Abstract
Dietary vitamin A deficiency causes eye disease in 40 million children each year and places 140 to 250 million at risk for health disorders. Many children in sub-Saharan Africa subsist on maize-based diets. Maize displays considerable natural variation for carotenoid composition, including vitamin A precursors α-carotene, β-carotene, and β-cryptoxanthin. Through association analysis, linkage mapping, expression analysis, and mutagenesis, we show that variation at the lycopene epsilon cyclase (lcyE) locus alters flux down α-carotene versus β-carotene branches of the carotenoid pathway. Four natural lcyE polymorphisms explained 58% of the variation in these two branches and a threefold difference in provitamin A compounds. Selection of favorable lcyE alleles with inexpensive molecular markers will now enable developing-country breeders to more effectively produce maize grain with higher provitamin A levels.
Maize is the dominant subsistence crop in much of sub-Saharan Africa and the Americas, where between 17 and 30% of children under age of 5 are vitamin A–deficient. This results in xerophthalmia (progressive blindness), increased infant morbidity and mortality, and depressed immunological responses (1). Vitamin A deficiency starts with inadequate provitamin A or vitamin A content or bioavailability in foods and is exacerbated by disease-induced malabsorption.
Diet diversification, food fortification, and supplementation (2–4) have all been used to combat dietary micronutrient deficiencies. Ideally, all children would have access to a varied diet rich in fruits and vegetables, but diet diversification is often limited by crop seasonality, expense, and low bioavailability of green leafy plant carotenoids (5, 6). Poor infrastructure in developing countries has limited widespread use of direct vitamin supplementation. Perhaps the most feasible approach to eradicating death and disease caused by dietary deficiencies is biofortification, a process by which staple crops are purposefully bred for higher nutritional density (7, 8). Although biofortified foods can potentially be an inexpensive, locally adaptable, and long-term solution to diet deficiencies, cultural preferences may limit their acceptance. This may be particularly true for those crops where transgenics are the only alternative to boost provitamin A content, given limited acceptance of genetically modified organisms in developing countries.
Carotenoids are derived from the isoprenoid biosynthetic pathway and are precursors of the plant hormone abscisic acid and of other apocarotenoids (9). The first committed step of this pathway [as recently revised (10)] is formation of phytoene from geranylgeranyl diphosphate by phytoene synthase (y1/psy1) (Fig. 1) (11). Recent studies in maize suggest that the psy1 locus has been the target of a selective sweep following selection for endosperm-accumulating carotenoids and shift from white to yellow kernels (12). The first branch point of this pathway (Fig. 1) occurs at cyclization of lycopene where action of lycopene beta cyclase (LCYB) at both ends of linear lycopene produces a molecule with two β rings. Alternatively, the coaction of LCYB and lycopene epsilon cyclase (LCYE) generates a β,ε-carotene that is a precursor to lutein (13). Relative activities of LCYB and LCYE are hypothesized to regulate the proportion of carotenes directed to each branch of this pathway (13–15). Indeed, transgenic manipulations of LCYE expression in Arabidopsis, potato, and Brassica increase the pool of β ring–containing carotenes and xanthophylls (13, 16–18).
Fig. 1.
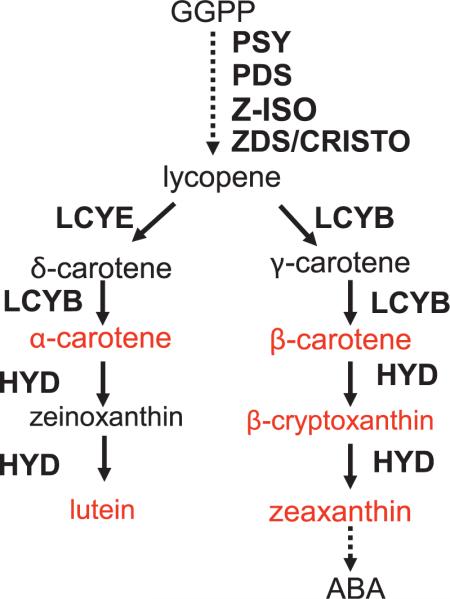
Simplified carotenoid biosynthetic pathway in plants (29). Enzymatic reactions are represented by arrows, dashed lines represent multiple enzymatic steps. Substrates in red were evaluated in this study. Compounds: GGPP, geranylgeranyl diphosphate; ABA, abscisic acid. Enzymes: PSY, phytoene synthase; PDS, phytoene desaturase; Z-ISO, 15-cis zetacarotene isomerase; ZDS, zetacarotene desaturase; CRTISO, carotene isomerase; HYD, carotene hydroxylase enzymes, which include ε- and β-ring hydroxylases.
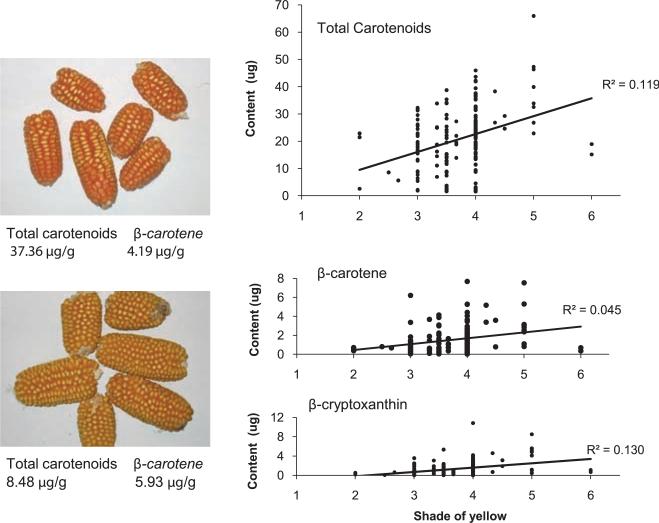
Maize exhibits considerable natural variation for kernel carotenoids, with some lines accumulating as much as 66 μg/g. The predominant carotenoids in maize kernels, in decreasing order of concentration, are lutein, zeaxanthin, β-carotene, β-cryptoxanthin, and α-carotene. β-Carotene contains two provitamin A structures (two nonhydroxylated β-ionone rings) and β-cryptoxanthin and α-carotene one each (single nonhydroxylated β-ionone ring). Among lines included in our diverse maize panel, β-carotene levels reached 13.6 μg/g. However, most yellow maize grown and consumed throughout the world has only 0.5 to 1.5 μg/g β-carotene. Comparisons between β-carotene and total carotenoids with grain color (scaled according to shade of yellow) revealed poor correlations with low R2 values (Fig. 2), which indicated that marker-assisted selection (MAS) may prove much more efficient than selection based on color alone.
