Abstract
Human noroviruses are difficult to study due to the lack of an efficient in vitro cell culture system or small animal model. Murine norovirus replicates in murine macrophages (MΦ) and dendritic cells (DCs), raising the possibility that human NoVs might replicate in such human cell types. To test this hypothesis, we evaluated DCs and MΦ derived from monocyte subsets and CD11c+ DCs isolated from peripheral blood mononuclear cells of individuals susceptible to Norwalk virus (NV) infection. These cells were exposed to NV and replication was evaluated by immunofluorescence and by quantitative RT-PCR. A few PBMC-derived DCs expressed NV proteins. However, NV RNA did not increase in any of the cells tested. These results demonstrate that NV does not replicate in human CD11c+ DCs, monocyte-derived DCs and MΦ, but abortive infection may occur in a few DCs. These results suggest that NV tropism is distinct from that of murine noroviruses.
Keywords: Norwalk virus, macrophages, dendritic cells, secretor-positive donors
Introduction
Norwalk virus (NV) is the prototype strain in the Norovirus (NoV) genus within the Caliciviridae family. The Norovirus genus is divided in 5 genogroups, with genogroups GI, GII, and GIV containing human strains, GIII containing bovine strains, and GV containing murine norovirus (MNV) strains. NoVs are noneveloped viruses with a positive-sense, polyadenylated, single-stranded RNA genome. The genome of ~7.7 kb encodes three open reading frames [(ORF)s 1, 2, and 3]. ORF1 encodes a nonstructural polyprotein, and ORF2 and ORF3 code for the two capsid proteins, VP1 and VP2, respectively (Glass et al., 2000; Jiang et al., 1992; Jiang et al., 1993). Human NoVs are the most common cause of outbreaks and sporadic non-bacterial gastroenteritis in the United States as well as the cause of significant disease in developing countries (Fankhauser et al., 1998; Mead et al., 1999; Patel et al., 2008). Symptoms include severe vomiting, watery diarrhea, nausea, abdominal cramps, fever and general malaise (Glass et al., 2009). Since its discovery in 1972, study of these highly contagious viruses has been difficult due to the lack of an in vitro cell culture system or a small animal model for virus replication and disease. Numerous attempts to develop a method for the cultivation of human NoVs have been unsuccessful (Duizer et al., 2004). Replication systems in cells have been described but these remain inefficient and cannot be used to study fully permissive virus replication (Asanaka et al., 2005; Katayama et al., 2006). A single report of cultivating human NoVs in a 3-dimensional, organoid model of human small intestinal epithelium has not been confirmed (Herbst-Kralovetz, 2009; Papafragkou, 2009; Straub et al., 2007). Gnotobiotic pigs and calves can replicate genogroup II (GII) human NoVs and viremia is detected (Cheetham et al., 2006; Souza et al., 2008), but currently no culture system supports efficient replication in vitro.
The MNV was originally isolated from immunodeficient mice (Karst et al., 2003) and can be propagated in cell culture (Wobus et al., 2004). MNV infects macrophages (MΦ) and dendritic cells (DC) in vivo in STAT-1−/− mice, cultured bone-marrow-derived MΦ and bone-marrow-derived DCs, as well as the several murine macrophage cell lines and a human-murine hybrid macrophage cell line (Changotra et al., 2009; Wobus et al., 2004). These results, together with data showing that NoVs can be detected in the serum of children with gastroenteritis (Takanashi et al., 2009), raise the possibility that human NoVs may replicate in cells of the hematopoietic lineage in humans. Studies with volunteers challenged with NV have shown that genetically determined factors are responsible for resistance to NV infection (Hutson et al., 2002; Hutson et al., 2005; Lindesmith et al., 2003; Marionneau et al., 2002; Parrino et al., 1977). In particular, secretor status (Se) of an individual, determined by the presence of functional α(1,2)fucosyltransferase (FUT2) gene products, regulates susceptibility to NV infection (Hutson et al., 2005; Lindesmith et al., 2003). The FUT2 enzyme produces H antigens on the surface of epithelial cells and in mucosal secretions (Kelly et al., 1995), and these glycans are thought to function as an initial receptor for NV (Guix et al., 2007). Thus, cells from histoblood group type O and Se positive (Se+) individuals bind to NV and are thought to be more susceptible to infection than those from Se negative (Se−) individuals (Marionneau et al., 2002). In this study, we tested the hypothesis that NV has a tropism for human monocyte-derived DCs and MΦ and circulating CD11c+ DCs obtained from PBMCs of blood group O, Se+ donors.
Results
Preparation of NV stocks for infectivity trials
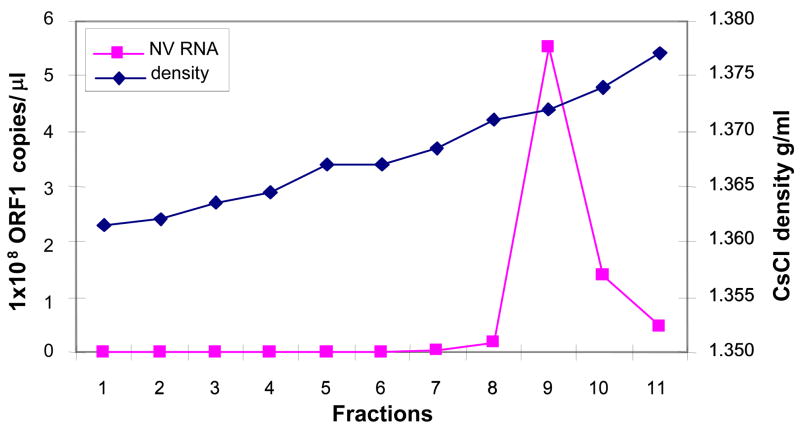
NV inocula to perform infectivity experiments in DCs and MΦ were prepared by purifying NV from stools containing high viral titers from a NV-infected individual as previously described (Guix et al., 2007). Briefly, NV was purified by CsCl gradients and fractions corresponding to CsCl densities of 1.37 to 1.39 g/ml were pooled and used as a viral stock (Fig. 1). The presence of NV particles in this pool was confirmed by real-time quantitative (q) reverse transcriptase (RT)-PCR and electron microscopy (data not shown). NV stocks purified in this gradient for infectivity studies had a viral titer ranging from 8 x 108 to 1.6 x 109 NV genome copies/μl, as estimated by immunomagnetic capture (IMC)/qRT-PCR assay.
Figure 1. NV quantification in fractions from NV purified from stools by isopycnic CsCl gradient ultracentrifugation.
Genomic NV RNA in each fraction of the CsCl density gradient was quantified by IMC/qRT-PCR, using primers specific for the ORF1 region of the NV genome (squares). The buoyant density of each fraction was also determined by measuring the refractive index (diamonds). Each square represents replicas of 3.
