Abstract
The Ras/mitogen-activated protein kinase (MAPK) pathway is considered to be a positive regulator of tumor initiation, progression, and maintenance. This study reports an opposite finding: we have found strong evidence that the MAPK pathway is inhibited in a subset of adenoid cystic carcinomas (ACCs) of the salivary glands. ACC tumors consistently overexpress the receptor tyrosine kinase (RTK) c-Kit, which has been considered a therapeutic target. We performed mutational analysis of the c-Kit gene (KIT in 17 cases of ACC and found that 2 cases of ACC had distinct missense mutations in KIT at both the genomic DNA and messenger RNA levels. These mutations caused G664R and R796G amino acid substitutions in the kinase domains. Surprisingly, the mutations were functionally inactive in cultured cells. We observed a significant reduction of MAPK (ERK1/2) activity in tumor cells, as assessed by immunohistochemistry. We performed further mutational analysis of the downstream effectors in the c-Kit pathway in the genes HRAS, KRAS, NRAS, BRAF, PIK3CA, and PTEN. This analysis revealed that two ACC tumors without KIT mutations had missense mutations in either KRAS or BRAF, causing S17N K-Ras and V590I B-Raf mutants, respectively. Our functional analysis showed that proteins with these mutations were also inactive in cultured cells. This is the first time that MAPK activity from the RTK signaling has been shown to be inhibited by gene mutations during tumor development. Because ACC seems to proliferate despite inactivation of the c-Kit signaling pathway, we suggest that selective inhibition of c-Kit is probably not a suitable treatment strategy for ACC.
Introduction
Adenoid cystic carcinoma (ACC) is the second most common malignant neoplasm of salivary glands [1–3]. It arises in the major and the minor salivary glands as well as in the seromucinous glands of the upper respiratory tract. ACC is composed of a bipopulation of duct-type epithelial cells and myoepithelial cells and forms three distinctive microscopic patterns. They are categorized as predominantly tubular, cribriform, or solid types. Among the three subtypes, the solid form often has the highest recurrence rate and the worst long-term prognosis.
ACC grows slowly with wide anatomic extension, although it infrequently metastasizes to regional lymph nodes. Advanced tumors can invade nerves, causing pain, numbness, and/or paralysis, with extension into vital structures, including the brain. Although short-term survival is high, almost half of patients develop distant metastases or succumb to local recurrences within 10 to 20 years of diagnosis. Metastases occur most commonly in the lungs and less commonly in the liver and bone. Unfortunately, even patients with good local control can develop distant metastases more than 10 years after initial therapy. As a result, ACC is considered by some to be a systemic disease with an unpredictable and unrelenting clinical course. Therapy is typically surgical resection with postoperative radiation [2,4,5]. Conventional chemotherapy has a poorly defined role in the treatment of ACC, and the lack of successful systemic therapy is a further obstacle to improving outcomes.
There is an increased interest in determining the molecular abnormalities that underpin ACC, with the hope that doing so will lead to the discovery of an effective targeted therapy [5]. One possibility involves transmembrane receptor tyrosine kinases (RTKs). RTKs play a critical role in initiation and development of many tumor types, including those arising in the head and neck, and offer potential as therapeutic targets in ACC [6].
C-Kit (also known as CD117) is an RTK encoded by the KIT gene. It is a member of the platelet-derived growth factor receptor (PDGFR) family that exhibits five Ig-like domains in its extracellular region [7,8]. C-Kit was originally identified as an oncoprotein encoded by a feline sarcoma virus. It is activated by receptor dimerization on stimulation by its ligand, stem cell factor (SCF) [9]. Once c-Kit is activated, diverse intracellular responses are induced through major signaling cascades such as the phosphoinositide-3 kinase (PI3K/Akt) and mitogen-activated protein kinase (MAPK) pathways [10–12]. The latter process is activated in the sequence of Ras, Raf (also known as MAPKKK), MEK1/2 (MAPKK), and ERK1/2 (classic MAPK) [13,14]. Thus, c-Kit regulates cell survival and growth control through the PI3K/Akt and MAPK signaling pathways and contributes to numerous life phenomena. For example, c-Kit cell signaling plays an important role in stimulating the formation of many types of blood and other cells, including melanocytes, germ cells, and the interstitial cells of Cajal, which mediate intestinal motility [6].
Recent studies have demonstrated that almost all ACCs show overexpression of c-Kit [2,3,15,16]. In contrast, c-Kit expression is seldom increased in other head and neck tumors. For this reason, c-Kit expression is often used as a diagnostic pathology aid for ACC. In addition, c-Kit might play important roles in the oncogenesis of ACC and in tumor maintenance. It could also be a therapeutic molecular target in ACC [5,17,18] and other conditions. For instance, in some leukemias and gastrointestinal tumors (GISTs), inhibiting c-Kit activity using imatinib mesylate (Gleevec) has proven to be clinically effective treatment modality [19].
Unfortunately, conflicting data exist regarding the impact of c-Kit inhibition on ACC. Two recent case reports have suggested that c-Kit targeted therapy inhibits the growth of ACC [20,21]. In contrast, a phase 2 clinical trial using imatinib found no objective responses in 27 patients with ACC despite high c-Kit expression levels in their tumors [5,17,18]. These results suggest that reducing c-Kit expression using imatinib may not be sufficient to achieve a clinical response in ACC. Moreover, sensitivity to the compound might depend on the presence of c-Kit mutations and/or the incidence of an autocrine-activating loop by its ligand SCF. The objective of this study was to determine the functional effects of c-Kit mutations in ACC.
Materials and Methods
Tumor Samples
Seventeen tumor samples of ACC were retrieved from the University of California, San Francisco (UCSF) Oral Pathology and Anatomic Pathology archives (Table 1) following UCSF guidelines for obtaining and handling human tissue. Slides were reviewed to determine the suitability of tissues for genomic analysis and for tumor grading into tubular, cribriform, and solid types [1].
Table 1.
Clinicopathologic Features of Sporadic ACC Samples Used in This Study.
| Case | Sex | Age (years) | Tumor Site | Histologic Diagnosis | c-Kit Expression |
|---|---|---|---|---|---|
| 1 | F | 61 | Palate | Cribriform | + |
| 2 | M | 51 | Palate | Cribriform | + |
| 3 | F | 74 | Palate | Tubular + cribriform | + |
| 4 | M | 57 | Palate | Tubular | + |
| 5 | F | 51 | Palate | Solid + cribriform | + |
| 6 | F | 45 | Maxilla | Solid | + |
| 7 | F | 72 | Maxilla | Tubular + cribriform | + |
| 8 | F | 43 | Parotid gland | Cribriform | + |
| 9 | F | 48 | Parotid gland | Tubular | + |
| 10 | F | 69 | Face | Cribriform | + |
| 11 | F | 75 | Floor of mouse | Tubular + cribriform | + |
| 12 | F | 73 | Lip | Cribriform | - |
| 13 | F | 52 | Paranasal sinus | Tubular + cribriform | - |
| 14 | M | 39 | Pterygopalatine fossa | Tubular | + |
| 15 | F | 77 | Submandibular gland | Cribriform | + |
| 16 | F | 62 | Tongue | Cribriform | + |
| 17 | F | 72 | Trachea | Tubular | + |
Immunohistochemistry
To examine c-Kit or active ERK1/2 protein expression in ACC tumors, immunohistochemistry (IHC) was performed on unstained sections using antibody-based staining kits for 104D2 c-Kit (Dako, Carpinteria, CA) or D13.14.4E phospho-p44/42 MAPK (Cell Signaling Technology, Danvers, MA [CST]) [16]. The detailed procedure for staining has been described [22]. Positive c-Kit protein expression was defined as membrane staining of at least 25% of tumor cells.