Fig. 2.
Grain color and carotenoid content. The graphs depict the low correlation between visual grain color and total carotenoids, β-carotene, and β-cryptoxanthin in diverse inbreds. In these kernels, the shade of yellow ranges from white (score of 1) to dark orange (score of 6). White kernels were excluded from the analysis. The difficulty in visual selection for β-carotene content is further exemplified by the images on the left, where the yellow maize below has higher β-carotene than the orange variety above. These correlations are across the diverse panel of 228 maize inbreds; correlations for grain color and total carotenoids are higher when scored across segregating populations and narrow ranges of germ plasm, but correlations for β-carotene and β-cryptoxanthin remain low.
To dissect the phenotypic diversity, we used an association-mapping approach that exploits the genetic diversity of maize to provide resolution within 2000 base pairs (bp) (19–21). In the context of plant breeding, this has the added advantage of identifying the most favorable allele within a diverse genetic background, which provides the necessary genotypic information to facilitate the design of efficient maize introgression and selection schemes throughout the world. We complemented the association mapping with linkage mapping to evaluate the effects in a genetically less complex background and with a mutagenesis program to isolate novel allelic variation within an elite near-isogenic background.
To evaluate functional diversity (Fig. 1), eight candidate genes representing select members of gene families encoding biosynthetic enzymes of the carotenoid pathway were sampled across a diverse panel of 288 maize lines, of which 204 were yellow. Subsets of yellow lines were grown in four different years and surveyed for whole-kernel carotenoids by high-performance liquid chromatography (HPLC). The yellow lines averaged 23 μg/g for total carotenoids (range 5.5 to 66.0 μg/g) and 1.7 μg/g for β-carotene (range 0.06 to 13.6 μg/g).
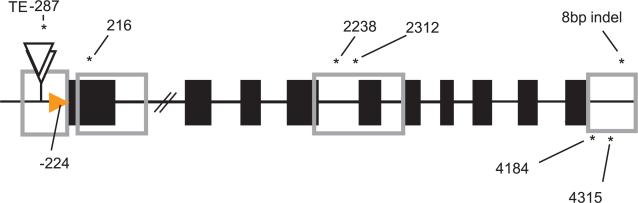
For association analysis, we used a mixed-model approach that controlled for complex population and pedigree relationships (22). Among our current sampling of candidate genes, lycopene epsilon cyclase (lcyE) (14) had the largest effect on partitioning the two branches of carotenoids and, consequently, on β-carotene and β-cryptoxanthin content. In maize, the single-copy lcyE gene consists of 10 exons spanning 3640 bp (Fig. 3). After initial association and screening for polymorphisms in key haplotypes, four regions were selected and scored across the entire panel. On the basis of the position of LCYE in the biochemical pathway, we predicted that the ratio of the sum of kernel carotenoids from each pathway branch would form the strongest association. Indeed, this was confirmed (Table 1), with the strength of the association confirming that lcyE plays a key role in controlling this ratio. Correspondingly, levels of predominant provitamin A compounds bcarotene and β-cryptoxanthin were also highly associated with lcyE.
Fig. 3.
Schematic diagram of the maize lcyE. Putative transcription start sites are depicted with orange arrow, translated exons as black squares, and the sampled regions as gray boxes. Polymorphisms that significantly associated with changes in flux between the lutein and zeaxanthin branches of the pathway are labeled with asterisks. The 5′ transposable element insertion(s) are represented by the white triangles. Positions relative to the sequence alignment are indicated numerically above the polymorphisms.
Table 1.
lcyE associations across seasons. Association results for significant polymorphisms identified in the four regions sampled along the lcyE gene. Each polymorphism is labeled numerically by its position on the alignment relative to the exon 1 start codon. Followed by the favorable allele (bold)/unfavorable allele at the site. An initial scan for association using both β-carotene and the ratio of the two pathway branches was conducted using the mixed model incorporating population structure and kinship. Subsequently a simpler general linear model (GLM) was used to evaluate data sets from additional years, including population structure (Q), given the oligogenic behavior of the trait the change in flux estimates for 2003 do not include Q. Avg., average; n.c., nonconvergence; n.s., not significant.
|
lcyE association (P), mixed model of |
lcyE association as a ratio across environments (GLM) (P) | Fold change in flux | |||||
|---|---|---|---|---|---|---|---|
| β-Carotene/all | Ratio of branches | ||||||
| Environment (year) | (2003) | (2003) | (2002) | (2003) | (2004) | (2005) | (2003) |
| Avg. observation no. | (157) | (154) | (44) | (156) | (154) | (156) | |
| Polymorphic site | |||||||
| 5′ TE 1+4/2/3 | 5.42 × 10−4 | 3.96 × 10−11 | 0.024 | 8.05 × 10−11 | 0.008 | 8.61 × 10−9 | 6.5 |
| 216 G / T | n.c. | 1.35 × 10−10 | 0.059 | 1.24 × 10−10 | 0.003 | 2.93 × 10−10 | 2.8 |
| 2238 G / T | 1.22 × 10−4 | 1.69 × 10−9 | 0.008 | 2.12 × 10−10 | 0.023 | 1.08 × 10−9 | 2.7 |
| 2312 A / T | 1.70 × 10−3 | n.c. | n.s. | 6.84 × 10−4 | 0.026 | 0.005 | 2.9 |
| 4184 G / A | 3.06 × 10−4 | n.c. | 8.87 × 10−4 | 2.23 × 10−10 | 0.019 | 1.13 × 10−8 | 2.6 |
| 4315 C / G | 1.84 × 10−4 | 7.01 × 10−10 | 0.012 | 3.07 × 10−9 | 5.75 × 10−4 | 6.79 × 10−7 | 2.6 |
| 3′Indel 8/0 | 4.80 × 10−3 | 2.75 × 10−9 | n.s. | 1.46 × 10−8 | 8.97 × 10−4 | 4.13 × 10−6 | 3.5 |
Subsequent haplotype analysis revealed several probable causative polymorphisms for the ratio of α- and β-carotene branches for the 2003 field season (table S1). A large promoter indel and an amino acid substitution in exon 1 explain most of the variation (R2 =36%; n = 135; P = 1.27 × 10−12) with a 5.2-fold effect. A second indel in the 3′ UTR also has a significant 3.3-fold effect and contributes to variation not explained by the promoter polymorphism (type III SS; P =1.9 × 10−4). The fourth significant polymorphism at position 2238 in intron 4 was associated with a 2.5-fold effect (type III SS; P = 0.0003). The overall, four-term model explains 58% of the variation (P = 9.2 × 10−17). These significant polymorphisms exhibit some linkage disequilibrium (LD), and only nine haplotypic classes exist in our sample, which limits full differentiation of the effects of each polymorphism. Overall, there is a ninefold difference between two of the more differentiated haplotype classes, and sixfold between two more common haplotypes (table S2). There was a threefold increase in the proportion of β-carotene and β-cryptoxanthin between the common haplotypes. Verification of these results was provided by significant associations in subsequent field seasons (Table 1).