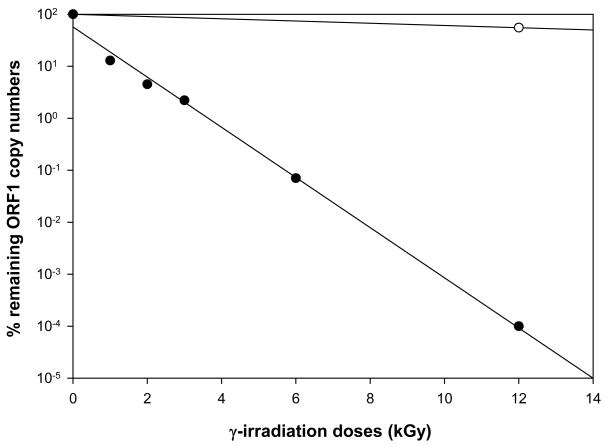
As a negative control, an aliquot of this NV stock was treated with γ-irradiation. This treatment inactivates virus infectivity while retaining residual, potentially toxic substances from the initial stool sample that could co-purify with virus particles. First, the γ-irradiation dose that significantly decreased NV RNA detection by IMC/qRT-PCR was determined (Fig. 2). Unexposed (mock-irradiated) NV, kept at room temperature (RT) for the same length of time as the sample exposed to the highest γ-irradiation dose, was used as a control. Progressively higher doses of γ-irradiation led to a log-linear decline in NV copy number (Fig. 2). Interpolation from this linear regression curve (r2=0.994) determined that 4 and 4.5 kGy were sufficient to decrease detection of the NV genome copies concentration by 99.4 and 99.6%, respectively, and were chosen to inactivate purified NV for use as a negative control for viral replication. Inactivation of NV viral infectivity with these doses of γ-irradiation was confirmed by complete abrogation of NV protein expression in NV RNA transfected Huh-7 cells, performed in similar manner as previously described (Guix et al., 2007) (data not shown).
Figure 2. Effect of γ-irradiation on NV genomic RNA qRT-PCR detection.
NV purified from stools was exposed to different γ-irradiation doses (black dots) expressed in kilo Gray units (kGy); or left unexposed at room temperature for equivalent periods of time (white dots). Quantitation of NV RNA was performed by IMC/qRT-PCR, using primers specific for the ORF1 region of the NV genome. The percentage of remaining ORF1 copy numbers is shown on a log10 scale. Lineal regression curves were calculated as r2=0.994 for γ-irradiated NV samples. Each dot represents 12 replicas.
Identification of the secretor status of type O group donors by FUT2 genotyping assay
All donors whose buffy coat cells were used in the study were blood type O and their secretor status was determined by FUT2 genotyping assay, which identifies the single nucleotide polymorphisms G428A (found in more than 95% of non-secretors of European and African descent) and A385T (found in non-secretors of Asian ancestry) that most commonly lead to dysfunction of the FUT2 gene product, as previously described (Hutson et al., 2005). Among 15 donors tested, twelve were Se+, and three were Se− as determined by the homozygosity of nucleotide A at position 428 (Table 1). Different cell types were obtained from the donors for use in infectivity trials, as summarized in Table 1. Cells from Se+ donors were used in most trials, and cells from Se− donors were included in some studies as an additional negative control.
Table 1.
Secretor status of donors and the different cell types used in this study, determined by the FUT2 genotyping assays
| Donor # | Cell type | Secretor status (Se) |
FUT2 gene SNPs |
|
|---|---|---|---|---|
| G428Aa | A385Ta | |||
| 1 | PBMC-DCs | + | GA | AA |
| 4 | PBMC-DCs | + | GA | AA |
| 30 | CD11c+DCs | + | GG | AA |
| 56 | CD14+/CD16+ DCs | − | AA | AA |
| 57 | CD14+/CD16+ DCs | − | AA | AA |
| 58 | CD14+/CD16+ DCs | + | GA | AA |
| 72 | CD14+/CD16+ MΦ | + | GG | AA |
| 103 | CD11c+DCs | + | GA | AA |
| 369 | PBMC-DCs | + | GA | AA |
| 453 | PBMC-DCs | + | GG | AA |
| 698 | PBMC-DCs | + | GA | AA |
| 748 | CD14+/CD16+ MΦ | + | GG | AA |
| 778 | PBMC-DCs | + | GA | AA |
| 829 | PBMC-DCs | + | GA | AA |
| 907 | CD11c+DCs | − | AA | AA |
Most common single nucleotide polymorphisms (SNP) of the FUT2 gene leading to lack of function. Homozygous AA at 428 or TT at 385 defines Se− genotype.
Few CX3CR1+ cells express viral antigens in NV-inoculated PBMC monocyte-derived DCs from type O/Se+ individuals
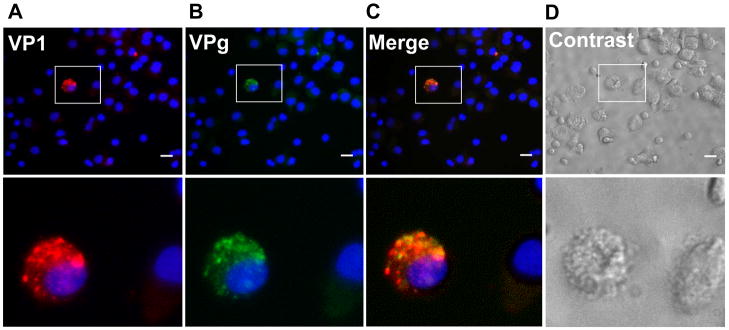
Characterization of PBMC monocyte-derived immature DCs (PBMC-DCs) obtained by the adherence method was performed by flow cytometry analysis after GM-CSF and IL-4 treatment for 6 days, as described in the materials and methods below. These cells displayed a typical profile of expression of surface markers for immature DCs: high expression of CD11c, CD1a, CD209, MHCI, MHCII, and low expression of CD14. Most of these cells did not show upregulated expression of mature DC markers e.g. CD80, CD83, CD86 and CD40 (data not shown). We also determined the percentage of cells expressing both CD11b+ and CD11c+ and CD11c+ cells expressing CX3CR1+ or CD103+ in this PBMC-DC population (Supplementary Fig. 1). The CX3CR1+ or CD103+ cell surface markers define two distinct DC populations in the intestinal lamina propria (Bogunovic et al., 2009); (Jaensson et al., 2008); (Varol et al., 2009). Most of the gated cells (86.8%), initially isolated using the adherence method, were positive for both CD11b+ and CD11c+ surface markers, typical findings for DCs. In addition, most (86.9%) of the CD11c+ cells expressed CX3CR1+ and only a small percentage (3.97%) of CD11c+ cells expressed CD103+ (Supplementary Fig. 1). To determine whether PBMC-DCs support NV replication, cells from several donors [1, 4, 369 and 698 (type O/Se+ individuals)], were inoculated with purified NV stocks at multiplicities of infection ranging from 275 to 1,941 NV genome copies/cell. Mock-inoculated (media only) and γ-irradiated NV (4 kGy or 4.5 kGy) were used as negative controls. Expression of viral protein and increases in viral RNA were analyzed by immunofluorescence (IF) and qRT-PCR, respectively. As determined by IF at 24 hours post-inoculation (hpi), the capsid protein VP1 and the nonstructural proteins VPg were co-expressed in 2–3 cells per 4 x104 cells from donor 1 (Fig. 3, A–C). Similar results were obtained with cells from donors 4 and 369 (data not shown). After 72 hpi, no increase in the number of cells expressing viral antigen was detected. Moreover, VP1 and the nonstructural protein polymerase were co-expressed in similar numbers of cells from donors 369 and 698 (data not shown). No fluorescence was observed in NV-exposed cells stained using a corresponding pre-immune serum as the primary antibody, or in mock exposed cells (data not shown). Finally, we infected these DCs with H1N1 (A/TX/36/91) and H3N2 (A/Brisbane/10/2007) influenza virus strains and observed significant cytopathic effect and positive IF staining for the nucleoprotein in the majority of cells (data not shown). These results indicate that the PBMC-DCs used in these experiments could support virus replication.