Mutation Analysis
Genomic DNA or messenger RNA (mRNA) from each case of ACC was isolated from formalin-fixed, paraffin-embedded (FFPE) tissue sections using a QIAampDNA FFPE Tissue kit (Qiagen, Valencia, CA) or a PureLink FFPE RNA isolation kit (Invitrogen, Carlsbad, CA). DNA samples were amplified by polymerase chain reaction (PCR) with the primer sets listed in Table 2 and proofreading capability platinum Taq DNA polymerase (Invitrogen). mRNA samples were used for first-strand complementary DNA (cDNA) synthesis after PCR amplification with the primer sets listed in Table 3, using an AccuScript high-fidelity RT-PCR kit (Agilent Technology, Santa Clara, CA). Direct sequencing of the PCR products was subsequently performed at the UCSF Genomics Core Facility using ABI BigDye v3.1 dye terminator sequencing chemistry (Applied Biosystems, Carlsbad, CA), an ABI PRISM 3730xl capillary DNA analyzer (Applied Biosystems) and Mutation Surveyor v2.5 (SoftGenetics, State College, PA). Each observed mutation was confirmed by at least three different sets of mutation analyses.
Table 2.
Oligonucleotide Primer Sequences for Detecting KIT, HRAS, KRAS, NRAS, BRAF, PIK3CA, and PTEN.
| Genes | Exon | Forward Primer Sequence (5′ to 3′) | Reverse Primer Sequence (5′ to 3 ′) | Expected PCR Product Size (bp) |
|---|---|---|---|---|
| KIT | 9 | TAAAAGTATGCCACATCCCAAG | CATGGTCAATGTTGGAATGAAC | 368 |
| KIT | 11 | TCCAGAGTGCTCTAATGAC | AGGTGGAACAAAACAAAGG | 292 |
| KIT | 13 | AAGATGCTCAAGCGTAAGTTC | AAGCAGTTTATAATCTAGCATTGCC | 302 |
| KIT | 17 | GTGAACATCATTCAAGGCGT | CCTTTGCAGGACTGTCAAGCA | 336 |
| HRAS | 1 | TCTAGAGGAAGCAGGAGACAGG | CTTTTCCCATCACTGGGTCAT | 515 |
| HRAS | 2 | GAGTCCTTCACCCGTTTGATCT | CCCTGTCTCCTGCTTCCTCTA | 519 |
| KRAS | 1 | GCACAGAGAGTGAACATCATGG | CATTTTTCTTAAGCGTCGATGG | 574 |
| KRAS | 2 | ACAGAAGGCTGTGGAGTCAAAC | TCATCTTTGGAGCAGGAACAAT | 574 |
| NRAS | 1 | GGACAGGTTTTAGAAACTTCAGCA | GGGAGTAATAGGAAGGGGGATT | 590 |
| NRAS | 2 | CCAAGTCATTCCCAGTAGCAAG | TTTGAGGGACAAACCAGATAGG | 568 |
| BRAF | 11 | TGAGGACTAGTTAACCTGGAGGA | TTTTTCCATCCTAATATTGACTTCAG | 581 |
| BRAF | 15 | TTTTTCAACAGGGTACACAGAACA | GCCCCAAAAATCTTAAAAGCAG | 745 |
| PIK3CA | 1 | GCCTAATCAAGTCAAACTATGGAAA | TCTAAGTCATCCCACAAATGACA | 769 |
| PIK3CA | 9 | TGGTTCTTTCCTGTCTCTGAAAA | GCATTTAATGTGCCAACTACCAA | 521 |
| PIK3CA | 20 | GCTTTGTCTACGAAAGCCTCTCT | GCAATAACAGCCTTTGTTGTGTC | 793 |
| PTEN | 2 | TCAATTTTGGGGTTTTGTTTTG | ACTGTATCCCCCTGAAGTCCAT | 563 |
| PTEN | 3 | TCTGAAAAGCTCTGGTTTTACTTCA | ATATGGGCTAGATGCCAAGTCA | 547 |
| PTEN | 4 | TGGCATCACAAGTTTTTAAGCA | AGACCAACTGCCTCAAATAGTAGG | 600 |
| PTEN | 5 | GGGGAAAATAATACCTGGCTTC | GAAGCTCTACTGGACAGCATAGTG | 798 |
| PTEN | 6 | CGCTGTTGTGACCTTTGAATAA | ATTGCTTTTGGCTTCTTTAGCC | 589 |
| PTEN | 7 | TTGAAGGTTCAAACTGGAGAAAA | AATGTCTCACCAATGCCAGAGT | 516 |
| PTEN | 8 | CATGTGAATGAAAATGCAACAGA | TGTTACTGCTACGTAAACACTGCTTC | 581 |
Table 3.
Oligonucleotide Primer Sequences for Detecting KIT mRNA.
| Genes | Targeted Codon | Forward Primer Sequence (5′ to 3′) | Reverse Primer Sequence (5′ to 3 ′) | Expected PCR Product Size (bp) |
|---|---|---|---|---|
| KIT | 664 | GAGGCAACTGCTTATGGCTTA | TGTCGGCCTTGGTTGGGACA | 400 |
| KIT | 796 | TGTCCCAACCAAGGCCGACA | TGCTTTCAGGTGCCATCCACTTCA | 329 |
DNA Plasmids and Site-Directed Mutagenesis
Wild-type human c-Kit cDNA (Origene, Rockville, MD) was subcloned into a pcDNA3.1 vector (Invitrogen). K-Ras cDNA (Origene) was subcloned into an N-terminal Myc epitope-tagged pcDNA3.1 expression vector [23]. The Myc epitope-tagged wild-type B-Raf pcDNA3.1 expression vector has been described [13]. C-Kit, K-Ras, and B-Raf mutants were generated with a QuickChange site-directed mutagenesis kit (Agilent Technologies). All mutations were verified by sequencing at the UCSF Genomics Core Facility.
Chemical, Cell Line, and Transfection
Human recombinant SCF was purchased from CST. Human embryonic kidney (HEK) 293T cells and African green monkey kidney fibroblast-like COS-7 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum (PAA, New Bedford, MA). They were maintained in a 5% CO2 incubator at 37 C. TransIT-LT1 (Mirus, Madison, WI) was used for transient transfections.