Expression analysis indicated that lcyE is preferentially expressed in the endosperm relative to the embryo (fig. S1). Expression profiling of kernels at 15 and 20 days after pollination (DAP) indicated expression levels correlated well with the ratio of carotenoids from each pathway branch, explaining 70 to 76% of the variance. Lines with transposon insertions near the start site had much lower expression levels [in 15 DAP and 20 DAP lower bya factor of 3.7 and 13, respectively (fig. S2)]. The 3′ indel may also have expression effects, but our statistical tests lacked the power to confirm this hypothesis. A quantitative trait locus (QTL) experiment that examined segregation of B73-Mo17 alleles in leaves found significant variation in the cis-regulation of lcyE expression, along with several other regions that also contribute to expression level control of lcyE (fig. S3).
In a previous study, three major QTL were identified for accumulation of carotenoids in maize (23). Two of these QTL colocalized with y1 and zeta carotene desaturase (zds); the third QTL mapped to a region without a candidate gene. We mapped lcyE to chromosome 8 bin 5, near marker bnlg1599, and it colocalized with this previously undetermined QTL. This QTL showed significant effects for modification of the ratio of α to β branch carotenoids [logarithm of the odds ratio for linkage (or lod) score of 34.05; R2 54.4%] and explained 31.7% of the variation for lutein (lod 16.5). The magnitude of effects was not as large as in association or mutagenesis analysis. However, this biparental QTL population only segregated for the amino substitution (at codon 216) and a modest promoter polymorphism and does not segregate for the 3′ polymorphism. Notably, this QTL was not significant for total carotenoids, which further supports the conclusion that variation within lcyE gene underlies this QTL for carotenoid composition.
To confirm association and QTL results, mutagenesis induced by ethane methyl sulfonate (EMS) was conducted to isolate additional alleles of lcyE. Two M2 ears of inbred Q×47 segregated for a distinct change in endosperm color from yellow to orange, with orange recessive to yellow (these color changes were apparent in the inbred isogenic background, but not in diverse breeding materials). HPLC analysis of orange and yellow kernels confirmed a shift in the zeaxanthin:lutein ratio in the direction of zeaxanthin. This orange endosperm mutation was backcrossed into the standard genetic inbred line B73, and lcyE was tested as a candidate gene, which revealed that the Q×47 lcyE haplotype cosegregates with orange endosperm and ratio of α-carotene versus β-carotene branch carotenoids (fig. S4).
The most favorable haplotype for higher βcarotene branch carotenoids included both the large promoter insertion and 3′ 8-bp insertion. In the diverse panel we tested, this haplotype occurs in 5% of temperate inbreds and 16% of tropical inbreds. MAS at this locus should be effective for several reasons: (i) The most favorable haplotype is found with at least modest frequency in different germ plasm sources and thus breeders can select donors from their relatively more adapted sources. (ii) The favorable haplotype has a large effect. (iii) Visual selection is ineffective for differentiating carotenoid composition and selecting provitamin A compounds. (iv) In comparison with HPLC analysis of carotenoids, polymerase chain reaction (PCR) scoring of the lcyE locusismuch less expensive (costing perhaps 1/1000th that of HPLC) and more accessible to developing countries with greatest need for provitamin A.
An approach that empowers local breeder involvement through inexpensive visual selection for darker yellow to orange kernels to enhance flux into carotenoid pathway, and also incorporates MAS for lcyE, should result in increased levels of provitamin A compounds. To expedite creation of improved germ plasm globally, we provide information on PCR-based markers (fig. S5). Donor inbreds and improved breeding lines derived at the International Maize and Wheat Improvement Center (CIMMYT) from synthetics of diverse panel inbreds with higher β-carotene are available by contacting T.R.R. This will facilitate selection worldwide of the most favorable lcyE alleles, which we have begun in our program. We are screening tropical breeding germ plasm collections in collaboration with CIMMYT.
To date, MAS for natural variation has been limited by resolution and scope (germ plasm diversity). Alleles have generally been characterized in the limited genetic background and resolution of biparental QTL studies, leaving in question their relevance to broader germ plasm (24), particularly for germ plasm outside of the temperate United States. As a result, the primary use for MAS is backcross breeding of transgenic traits. In contrast, the association mapping approach used here allows for rapid generation of selectable markers based on performance of diverse germ plasm. This provides markers more relevant in a broad genetic background, and that enables breeders to search for favorable alleles in their locally adapted germ plasm sources.
In ongoing studies, we are attempting to identify alleles for other genes in the pathway that increase total carotenoids and that slow the conversion of β-carotene to β-cryptoxanthin and zeaxanthin, to exploit more fully the natural genetic variation potential in provitamin A biofortification of maize. These results will then be further incorporated in breeding efforts to create a healthier maize crop for the world's poorest people.
