Abstract
We report the development of universal primers for the reverse-transcription polymerase chain reaction (RT-PCR) amplification and nucleotide sequence analysis of actin cDNAs from taxonomically diverse mosquito species. Primers specific to conserved regions of the invertebrate actin-1 gene were designed after actin cDNA sequences of Anopheles gambiae, Bombyx mori, Drosophila melanogaster, and Caenorhabditis elegans. The efficacy of these primers was determined by RT-PCR with the use of total RNA from mosquitoes belonging to 30 species and 8 genera (Aedes, Anopheles, Culex, Deinocerites, Mansonia, Psorophora, Toxorhynchites, and Wyeomyia). The RT-PCR products were sequenced, and sequence data were used to design additional primers. One primer pair, denoted as Act-2F (5′-ATGGTCGGYATGGGNCAGAAGGACTC-3′) and Act-8R (5′-GATTCCATACCCAGGAAG-GADGG-3′), successfully amplified an RT-PCR product of the expected size (683-nt) in all mosquito spp. tested. We propose that this primer pair can be used as an internal control to test the quality of RNA from mosquitoes collected in vector surveillance studies. These primers can also be used in molecular experiments in which the detection, amplification or silencing of a ubiquitously expressed mosquito housekeeping gene is necessary. Sequence and phylogenetic data are also presented in this report.
Keywords: Mosquito, actin, universal primers, reverse transcription-polymerase chain reaction, sequence
All mosquitoes are members of the family Culicidae (Knight and Stone 1977). This family contains of over 3,500 species and is divided into 3 subfamilies: Anophelinae, Culicinae, and Toxorhynchitinae. The subfamily Anophelinae is further divided into 3 genera: Anopheles, Bironella and Chagasia (Gwadz and Collins 1996). The genus Anopheles is of particular relevance to human health because all human malarial parasites are transmitted by Anopheles spp. Malaria is an enormous public health problem with an estimated 300–500 million cases and 1–3 million deaths occurring worldwide each year (Gwadz and Collins 1996, Snow et al. 2005). Anopheline mosquitoes are also important vectors of human filarial parasites and O’nyongnyong virus (Johnson 1988, Bartholomay and Christensen 2002).
The subfamily Culicinae contains 29 genera (Nasci and Miller 1996), 6 of which are represented in this study: Aedes, Culex, Deinocerites, Mansonia, Psorophora, and Wyeomyia. Mosquitoes in the genera Aedes, Culex, and Mansonia are important vectors of human filarial parasites (Bartholomay and Christensen 2002). Aedes spp. are also important vectors of many medically significant arboviruses; for example: dengue, yellow fever, chikungunya, Rift Valley fever, and La Crosse viruses (Weaver 2005, Gubler et al. 2007, Schmaljohn and Nichol 2007). Dengue virus has a particularly devastating impact on human health with an estimated 50 million–100 million cases of dengue fever and 500,000 cases of life-threatening dengue hemorrhagic fever occurring annually in tropical regions of the world (Gubler 2006). Many viruses that cause morbidity and mortality in humans are also transmitted by Culex spp.; for example: Japanese encephalitis, West Nile, St. Louis encephalitis, Western equine encephalitis, and Venezuelan equine encephalitis viruses (Weaver 2005, Gubler et al. 2007). Human pathogens, such as WNV, have been isolated from Deinocerites and Psorophora spp. mosquitoes (Granwehr et al. 2004).
The subfamily Toxorhynchitinae contains a single genus: Toxorhynchites (Nasci and Miller 1996). Toxorhynchites spp. are among the few mosquito species that do not take blood meals; adults feed primarily on plant nectar. Thus, Toxorhynchites spp. do not transmit infectious agents to humans.