Western Blot Analysis
Total cellular protein was prepared using cell lysis buffer (CST). Equal amounts of total protein were resolved by SDS-PAGE. Western blots were developed by enhanced chemiluminescence (GE Healthcare Life Sciences, Piscataway, NJ). The following primary and secondary antibodies were used: phospho-c-Kit tyrosine 721 (3391; CST), c-Kit (AB81; CST), phospho-C-Raf serine 289/296/301 (9431; CST), C-Raf (610151; BD Biosciences, San Jose, CA), phospho-MEK1/2 serine 218/222 or serine 222/226 (9121; CST), MEK1/2 (9122; CST), phospho-ERK1/2 (E-4; Santa Cruz Biotech [SCB], Santa Cruz, CA), ERK1/2 (K-23; SCB), Myc (A-14 HRP-conjugated; SCB), β-actin-HRP (Sigma, St. Louis, MO), antimouse IgG HRP (7076; CST), and antirabbit IgG HRP (7074; CST).
Results
Characteristics of Tumors
We identified 17 tumor samples that fulfilled the criteria for ACC in the UCSF Oral Pathology and Anatomic Pathology archives. Table 1 summarizes the clinical attributes of their donor patients. All tumors had arisen sporadically. Fourteen tumors occurred in women, and three tumors occurred in men. The mean age at presentation was 60 years (range, 39–77 years). All tumors arose at the following sites: palate (five tumors), maxilla (two tumors), parotid gland (two tumors), and one each for the following sites: face, floor of mouth, lip, paranasal sinus, pterygopalatine fossa, submandibular gland, tongue, and trachea. The tumors were graded into the following morphologic patterns: cribriform (seven tumors), tubular (four tumors), solid (one tumor), combined cribriform and tubular (four tumors), and combined solid and tubular (one tumor).
Expression of c-Kit Was Elevated in Sporadic ACC Tumors
Given that recent studies have demonstrated that almost all ACC tumors show increased expression of c-Kit, we performed IHC for this protein on tumor sample sections using a c-Kit/CD117 antibody. IHC was assessed by scoring the presence or absence of positively stained cells. Positive cases were defined as membrane staining in at least 25% of tumor cells. Table 1 shows the results of this study. We identified expression of c-Kit in 15 (88%) of 17 tumors. No immunoreactivity was observed in adjacent normal salivary gland tissue, suggesting that c-Kit immunoreaction was specific.
Two ACC Tumors Demonstrated Heterogeneous KIT Missense Mutations That Caused Distinct Amino Acid Changes in the c-Kit Protein
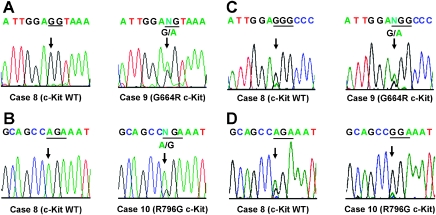
We searched for genomic alterations in KIT in our 17 ACC tumor samples. Similar investigations have been conducted by others specifically in exons 11 and 17 of c-Kit in the ACC tumors [15]. We included other important functional domains such as exons 9 and 13 within c-Kit in our mutation analysis. Genomic DNA was isolated from FFPE tissue sections. DNA samples were amplified by PCR with primer sets independently designed to amplify specific sized products of exons 9, 11, 13, and 17 of KIT (Table 2). These exons encode domains for dimerization (exon 9), the juxtamembrane region (exon 11), or protein kinase activity (exons 13 and 17) [24]. We chose them because gain-of-function mutations are frequently found in these regions of KIT in other neoplasms, such as in acute myeloid leukemia, GIST, mastocytosis, and seminomas [25]. We performed direct sequencing of each exon's PCR product. Each observed mutation was confirmed by at least three different sets of mutation analyses. We identified two heterogeneous missense mutations in KIT (Table 4). Mutations were identified at a lower frequency than in other neoplasms, but they were specific and nonrandom: no frameshift, nonsense, synonymous missense, or splice mutations were detected in the cohort of sporadic ACC tumor specimens. Two ACC tumors (cases 9 and 10) had nucleotide (nt) switches. Specifically, they were nt1990G → A in exon 13 in case 9 (Figure 1A) and nt2386A → G in exon 17 in case 10 (Figure 1B). The predicted missense substitutions were arginine for glycine 664 (G664R) and glycine for arginine 796 (R796G) in the protein kinase domain. These mutations were found in highly conserved residues in the intracellular kinase domain of the gene, suggesting that they should affect the protein kinase activities of the ACC c-Kit mutants.
Table 4.
Summary of Mutation Analysis in Sporadic ACC.
| Case | Genes | ||||||
|---|---|---|---|---|---|---|---|
| KIT | HRAS | KRAS | NRAS | BRAF | PIK3CA | PTEN | |
| 1 | — | — | — | — | N/A | — | — |
| 2 | — | nt81T → C (H27H) | nt50G → A (S17N) | — | — | — | — |
| 3 | — | — | — | — | — | — | — |
| 4 | — | — | — | — | — | nt2994C → T (F998F) | — |
| 5 | — | nt81T → C (H27H) | — | — | — | — | — |
| 6 | — | nt81T → C (H27H) | — | — | — | — | — |
| 7 | — | — | — | — | — | — | — |
| 8 | — | — | — | — | — | — | — |
| 9 | nt1990G → A (G664R) | — | — | — | — | — | — |
| 10 | nt2386A → G (R796G) | — | — | N/A | N/A | N/A | N/A |
| 11 | — | nt81T → C (H27H) | — | — | nt1768G → A (V590I) | — | — |
| 12 | — | nt81T → C (H27H) | — | N/A | — | nt318C → A (G106G) | — |
| 13 | — | — | — | — | — | — | — |
| 14 | — | nt81T → C (H27H) | — | — | — | — | — |
| 15 | — | — | — | nt57G → A (L19L) | — | — | — |
| 16 | — | nt81T → C (H27H) | — | — | — | N/A | — |
| 17 | — | nt81T → C (H27H) | — | — | — | — | — |
Four missense mutations in three different genes within the c-Kit RTK signaling pathway, namely KIT, KRAS, and BRAF, were found. All of these ACC mutations caused amino acid changes in the resultant proteins.
Figure 1.