Figure 3. Detection of NV proteins by immunofluorescence in a few PBMC monocyte-derived DCs inoculated with NV.
NV inoculated PBMC-DCs from a type O/Se+ individual (donor 1) were dual labeled with antibodies specific for VP1 (NV capsid protein) and for VPg (NV nonstructural protein). A few cells (2–3 out of 4 x 104 cells) express VP1 and VPg proteins (A, B and C). Antibodies were visualized with species-specific IgG conjugated to Alexa Fluor 594 and 488, respectively. DAPI staining specific for nucleus was also performed (A, B and C). Phase contrast image of the stained cells (D). White scale bar represents 20 μm (upper panel); an enlargement of the boxed area (lower panel).
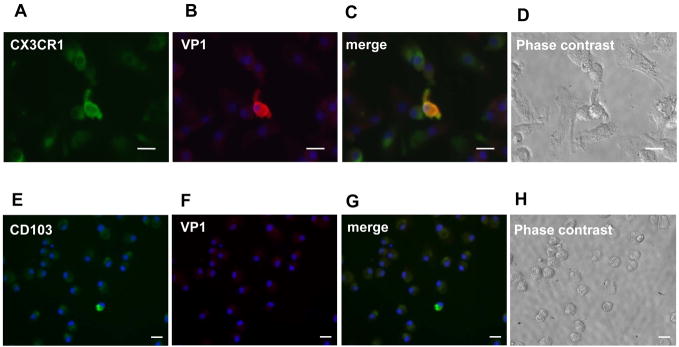
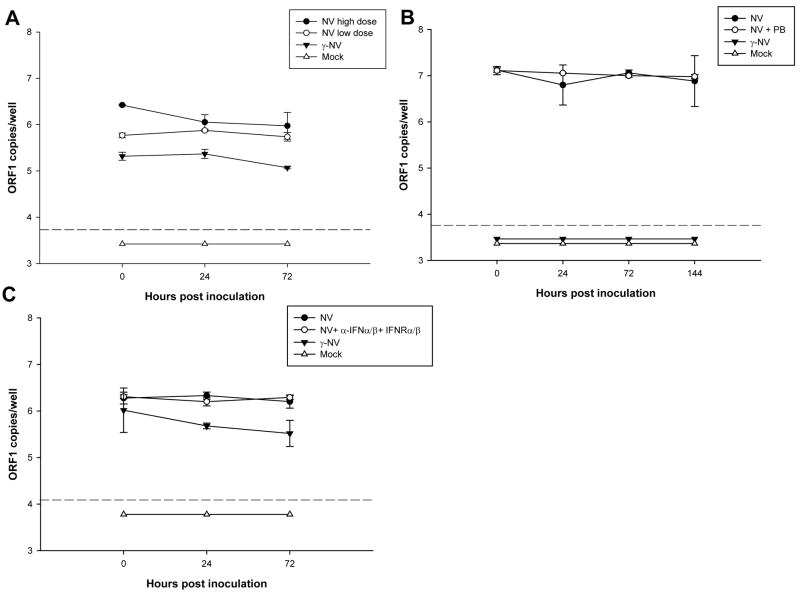
To identify what kind of DCs were expressing NV proteins, we also dual labeled cells with antibodies specific for CX3CR1 and for CD103 surface cell markers. We found that VP1 expression in a few (0.5%) CX3CR1+ cells (Fig. 4A–D), but not in CD103+ cells (Fig. 4E–H). Despite this finding, the titer of NV genome copies in total pooled RNA extracted from both the culture supernatant and cells of the inoculated PBMC-DCs did not increase from time 0 to 24–72 hpi (Fig. 5A). Similar results were obtained in three separate inoculation experiments using cells from donor 4 (data not shown). Additionally, no increase in NV RNA was observed up to 144 hpi. (Fig. 5B). These results indicate that efficient, productive replication did not occur in human PBMC-DCs inoculated with NV, and even though a few isolated CX3CR1+ DCs expressed viral antigens,
Figure 4. NV protein expressing cells co-localize with some CX3CR1+ cells, but not with CD103+ cells by immunofluorescence.
NV-inoculated PBMC-DCs from a type O/Se+ individual (donor 698) were stained with antibodies specific for CX3CR1 cell surface marker (A) and for capsid proteinVP1 (B). Antibodies were visualized with species-specific IgG conjugated to Alexa Fluor 488 and 594, respectively. Similar staining was performed with antibodies specific for CD103 surface marker (E) and for VP1 (F). Merge images from the respective co-staining are shown in C and G. DAPI staining specific for nucleus was also performed (A, B, C, E, F and G). Phase contrast image of the stained cells (D and H). White scale bar represents 20 μm.
Figure 5. Detection of genomic NV RNA in PBMC monocyte-derived DCs inoculated with NV by qRT-PCR.
Total RNA combined from the cells and the supernatant of PBMC-DCs from a type O/Se+ individual (donor 1) exposed to mock (white triangles), or γ-irradiated NV (black triangles) or 275 NV genome copies/cell (white circles) or 1,800 NV genome copies/cell (black circles) of NV purified from stool, was analyzed by qRT-PCR, using primers specific to ORF1, at 0, 24 and 72 hpi (A). Total RNA from PBMC-DCs inoculated with NV compared to NV inoculated PBMC-DCs (donor 778) treated with 10 μg/ml polymixin B (B); or NV inoculated PBMC-DCs (donor 453) treated with a cocktail of antibodies against IFN-α, IFN-β and IFN-α/β receptor (C) were analyzed by qRT-PCR at different times. Total ORF1 copy numbers per well are represented on a log10 scale. Each well contained from 4–7 x105 cells. The data plotted represent the means ± standard deviation of triplicate wells. Dotted lines represent the detection limit of the assay. All points below the limit of detection had the same value but were offset for visualization.