Although the genetic results and strategy presented here are encouraging, they need to be placed in context as part of an overall biofortification effort encompassing breeding infrastructure, seed distribution, societal acceptance, dietary habits, and nutritional impact. Information now available on some of these issues is encouraging. Results from an animal model for human vitamin A metabolism indicated vitamin A activity of provitamin A in orange maize was greater than assumed by a factor of about four (25). A successful intervention to introduce βcarotene–rich, orange sweet potato in Mozambique, where only white sweet potato was previously cultivated, suggests that orange-colored staple foods can be acceptable, and their regular consumption results in improved vitamin A status (26). Related follow-up acceptance studies of yellow and orange maize in Mozambique and Zimbabwe are in progress with initial results encouraging (27). The dietary habits of many Africans, in which maize is consumed for all three meals a day, indicates that maize is a good target for biofortification (28). The recent positive nutritional and acceptance results will need to be coordinated with comprehensive breeding and seed distribution efforts to realize the potential of provitamin A–biofortified maize, as, for example, is coordinated by the HarvestPlus Global Challenge Program.
Supplementary Material
Supporting Online Material for
Natural Genetic Variation in Lycopene Epsilon Cyclase Tapped for Maize Biofortification
Carlos E. Harjes, Torbert R. Rocheford,* Ling Bai, Thomas P. Brutnell, Catherine Bermudez Kandianis, Stephen G. Sowinski, Ann E. Stapleton, Ratnakar Vallabhaneni, Mark Williams, Eleanore T. Wurtzel, Jianbing Yan, Edward S. Buckler*
*To whom correspondence should be addressed. esb33@cornell.edu (E.S.B); trochefo@uiuc.edu (T.R.R.)
Published 18 January 2008, Science 319, 330 (2008) DOI: 10.1126/science.1150255
This PDF file includes
Materials and Methods
Figs. S1 to S8
Tables S1 to S5
References
Supporting Online Material
MATERIALS AND METHODS
Germplasm Evaluation: A diverse association panel of 282 lines(S1) was phenotyped using HPLC analysis for carotenoid content and genotyped using SSR and SNP markers. The inbreds were grown in one-row plots in a randomized complete block design in Champaign-Urbana, Illinois during the summers of 2002, 2003, 2004, and 2005. HPLC analysis was performed on balanced bulks of a few to several self-pollinated ears of seed from each plot.
Carotenoid Quantification: Quantification of carotenoids was accomplished by standard regression with external standards (S2, 3). Evaluations were done on dry kernels, and values are quantified relative dry weight. The HPLC system consisted of a Waters Alliance 2690 separation module (system includes solvent delivery system, inline degasser, column heater, and sample cooler) attached to a Waters 996 Photodiode Array detector (PDA) and this system was managed by Waters Millennium 2001 (v 3.2). The column used for quantification was a reverse phase YMC carotenoid C30 column (5 μm, 4.6 × 100 mm) connected to a YMC C30 filter insert that acted as a guard column. The mobile phase was Acetonitrile, methanol, methylene chloride (75:20:5 v/v/v) with triethylamine (.05% of total volume) and BHT, mixed by the Alliance 2690 solvent delivery system to help reduce composition differences between runs. Samples were loaded into amber glass vials with 50 μl limited volume inserts (Alltech and Associates, Deerfield, IL). All samples were run immediately after extraction. Samples were stored at 4°C if they were unable to be loaded into the HPLC immediately. Flow rate was 1.8 ml min.-1, with each run taking 20 – 23 minutes. To control fluctuations in retention time due to temperature differences, column temperature was set at 30°C. Sample temperature was set at 4°C to control degradation of samples by heat. Carotenoids were detected at 450 nm, and were quantified by external standards as described previously (S2, 3).
The ratio of the two branches was evaluated as natural logarithm of (α-carotene + lutein) / (β-carotene + β-cryptoxanthin + zeaxanthin). Evaluations of individual compounds generally compared the proportion of individual compound relative to the total carotenoid levels.
Mutagenesis: EMS pollen mutagenesis was conducted following the protocol of Neuffer(S4) on the DuPont inbred line Q×47. Carotenoid analysis of maize kernels was conducted by Craft Technologies, Inc (Wilson, NC, USA). Two self-pollinated M2 ears derived from the EMS pollen mutagenesis of the inbred line Q×47 segregated for a distinct change in endosperm color from yellow to orange. These putative mutants were designated #26 and #28. M2 kernels of each type were grown and self-pollinated. All of the orange M2 kernels gave uniformly orange M3 ears whereas yellow M2 kernels produced both uniformly yellow and yellow/orange segregating ears, indicating dominance of yellow over orange. The #28 putative mutant was backcrossed 4 times into the public inbred B73 and selfed. Orange kernels were crossed to yellow kernels from the last backcross, and a 1:1 yellow:orange segregation was observed on 50% of the ears, again consistent with a single, Mendelian recessive gene. A SNP identified in lcyE between B73 and Q×47, which confers a HinfI restriction site in Q×47 and not in B73, allowed a co-segregation analysis on the backcross population described above. Following 4 backcrosses to B73, all of the orange kernels (n=47) were homozygous for the Q×47 (mutant #28) lcyE haplotype, and all of the yellow kernels (n=48) were heterozygous.
Genotyping: Candidate genes were selected based on previous characterization in maize or by homology with tomato, Arabidopsis, or rice genes (S5, 6). Given the ancient duplication in the maize genome, active site homology was used to select homologs. Robust PCR primers for the diverse germplasm were designed based on MAGI (Maize Assembled Genomic Islands)(S7) or the TIGR Maize Database(S8) and refined by prescreening on a diverse panel of 32 lines (Genbank numbers: BV709178-BV709204, BV709242-BV709256, BV709298-BV709330, BV709398-BV709439, BV709481-BV709506, BV727024-BV727770). The selected amplicons with an average span of 500bp were then sequenced across the full panel of maize lines. One to two regions were sampled from each candidate gene. Given the rapid LD decay in maize we cannot rule out genes that showed no evidence of association (ie. we may have sampled the wrong region or paralogue). Once the lcyE association was identified the gene was sequenced from 12 diverse haplotypes. Three more regions were sampled across entire panel based on putative functional SNPs suggested by sequencing. Promoter haplotypes with multiple TE were scored by amplicon length, using PCR markers provided for maker assisted selection of these alleles.