KIT mutations in tumor samples from sporadic ACC. (A) Genomic DNA electropherograms of case 8 (left) and case 9 (right) are shown. They identify the KIT missense mutation in exon 13. Case 8 has a wild-type (WT) sequence. Case 9 has nucleotide (nt) switches, specifically nt1990G → A in exon 13, with a predicted missense substitution of arginine for glycine 664 (G664R). The first and second nucleotides for codon 664 are highlighted with underlines. The third nucleotide is located in a different exon. (B) Genomic DNA electropherograms of case 8 (left) and case 10 (right) are shown, identifying the KIT missense mutation in exon 17. Case 8 has a WT sequence. Case 10 has an nt2386A → G transition in exon 17, with a predicted missense substitution of glycine for arginine 796 (R796G). Triplet nucleotides at codon 796 are marked with underlines. (C) RT-PCR products electropherograms of case 8 (left) and case 9 (right). Case 8 has a wild-type (WT) sequence. Case 9 has the KIT missense mutation in codon 664. Triplet nucleotides for codon 664 are highlighted with underlines. (D) RT-PCR products electropherograms of case 8 (left) and case 10 (right) RT-PCR products are shown. Case 8 has a wild-type (WT) sequence. Case 10 has the KIT missense mutation in codon 796. Triplet nucleotides for codon 796 are highlighted with underlines.
Because cases 9 and 10 were heterozygous for these c-Kit mutations at the genomic DNA level (Figure 1, A and B), we investigated whether the c-Kit protein expression noted in Table 1 was from the mutant allele. We performed reverse transcription (RT)-PCR and subsequent direct sequencing of the products to address the question (Figure 1, C and D). mRNA was isolated from FFPE tissue sections. Next, first-strand cDNA was synthesized from each sample. Specific-sized products of the c-Kit gene, which contained either codon 664 or 796, were independently amplified by PCR with the primer sets indicated in Table 3. We performed direct sequencing of these PCR products. Each observed mutation was confirmed by at least three different RT-PCR assays. Case 9 expressed both the wild-type and G664R mutant forms of c-Kit (Figure 1C), suggesting that tumor cells in this sample might consist of a heterogenous population. In contrast, we found that case 10 expressed the R796G mutant form but not the wild-type form (Figure 1D), indicating that most of the cells were homogeneous and had the same R786G c-Kit mutation.
Taken together, we concluded that the elevated c-Kit protein expression found in cases 9 and 10 was indeed from the mutant allele. We reasoned that, if our ACC c-Kit mutants functioned actively, c-Kit might play a role in the development of at least some subsets of ACC and that patients may benefit from c-Kit targeted therapy with imatinib.
ACC c-Kit Mutations Are Inactive
To explore the functional consequences of these mutations, we introduced expression vectors containing G664R and R796G mutant forms of c-Kit into HEK 293T cells. HEK 293T cells were chosen because they barely express endogenous c-Kit and because widely used cell lines thought to be ACC cells have recently been shown to have been replaced with other cells because of cross-contamination and misidentification [3].
Twenty-four hours later, we assessed the cells' ability to autophosphorylate at tyrosine 721 [13,25]. Tyrosine 721 is located in the kinase insert region of the catalytic domain and its phosphorylation is essential for c-Kit activation [25]. Phosphorylation status was determined by Western blot analysis (Figure 2A).
Figure 2.
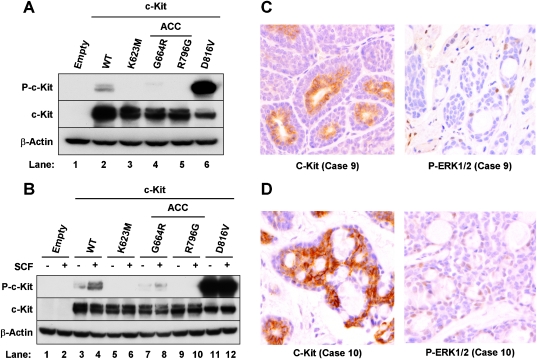
ACC c-Kit mutations are inactive and caused substantial reduction of MAPK activity in tumor cells. (A, B) HEK 293T cells were plated into six-well plates. Twenty-four hours later, subconfluent cells were transfected with 2 µg each of expression vectors containing the various c-Kit mutants. (A) After 24 hours of incubation, the cells were harvested. (B) After 24 hours of incubation, the cells were serum-starved for 16 hours with medium containing 0.5% fetal bovine serum. Cultures were incubated for 15 minutes in the presence (+) or absence (-) of 50 ng/ml SCF before harvesting. Western blot analysis was performed using antibodies against phosphorylation specific c-Kit (P-c-Kit), c-Kit specific for the c-Kit variants, and β-actin as a loading control. The following c-Kit vectors were independently transfected: Empty (pcDNA 3.1) wild-type, K623M (catalytically inactive), D816V (constitutively active), and G664R and R796G (found in ACC). (C, D) IHC was performed on unstained sections using antibodies to Kit (left panels) or phospho-p44/42 MAPK (right panels). Phosphorylated forms of ERK1/2 (right panels) were barely detectable in both cases despite strong expression of c-Kit (left panels).
We compared the activities of our ACC mutants with wild-type c-Kit and two other variants. The variant forms included K623M (methionine substitution for the catalytic lysine at residue 623) and D816V (substitution of valine for aspartic acid at residue 816). K623M is catalytically inactive, and D816V is constitutively active. D816V c-Kit is often found in different types of human malignancies, including acute myeloid leukemia, mastocytosis, seminomas, lymphomas, GIST, and teratomas [26,27]. We observed significant autophosphorylation at tyrosine 721 in the cells with D816V c-Kit (Figure 2A, lane 6; P-c-Kit). In contrast, K623M c-Kit was not phosphorylated at tyrosine 721, as expected (lane 3). Surprisingly, the phosphorylation of both ACC mutants was much lower than wild-type, and they seemed to be kinase-inactivated (lanes 4 and 5). Similar profiles were obtained when the experiments were performed in COS-7 cells (not shown).
We next examined whether our ACC c-Kit mutants could be activated in response to stimulation with SCF, the c-Kit ligand (Figure 2B). SCF is the sole activator of c-Kit, and the ACC mutants might be activated in the presence of an autocrine-activating loop by SCF. Cells were serum-starved for 16 hours after transfection and then cultured with (+) or without (-) SCF for 15 minutes before harvesting [28]. We found constitutive phosphorylation of c-Kit in the cells transfected with D816V (lanes 11 and 12). Autophosphorylation was observed with or without SCF. No activation was observed in the samples transfected with K623M c-Kit (lanes 5 and 6). SCF increased phosphorylation of wild-type c-Kit (lanes 3 and 4). In contrast, we did not see an increase of phosphorylated c-Kit protein in the samples transfected with the ACC mutant R796G (lanes 9 and 10). There was a slight increase in the other mutant form (G664R; lanes 7 and 8), but the level of activation was much lower than that of wild-type c-Kit. Similar data were obtained in COS-7 cells (not shown).
These results suggested that ACC c-Kit mutants had lost most (G664R) or all (R796G) protein kinase activity. Taken together, we concluded that c-Kit is inactive in the ACC mutants.
ACC c-Kit Mutations Reduced MAPK Activity in Tumor Cells
To determine the biological significance of these c-Kit mutations in downstream effectors—particularly in the MAPK signaling pathway in ACC tumors—we performed IHC on tumor sample sections using a phosphorylation-specific ERK1/2 antibody (Figure 2, C and D).