Because of the significant burden that Anophelinae and Culicinae mosquitoes have on human health, numerous mosquito surveillance and control programs have been established throughout the world. Mosquito surveillance often involves the use of reverse-transcription polymerase chain reaction (RT-PCR) to test field-caught mosquitoes for specific pathogens. Often, however, only a small proportion of mosquitoes are positive. Because of the often small proportion of positive specimens, it is useful to amplify a reference or housekeeping gene to be sure that the RNA isolation and cDNA generation were successful. This is especially true when mosquitoes are collected in remote study sites and cannot be transported to the laboratory in a timely manner. Thus, we have developed generic RT-PCR primers for the detection of actin-1 in mosquitoes from all 3 Culicidae subfamilies.
To facilitate the design of universal actin-specific primers, the actin-1 cDNA sequences of Anopheles gambiae (Giles), Bombyx mori (Linnaeus), Drosophila melanogaster (Meigen), and Caenorhabditis elegans were downloaded from the Genbank database (accession numbers U02933. 1, NM_001126255.1, NM_079643.1, and NM_073418.4, respectively) and aligned. Highly conserved regions were identified and used to design 10 actin-specific primers, including Act-2F (5′-ATGGTCGGYATGGGNCAGAAG-GACTC-3′), Act-2R (5′-TCGCACTTCAT-GATSGAGTTGTA-3′), and Act-3R (5′-CCNGGGTACATGGTGGTACCNCCGGA-3′). Act-2F is a forward primer that aligns to nucleotides 269–294 of the actin cDNA sequence of An. gambiae ( Ge nB a nk ac ce ss i on number U02933.1). Act-2R and Actin-3R are reverse primers that align to nucleotides 974–996 and 1037–1062, respectively of the aforementioned sequence.
The efficacy of various primer combinations was evaluated by RT-PCR with the use of 30 species of mosquitoes from 8 genera and 3 subfamilies (Table 1). Twenty-three mosquito species were collected in northern Mexico or the Yucatan Peninsula of Mexico as part of our ongoing arbovirus surveillance studies. Seven species were obtained from the insectaries at Iowa State University. Detailed protocols for the collection, identification, and homogenization of mosquitoes have been provided elsewhere (Far-fan-Ale et al. 2009). Total RNA was extracted from mosquito homogenates with the use of the QIAamp viral RNA extraction kit (QIAGEN, Valencia, CA). Complementary DNAs were generated with the use of Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA), and PCRs were performed with the use of Taq polymerase (Invitrogen).
Table 1.
Mosquito species used in this study, reactivities of actin-specific primers by RT-PCR, and GenBank accession numbers.
| Species | Subfamily | Source of mosquitoes | Primer pair1 |
GenBank accession no. | ||
|---|---|---|---|---|---|---|
| Act-2F + 2R | Act-2F + 3R | Act-2F + 8R | ||||
| Aedes aegypti | Culicinae | YP2 | +3 | + | + | GQ981439 |
| Ae. albopictus | Culicinae | ISU4 | + | + | + | GQ981440 |
| Ae. caspius | Culicinae | ISU | + | + | + | GQ981456 |
| Ae. scapularis | Culicinae | NM5 | + | + | + | GQ981441 |
| Ae. taeniorhynchus | Culicinae | NM | + | + | + | GQ981457 |
| Ae. triseriatus | Culicinae | ISU | + | + | + | GQ981442 |
| Ae. trivittatus | Culicinae | ISU | + | + | + | GQ981443 |
| Anopheles albimanus | Anophelinae | YP | + | + | + | GQ981444 |
| An. crucians | Anophelinae | NM | + | + | + | GQ981467 |
| An. pseudopunctipennis | Anophelinae | NM | + | + | + | GQ981445 |
| An. quadrimaculatus | Anophelinae | NM | + | + | + | GQ981446 |
| An. stephensi | Anophelinae | ISU | + | + | + | GQ981447 |
| An. vestitipennis | Anophelinae | YP | + | + | + | GQ981448 |
| Culex coronator | Culicinae | NM | + | + | + | GQ981449 |
