Abstract
Previously, we reported a high prevalence of Culex flavivirus (CxFV) in Culex quinquefasciatus (Say) in the Yucatan Peninsula of Mexico. To determine whether other Culex spp. mosquitoes in this region are susceptible to natural CxFV infection, Cx. bahamensis (Dyar and Knab), Cx. coronator (Dyar and Knab), Cx. interrogator (Dyar and Knab), Cx. nigripalpus (Theobald) and Cx. opisthopus (Komp) in the Yucatan Peninsula of Mexico were tested for CxFV. Two pools of Cx. interrogator were positive. The envelope protein genes of these isolates and 16 isolates from Cx. quinquefasciatus were sequenced and shown to have ≥99.2% nucleotide identity. These data suggest that there is limited genetic diversity among CxFV isolates in the Yucatan Peninsula of Mexico.
Keywords: Flavivirus, Culex flavivirus, Mosquito, Phylogeny, Mexico
Culex flavivirus (family Flaviviridae, genus Flavivirus) is an insect-specific virus that was first isolated from Cx. pipiens (Linnaeus), Cx. quinquefasciatus (Say) and Cx. tritaeniorhynchus (Giles) collected in Japan and Indonesia in 2003 and 2004 [13]. More recently, CxFV was detected in Cx. quinquefasciatus in Guatemala [20], the Yucatan Peninsula of Mexico [10], Trinidad [17] and Uganda [4], as well as Cx. quinquefasciatus and Cx. restuans (Theobald) in Texas [17], and Cx. pipiens and Cx. tarsalis (Coquillet) in Iowa [2]. CxFV replicates in Aedes albopictus (C6/36) cells but not in African green monkey kidney (Vero) cells, baby hamster kidney cells or intracerebrally inoculated newborn mice, which suggests that this virus is insect-specific.
Eight other insect-specific flaviviruses have been isolated from mosquitoes: cell fusing agent virus [3, 16, 26], Kamiti River virus [6, 25], Quang Binh virus [5], Aedes flavivirus [12], Nakiwogo virus [4], Lammi virus [14], Nouname virus [15] and Calbertado virus (M. A. Drebot, personal communication). A potentially novel insect-specific flavivirus (es) has also been identified in Phlebotomine sandflies [21, 24]. In addition, flavivirus-related DNA known as cell silent agent is integrated into the genomes of some Aedes spp. mosquitoes [7, 22].
There is a current lack of information of the host range and genetic diversity of CxFV in Mexico. The prototype Mexican strain of CxFV (designated CxFV-Mex07), which was isolated from Cx. quinquefasciatus in the Yucatan Peninsula of Mexico in 2007, represents the only CxFV isolate from Mexico for which sequence data are available [10]. Furthermore, Cx. quinquefasciatus is the only mosquito species from which the Mexican strain of CxFV has been isolated [10, 9]. In this report, all Culex spp. mosquitoes (with the exception of Cx. quinquefasciatus) that had been collected in our mosquito-based virus surveillance in the Yucatan Peninsula of Mexico in 2008 [9] were assayed by reverse transcription-polymerase chain reaction (RT-PCR) using flavivirus-and CxFV-specific primers. All of these mosquitoes had previously been tested for cytopathic virus by virus isolation in Vero cells. The purpose of our present study was to identify other insect-specific flaviviruses that may be present in the Yucatan Peninsula of Mexico and to increase our knowledge on the host range and genetic diversity of CxFV in this region.