Transcript profiling: Maize inbreds used for LCYE transcript profiles were grown at the Lehman College field station, Bronx, New York, and developmentally staged endosperms dissected and stored at -80°C until use. RNA Isolation and Reverse transcription: Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen Sciences, Maryland), and DNase I-treated (Invitrogen, Carlsbad, CA) prior to first strand cDNA synthesis using oligo (dT) as a primer and SuperscriptTM III RT (Invitrogen, Carlsbad, CA). 1ul of 50μM oligo (dT)20’ and 1ul of 10mM dNTP mix were mixed with 8μl of DNase I treated total RNA (~ 1 μg) and incubated at 65°C for 5min, and left on ice for at least 1 min. 10μl of cDNA synthesis mix (2μl of 10X RT buffer, 4μl of 25mM MgCl2, 2μl of 0.1M DTT, 1μl of RNaseOUT™ (40 U/μl), 1ul of Superscript™ III RT (200 U/μl) was added and incubated for 50min at 50°C and reactions terminated at 85°C for 5min. Samples were collected by brief centrifugation and 1μl of RNase H added and incubated for 20min at 37°C. Quantitative RT-PCR: cDNA samples were amplified on the MyIQ Single-Color Real-Time PCR detection system (Bio-Rad, Hercules, CA), using iQTM SYBR Green Supermix (Bio-Rad, Hercules, CA). 2μl (5ng/μl) of cDNA; 15μl of 2X iQTM SYBR Green Supermix; 11μl of Nuclease-Free water; 1μl (20ρm/μl) of each primer were used in a 30μl reaction volume. Thermal cycling conditions included an initial incubation at 94°C for 10s, followed by 35 cycles of 95°C for 10s, 58°C for 35s, and 72°C 10s. Melt curve analysis was performed verify primer specificity, and PCR products were confirmed by sequencing. The relative quantity of the transcripts was calculated by using the comparative threshold cycle (CT) method. Actin mRNA was amplified simultaneously for normalization between samples. Primers were designed to flank introns (lcyE: TTTACGTGCAAATGCAGTCAA, TGACTCTGAAGCTAGAGAAAG; Actin: CGATTGAGCATGGCATTGTCA, CCCACTAGCGTACAACGAA).
eQTL in maize leaf tissue: Experimental design, plant material, and array methods are described in detail in the Maize Oligonucleotide Array project database, which is publicly available from www.maizearray.org; this work is Study ID 21, Investigator Stapleton. Array element median spot value from the maizearray spreadsheet page was log-transformed and averaged within treatment group (UV and control). The mean values were merged with marker data for the recombinant inbred lines; the marker data was retrieved from MaizeGDB (www.maizegdb.org); with Iowa combined markers and Genoplante markers merged into the list for a total of 4751 markers. SAS Proc GLM was used to test for significant marker association with expression level for genes TM00018798 and TM00050085 and for treatment interaction for each gene.
Statistical Analysis: Association analysis was conducted using a mixed model incorporating kinship and population structure(S9) as implemented in TASSEL (www.maizegenetics.net)(S10) and SAS/STAT (Version 9.1, SAS Institute Inc., Cary, NC, USA). This approach simultaneously accounts for the multiple levels of relatedness based on random genetic markers that are used to establish population structure and a kinship matrix. The method has very good control of Type I error rates when a trait is polygenic. When examining the ratio of the two branches, we used a standard GLM model as this trait is either a mono- or oligo-genic trait.
Figure S1. Relative gene expression of LCYB and LCYE in W22 maize seeds by quantitative RT-PCR.
• Figure S2. LCYE Quantitative RT-PCR for endosperm at 15 days after pollination (top) and 20 days after pollination (bottom). Samples surrounded by green box have transposon insertions near the start site.
Figure S3. Significant markers at a FDR of 10% are shown; P values are from SAS GLM analysis. Expression levels of probe TM00050085 designed from (lcyE sequence based AZM_584852) in IBM (B73 × Mo17) mapping lines were calculated by averaging the expression levels by allele at each of these marker loci. Probe TM00050085 was used to estimate expression. The second most significant marker is umc1316, which is on the same physical contig as lcyE. Although these lines differ for a significant promoter haplotypes, they do not segregate for the large transposon promoter differences.
Figure S4. Comparison of ( and ( carotenoid levels for the EMS mutagenized lcyE allele (Orange allele is the #28 EMS mutation of the Q×47 allele; Y is wild-type Q×47 allele). This is the average of four samples for each endosperm genotype (endosperm tissue is triploid). The error bars are standard errors of the mean.
Figure S5. PCR assay for target polymorphisms. PCR assay for key associated polymorphisms haplotypes observed across the inbred line panel and diagram of primer annealing positions, black triangles represent indels (not to scale), colored lines represent the PCR amplicons corresponding to bands on gel identified by color. Assays are designed to incorporate the use of all indicated primers in a single reaction; the haplotype assignment (listed in each depicted lane) will result from one to multiple amplicons. For the 5’ indels, class 1 and class 4 both share the promoter transposon; they were statistically the same in their effect and were fused for analysis.
Figure S6. Details for the lcyE 5’ Indel PCR for Marker-Assisted Breeding
Figure S7. Details for the lcyE SNP216 PCR for Marker-Assisted Breeding
Figure S8: Details for the lcyE 3’Indel PCR for Marker-Assisted Breeding
Table S1. Haplotype estimated effects. Count of haplotypic classes and estimated effects on the ratio of the pathway braches ln(lutein + α-carotene) / (zeaxanthin + β-carotene + β-cryptoxanthin)
Table S2. lcyE gene structure. Based on maize contig ZmGSStuc11-12-04.976.1. Predicted exons are in yellow, predicted in other genes features in green, polymorphisms of interest in red (number next to them are the aligned sequence position used in the text), and primers to score key polymorphisms are in blue.
Table S3. Association for other candidate genes with lcyE effects included as a covariate. NC is noncovergence of the mixed model often caused by non-normal trait distributions or low frequency SNPs. Please note this table includes over 600 tests, and these P values have not been multiple test corrected. Except for a few false very significant results (P < 0.002) most of these are false positives. For contrast lcyE's initial association was P = 2.23×10-10.
Since the completion of the study of the lcyE, we surveyed the numerous hydroxylase paralogues in maize and we have found an additional important modifier.
Table S4. HPLC carotenoid values for the 2003 field in IL.
Table S5. PCR scores for the diverse maize association panel.
Supplementary References
S1. S. A. Flint-Garcia et al., Plant J 44, 1054 (2005).