The type I IFN pathway does not affect NV replication in PBMC-DCs from type O Se+ individuals
Since the NV inoculum was purified from stools from a NV-infected individual, we evaluated the viral inoculum for the presence of endotoxin (LPS). We determined that NV and γ-NV contained 200 ng/ml and 20 ng/ml of LPS, respectively. We next examined whether the NV inoculum might induce upregulation of the type I IFN pathway, a process that could lead to inhibition of NV replication in PBMC-DCs. The NV inoculum (1,941 genomic copies/cell), which contained only 1.6 ng/ml of LPS, did not significantly induce IFN-β mRNA production in PBMC-DCs (donor 829) at 10 hpi as measured by relative quantification (RQ) qRT-PCR compared to mock exposed cells (data not shown), while LPS (1 μg/ml) and poly-IC (50 μg/mL) did significantly increase IFN-β mRNA production 16- and 32- fold, respectively (data not shown). To further assess whether upregulation of the type I IFN pathway by the NV inoculum may have had an effect on NV replication in PBMC-DCs, we inoculated PBMC-DCs (donor 778) with NV treated with 10 μg/ml polymixin B, an antagonist of LPS effect (Inden et al., 2009). No increase in NV RNA was observed in these cells at 72 hpi (Fig. 5B). In addition, NV inoculated PBMC-DCs (donor 453) treated with a cocktail of antibodies against IFN-α, IFN-β and IFN-α/β receptor (anti-IFNs Abs), as previously reported (Hanabuchi et al., 2006), did not result in an increase in levels of NV RNA at 72 hpi (Fig. 5C). Similarly, no increase in the number of VP1 positive cells was observed by IF in NV-inoculated DCs treated with polymixin B compared to untreated, NV-inoculated cells (data not shown). Taken together, these results indicate that the replication of NV in these cells was not inhibited by the induction of the type I IFN pathway by the NV inoculum.
NV does not replicate in human CD14+ and CD16+ monocyte-derived DCs, and CD11c+ blood DCs from type O/Se+ individuals
Since positive staining for NV proteins was observed in a few CX3CR1 cells from PBMC-DCs, we determined whether NV might have a tropism for either one or both types of DCs derived by in vitro differentiation from the two principal subsets (CD16high and CD14low, and CD14high and CD16low) of human monocytes (Passlick et al., 1989), that are reported to both express CX3CR1 (Seidler et al., 2010). In addition, we also evaluated whether NV replicates in a directly sorted, purified population of CD11c+ DCs, which are primary committed-DCs circulating in the peripheral blood (Ito et al., 2005). To test these hypotheses, first human CD14+ and CD16+ monocyte subsets were isolated from PBMCs of type O/Se+ or Se− individuals and differentiated into immature DCs. Characterization of the two isolated monocyte populations and monocye-derived DCs was performed by flow cytometry analysis at day 0 and 8, after GM-CSF and IL-4 treatment (Supplementary Fig. 2). At day 0, the purity of CD16+ and CD14+ monocytes was 77.9% and 78.4%, respectively (Supplementary Fig. 2, A and B, respectively). CD3 and CD56 expression was absent in both populations, indicating that no contamination with T cells or NK cells in either of the two isolated monocyte populations (data not shown). CD11c (highly expressed in cells of myeloid origin) expression corresponded to 98% and 95% in CD16+ and CD14+ monocyte subsets, respectively (Supplementary Fig. 2, A and B, respectively). Analysis at day 8 showed that both populations of CD16+ and CD14+ monocytes differentiated to immature DCs (CD16+ DCs and CD14+ DCs, respectively), as indicated by the upregulation of a major DC marker CD1a, moderate and low expression of the co-stimulatory molecules CD80 and CD40, and low expression of CD16+ and CD14+ monocyte markers, respectively. High expression of MHC-I and MHC-II markers present in both monocyte subsets at day 0 and monocyte-derived DCs at day 8 are consistent with the antigen presenting properties of these cells (Supplementary Fig. 2, A and B). CD11c+ DCs were isolated from PBMCs and characterized as previously reported (Reche et al., 2001).
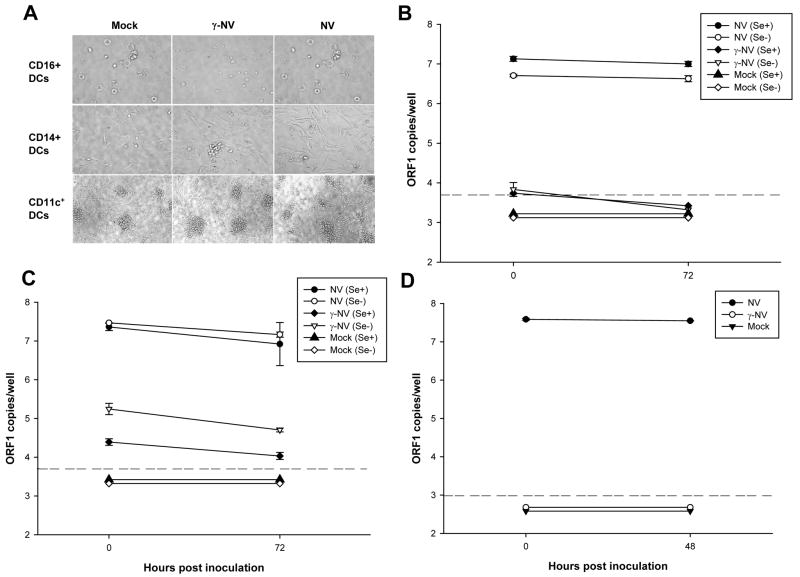
The CD16+ DC, CD14+ DC subsets and CD11c+ DCs were exposed to NV, to γ-irradiated NV, or to media only (mock). Differences in cell morphology were observed among the different DC subsets by light microscopy, but no morphological changes were observed between cells exposed to NV compared to the negative controls, indicating that no cytopathic effect occurred (Fig. 6A). Positive IF staining for VP1 and VPg NV proteins was observed only in a few (2 cells per 4x104 cells) CD14+ DCs cells from a type O/Se+ (donor 1). No fluorescence was observed in γ-NV exposed or in NV-exposed cells stained using a corresponding pre-immune serum as the primary antibody (data not shown). In addition, levels of viral RNA measured by qRT-PCR did not increase from 0 to 72 hpi in any of the trials performed with the NV-inoculated CD14+ DCs (Fig. 6B) and the NV-inoculated CD16+ DCs (Fig. 6C) from type O/Se+ (donor 58) and Se− (donor 56) individuals. Finally, no increase was observed either in levels of total viral RNA in CD11c+ DCs from a type O/Se+ individual (donor 103) or from a Se− individual (donor 907, data not shown) exposed to NV for 48 h (Fig. 6D). These results demonstrate that NV does not replicate in CD16+ DCs, CD14+ DCs and CD11c+ DCs from peripheral blood, indicating that NV does not productively replicate in CD11c+ DCs isolated directly from PBMCs and DCs derived from the two main subsets of circulating monocytes from type O/Se+ individuals.
Figure 6. NV does not replicate in CD16+, CD14+ monocyte-derived DCs or CD11c+ blood DCs.
Analysis of cell morphology of CD16+ DCs, CD14+ DCs, and CD11c+ DCs exposed to mock, γ-NV and NV after 48 hpi by phase contrast microscopy (A). The titer of NV RNA in each sample: mock, γ-NV and NV exposed CD16+ DCs (B) and CD14+ DCs (C), from type O/Se+ (donor 58) and type O/Se− (donor 56) individuals; and CD11c+ DCs (D) from a type O/Se+ (donor 103) individual, was measured by qRT-PCR at 0, 48 or 72 hpi. Primers targeting the ORF1 region of the NV genomic RNA were used. Total ORF1 copy numbers per well are represented on a log10 scale. Each well contained from 2–4 x105 cells. The data plotted represent the means ± standard deviation of 6 (B and C) or 4 (D) replicate wells. Dotted lines represent the detection limit of the assay. All points below the limit of detection had the same value but were offset for visualization.