We included a positive control from a breast cancer sample to confirm whether our protocol and procedure worked properly (data not shown). As expected, phosphorylated forms of ERK1/2 were barely detectable in both cases (right panels), despite the fact that both tumors strongly expressed c-Kit (left panels). Positive staining was found in only 1% of the cells in case 9, whereas almost no immunoreactivity was observed in case 10: some cells stained weakly, but no strong staining was observed. We concluded that c-Kit mutations caused substantially reduced MAPK activity in ACC tumor cells.
Two ACC Tumors without KIT Mutations Had Missense Mutations in KRAS and BRAF, Downstream Effectors of c-Kit
We performed further mutation analysis in the components of the c-Kit downstream signaling pathways, including the Ras/MAPK and PI3K/Akt cascades. The PI3K/Akt and MAPK signaling pathways are important for tumor progression in several human cancers [13,14]. In fact, mutations in the downstream signaling elements often amplify the transmission of messages from RTKs.
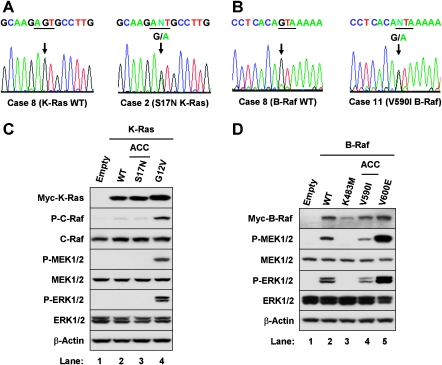
Because oncogenic mutations are frequently found in HRAS, KRAS, NRAS, and BRAF in the MAPK pathway as well as in PIK3CA and PTEN in the PI3K/Akt cascade, we searched for mutations in these genes [14]. Table 2 shows primer sets used to detect oncogenic mutations in these genes and our exons of focus. These exons were chosen because most gain-of-function mutations in HRAS, KRAS, NRAS, BRAF, and PIK3CA are frequently found in them in a variety of neoplasms, as are loss-of-function mutations in PTEN [29–31]. Table 4 summarizes our results. There were amino acid changes in two ACC tumors: case 2 (K-Ras) and case 11 (B-Raf). There was a missense substitution of asparagine for serine 17 (S17N) of K-Ras (nt50G → A transition in exon 1; Figure 3A) and an isoleucine-for-valine 590 substitution (V590I) in the protein kinase domain of B-Raf (nt1768A → G in exon 15; Figure 3B).
Figure 3.
ACC K-Ras and B-Raf mutations were inactive or kinase-impaired. (A and B) KRAS and BRAF mutations were detected in sporadic ACC. (A) Genomic DNA electropherograms of case 8 (left) and case 2 (right) identify the KRAS missense mutation in exon 1. Case 8 has a wild-type (WT) sequence. In contrast, case 2 has a nt50G → A transition with a predicted missense substitution of asparagine for serine 17 (S17N). Triplet nucleotides for codon 17 are highlighted with underlines. (B) Genomic DNA electropherograms of case 8 (left) and case 11 (right) identify the BRAF missense mutation in exon 15. Case 8 shows normal wild-type (WT) sequence. In contrast, case 11 has a nt1768A → G change with a predicted missense substitution of isoleucine for valine 590 (V590I). Triplet nucleotides cording codon 590 are marked with underlines. (C and D) HEK 293T cells were plated into six-well plates. Twenty-four hours later, subconfluent cells were transfected with 2 µg each of expression vectors containing the various K-Ras mutants (C) or 0.25 µg each of expression vectors containing the various B-Raf mutants (D). After 24 hours of incubation, the cells were harvested. Western blot analysis was performed using antibodies against phosphorylation-specific C-Raf (P-C-Raf), MEK1/2 (P-MEK1/2), p44 ERK1 and p42 ERK2 (P-ERK1/2), total C-Raf, total MEK1/2, total ERK1/2, and β-actin. The following vectors were independently transfected: (C) Empty (pcDNA 3.1), wild-type, G12V (constitutively active), and S17N K-Ras (found in ACC). In (D), the vectors were Empty (pcDNA 3.1), wild-type, K483M (catalytically inactive), V600E (constitutively active), and V590I (found in ACC).
No amino acid change, frameshift, nonsense, or splice mutations were detected in HRAS, NRAS, PIK3CA, and PTEN in the cohort of ACC tumors. Eight ACC tumors, however, had a synonymous single nucleotide polymorphism (SNP) in HRAS. This SNP was rs12628 (nt81T → C); it was found in patients 2, 5, 6, 11, 12, 14, 16, and 17. There was one synonymous SNP (nt57G → A) in NRAS in case 15, and there were two synonymous SNPs (nt2994C → T and nt318C → A) in PIK3CA in cases 4 and 12. These synonymous mutations do not change the amino acid sequence of the gene products of HRAS, NRAS, or PIK3CA, and they are therefore functionally identical to their wildtype counterparts.
Because c-Kit protein was expressed in the tumors harboring K-Ras and B-Raf mutations (cases 2 and 11 in Table 1), we hypothesized that these mutants must render c-Kit-MAPK signaling by their loss or reduced function. Alternatively, they might antagonize the gene product of the wild-type allele. Thus, we expected that S17N K-Ras and V590I B-Raf would have similar functional consequences in the signaling pathway.
ACC K-Ras and B-Raf Mutations Are Inactive or Kinase-Impaired
The ACC K-Ras mutant form S17N was particularly interesting because it has not been reported in human cancer and genetic disease. Nonetheless, it is well known as a dominant-negative form of the protein, and has been widely used as a tool to study the Ras/MAPK signaling pathway [32,33]. The asparagine substitution for serine 17 dramatically reduces Ras's affinity for GTP without affecting its affinity for GDP. Thus, expression of S17N K-Ras can block exchange factor. mediated activation of endogenous K-Ras.
The functional significance of the ACC B-Raf mutant V590I is unknown, although it has been found in one case of childhood acute lymphoblastic leukemia [34]. This leukemia patient simultaneously carried an N-Ras activated mutant.
We examined the functional consequences of the mutations in KRas and B-Raf in the ACC samples (Figure 3, C and D). To do so, we introduced expression vectors containing S17N K-Ras and V590I B-Raf into HEK 293T cells and assessed the effects of endogenous activation within the MAPK signaling pathway [13]. Activation was assessed by Western blot analysis of the phosphorylation status of the molecules (see P-C-Raf in Figure 3C and P-MEK1/2, P-ERK1/2 in Figure 3, C and D).
We compared the activities of S17N K-Ras with wild-type and G12V (substitution of valine for glycine 12 of K-Ras). We also compared the activity of V590I B-Raf with wild-type B-Raf, K483M (methionine substitution for the catalytic lysine 483) and V600E (substation of glutamine for valine 600). G12V K-Ras and V600E B-Raf are catalytically active; K483M B-Raf is inactive. K-Ras is commonly mutated in pancreatic and colorectal cancers, and the G12V mutation is its most frequently observed mutation in these tumors [29,35,36]. Conversely, the V600E B-Raf mutation occurs at a high rate in numerous cancers. This mutation accounts for more than 90% of BRAF mutations [37].