S2. A. C. Kurilich, J. A. Juvik, Journal of Agricultural and Food Chemistry 47, 1948 (1999).
S3. A. C. Kurilich, J. A. Juvik, Journal of Liquid Chromatography & Related Technologies 22, 2925 (1999).
S4. M. G. Neuffer, in The Maize Handbook M. Freeling, V. Walbot, Eds. (Springer-Verlag, New York, 1994) pp. 212-219.
S5. P. D. Fraser, P. M. Bramley, Prog Lipid Res 43, 228 (2004).
S6. J. Hirschberg, Current Opinion in Plant Biology 4, 210 (2001).
S7. Y. Fu et al., Proc Natl Acad Sci U S A 102, 12282 (2005).
S8. A. P. Chan et al., Nucl. Acids Res. 34, D771 (2006).
S9. J. M. Yu et al., Nature Genet. 38, 203 (2006).
S10. P. J. Bradbury et al., Bioinformatics 23, 2633 (2007).
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/319/5861/330/DC1
Materials and Methods
Figs. S1 to S9
Tables S1 to S5
References
References and Notes
- 1.Underwood BA. J. Nutr. 2004;134:231S. doi: 10.1093/jn/134.1.231S. [DOI] [PubMed] [Google Scholar]
- 2.Mora JO. J. Nutr. 2003;133:2990S. doi: 10.1093/jn/133.9.2990S. [DOI] [PubMed] [Google Scholar]
- 3.West CE. Nutr. Rev. 2000;58:341. doi: 10.1111/j.1753-4887.2000.tb01831.x. [DOI] [PubMed] [Google Scholar]
- 4.Dary O, Mora JO. J. Nutr. 2002;132:2927S. doi: 10.1093/jn/132.9.2927S. [DOI] [PubMed] [Google Scholar]
- 5.West CE, Eilander A, van Lieshout M. J. Nutr. 2002;132:2920S. doi: 10.1093/jn/132.9.2920S. [DOI] [PubMed] [Google Scholar]
- 6.van Lieshout M, de Pee S. Am. J. Clin. Nutr. 2005;81:943. doi: 10.1093/ajcn/81.4.943. [DOI] [PubMed] [Google Scholar]
- 7.Graham RD, Welch RM, Bouis HE. Adv. Agron. 2001;70:77. [Google Scholar]
- 8.Fraser PD, Bramley PM. Prog. Lipid Res. 2004;43:228. doi: 10.1016/j.plipres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 9.DellaPenna D, Pogson BJ. Annu. Rev. Plant Biol. 2006;57:711. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- 10.Li F, Murillo C, Wurtzel ET. Plant Physiol. 2007;144:1181. doi: 10.1104/pp.107.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckner B, Miguel PS, Janick-Buckner D, Bennetzen JL. Genetics. 1996;143:479. doi: 10.1093/genetics/143.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palaisa K, Morgante M, Tingey S, Rafalski A. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9885. doi: 10.1073/pnas.0307839101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D. Plant Cell. 1996;8:1627. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham FX, Jr., et al. Plant Cell. 1996;8:1613. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecker I, Gabbay R, Cunningham FX, Jr., Hirschberg J. Plant Mol. Biol. 1996;30:807. doi: 10.1007/BF00019013. [DOI] [PubMed] [Google Scholar]
- 16.Yu B, Lydiate D, Young L, Schäfer U, Hannoufa A. Transgenic Res. 2007 doi: 10.1007/s11248-007-9131-x. 10.1007/s11248. [DOI] [PubMed] [Google Scholar]
- 17.Diretto G, et al. BMC Plant Biol. 2006;6:13. doi: 10.1186/1471-2229-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogson BJ, Rissler HM. Philos. Trans. R. Soc. London B Biol. Sci. 2000;355:1395. doi: 10.1098/rstb.2000.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint-Garcia SA, et al. Plant J. 2005;44:1054. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 20.Remington DL, et al. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11479. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Materials and methods are available as supporting material on Science Online.
- 22.Yu J, et al. Nat. Genet. 2006;38:203. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 23.Wong JC, Lambert RJ, Wurtzel ET, Rocheford TR. Theor. Appl. Genet. 2004;108:349. doi: 10.1007/s00122-003-1436-4. [DOI] [PubMed] [Google Scholar]
- 24.Asíns MJ. Plant Breed. Rev. 2002;121:281. [Google Scholar]
- 25.Howe JA, Tanumihardjo SA. J. Nutr. 2006;136:2562. doi: 10.1093/jn/136.10.2562. [DOI] [PubMed] [Google Scholar]
- 26.Low JW, et al. J. Nutr. 2007;137:1320. doi: 10.1093/jn/137.5.1320. [DOI] [PubMed] [Google Scholar]
- 27.Stevens RA, Winter-Nelson A. Food Policy. in press. [Google Scholar]
- 28.Li S, Tayie FAK, Young MF, Rocheford T, White WS, Agric J. Food Chem. 2007;55:10744. doi: 10.1021/jf071815v. [DOI] [PubMed] [Google Scholar]
- 29.Matthews PD, Wurtzel ET. In: Food Colorants: Chemical and Functional Properties. Socaciu C, editor. CRC Press; Boca Raton, FL: 2007. pp. 347–398. [Google Scholar]
- 30.We thank S. Islam, C. Paul, W. Liu, and W. White for running HPLC on samples; H. Yates for support of molecular genetics; and N. Stevens for technical editing of the manuscript. This work was supported by NSF DBI-0321467 (to E.S.B.), U.S. Agency for International Development (to T.R.R.), HarvestPlus (to T.R.R.), NIH (S06-GM08225 to E.T.W.), Professional Staff Congress—CUNY research award (to E.T.W.), New York State (to E.T.W.), NSF DBI-0604923 (to T.R.R), TRIAD Foundation (L.B. and T.P.B.), U.S. Department of Agriculture (USDA) Cooperative State Research, Education, and Extension Service (CSREES), National Research Initiative grant 2003-00745 (to A.E.S.), and USDA—Agricultural Research Service (to E.S.B.). Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Online Material for
Natural Genetic Variation in Lycopene Epsilon Cyclase Tapped for Maize Biofortification
Carlos E. Harjes, Torbert R. Rocheford,* Ling Bai, Thomas P. Brutnell, Catherine Bermudez Kandianis, Stephen G. Sowinski, Ann E. Stapleton, Ratnakar Vallabhaneni, Mark Williams, Eleanore T. Wurtzel, Jianbing Yan, Edward S. Buckler*
*To whom correspondence should be addressed. esb33@cornell.edu (E.S.B); trochefo@uiuc.edu (T.R.R.)