| Cx. erraticus | Culicinae | NM | + | + | + | GQ981450 |
| Cx. interrogator | Culicinae | YP | + | + | + | GQ981451 |
| Cx. nigripalpus | Culicinae | YP | + | + | + | GQ981452 |
| Cx. pipiens | Culicinae | ISU | + | + | + | GQ981453 |
| Cx. quinquefasciatus | Culicinae | NM | + | + | + | GQ981468 |
| Deinocerites cancer | Culicinae | YP | + | + | + | GQ981454 |
| Mansonia titillans | Culicinae | YP | + | + | + | GQ981455 |
| Psorophora albipes | Culicinae | YP | + | − | + | GQ981458 |
| Ps. cyanescens | Culicinae | YP | + | + | + | GQ981459 |
| Ps. ciliata | Culicinae | YP | + | + | + | GQ981460 |
| Ps. columbiae | Culicinae | NM | + | + | + | GQ981461 |
| Ps. confinnis | Culicinae | YP | −6 | + | + | GQ981462 |
| Ps. ferox | Culicinae | YP | + | + | + | GQ981463 |
| Ps. howardii | Culicinae | YP | + | + | + | GQ981464 |
| Toxorhynchites amboinensis | Toxorhynchitinae | ISU | + | + | + | GQ981465 |
| Wyeomyia spp.7 | Culicinae | YP | − | + | + | GQ981466 |
Sizes of RT-PCR products generated by primer pairs Act-2F + 2R, Act-2F + 3R and Act-2F + 8R are 728, 794, and 683 nt, respectively.
Yucatan Peninsula of Mexico.
Positive.
Insectaries at Iowa State University.
Northern Mexico.
Negative.
Mosquitoes were identified to the genus but not species level.
Act-2F and Act-3R was the most consistent and efficient primer combination; RT-PCR products were detected in 29 of the 30 species tested (Table 1), although a nonspecific band of approximately 400 nt was detected with several samples. Primers Act-2F and Act-2R recognized actin-1 from 28 species, and no nonspecific bands were detected. The longest RT-PCR product of the expected size from each species was purified using the Purelink Gel Extraction Kit (Invitrogen) and sequenced using a 3730×1 DNA sequencer (Applied Biosystems, Foster City, CA). The resulting sequences were aligned and used to design additional primers including Act-8R (5′-GATTCCATACCCAGGAAGGADGG-3′). Act-8R is a reverse primer that aligns to nucleotides 929–951 of the actin cDNA sequence of An. gambiae (GenBank accession number U02933.1). When primers Act-2F and Act-8R were used in combination, RT-PCR products of the expected size (683 nt) were detected in all mosquito species. Nonspecific bands were not observed in any samples. All RT-PCR products generated with the use of Act-2F and Act-8R were sequenced with the use of these same primers in the sequencing reactions. Resulting sequences were aligned and in most instances the forward and reverse sequences did not overlap at the 5′ and 3′ ends. Thus, additional primers were designed and used in subsequent sequencing reactions. The resulting sequences were once again aligned and approximately 20 nts were trimmed from the 5′ and 3′ ends to confidently yield sequences of 641 nt (GenBank accession numbers GQ981439 to GQ981468).
Alignment of the 641-nt region of actin-1 from all the mosquito spp. examined in this study using the CLUSTAL W algorithm (version 2) (Higgins and Sharp 1988, Larkin et al. 2007) revealed that this region is highly conserved. Highest (99.2%) nucleotide identity occurs between the actin cDNAs of Ae. aegypti (Linnaeus) and Ae. albopictus (Skuse), and identity was lowest (85.3%) between the actin cDNAs of De. cancer (Theobald) and Mn. titillans (Walker). A total of 465 (72.5%) nucleotide positions are strictly conserved among all mosquito species. This region also has striking identity to the homologous regions of actin cDNAs of other invertebrate species. For instance, alignment of the 641-nt regions of actin-1 from C. elegans and all mosquito species revealed that there are 441 (68.8%) strictly conserved nucleotide positions.