Figure 3C shows constitutive activation of C-Raf and MEK1/2 with consequent induction of ERK1/2 activities in the sample transfected with G12V K-Ras (lane 4). In contrast, wild-type and S17N K-Ras failed to stimulate the MAPK signaling cascade (lanes 2 and 3), suggesting that the S17N is inactive. Similar profiles were obtained when the experiments were performed in COS-7 cells (not shown).
Likewise, we observed that K483M B-Raf hardly activated its downstream effectors MEK1/2 and ERK1/2 (Figure 3D, lane 3). The V600E B-Raf mutant significantly increased activation of MEK1/2 and ERK1/2 compared with B-Raf wild-type (lanes 2 and 5). However, the activity of our ACC B-Raf mutant V590I was lower than that of the wild-type (lanes 2 and 4) [38]. Similar data were obtained in COS-7 cells (not shown). We concluded that ACC K-Ras and B-Raf mutants are inactive or kinase-impaired.
Discussion
We performed a functional genomic study of the RTK c-Kit gene (KIT) using 17 sporadic ACC tumor specimens. We found that two ACC cases had distinct missense mutations in KIT (Figure 4). We then explored the functional consequences of these mutations. Surprisingly, they were inactive, and we observed a significant reduction of MAPK activity in tumor cells harboring them. In addition, we observed that two ACC tumors without KIT mutations had missense mutations in the downstream effectors KRAS or BRAF, causing kinase-inactive mutant forms. This is the first time that a study has shown that MAPK activity from RTK signaling is inhibited by gene mutations during tumor development. Our results suggest that ACC is able to proliferate despite inactivation of the RTK c-Kit signaling pathway. This mechanism may explain why no objective responses were observed in the phase 2 clinical trial of imatinib for 27 ACC patients.
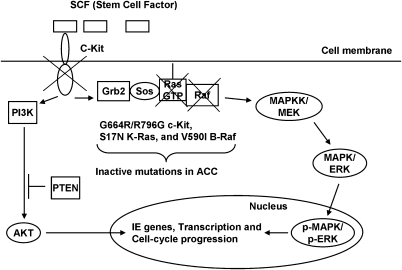
Figure 4.
Cell signaling from the RTK c-Kit is disrupted during tumor development in a subset of salivary gland ACCs. C-Kit regulates cell survival and growth control through the PI3K/Akt and MAPK signaling pathways. We found inactivating mutations in KIT, KRAS, and BRAF. Because ACC can proliferate despite inactivation of c-Kit cell signaling, c-Kit must be dispensable for maintaining established ACC tumors. This is the first time that inactivation of MAPK accompanied tumorigenesis. Our results also suggest that selective inhibition of c-Kit is not a promising strategy for ACC therapeutic development.
Tumors generally have a number of genomic aberrations, and no single genetic alteration is thought to be enough to cause tumor progression. Some mutations are necessary for initiating and/or developing tumors but are not necessary for late-stage tumor maintenance [39].
Only a few of the numerous defective proteins responsible for tumor maintenance might be attractive targets for drug development [19]. In addition, anticancer drugs, which are usually low-molecular weight organic compounds, inhibit biochemical functions rather than enhance them. Thus, target molecules should be activated in cancer. Recent data from mouse models of cancer have shown that even the brief interruptions of the activities of a single oncogene such as H-Ras, K-Ras, c-Myc, and EGFR can reverse cancer [39]. There have been no reports of reversibility of c-Kit-induced cancer in mice. Activation of c-Kit might play an important role at some stage of ACC tumor progression. However, its expression is probably not necessary for ACC tumor maintenance at the late stage because at least a subset of ACC cells can proliferate despite inhibition of the c-Kit signaling pathway.
Thus, c-Kit does not seem to be a suitable therapeutic target in ACC. Partial or complete loss of c-Kit function is associated with other diseases. For example, piebaldism (also known as piebald trait; OMIM 172800) is an autosomal-dominant genetic disorder of melanocyte development. It is characterized by white patches of skin and hair [40]. Piebaldism is a disorder of haploinsufficiency, in which there is loss of function in one allele of the c-Kit proto-oncogene. Some subsets of piebaldism, however, are attributed to distinct mutations in the zinc finger transcription factor SNAI2 [41].
A disorder similar to human piebaldism also occurs in mice [42,43]. Spontaneous mutations at the dominant white (W) locus of the laboratory mouse are associated with defects of pigmentation (albinism; white spotting of the skin), hematopoiesis (anemia), and germ cell development (sterility). Affected mice have partial or complete loss of c-Kit function. This fact highlights a significant function of c-Kit in normal stem and progenitor cells for hematopoiesis and pigmentation.
In this study, we examined the biochemical functions of two ACC c-Kit mutants: G664R and R796G. Interestingly, both mutations have been reported in patients with piebaldism [40,44]. In particular, a patient harboring the R796G c-Kit mutation had severe piebaldism and profound sensorineural deafness [44]. The symptom seemed much worse than in other cases of piebaldism, suggesting that the R796G mutant must completely abolish c-Kit function and that the added severity may be attributed to a dominant-negative effect [6]. This observation is in accordance with our findings. The R796G c-Kit could not be activated by SCF stimulation. Piebaldism patients do not have a higher incidence of ACC, even if they have completely lost c-Kit function through a R796G mutation. Thus, loss-of-function mutations in c-Kit are probably not sufficient to cause ACC.
Gain-of-function mutations of c-Kit are frequently found in neoplasms, including GIST (OMIM 606764). GISTs are mesenchymal tumors found in the gastrointestinal tract. They originate from the interstitial cells of Cajal in the intestines [25]. Approximately 50% to 90% of GISTs are caused by mutations in KIT, and 10% to 35% of GIST subsets arise due to the mutations in PDGFRA (PDGFRα) [25,45]. Mutations in KIT and PDGFRA are mutually exclusive. Familial cases of GIST have germ line mutations, whereas patients with sporadic disease have somatic mutations in tumor tissue. In contrast to piebaldism, familial GIST patients often present with hyperpigmentation and mast cell hyperplasia [46].
GISTs are sensitive to c-Kit-targeted therapy with imatinib. Most GISTs, however, develop resistance to this compound. After 2.5 years of initial treatments, ∼75% of tumors did not respond to further treatment with the drug [19,47]. This fact suggests that long-term therapy with c-Kit targeted therapy may not be effective. To date, there have been no reports of familial ACC or other salivary gland tumors in GIST patients, indicating that gain-of-function mutations in c-Kit play little role in ACC. In fact, our ACC c-Kit mutations demonstrate loss of function.