Published 18 January 2008, Science 319, 330 (2008) DOI: 10.1126/science.1150255
This PDF file includes
Materials and Methods
Figs. S1 to S8
Tables S1 to S5
References
Supporting Online Material
MATERIALS AND METHODS
Germplasm Evaluation: A diverse association panel of 282 lines(S1) was phenotyped using HPLC analysis for carotenoid content and genotyped using SSR and SNP markers. The inbreds were grown in one-row plots in a randomized complete block design in Champaign-Urbana, Illinois during the summers of 2002, 2003, 2004, and 2005. HPLC analysis was performed on balanced bulks of a few to several self-pollinated ears of seed from each plot.
Carotenoid Quantification: Quantification of carotenoids was accomplished by standard regression with external standards (S2, 3). Evaluations were done on dry kernels, and values are quantified relative dry weight. The HPLC system consisted of a Waters Alliance 2690 separation module (system includes solvent delivery system, inline degasser, column heater, and sample cooler) attached to a Waters 996 Photodiode Array detector (PDA) and this system was managed by Waters Millennium 2001 (v 3.2). The column used for quantification was a reverse phase YMC carotenoid C30 column (5 μm, 4.6 × 100 mm) connected to a YMC C30 filter insert that acted as a guard column. The mobile phase was Acetonitrile, methanol, methylene chloride (75:20:5 v/v/v) with triethylamine (.05% of total volume) and BHT, mixed by the Alliance 2690 solvent delivery system to help reduce composition differences between runs. Samples were loaded into amber glass vials with 50 μl limited volume inserts (Alltech and Associates, Deerfield, IL). All samples were run immediately after extraction. Samples were stored at 4°C if they were unable to be loaded into the HPLC immediately. Flow rate was 1.8 ml min.-1, with each run taking 20 – 23 minutes. To control fluctuations in retention time due to temperature differences, column temperature was set at 30°C. Sample temperature was set at 4°C to control degradation of samples by heat. Carotenoids were detected at 450 nm, and were quantified by external standards as described previously (S2, 3).
The ratio of the two branches was evaluated as natural logarithm of (α-carotene + lutein) / (β-carotene + β-cryptoxanthin + zeaxanthin). Evaluations of individual compounds generally compared the proportion of individual compound relative to the total carotenoid levels.
Mutagenesis: EMS pollen mutagenesis was conducted following the protocol of Neuffer(S4) on the DuPont inbred line Q×47. Carotenoid analysis of maize kernels was conducted by Craft Technologies, Inc (Wilson, NC, USA). Two self-pollinated M2 ears derived from the EMS pollen mutagenesis of the inbred line Q×47 segregated for a distinct change in endosperm color from yellow to orange. These putative mutants were designated #26 and #28. M2 kernels of each type were grown and self-pollinated. All of the orange M2 kernels gave uniformly orange M3 ears whereas yellow M2 kernels produced both uniformly yellow and yellow/orange segregating ears, indicating dominance of yellow over orange. The #28 putative mutant was backcrossed 4 times into the public inbred B73 and selfed. Orange kernels were crossed to yellow kernels from the last backcross, and a 1:1 yellow:orange segregation was observed on 50% of the ears, again consistent with a single, Mendelian recessive gene. A SNP identified in lcyE between B73 and Q×47, which confers a HinfI restriction site in Q×47 and not in B73, allowed a co-segregation analysis on the backcross population described above. Following 4 backcrosses to B73, all of the orange kernels (n=47) were homozygous for the Q×47 (mutant #28) lcyE haplotype, and all of the yellow kernels (n=48) were heterozygous.
Genotyping: Candidate genes were selected based on previous characterization in maize or by homology with tomato, Arabidopsis, or rice genes (S5, 6). Given the ancient duplication in the maize genome, active site homology was used to select homologs. Robust PCR primers for the diverse germplasm were designed based on MAGI (Maize Assembled Genomic Islands)(S7) or the TIGR Maize Database(S8) and refined by prescreening on a diverse panel of 32 lines (Genbank numbers: BV709178-BV709204, BV709242-BV709256, BV709298-BV709330, BV709398-BV709439, BV709481-BV709506, BV727024-BV727770). The selected amplicons with an average span of 500bp were then sequenced across the full panel of maize lines. One to two regions were sampled from each candidate gene. Given the rapid LD decay in maize we cannot rule out genes that showed no evidence of association (ie. we may have sampled the wrong region or paralogue). Once the lcyE association was identified the gene was sequenced from 12 diverse haplotypes. Three more regions were sampled across entire panel based on putative functional SNPs suggested by sequencing. Promoter haplotypes with multiple TE were scored by amplicon length, using PCR markers provided for maker assisted selection of these alleles.
Transcript profiling: Maize inbreds used for LCYE transcript profiles were grown at the Lehman College field station, Bronx, New York, and developmentally staged endosperms dissected and stored at -80°C until use. RNA Isolation and Reverse transcription: Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen Sciences, Maryland), and DNase I-treated (Invitrogen, Carlsbad, CA) prior to first strand cDNA synthesis using oligo (dT) as a primer and SuperscriptTM III RT (Invitrogen, Carlsbad, CA). 1ul of 50μM oligo (dT)20’ and 1ul of 10mM dNTP mix were mixed with 8μl of DNase I treated total RNA (~ 1 μg) and incubated at 65°C for 5min, and left on ice for at least 1 min. 10μl of cDNA synthesis mix (2μl of 10X RT buffer, 4μl of 25mM MgCl2, 2μl of 0.1M DTT, 1μl of RNaseOUT™ (40 U/μl), 1ul of Superscript™ III RT (200 U/μl) was added and incubated for 50min at 50°C and reactions terminated at 85°C for 5min. Samples were collected by brief centrifugation and 1μl of RNase H added and incubated for 20min at 37°C. Quantitative RT-PCR: cDNA samples were amplified on the MyIQ Single-Color Real-Time PCR detection system (Bio-Rad, Hercules, CA), using iQTM SYBR Green Supermix (Bio-Rad, Hercules, CA). 2μl (5ng/μl) of cDNA; 15μl of 2X iQTM SYBR Green Supermix; 11μl of Nuclease-Free water; 1μl (20ρm/μl) of each primer were used in a 30μl reaction volume. Thermal cycling conditions included an initial incubation at 94°C for 10s, followed by 35 cycles of 95°C for 10s, 58°C for 35s, and 72°C 10s. Melt curve analysis was performed verify primer specificity, and PCR products were confirmed by sequencing. The relative quantity of the transcripts was calculated by using the comparative threshold cycle (CT) method. Actin mRNA was amplified simultaneously for normalization between samples. Primers were designed to flank introns (lcyE: TTTACGTGCAAATGCAGTCAA, TGACTCTGAAGCTAGAGAAAG; Actin: CGATTGAGCATGGCATTGTCA, CCCACTAGCGTACAACGAA).
