J Primatol. 2007;28:673–703. [Google Scholar]
- 106.Harris TR, Chapman CA. Variation in diet and ranging of black and white colobus monkeys in Kibale National Park, Uganda. Primates. 2007;48:208–221. doi: 10.1007/s10329-006-0036-8. [DOI] [PubMed] [Google Scholar]
- 107.Lazaro-Perea C. Intergroup interactions in wild common marmosets, Callithrix jacchus: territorial defence and assessment of neighbours. Anim Behav. 2001;62:11–21. [Google Scholar]
- 108.Mitchell CL, et al. Competitive regimes and female bonding in 2 species of squirrel monkeys (Saimiri oerstedi and S sciureus) Behav Ecol Sociobiol. 1991;28:55–60. [Google Scholar]
- 109.Boinski S, et al. Dispersal patterns among three species of squirrel monkeys (Saimiri oerstedii S boliviensis and S sciureus): I. Divergent costs and benefits. Behaviour. 2005;142:525–632. [Google Scholar]
- 110.Wright PC. Female Primates: Studies by Women Primatologists. Alan R. Liss; 1984. Biparental care in Aotus trivirgatus and Callicebus moloch; pp. 59–75. [Google Scholar]
- 111.Schmid J, Kappeler PM. Fluctuating sexual dimorphism and differential hibernation by sex in a primate, the gray mouse lemur (Microcebus murinus) Behav Ecol Sociobiol. 1998;43:125–132. [Google Scholar]
- 112.Nash LT, Harcourt CS. Social organization of galagos in Kenyan coastal forests. 2. Galago garnettii. Am J Primatol. 1986;10:357–369. doi: 10.1002/ajp.1350100407. [DOI] [PubMed] [Google Scholar]
- 113.Mitani JC. Demography of Agile Gibbons (Hylobates agilis) Int J Primatol. 1990;11:411–424. [Google Scholar]
- 114.Palombit RA. Pair bonds in monogamous apes: A comparison of the siamang Hylobates syndactylus and the white-handed gibbon Hylobates lar. Behaviour. 1996;133:321–356. [Google Scholar]
- 115.Brockelman WY, et al. Dispersal, pair formation and social structure in gibbons (Hylobates lar) Behav Ecol Sociobiol. 1998;42:329–339. [Google Scholar]