The mechanism of c-Kit overexpression in ACC is not yet elucidated. In the melanocyte, c-Kit is transcriptionally regulated by the basic-helix-loop-helix-leucine zipper protein family of transcription factor MITF [48]. We speculate that MITF might contribute to c-Kit expression in ACC and could play a role for tumor progression in a similar way in pigment precursor cells. The MITF is encoded at microphthalmia (mi) locus in mice [49]. Mutant mice have small eyes, show loss of pigmentation in the eye, inner ear, and skin, and early-onset sensorineural deafness (OMIM 156845). The phenotype apparently associates mi mutant animals with W locus-deletion mice. The association of MITF with c-Kit is mutual. MITF must be phosphorylated at serine 73 and serine 409 to get full transcriptional activity in melanocytes. This phosphorylation is mediated by an upstream signaling pathway from the RTK c-Kit [50]. Despite their strong correlation in the progenitor cell melanocyte and frequent MITF locus amplification particularly in advanced melanomas, c-Kit expression is often lost in malignant melanoma [51]. Further studies are necessary, but c-Kit V600E B-Raf mutations and p16Ink4a inactivation may reduce the c-Kit requirement for MITF activation in melanoma cells. Otherwise, c-Kit may have the properties of a tumor-suppressor gene product in some contexts [52].
We conclude that c-Kit must be dispensable for tumor maintenance in ACC and that selective c-Kit inhibition may not be a promising strategy for ACC therapeutic development. C-Kit activity may be necessary for tumor initiation and at some stage of tumor progression, although further studies are required to answer this question.
Acknowledgments
The authors gratefully acknowledge Frank McCormick for his valuable suggestions and critical discussion. The authors thank Jeffrey Kaufmann and Valerie Natale for their helpful comments. The authors also thank Jonathan M. Woo, Rick Baehner, Loretta Chan, and the UCSF Genomics and UCSF Comprehensive Cancer Center Immunohistochemistry and Molecular Pathology Core Facilities for their support of mutation analyses and immunohistochemistry.
Footnotes
This work was supported by grants from the University of California Cancer Research Coordinating Committee, Hearing Research, Inc, and American Head and Neck Society to O.T. and by grants DE017249-01, U10 CA21661, and 1T32DE019096 from the National Institutes of Health to R.C.K.J.
References
- 1.Spiro RH, Huvos AG, Strong EW. Adenoid cystic carcinoma of salivary origin. A clinicopathologic study of 242 cases. Am J Surg. 1974;128:512–520. doi: 10.1016/0002-9610(74)90265-7. [DOI] [PubMed] [Google Scholar]
- 2.Carlson ER, Kim J, Mirowski GW, Stewart JCB, Zarbo RJ. Salivary gland diseases. In: Regezi JA, Sciubba JJ, Jordan RCK, editors. Oral Pathology: Clinical Pathology Correlations. St Louis, MO: Saunders; 2008. pp. 179–215. [Google Scholar]
- 3.Phuchareon J, Ohta Y, Woo JM, Eisele DW, Tetsu O. Genetic profiling reveals cross-contamination and misidentification of 6 adenoid cystic carcinoma cell lines: ACC2, ACC3, ACCM, ACCNS, ACCS and CAC2. PLoS One. 2009;4:e6040–e6040. doi: 10.1371/journal.pone.0006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen AM, Garcia J, Lee NY, Bucci MK, Eisele DW. Patterns of nodal relapse after surgery and postoperative radiation therapy for carcinomas of the major and minor salivary glands: what is the role of elective neck irradiation? Int J Radiat Oncol Biol Phys. 2007;67:988–994. doi: 10.1016/j.ijrobp.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 5.Laurie SA, Licitra L. Systemic therapy in the palliative management of advanced salivary gland cancers. J Clin Oncol. 2006;24:2673–2678. doi: 10.1200/JCO.2005.05.3025. [DOI] [PubMed] [Google Scholar]
- 6.Robertson SC, Tynan JA, Donoghue DJ. RTK mutations and human syndromes when good receptors turn bad. Trends. 2000;Genet 16:265–271. doi: 10.1016/s0168-9525(00)02077-1. [DOI] [PubMed] [Google Scholar]
- 7.Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, Ullrich A. Human proto-oncogene c-Kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin FH, Suggs SV, Langley KE, Lu HS, Ting J, Okino KH, Morris CF, McNiece IK, Jacobsen FW, Mendiaz EA, et al. Primary structure and functional expression of rat and human stem cell factor DNAs. Cell. 1990;63:203–211. doi: 10.1016/0092-8674(90)90301-t. [DOI] [PubMed] [Google Scholar]
- 9.Besmer P, Murphy JE, George PC, Qiu FH, Bergold PJ, Lederman L, Snyder HW, Jr, Brodeur D, Zuckerman EE, Hardy WD. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature. 1986;320:415–421. doi: 10.1038/320415a0. [DOI] [PubMed] [Google Scholar]
- 10.Sattler M, Salgia R, Shrikhande G, Verma S, Pisick E, Prasad KV, Griffin JD. Steel factor induces tyrosine phosphorylation of CRKL and binding of CRKL to a complex containing c-Kit, phosphatidylinositol 3-kinase, and p120 (CBL) J Biol Chem. 1997;272:10248–10253. doi: 10.1074/jbc.272.15.10248. [DOI] [PubMed] [Google Scholar]
- 11.Blume-Jensen P, Jiang G, Hyman R, Lee KF, O'Gorman S, Hunter T. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3′-kinase is essential for male fertility. Nat Genet. 2000;24:157–162. doi: 10.1038/72814. [DOI] [PubMed] [Google Scholar]
- 12.Lennartsson J, Blume-Jensen P, Hermanson M, Ponten E, Carlberg M, Rönnstrand L. Phosphorylation of Shc by Src family kinases is necessary for stem cell factor receptor/c-Kit mediated activation of the Ras/MAP kinase pathway and c-fos induction. Oncogene. 1999;18:5546–5553. doi: 10.1038/sj.onc.1202929. [DOI] [PubMed] [Google Scholar]
- 13.Senawong T, Phuchareon J, Ohara O, McCormick F, Rauen KA, Tetsu O. Germline mutations of MEK in cardio-facio-cutaneous syndrome are sensitive to MEK and Raf inhibition: implications for therapeutic options. Hum Mol Genet. 2008;17:419–430. doi: 10.1093/hmg/ddm319. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Viciana P, Tetsu O, Oda K, Okada J, Rauen K, McCormick F. Cancer targets in the RAS pathway. Cold Spring Harb Symp Quant Biol. 2005;70:461–467. doi: 10.1101/sqb.2005.70.044. [DOI] [PubMed] [Google Scholar]
- 15.Holst VA, Marshall CE, Moskaluk CA, Frierson HF., Jr KIT protein expression and analysis of c-Kit gene mutation in adenoid cystic carcinoma. Mod Pathol. 1999;12:956–960. [PubMed] [Google Scholar]
- 16.Mino M, Pilch BZ, Faquin WC. Expression of KIT (CD117) in neoplasms of the head and neck: an ancillary marker for adenoid cystic carcinoma. Mod Pathol. 2003;16:1224–1231. doi: 10.1097/01.MP.0000096046.42833.C7. [DOI] [PubMed] [Google Scholar]