NV does not replicate in human CD14+ and CD16+ monocyte derived-MΦ from type O/Se+ individuals
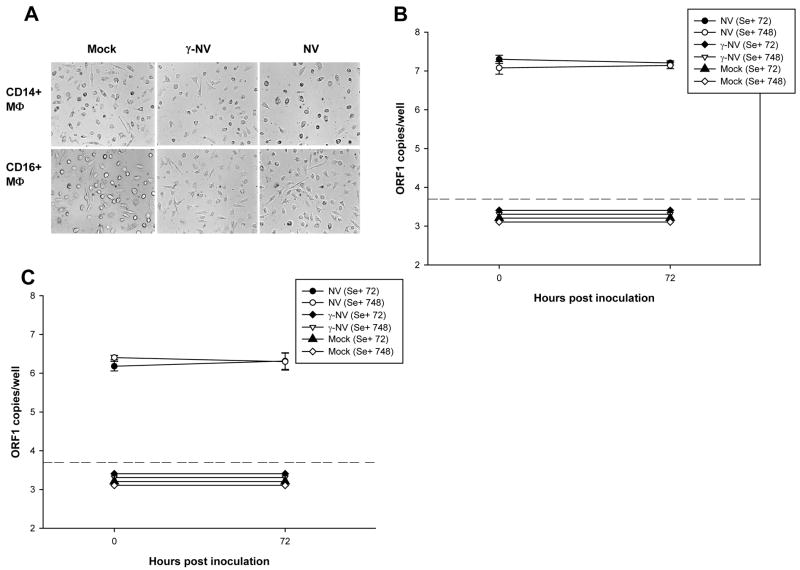
We also tested whether NV has a tropism for either one or both of the two MΦ subsets derived from human CD14+ and CD16+ monocytes. After 7 days of treatment with M-CSF, CD14+ and CD16+ monocyte-derived MΦ subsets (CD14+ MΦ and CD16+ MΦ, respectively) were exposed to NV, to γ-irradiated NV, or to media only (mock). No morphological changes were observed in these different MΦ subsets exposed to NV compared to the negative controls (Fig. 8A). As expected, levels of viral RNA measured by qRT-PCR in γ-NV-inoculated CD14+ MΦ (Fig. 7B) and γ-NV-inoculated CD16+ MΦ (Fig.7C) did not increase over time. In addition, viral RNA did not increase in either NV-inoculated CD14+ MΦ (Fig. 7B) or NV-inoculated CD16+ MΦ (Fig. 7C) from type O/Se+ (donors 72 and 748) individuals after 72 hpi. These results indicate that NV does not replicate in these two subsets of monocyte-derived MΦ from type O/Se+ individuals.
Figure 7. NV does not replicate in CD14+ or CD16+ monocyte-derived MΦ.
Analysis of cell morphology observed by phase contrast microscopy in CD14+ MΦ and CD16+ MΦ exposed to mock, γ-NV and NV after 48 hpi (A). The titer of NV RNA in each sample: mock, γ-NV and NV exposed CD14+ MΦ (B), and CD16+ MΦ (C) from type O/Se+ individuals (donors 72 and 748), was measured by qRT. PCR at 0 and 72 hpi. Primers targeting the ORF1 region of the NV genomic RNA were used. Total ORF1 copy numbers per well are represented on a log10 scale. Each well contained 2 x105 cells. The data plotted represent the means ± standard deviation of 4 (B) or 3 (C) replicate wells. Dotted lines represent the detection limit of the assay. All points below the limit of detection had the same value but were offset for visualization.
Discussion
The major hurdle to working with human NoVs is the inability to efficiently cultivate these viruses in cultured cells. This hurdle prevents complete studies of the virus replication cycle or studies to assess the public health hazard of detecting viral nucleic acid in food and environmental samples. A single report of replication in a 3-dimensional organoid model of human small intestinal epithelium has not been confirmed (Herbst-Kralovetz, 2009; Papafragkou, 2009; Straub et al., 2007). Our laboratory has reported previous unsuccessful attempts to cultivate human NoVs in a variety of cell lines (Duizer et al., 2004). The present paper reports the results of inoculation of cells of hematopoietic origin, studies that were performed because a murine norovirus that causes systemic infection of mice and is proposed as a model to study human NoVs (Wobus et al., 2006) is cultivatable in murine MΦ and DCs (Wobus et al., 2004). In addition, human NoVs have been detected in serum samples from children with gastroenteritis and in gnotobiotic calves infected with human NoVs, (Souza et al., 2008; Takanashi et al., 2009). These are interesting reports that are reminiscent of the recent recognition that rotaviruses cause a viremia in children and animals (Blutt and Conner, 2007) and that PBMCs from adults are susceptible to infection with purified rotavirus (Mesa et al., 2007). Furthermore, using a biopsy of a NV-infected individual after 24 hpi, viral capsid expression has been detected in mononuclear cells in the intestinal lamina propria (personal communication, T. Tanaka, D.Y. Graham, M.K. Estes), suggesting that cells in the lamina propria, including DCs and MΦ, might be infected with NV. The precursors of these cells are likely to be monocytes or other precursors of intestinal DCs circulating in the blood (Bogunovic et al., 2009; Geissmann et al., 2003; Varol et al., 2007; Varol et al., 2009).
We did not detect productive replication of NV in CD11c+ DCs isolated directly from PBMCs, or in different subsets of PBMC monocyte-derived DCs or MΦ for up to 144 hpi. This time frame was chosen for our studies because recent results, using RNA extracted from NV purified from stool samples of infected volunteers, show that within this period of time the viral RNA transfected into cultured cells replicates to high titers that are detectable even though the infection does not spread beyond the initially RNA-transfected cells (Guix et al., 2007). In these previous transfection studies, we estimated that ~ 20,000 genomic copies of RNA were produced per transfected cell. It is possible that we were not able to detect a low level of NV replication from the 2 or 3 antigen positive cells (estimated total virus yield <105 genomic copies) per 103–104 inoculated cells due to the high levels of NV inoculum used. However, efficient replication as seen in the murine norovirus system was not observed in our studies with NV. It remains to be seen if other NoV strains might replicate in such cells.
Several approaches were used in our current studies to allow evaluation of the specificity of any potential replication. First, we used cells from donors characterized to be blood type O, Se+ who are known to be genetically susceptible to NV infection (Hutson et al., 2005; Lindesmith et al., 2003), and cells from genetically-resistant Se− donors were used as negative controls. In addition, aliquots of the live virus inoculum were inactivated with γ-irradiation to allow assessment of the effects of any residual stool components that might be present in the fractions of virus purified from stool. The results in all the cells tested indicate that NV does not replicate in CD11c+ DCs, DCs and MΦ derived from the two main monocyte subsets from PBMCs, and no differences were observed between Se+ and Se− individuals. One might question if the purified virus used for these experiments is truly infectious. Until positive results are obtained in cultured cells, this question cannot be fully answered. However, the RNA extracted from similar preparations of virus is infectious in Huh-7 cells (Guix et al., 2007), and purification and characterization of recombinant virus-like particles (VLPs), using the same purification methods, results in particles that are antigenically and morphologically identical to authentic virus (Green et al., 1993; Jiang et al., 1992). Moreover, VLPs also bind and enter cells that express the needed histoblood group antigens (Guix et al., 2007; Marionneau et al., 2002; White et al., 1996). The DCs from the Se+, but not the Se− individuals, used in the current studies expressed histoblood group antigens as shown by positive binding of NV VLPs (data not shown). Murine norovirus purified in a similar manner also retains its infectivity (Changotra et al., 2009).