Culex spp. mosquitoes tested in this study were collected at study sites in Isla Mujeres, Merida, Sian Ka’an and Tixkokob in the Yucatan Peninsula of Mexico. Detailed descriptions of these study sites and the protocols used for the collection, identification and homogenization of mosquitoes have been provided elsewhere [8–10]. Total RNA was extracted from mosquito homogenates using a QIAamp viral RNA extraction kit (QIAGEN, Valencia, CA). Complementary DNAs were generated using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA), and PCRs were performed using Taq polymerase (Invitrogen, Carlsbad, CA, USA) and flavivirus- and CxFV-specific primers. The flavivirus-specific primers, FU2 and cFD3, target a 845-nt region of the NS5 gene [18]. The CxFV-specific primers, CxFV(E)-PCR-F (5′-ACTGGTGACGTTCAAGGCCATAAG-3′) and CxFV (E)-PCR-R (5′-GCCGTGATCAGGTGCTGGTCATCG-3′), target a 1,551 nt region of the CxFV genome that includes the entire (1,281 nt) envelope (E) protein gene. RT-PCR products were purified using a Purelink Gel Extraction Kit (Invitrogen, Carlsbad, CA, USA) and sequenced using a 3730×1 DNA sequencer (Applied Biosystems, Foster City, CA, USA). Sequencing reactions were performed using 12 CxFV-specific primers, and primer sequences are available upon request.
A total of 1,856 Culex spp. mosquitoes in 121 pools were tested by RT-PCR. The mosquitoes belong to five species: Cx. bahamensis (Dyar and Knab) (n = 3), Cx. coronator (Dyar and Knab) (n = 154), Cx. interrogator (Dyar and Knab) (n = 766), Cx. nigripalpus (Theobald) (n = 235) and Cx. opisthopus (Komp) (n = 698). Of these, 1,805 (97.3%) were identified as female and 51 (2.7%) as male. Two pools of female Cx. interrogator were positive by RT-PCR using flavivirus- and CxFV-specific primers. All other mosquitoes were negative with both primer pairs. However, due to the small numbers of Cx. bahamensis, Cx. coronator, Cx. nigripalpus and Cx. opisthopus available for testing, it would be premature to conclude that these species are not natural hosts of CxFV until more research is done to address this issue. The CxFV minimal infection rate (MIR), expressed as the number of positive mosquito pools per 1,000 mosquitoes tested, in Cx. interrogator was 2.6. Because of the considerable variation in pool sizes, bias-corrected maximum likelihood estimation (MLE) values were also calculated using the PooledInfRate statistical software package [1]. The MLE value (and 95% confidence interval) for CxFV in Cx. interrogator was 2.6 (0.48–8.42). One pool positive for CxFV RNA was collected in Tixkokob in January 2008; the other pool was collected in Merida in June 2008. Previously, we reported a much higher CxFV MIR in Cx. quinquefasciatus in the Yucatan Peninsula of Mexico [9, 10]. The CxFV MIRs in Cx. quinquefasciatus collected in Merida and Tixkokob from June through August 2007 were 10.9 and 26.0, respectively. The overall CxFV MIR in Cx. quinquefasciatus collected in Merida from January to December 2008 was 7.7, and the monthly MIRs ranged from 4.3 to 16.6. Previously, we also reported that CxFV RNA was not detected by RT-PCR in any Aedes, Anopheles, Ochlerotatus, Mansonia and Psorophora spp. mosquitoes tested [9].
The two mosquito homogenates that were positive for CxFV RNA were tested for virus by virus isolation in C6/36 cells as described previously [10]. CxFV was isolated from both homogenates. The isolate from Tixkokob has been designated T-955, and the isolate from Merida have been designated M-2168. Twenty homogenates that had previously tested positive for CxFV RNA by RT-PCR (Farfan-Ale et al. 2010) were also processed by virus isolation in C6/36 cells. Sixteen isolates were obtained, and all are from Cx. quinquefasciatus collected in Merida in 2008.
The E protein genes of the 2 CxFV isolates from Cx. interrogator as well as the 16 CxFV isolates from Cx. quinquefasciatus were sequenced (GenBank accession nos. GU289683 to GU289700). Pairwise alignments of the nucleotide sequences using the CLUSTAL W algorithm (version 2) [11, 19] revealed that they have 99.2 to 100% identity. Of the 1280 nucleotides that comprise the E protein gene, 1245 (97.3%) are strictly conserved between all isolates. The deduced amino acid sequences have 99.5 to 100% identity, and 99.5 to 100% similarity. In total, 419 of the 426 (98.4%) amino acid positions are strictly conserved between these isolates. The nucleotide and deduced amino acid sequences of these isolates were also aligned to the homologous region of the prototype Mexican strain of this virus, CxFV-Mex07, which was isolated from Cx. quinquefasciatus in Tixkokob in 2007 [10] (GenBank accession number EU879060). The E protein gene of CxFV-Mex07 has at least 99.5% nucleotide identity, 99.5% amino acid identity and 99.8% amino acid similarity to the 18 E protein gene sequences from in this study. These findings suggest that there is minimal genetic diversity between CxFV isolates in the Yucatan Peninsula of Mexico.