- 17.Hotte SJ, Winquist EW, Lamont E, MacKenzie M, Vokes E, Chen EX, Brown S, Pond GR, Murgo A, Siu LL. Imatinib mesylate in patients with adenoid cystic cancers of the salivary glands expressing c-Kit: a Princess Margaret Hospital phase II consortium study. J Clin Oncol. 2005;23:585–590. doi: 10.1200/JCO.2005.06.125. [DOI] [PubMed] [Google Scholar]
- 18.Bruce IA, Slevin NJ, Homer JJ, McGown AT, Ward TH. Synergistic effects of imatinib (STI 571) in combination with chemotherapeutic drugs in head and neck cancer. Anticancer Drugs. 2005;16:719–726. doi: 10.1097/01.cad.0000168392.04676.bb. [DOI] [PubMed] [Google Scholar]
- 19.Weinberg RA. The rational treatment of cancer. In: Weinberg RA, editor. The Biology of Cancer. New York, NY: Garland Science; 2007. pp. 725–796. [Google Scholar]
- 20.Alcedo JC, Fabrega JM, Arosemena JR, Urrutia A. Imatinib mesylate as treatment for adenoid cystic carcinoma of the salivary glands: report of two successfully treated cases. Head Neck. 2004;26:829–831. doi: 10.1002/hed.20094. [DOI] [PubMed] [Google Scholar]
- 21.Faivre S, Raymond E, Casiraghi O, Temam S, Berthaud P. Imatinib mesylate can induce objective response in progressing, highly expressing KIT adenoid cystic carcinoma of the salivary glands. J Clin Oncol. 2005;23:6271–6273. doi: 10.1200/JCO.2005.01.6055. [DOI] [PubMed] [Google Scholar]
- 22.Solar AA, Schmidt BL, Jordan RC. Hyalinizing clear cell carcinoma: case series and comprehensive review of the literature. Cancer. 2009;115:75–83. doi: 10.1002/cncr.23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tetsu O, McCormick F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 24.Weinberg RA. Growth factors, receptors, and cancer. In: Weinberg RA, editor. The Biology of Cancer. New York, NY: Garland Science; 2007. pp. 119–158. [Google Scholar]
- 25.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, et al. Gain-of-function mutations of c-Kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y, Zeng S, Metcalfe DD, Akin C, Dimitrijevic S, Butterfield JH, McMahon G, Longley BJ. The c-Kit mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors; kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood. 2002;99:1741–1744. doi: 10.1182/blood.v99.5.1741. [DOI] [PubMed] [Google Scholar]
- 27.Foster R, Griffith R, Ferrao P, Ashman L. Molecular basis of the constitutive activity and STI571 resistance of Asp816Val mutant KIT receptor tyrosine kinase. J Mol Graph Model. 2004;23:139–152. doi: 10.1016/j.jmgm.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Nakagomi N, Hirota S. Juxtamembrane-type c-Kit gene mutation found in aggressive systemic mastocytosis induces imatinib-resistant constitutive KIT activation. Lab Invest. 2007;87:365–371. doi: 10.1038/labinvest.3700524. [DOI] [PubMed] [Google Scholar]
- 29.Karnoub AE, Weinberg RA. RAS oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554–554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 31.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 32.Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matallanas D, Arozarena I, Berciano MT, Aaronson DS, Pellicer A, Lafarga M, Crespo P. Differences on the inhibitory specificities of H-Ras, K-Ras, and N-Ras (N17) dominant negative mutants are related to their membrane microlocalization. J Biol Chem. 2003;278:4572–4581. doi: 10.1074/jbc.M209807200. [DOI] [PubMed] [Google Scholar]
- 34.Gustafsson B, Angelini S, Sander B, Christensson B, Hemminki K, Kumar R. Mutations in the B-Raf and N-Ras genes in childhood acute lymphoblastic leukaemia. Leukemia. 2005;19:310–312. doi: 10.1038/sj.leu.2403589. [DOI] [PubMed] [Google Scholar]
- 35.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-Ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 36.Smit VT, Boot AJ, Smits AM, Fleuren GJ, Cornelisse CJ, Bos JL. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res. 1988;16:7773–7782. doi: 10.1093/nar/16.16.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 38.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Mechanism of activation of the Raf-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 39.Felsher DW. Reversibility of oncogene-induced cancer. Curr Opin Genet Dev. 2004;14:37–42. doi: 10.1016/j.gde.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Giebel LB, Spritz RA. Mutation of the KIT (mast/stem cell growth factor receptor) protooncogene in human piebaldism. Proc Natl Acad Sci USA. 1991;88:8696–8699. doi: 10.1073/pnas.88.19.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez-Martín M, Pérez-Losada J, Rodríguez-García A, González-Sánchez B, Korf BR, Kuster W, Moss C, Spritz RA, Sánchez-García I. Deletion of the SLUG (SNAI2) gene results in human piebaldism. Am J Med Genet A. 2003;122A:125–132. doi: 10.1002/ajmg.a.20345. [DOI] [PubMed] [Google Scholar]
- 42.Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-Kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335:88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 43.Geissler EN, Ryan MA, Housman DE. The dominant-white spotting (W) locus of the mouse encodes the c-Kit proto-oncogene. Cell. 1988;55:185–192. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 44.Spritz RA, Beighton P. Piebaldism with deafness: molecular evidence for an expanded syndrome. Am J Med Genet. 1998;75:101–103. doi: 10.1002/(sici)1096-8628(19980106)75:1<101::aid-ajmg20>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 45.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 46.Nishida T, Hirota S, Taniguchi M, Hashimoto K, Isozaki K, Nakamura H, Kanakura Y, Tanaka T, Takabayashi A, Matsuda H, et al. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet. 1998;19:323–324. doi: 10.1038/1209. [DOI] [PubMed] [Google Scholar]
- 47.Demetri GD. Identification and treatment of chemoresistant inoperable or metastatic GIST: experience with the selective tyrosine kinase inhibitor imatinib mesylate (STI571) Eur J Cancer. 2002;38(suppl 5):S52–S59. doi: 10.1016/s0959-8049(02)80603-7. [DOI] [PubMed] [Google Scholar]
- 48.Tsujimura T, Morii E, Nozaki M, Hashimoto K, Moriyama Y, Takebayashi K, Kondo T, Kanakura Y, Kitamura Y. Involvement of transcription factor encoded by the mi locus in the expression of c-Kit receptor tyrosine kinase in cultured mast cells of mice. Blood. 1996;88:1225–1233. [PubMed] [Google Scholar]
- 49.Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 50.Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- 51.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 52.Carl-Henrik H, Ronnstrand L. Growth factor receptors in cell transformation. In: Peters G, Vousden KH, editors. Oncogenes and Tumor Suppressors. Oxford, UK: Oxford University Press; 1997. pp. 55–85. [Google Scholar]