While NV RNA does not efficiently replicate in these cells of myeloid origin inoculated with virus, rare PBMC-derived DCs were found to be positive for both the NV VP1 and VPg proteins by IF at 24 and 72 hpi (data not shown). These results suggest that, in a subset of cells, abortive expression of the viral proteins or limited virus replication may occur. We also tried to identify these cells using two cell surface markers, which define two distinct (CX3CR1 and CD103) DC populations in the intestinal lamina propria (Bogunovic et al., 2009); (Jaensson et al., 2008); (Varol et al., 2009); VP1 expression was detected in a few CX3CR1+ cells, but not in CD103+ cells. One possibility is that these few permissive cells might represent DCs derived from CD14+ or CD16+ monocytes, which both are known to express CX3CR1 (Seidler et al., 2010), present in the peripheral blood and are suggested to seed the lamina propria for renewal of DCs or MΦ (Geissmann et al., 2003; Niess et al., 2005; Varol et al., 2007). However, our results failed to detect efficient replication in either type of these subsets of monocytes present in the peripheral blood of individuals susceptible to NV infection.
Our negative culture results cannot provide an explanation for the NV capsid-specific staining observed in cells of the intestinal lamina propria in the above-mentioned biopsy. It remains possible that NV might replicate in intestinal DC or MΦ that, under the local microenvironment that influences the phenotype of these cells (Niess, 2008; Smith et al., 2005), may have different properties from those of PBMC-derived cells. Another possibility is that these cells may come from precursors, which are different from the two main subsets of monocytes present in the peripheral blood. A third possibility is that replication was not occurring in these cells but, instead, these cells took up viral capsid antigen by phagocytosis. Further studies will be required to test human DCs or MΦ already differentiated in the lamina propria of the intestine to see if they might support the replication of NV or other human NoVs.
Our results suggest that the human NV possesses distinct biological properties from the only norovirus, MNV, that replicates efficiently in cultured primary, bone marrow-derived murine DCs and MΦ as well as several murine macrophage cell lines and a human-murine hybrid macrophage cell line in vitro (Changotra et al., 2009; Wobus et al., 2004). In fact, to date there are no reports of MNVs failing to replicate in any macrophage cell line of murine origin. Thus, if the lack of efficient replication of NV in human blood-derived MΦ or DCs and CD11c+ DCs was due to the source of these cells (i.e., not derived from the bone marrow and differentiated in vitro), this would contrast with the apparently more diverse tropism of MNVs for murine macrophage cell lines. Another difference between human NV and murine strains is that MNV-1 causes extraintestinal infection, including encephalitis, in immunocompromised mice (Karst et al., 2003) and extraintestinal infection in immunocompetent mice, but it does not cause frank gastrointestinal disease or obvious detrimental effects in immunocompetent mice (Henderson, 2008; Mumphrey et al., 2007; Wobus et al., 2006). In addition, human NoVs bind to histoblood group antigens likely as initial receptors (Marionneau et al., 2002), and these antigens are host susceptibility factors for human infection. In contrast, MNV is reported to bind to a ganglioside-linked sialic acids on RAW 264.7 cells (Taube et al., 2009) and no genetic host susceptibility factors have been identified in immunocompetent mice, although immunocompromised interferon (IFN)-αβγR and STAT-1 knock-out mice are highly susceptible to infection and disease (Karst et al., 2003; Mumphrey et al., 2007). Thus, the cell tropisms for the MNVs and human NoVs in their respective hosts appear to be quite distinct.
Resistance to NV replication of the DCs and MΦ tested in this study might have been due to strong IFN responses produced in these cells. IFN-α and IFN-γ have been recently reported to play a pivotal role in controlling replication of a NV replicon in human hepatoma cells (Huh-7 cells) or hamster kidney cells (BHK21 cells) (Chang et al., 2006; Chang and George, 2007), suggesting that IFN production is an efficient antiviral host mechanism against NV infection. Moreover, NV replication in Huh-7 cells seen after transfecting genomic NV RNA (Guix et al., 2007) is inhibited if cells are pre-treated with the supernatant from cells that have been transfected with Poly(I:C) to induce IFN-α/β production (Guix and Estes, 2009). Finally, permissive NV genome replication after transfecting NV RNA in Huh-7 cells does not induce a strong type I IFN response (Guix and Estes, 2009). However, Huh-7 cells lack some key components of the type I IFN pathway (Li et al., 2005). Thus, it was worthwhile to test whether PBMC-DCs might produce type I IFN responses following exposure to the NV inoculum, since we found few of these cells expressing NV proteins. The NV inoculum contained small amounts of residual endotoxin due to its derivation from stool, so we analyzed whether LPS present in this inoculum might preclude NV replication in cells by inducing the type I IFN pathway using three different approaches. No significant induction of IFN-β mRNA was identified 10 hpi and the use of either polymixin B (to antagonize possible IFN-β induction by LPS) or antibodies targeting IFN-α and IFN-β and their receptors did not lead to increases in NV RNA production following inoculation of cells. Thus, other cellular barriers are preventing NV replication in these cells.
In conclusion, attempts to determine if NV grows in human CD11c+ DCs, monocyte-derived DCs and MΦ, cells of the hematopoietic lineage, were unsuccessful, and cultivatable NV was not recovered. These results raise the question of whether NoV nucleic acid detected in the serum of children with gastroenteritis (Takanashi et al., 2009) represents free virus in serum or virus released from another subset of infected cells. Future studies could examine this question in volunteers infected with NV. Further studies are needed to elucidate NV tropism and to fully understand NoV pathogenesis in humans. Such studies may facilitate the development of a fully permissive cell culture system for human NoV, which would then lead to the design of new therapeutic treatments for these fastidious viruses.
Materials and Methods
NV purification from stools from a NV infected volunteer
NV was purified from stool as previously described (Guix et al., 2007). In brief, stools obtained 3 days post-NV inoculation from volunteer #732 (Atmar et al., 2008) were prepared as ten percent suspensions in 0.1 M phosphate-buffered saline (PBS) −0.5% Zwittergent detergent (Calbiochem, Gibbstown, NJ), extracted with Vertrel XF (Miller-Stephenson, Sylmar, CA) and clarified by centrifugation for 10 min at 12,400 x g. Virus in the supernatant was precipitated with 8% polyethylene glycol and 40 mM NaCl solution, and after centrifugation, the pellet was suspended in 0.1 M PBS. The virus suspension was centrifuged through a 30% sucrose cushion for 3 h at 124,000 × g and further purified by isopycnic cesium chloride (CsCl) gradient centrifugation for 24 h at 150,000 × g. Fractions were collected, and densities were determined by measuring the refractive index (RI) of each fraction. CsCl was removed from each fraction and suspended into 120 μl diethylpyrocarbonate-treated (DEPC) water. The titer of NV genome copies in each fraction was determined by IMC/qRT-PCR assay as described below. Endotoxin levels in CsCl purified NV and γ-NV (see below), respectively, were determined by using the Pyrotell Gel-Clot method (Associates of Cape Cod Incorporated), following the manufacturer’s protocol.