A phylogenetic tree was constructed with Bayesian methods using the E protein gene sequences of 34 CxFV isolates, including the 18 isolates obtained in this study (Fig. 1). The analysis revealed that all isolates from this study have a close phylogenetic relationship with CxFV-Mex07, in addition to CxFV isolates from Guatemala, Trinidad and Uganda. These isolates comprise a distinct clade (denoted as clade 1). CxFV isolates from the United States and Asia comprise a second clade (denoted as clade 2). The posterior support for the branch separating these two clades is 100%. The Mexican isolates form a monophyletic group within clade 1, and the posterior support for this topological arrangement is 84%. Phylogenetic trees were also generated using neighbor-joining, maximum parsimony and maximum likelihood methods, and all of these trees showed the same topological features as the Bayesian tree (data not shown).
Fig. 1.
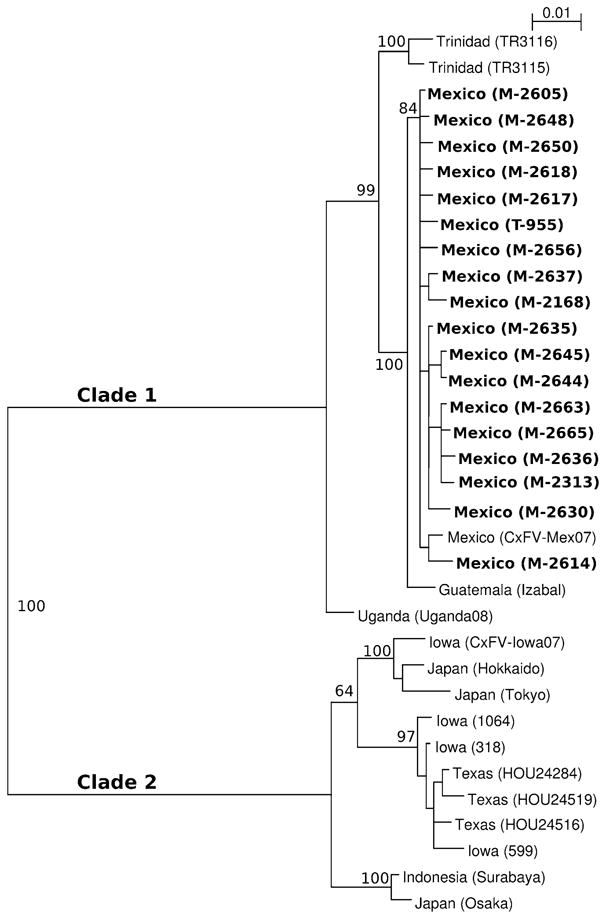
Phylogenetic analysis of the envelope protein gene of 18 CxFV isolates obtained in this study and 16 other CxFV isolates. The envelope protein gene encompasses nucleotide positions 938–2,217 relative to the genomic sequence of the prototype CxFV strain (Tokyo). The displayed phylogeny was estimated by using the program MRBAYES, version 3.1 [23]. Posterior support (out of 100) for selected branches is indicated. An unrooted tree was inferred but is shown rooted using the midpoint method. CxFV isolates obtained in the present study are denoted in bold. GenBank accession numbers for sequences used in the phylogenetic analysis are: M-2168 (GU289684), M-2313 (GU289685), M-2605 (GU289686), M-2614 (GU289687), M-2617 (GU289688), M-2618 (GU289689), M-2630 (GU289690), M-2635 (GU289691), M-2636 (GU289692), M-2637 (GU289693), M-2644 (GU289694), M-2645 (GU289695), M-2648 (GU289696), M-2650 (GU289697), M-2656 (GU289698), M-2663 (GU289699), M-2665 (GU289700), T-955 (GU289683), Mex07 (EU879060.1), Guatemala (EU805805.1), Iowa07 (FJ663034.1), Iowa 1064 (FJ663026.1), Iowa 318 (FJ663030.1), Iowa 599 (FJ663032.1), HOU24284 (FJ502997.1), HOU24519 (FJ502996.1), HOU24516 (FJ502999.1), Hokkaido (AB262762.1), Osaka (AB262763.1), Sura-baya (AB262766.1), Tokyo (AB262759.2), TR3115 (FJ503002.1), TR3116 (FJ503003.1) and Uganda (GQ165808.1)
In summary, we demonstrate that the host-range of CxFV in the Yucatan Peninsula of Mexico is not restricted to Cx. quinquefasciatus and provide evidence of limited genetic and phylogenetic diversity between CxFV isolates in this region. Comparative studies between insect-specific viruses and mosquito-vertebrate flaviviruses will provide important insight into flavivirus evolution and will help us understand why some flaviviruses possess the capacity to replicate and cause disease in humans and vertebrate animals while others do not.