The predicted amino acid sequences of actin-1 from all the mosquito species evaluated in this study were aligned, and amino acid identities and similarities were calculated as described by Altschul et al. 2005. This analysis revealed that the sequences are 94.4–100% identical and 96.2–100% similar. A total of 186 (87.3%) amino acid residues are strictly conserved between all species. This region also has striking identity and similarity to the homologous region of actin-1 of other invertebrate species. A total of 181 (85.0%) amino acid residues are strictly conserved in actin-1 of C. elegans and all the mosquito spp.
Because the nucleotide and amino acid sequence alignments revealed that there is limited sequence diversity between actin-1 from different mosquito species, the universal primers reported here can not be used for RT-PCR sequencing to determine the identity of field mosquitoes that could not be classified based on morphological characteristics. In contrast, the D2 region of 28S ribosomal DNA provides a suitable target in multiplex PCR assays for the differentiation of different members of the An. culicifacies complex (Raghavendra et al. 2009). The ribosomal internal transcribed spacer 1 region provides a reliable target to differentiate Ae. aegypti, Ae. albopictus, and Ae. scutellaris (Walker) in real-time RT-PCR assays (Hill et al. 2008).
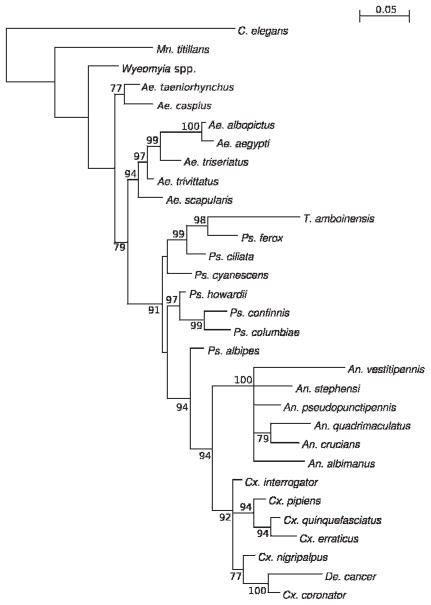
A phylogenetic tree was constructed with the use of the MrBayes GTR+I+G model applied to the 641-nt actin region of 30 mosquito species (Fig. 1). Phylogenetic trees were also generated with the use of neighbor-joining (NJ) and maximum-likelihood (ML) methods (data not shown). To our surprise, the mosquitoes did not separate according to subfamily; no tree has the 3 expected clades corresponding to Anophelinae, Culicinae, or Toxorhynchitinae species mosquitoes. Toxorhynchites amboinensis (Doleschall), Wyeomyia, and Mn. titillans are positioned with the greatest uncertainty and disparity among the phylogenetic trees. In the Bayesian and NJ trees, Wyeomyia and Mn. titillans are basal to all other mosquito sequences, but cluster with the Aedes sequences in the ML tree. Toxorhynchites amboinensis is the most basal sequence in the ML tree, but clusters with some Psorophora spp. in the Bayesian and NJ trees. The Psorophora spp. do not appear as a monophyletic clade in any tree, but instead appear basal to Culex and Anopheles sequences. In particular, Ps. albipes (Theobald) is significantly clustered in the Bayesian tree with Culex and Anopheles spp. rather than other Psorophora spp. Because of the considerable amount of disagreement in the topological arrangements of the mosquito sequences in the Bayesian, NJ, and ML trees and because the mosquitoes did not separate according to subfamily, these data indicate that actin sequences are not suitable for the study of mosquito phylogeny.
Fig. 1.
Phylogenetic analysis of a 641 nucleotide region of the actin-1 cDNA sequence of 30 species of mosquitoes. The displayed phylogeny was estimated by using the program MRBAYES, version 3.1 (Ronquist and Huelsenbeck 2003). Posterior support (out of 100) for selected branches is indicated. All trees were midpoint rooted; the midpoint was on the branch to the homologous region of actin-1 of C. elegans (GenBank accession number NM_079643.1).