NV inactivation by gamma (γ)-irradiation
NV was inactivated by γ-irradiation for use as a negative control to monitor NV replication in cells. Initially, 5 μl of the NV purified from stool was added into a 2 ml cryovial ‘o’ ring tube and was exposed to γ-irradiation doses of 1, 2, 3, 6, or 12 kilo Grays (kGy) at RT using a Gammacell-1000 Irradiator (Atomic Energy of Canada Ltd., Ontario) with a Cesium-137 source, at a dose rate of 0.5 kGy/h. Unexposed NV was left at RT for the same length of time as the exposed samples to determine the effect of γ-inactivation on NV genomic RNA.
Immunomagnetic capture/real-time quantitative RT-PCR (IMC/qRT-PCR) assay
NV RNA titers were measured in samples either from gradient fractions after CsCl ultracentrifigation or from purified NV exposed to different doses of γ-irradiation by IMC/qRT-PCR assay as previously described with some modifications (Gilpatrick et al., 2000). In brief, 10 μl of a 1:1,000 dilution of each gradient sample, or 10 μl of undiluted, 1:10, 1:100, or 1:000 dilutions from γ-irradiated samples, were mixed with 1.2 mL of PBS containing 0.1% bovine serum albumin (BSA, Sigma, Atlanta, GA) and 50 μl of polyclonal rabbit anti-rNV IgG coupled to magnetic beads (1 μg/μl BioMag goat anti-rabbit IgG, Qiagen, Carol Stream, IL), in RNAse-free 2 ml tubes and incubated on a rotating platform for 2 h at RT. Each sample was washed 3X with 0.1% BSA/PBS. After the last wash, 35 μl of DEPC-treated water (Invitrogen, Chicago, IL) was added to each sample and the suspended sample was transferred into a RNAse-free 0.5 ml tube. RNA was released from the NV particles by incubating each tube for 5 min at 95°C. RNA was analyzed by qRT-PCR with primers targeting the ORF1 region of NV as previously described (Guix et al., 2007).
Fucosyltransferase 2 (FUT2) genotyping assay
DNA was extracted from 200 μl of buffy coat of each donor using the QIAamp DNA Blood Mini Kit (QIAGEN). A region of the FUT2 gene was amplified using the following primers: forward 5′-CCCATCTTCAGAATCACCCTGCCGGTGCTG-3′, and reverse 5′-TCGGCCGGCCCGTGGAAACATCCCCAGGTA-3′, which anneal at nucleotide (nt) positions 280–309 and 535–564, respectively, as previously described (Hutson et al., 2005). Reactions were carried out in a final volume of 50 μl using 2.5 U AmpliTaq (Invitrogen) with a thermal program that consisted of an initial denaturation step for 2 min at 95°C, followed by 35 cycles for 30 sec at 95°C, 30 sec at 65°C, 1 min at 72°C and a final elongation for 10 min at 72°C. Detection of the single nucleotide polymorphism of G428A (SNP) was performed on 2 μl of the PCR product, the Custom TaqMan SNP Genotyping Assay for the 428 SNP of the FUT2 gene designed by Dr. Le Pendu, France (Applied Biosystems, Chicago, IL) and the TaqMan Genotyping Master Mix (Applied Biosystems), in a final reaction volume of 20 μl. The thermal program consisted of 40 cycles of amplification 95°C 10 min, 92°C 15 sec, and 60°C 1 min, using the Applied Biosystems 7500 real-time PCR system, and applying the Allelic Discrimination program (Applied Biosystems). Samples were tested in triplicate wells. To assay for the A385T polymorphism, a fragment of the FUT2 gene was amplified using the forward primer 5′–AACGACTGGATGGAGGAGGAATA-3′ and the reverse primer 5′ –CTCATGCAGGCGAAGTGGC-3′ that span nt 355–412.
Reactions were carried out as described above, and detection of the SNP was performed as above using the Custom TaqMan SNP Genotyping Assay for the 385 SNP of the FUT2 gene (Applied Biosystems) with the following fluorescent probes both at nt position 378–391: 5′-CCGCCACATCCCGG-3′ and 5′-CCGCCACTTCCCGG-3′. Results were analyzed as described previously (Hutson et al., 2005).
Isolation and culturing of cell subsets from buffy coats of adult donors
DCs were derived from monocytes isolated from peripheral blood mononuclear cells by the adherence method (PBMC-DCs). Buffy coats of blood samples from healthy adult donors, blood group type O, were purchased from the Gulf Coast Regional Blood Center (Houston, TX), and PBMCs were isolated by density gradient centrifugation using Ficoll-Paque PLUS following the manufacturer’s procedure (GE Healthcare, Pittsburgh, NJ). Following suspension of PBMCs into RPMI 1640 media (Invitrogen) supplemented with 1% heat inactivated fetal bovine serum (FBS) (Invitrogen) and incubated for 2 h at 37°C and 5% CO2, adherent cells (mostly CD14+ monocytes) were washed 3X with media by removing floating cells. Adherent cells were suspended in DC media [RPMI 1640 media supplemented with 10% heat inactivated FBS (Invitrogen), 100 U/ml Penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM Hepes, 1X β-mercaptoethanol (Invitrogen), and 500 U/ml granulocyte macrophage-colony stimulator factor (GM-CSF) and 500 U/ml interleukin-4 (IL-4) (R&D Systems, Minneapolis, MN)] and incubated for at least 6 days at 37°C in 5% CO2. DCs and MΦ were also derived from CD16+ and CD14+ monocyte subsets isolated from PBMCs. CD16+ monocytes were isolated by using the CD16+ Monocyte Isolation Kit (Milteny Biotec, San Francisco, CA), and CD14+ monocytes were isolated from the flow through fraction using CD14 microbeads following the manufacturer’s protocol (Milteny Biotec). The CD16+ and CD14+ monocyte subsets were incubated at 37°C in 5% CO2, for at least 6 days, in DC media to give rise to immature CD16+ DCs and CD14+ DCs, respectively. In some cases, monocyte subsets were incubated in MΦ media, which consists of RPMI 1640 media supplemented with 10% heat inactivated FBS, 100 U/ml Penicillin, 100 μg/ml streptomycin (Invitrogen) plus 10 ng/ml of human rM-CSF (R&D Systems), to give rise to CD16+ MΦ and CD14+ MΦ, respectively.