Acknowledgments
This study was supported by grant 5R21AI067281-02 from the National Institutes of Health.
Contributor Information
Rungrat Saiyasombat, Department of Veterinary Microbiology and Preventive Medicine, College of Veterinary Medicine, Iowa State University, 2116 Veterinary Medicine, Ames, IA 50011, USA.
Karin S. Dorman, Department of Statistics and Genetics, Development and Cell Biology, College of Agriculture and Life Sciences, Iowa State University, Ames, IA, USA
Julian E. Garcia-Rejon, Laboratorio de Arbovirologia, The Universidad Autonoma de Yucatan, Merida, Yucatan, Mexico
Maria A. Loroño-Pino, Laboratorio de Arbovirologia, The Universidad Autonoma de Yucatan, Merida, Yucatan, Mexico
Jose A. Farfan-Ale, Laboratorio de Arbovirologia, The Universidad Autonoma de Yucatan, Merida, Yucatan, Mexico
Bradley J. Blitvich, Email: blitvich@iastate.edu, Department of Veterinary Microbiology and Preventive Medicine, College of Veterinary Medicine, Iowa State University, 2116 Veterinary Medicine, Ames, IA 50011, USA
References
- 1.Biggerstaff B. Software for mosquito surveillance (cited February 15, 2010) Centers for Disease Control and Prevention; Fort Collins, Colorado: 2006. Available from http://www.cdc.gov/ncidod/dvbid/westnile/software.htm. [Google Scholar]
- 2.Blitvich BJ, Lin M, Dorman KS, Soto V, Hovav E, Tucker BJ, Staley M, Platt KB, Bartholomay LC. Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa. J Med Entomol. 2009;46:934–941. doi: 10.1603/033.046.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballerie X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol. 2006;87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- 4.Cook S, Moureau G, Harbach R, Mukwaya L, Goodger K, Ssenfuka F, Gould E, Holmes EC, de Lamballerie X. Isolation of a new species of flavivirus and a novel strain of Culex flavivirus (Flaviviridae), from a natural mosquito population in Uganda. J Gen Virol. 2009;90:2669–2678. doi: 10.1099/vir.0.014183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabtree MB, Nga PT, Miller BR. Isolation and characterization of a new mosquito flavivirus, Quang Binh virus, from Vietnam. Arch Virol. 2009;154:857–860. doi: 10.1007/s00705-009-0373-1. [DOI] [PubMed] [Google Scholar]
- 6.Crabtree MB, Sang RC, Stollar V, Dunster LM, Miller BR. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch Virol. 2003;148:1095–1118. doi: 10.1007/s00705-003-0019-7. [DOI] [PubMed] [Google Scholar]
- 7.Crochu S, Cook S, Attoui H, Charrel RN, De Chesse R, Belhouchet M, Lemasson JJ, de Micco P, de Lamballerie X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 2004;85:1971–1980. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- 8.Darsie RF., Jr A survey and bibliography of the mosquito fauna of Mexico (Diptera: Culicidae) J Am Mosq Control Assoc. 1996;12:298–306. [PubMed] [Google Scholar]