In summary, we report the development of primers that detected by RT-PCR a 683-nt region of actin-1 in all mosquito species tested. Because these studies were performed with the use of a taxonomically diverse group of mosquitoes, with representatives from all 3 subfamilies, it is likely that these primers can amplify actin RNA in all mosquito species. Thus, we propose that these primers can be used to monitor the integrity of mosquito samples collected in the field. These primers, in addition to the sequence data presented here, can also be used to facilitate molecular experiments in which the detection, expression, or silencing of a mosquito housekeeping gene is required as a positive or negative control, for example, in Northern blots and RNA interference experiments. The mosquito actin sequences could also be used to design smaller primer sets for quantitative RT-PCR assays if the abundance of a target gene needs to be normalized against a ubiquitously expressed housekeeping gene.
Acknowledgments
This study was supported by Grant 5R21AI0-67281-02 from the National Institutes of Health.
REFERENCES CITED
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu Y. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomay LC, Christensen BM. Vector–parasite interactions in mosquito-borne filariasis. In: Klei TR, Rajan TV, editors. The Filaria. Dordrecht, Netherlands: Kluwer Academic; 2002. pp. 9–20. [Google Scholar]
- Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, Lin M, Dorman KS, Platt KB, Bartholomay LC, Soto V, Beaty BJ, Lanciotti RS, Blitvich BJ. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80:85–95. [PMC free article] [PubMed] [Google Scholar]
- Granwehr BP, Lillibridge KM, Higgs S, Mason PW, Aronson JF, Campbell GA, Barrett AD. West Nile virus: where are we now? Lancet Infect Dis. 2004;4:547–556. doi: 10.1016/S1473-3099(04)01128-4. [DOI] [PubMed] [Google Scholar]
- Gubler DJ. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp. 2006;277:3–16. doi: 10.1002/0470058005.ch2. discussion 16–22, 71–73, 251–253. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. pp. 1153–1252. [Google Scholar]
- Gwadz R, Collins FH. Anopheline mosquitoes and the agents they transmit. In: Beaty BJ, Marquardt WC, editors. The biology of disease vectors. Niwot, CO: University Press of Colorado; 1996. pp. 73–84. [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hill LA, Davis JB, Hapgood G, Whelan PI, Smith GA, Ritchie SA, Cooper RD, van den Hurk AF. Rapid identification of Aedes albopictus, Aedes scutellaris, and Aedes aegypti life stages using real-time polymerase chain reaction assays. Am J Trop Med Hyg. 2008;79:866–875. [PubMed] [Google Scholar]
- Johnson BK. O’nyongnyong virus disease. In: Monath TP, editor. The arboviruses: epidemiology and ecology. Boca Raton, FL: CRC Press; 1988. pp. 217–223. [Google Scholar]
- Knight KL, Stone A. A catalogue of the mosquitoes of the world (Diptera: Culicidae) College Park, MD: Thomas Say Foundation and Entomological Society of America; 1977. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Nasci RS, Miller BR. Culicine mosquitoes and the agents they transmit. In: Beaty BJ, Marquardt WC, editors. The biology of disease vectors. Niwot, CO: University Press of Colorado; 1996. pp. 85–97. [Google Scholar]
- Raghavendra K, Cornel AJ, Reddy BP, Collins FH, Nanda N, Chandra D, Verma V, Dash AP, Subbarao SK. Multiplex PCR assay and phylogenetic analysis of sequences derived from D2 domain of 28S rDNA distinguished members of the Anopheles culicifacies complex into two groups, A/D and B/C/E. Infect Genet Evol. 2009;9:271–277. doi: 10.1016/j.meegid.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schmaljohn CS, Nichol ST. Bunyaviridae. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. pp. 1741–1789. [Google Scholar]
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC. Host range, amplification and arboviral disease emergence. Arch Virol Suppl. 2005;19:33–44. doi: 10.1007/3-211-29981-5_4. [DOI] [PubMed] [Google Scholar]