eQTL in maize leaf tissue: Experimental design, plant material, and array methods are described in detail in the Maize Oligonucleotide Array project database, which is publicly available from www.maizearray.org; this work is Study ID 21, Investigator Stapleton. Array element median spot value from the maizearray spreadsheet page was log-transformed and averaged within treatment group (UV and control). The mean values were merged with marker data for the recombinant inbred lines; the marker data was retrieved from MaizeGDB (www.maizegdb.org); with Iowa combined markers and Genoplante markers merged into the list for a total of 4751 markers. SAS Proc GLM was used to test for significant marker association with expression level for genes TM00018798 and TM00050085 and for treatment interaction for each gene.
Statistical Analysis: Association analysis was conducted using a mixed model incorporating kinship and population structure(S9) as implemented in TASSEL (www.maizegenetics.net)(S10) and SAS/STAT (Version 9.1, SAS Institute Inc., Cary, NC, USA). This approach simultaneously accounts for the multiple levels of relatedness based on random genetic markers that are used to establish population structure and a kinship matrix. The method has very good control of Type I error rates when a trait is polygenic. When examining the ratio of the two branches, we used a standard GLM model as this trait is either a mono- or oligo-genic trait.
Figure S1. Relative gene expression of LCYB and LCYE in W22 maize seeds by quantitative RT-PCR.
• Figure S2. LCYE Quantitative RT-PCR for endosperm at 15 days after pollination (top) and 20 days after pollination (bottom). Samples surrounded by green box have transposon insertions near the start site.
Figure S3. Significant markers at a FDR of 10% are shown; P values are from SAS GLM analysis. Expression levels of probe TM00050085 designed from (lcyE sequence based AZM_584852) in IBM (B73 × Mo17) mapping lines were calculated by averaging the expression levels by allele at each of these marker loci. Probe TM00050085 was used to estimate expression. The second most significant marker is umc1316, which is on the same physical contig as lcyE. Although these lines differ for a significant promoter haplotypes, they do not segregate for the large transposon promoter differences.
Figure S4. Comparison of ( and ( carotenoid levels for the EMS mutagenized lcyE allele (Orange allele is the #28 EMS mutation of the Q×47 allele; Y is wild-type Q×47 allele). This is the average of four samples for each endosperm genotype (endosperm tissue is triploid). The error bars are standard errors of the mean.
Figure S5. PCR assay for target polymorphisms. PCR assay for key associated polymorphisms haplotypes observed across the inbred line panel and diagram of primer annealing positions, black triangles represent indels (not to scale), colored lines represent the PCR amplicons corresponding to bands on gel identified by color. Assays are designed to incorporate the use of all indicated primers in a single reaction; the haplotype assignment (listed in each depicted lane) will result from one to multiple amplicons. For the 5’ indels, class 1 and class 4 both share the promoter transposon; they were statistically the same in their effect and were fused for analysis.
Figure S6. Details for the lcyE 5’ Indel PCR for Marker-Assisted Breeding
Figure S7. Details for the lcyE SNP216 PCR for Marker-Assisted Breeding
Figure S8: Details for the lcyE 3’Indel PCR for Marker-Assisted Breeding
Table S1. Haplotype estimated effects. Count of haplotypic classes and estimated effects on the ratio of the pathway braches ln(lutein + α-carotene) / (zeaxanthin + β-carotene + β-cryptoxanthin)
Table S2. lcyE gene structure. Based on maize contig ZmGSStuc11-12-04.976.1. Predicted exons are in yellow, predicted in other genes features in green, polymorphisms of interest in red (number next to them are the aligned sequence position used in the text), and primers to score key polymorphisms are in blue.
Table S3. Association for other candidate genes with lcyE effects included as a covariate. NC is noncovergence of the mixed model often caused by non-normal trait distributions or low frequency SNPs. Please note this table includes over 600 tests, and these P values have not been multiple test corrected. Except for a few false very significant results (P < 0.002) most of these are false positives. For contrast lcyE's initial association was P = 2.23×10-10.
Since the completion of the study of the lcyE, we surveyed the numerous hydroxylase paralogues in maize and we have found an additional important modifier.
Table S4. HPLC carotenoid values for the 2003 field in IL.
Table S5. PCR scores for the diverse maize association panel.
Supplementary References
S1. S. A. Flint-Garcia et al., Plant J 44, 1054 (2005).
S2. A. C. Kurilich, J. A. Juvik, Journal of Agricultural and Food Chemistry 47, 1948 (1999).
S3. A. C. Kurilich, J. A. Juvik, Journal of Liquid Chromatography & Related Technologies 22, 2925 (1999).
S4. M. G. Neuffer, in The Maize Handbook M. Freeling, V. Walbot, Eds. (Springer-Verlag, New York, 1994) pp. 212-219.
S5. P. D. Fraser, P. M. Bramley, Prog Lipid Res 43, 228 (2004).
S6. J. Hirschberg, Current Opinion in Plant Biology 4, 210 (2001).
S7. Y. Fu et al., Proc Natl Acad Sci U S A 102, 12282 (2005).
S8. A. P. Chan et al., Nucl. Acids Res. 34, D771 (2006).
S9. J. M. Yu et al., Nature Genet. 38, 203 (2006).
S10. P. J. Bradbury et al., Bioinformatics 23, 2633 (2007).