- 9.Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Soto V, Lin M, Staley M, Dorman KS, Bartholomay LC, Hovav E, Blitvich BJ. Detection of flaviviruses and orthobunyaviruses in mosquitoes in the Yucatan Peninsula of Mexico in 2008. Vector Borne Zoonotic Dis. 2010 doi: 10.1089/vbz.2009.0196. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, Lin M, Dorman KS, Platt KB, Bartholomay LC, Soto V, Beaty BJ, Lanciotti RS, Blitvich BJ. Detection of RNA from a Novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80:85–95. [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 12.Hoshino K, Isawa H, Tsuda Y, Sawabe K, Kobayashi M. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology. 2009;391:119–129. doi: 10.1016/j.virol.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, Yuda M, Takasaki T, Kobayashi M, Sawabe K. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–414. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 14.Huhtamo E, Putkuri N, Kurkela S, Manni T, Vaheri A, Vapalahti O, Uzcategui NY. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J Virol. 2009;83:9532–9540. doi: 10.1128/JVI.00529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junglen S, Kopp A, Kurth A, Pauli G, Ellerbrok H, Leendertz FH. A new flavivirus and a new vector: characterization of a novel flavivirus isolated from uranotaenia mosquitoes from a tropical rain forest. J Virol. 2009;83:4462–4468. doi: 10.1128/JVI.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kihara Y, Satho T, Eshita Y, Sakai K, Kotaki A, Takasaki T, Rongsriyam Y, Komalamisra N, Srisawat R, Lapcharoen P, Sumroiphon S, Iwanaga S, Ushijima H, Endoh D, Miyata T, Sakata A, Kashige N, Miake F, Fukushi S, Saijo M, Kurane I, Morikawa S, Mizutani T. Rapid determination of viral RNA sequences in mosquitoes collected in the field. J Virol Methods. 2007;146:372–374. doi: 10.1016/j.jviromet.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Kim DY, Guzman H, Bueno R, Jr, Dennett JA, Auguste AJ, Carrington CV, Popov VL, Weaver SC, Beasley DW, Tesh RB. Characterization of Culex Flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology. 2009;386:154–159. doi: 10.1016/j.virol.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 18.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Morales-Betoulle ME, Monzon Pineda ML, Sosa SM, Panella N, Lopez MR, Cordon-Rosales C, Komar N, Powers A, Johnson BW. Culex flavivirus isolates from mosquitoes in Guatemala. J Med Entomol. 2008;45:1187–1190. doi: 10.1603/0022-2585(2008)45[1187:cfifmi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 21.Moureau GL, Ninove A, Izri S, Cook X, De Lamballerie, Charrel RN. Flavivirus RNA in Phlebotomine Sandflies. Vector Borne Zoonotic Dis. 2010;10:195–197. doi: 10.1089/vbz.2008.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roiz D, Vazquez A, Seco MP, Tenorio A, Rizzoli A. Detection of novel insect flavivirus sequences integrated in Aedes albopictus (Diptera: Culicidae) in Northern Italy. Virol J. 2009;6:93. doi: 10.1186/1743-422X-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Seco MP, Vazquez A, Collao X, Hernandez L, Aranda C, Ruiz S, Escosa R, Marques E, Bustillo MA, Molero F, Tenorio A. Surveillance of Arboviruses in Spanish Wetlands: detection of new Flavi- and Phleboviruses. Vector Borne Zoonotic Dis. 2009 doi: 10.1089/vbz.2008.0188. [DOI] [PubMed] [Google Scholar]
- 25.Sang RC, Gichogo A, Gachoya J, Dunster MD, Ofula V, Hunt AR, Crabtree MB, Miller BR, Dunster LM. Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch Virol. 2003;148:1085–1093. doi: 10.1007/s00705-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 26.Stollar V, Thomas VL. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology. 1975;64:367–377. doi: 10.1016/0042-6822(75)90113-0. [DOI] [PubMed] [Google Scholar]