Medium and reagents were changed every 3 days. CD11c+ blood DCs (CD11c+ DCs) were also isolated from buffy coats by fluorescent activated cell sorting (FACS) as previously described (Reche et al., 2001). In brief, the DC-enriched population (lineage negative cells) was obtained from PBMCs by negative selection using a mixture of monoclonal antibodies (mAbs) against the lineage markers CD3 (OKT3), CD14 (M5E2), CD16 (HB78), CD20 (L27), CD56 (B159), and CD235a (10F7MN) (BD Biosciences, San Jose, CA), followed by the use of magnetic beads (Invitrogen and Miltenyi Biotec). The DC-enriched population was subsequently stained with a mixture of fluorescein isothiocyanate (FITC)-labeled mAbs against lineage markers including CD3 (SK7), CD14 (MØP9), CD16 (3G8), CD19 (HIB19) and CD56 (NCAM16.2), APC-labeled anti-CD11c (Bly6) (BD Biosciences) and pacific blue-labeled anti-CD4 (OKT4; eBioscience, San Diego, CA). The lineage negative and CD11c, CD4 double positive cells were isolated by a FACS-Aria™ cell sorter system (BD Biosciences) to reach >99% purity. Viability of the CD11c+ DCs was determined by the standard trypan blue dye-exclusion method. The CD11c+ DCs were cultured in RPMI containing 10% FBS by seeding at a density of 1–3×105 cells/well in 200 μl volume in flat-bottom 96-well plates.
Cell surface phenotyping analysis of PBMC and isolated monocyte-derived DCs
Phenotypic analysis of isolated monocytes and immature DCs was performed by flow cytometry. At day 0 (monocytes), days 6 (PBMC-DCs) or 8 (CD16+DCs and CD14+DCs) of GM-CSF and IL-4 treatment, cells were harvested and washed with cold 0.1 M PBS containing 2% FBS (PBS/2% FBS). Subsequently, cells were stained in 100 μl of PBS/2% FBS for 30 minutes at 4°C with phycoerythrin (PE) or FITC-conjugated mouse anti-human antibodies to: CD14, CD16, CD11c, CD1a, CD80, CD83, CD3, CD40, CD31, CD56, HLA-A,B,C (MHC class I); and HLA-DR,DP,DQ (MHC class II) and in some cases, to CD209 (BD Biosciences). FITC or PE-conjugated mouse anti-human IgG1 antibodies were included as isotype controls. Cells were washed and suspended in 200 μl of cold PBS/2% FBS and fixed with 2% paraformaldehyde. Stained cells were analyzed (5,000 events per sample) for single or double color immunofluorescence with a BD FACSCalibur system–dual-laser flow cytometer (BD Biosciences). Data were acquired and analyzed using CellQuest software (Becton Dickinson). For analysis of CD103 and CX3CR1 expression in PBMC-DCs, a similar staining procedure was performed including: FITC-labeled anti-human CD11c (Clone 3.9, mouse IgG1, eBiosciences), PE-labeled anti- human CD103 (Clone B-Ly7, mouse IgG1, eBiosciences), tri-color (TC)-labeled anti-human CD3 (Clone S4.1, mouse IgG2a, Caltag Laboratories), APC-Cy7 labeled anti-human CD11b (Clone ICRF44, mouse IgG1, BD Biosciences) and Alexa Fluor 647 labeled anti-human CX3CR1 (Clone2A9-1 Rat IgG2b, Biolegend). The stained cells were acquired on a Cyan ADP (Dako, Carpinteria, CA), and data were analyzed by using Flowjow software (Tree Star, Ashland, OR).
NV inoculation of PBMC monocyte-derived DCs/MΦ and CD11c+ DCs
After at least 6 days of culture, PBMC-DCs, CD16+ DCs and CD14+ DCs were seeded at 3–7x105 cells/well. CD16+ MΦ, CD14+MΦ, and sorted CD11c+ DCs were seeded at 2x105 cells/well. Seeded cells were exposed to media only (mock), to NV (viral doses were in the range of 275 to 1,941 NV genome copies/cell) or to γ-irradiated NV (4 or 4.5 kGy). Treated cells were incubated for 2 hours at 37°C. Supernatants were removed and CD11c+ DCs were suspended into RPMI containing 10% FBS. After 3 washes with RPMI media, PBMC-DCs, CD16+ DCs or CD14+ DCs were suspended into DC media. CD16+ MΦ or CD14+ MΦ were suspended into MΦ media. In some experiments, PBMC-DCs were also treated with 10 μg/ml polymyxin B (PB, SIGMA-ALDRICH, St. Louis, MO); 1μg/ml LPS (SIGMA-ALDRICH); 50μg/ml -polynosinic polycytidylic acid sodium (Poly [I:C], SIGMA-ALDRICH); or a cocktail of rabbit polyclonal anti–IFN-α Abs (2000 neutralizing U/mL), rabbit polyclonal anti–IFN-β Abs (1000 neutralizing U/mL), and mouse anti–IFN-α/β receptor mAb (10 μg/mL, MMHAR-2; PBL Biomedical Laboratories). These treatments were added at the time of inoculation and kept in the media after the inoculum was removed. Treated cells were incubated at 37°C in 5% CO2 and harvested for viral RNA amplification analysis by qRT-PCR at 0, 24, 48, 72 or 144 hours post-inoculation (hpi) and protein expression analysis by IF at 24 and 72 hpi. Inoculation of cells from each donor was performed in triplicate.
Detection of NV proteins and cell markers in inoculated PBMC-DCs by IF assays
Immunofluorescence (IF) analysis was performed as previously reported (Guix et al., 2007), except for the following modifications: For dual-staining for NV structural and non-structural proteins at 24 or 72 hpi, cells were incubated with primary antibodies and added to the wells at the appropriate dilution in 0.1 M PBS-5% BSA (1:1,000 dilution for guinea pig polyclonal anti-VP1 with 1:1,000 rabbit polyclonal anti-VPg; or guinea pig polyclonal anti-VP1 with 1:2,000 for mouse polyclonal anti-polymerase serum. The same dilutions of their respective pre-immune sera were used as controls). For dual-staining of VP1 and CD103, anti-VP1 antibody and 1:200 dilution of a rabbit polyclonal anti-CD103 antibody (Integrin aE [H-260], Santa Cruz Biotechnology, Santa Cruz, CA) in 0.1 M PBS-5% BSA was added. Samples were incubated overnight at 4°C. Cells were washed 3X with 0.1 M PBS and incubated for 2 h at RT in a 1:1,000 dilution of the corresponding secondary antibodies conjugated with Alexa 488, or Alexa 594 (Invitrogen). Cells were rinsed 3X with 0.1 M PBS. Nuclei were stained with 300 nM 4′,6′-diamidino-2-phenylindole (DAPI, Invitrogen) for 5 min at RT. For dual-staining of VP1 and CX3CR1, cells were blocked at RT for 2 hours in 0.1 M PBS-5% BSA. Rabbit polyclonal anti-CX3CR1 antibody (Abcam) was added to the wells at 1:400 dilution in 0.1 M PBS-5% BSA and incubated overnight at 4°C. After washes, cells were incubated for 2 h at RT with the respective secondary antibody conjugated with Alexa 488 in 0.1 M PBS-5% BSA. After washing, cells were permeabilized with 0.5% Triton X-100-0.1 M PBS for 20 min at RT. Cells were stained with primary antibody for VP1 staining as shown above. IF and cell morphology analyses were performed using an Olympus IX-70 inverted microscope and Microfire digital camera.
RNA isolation of inoculated DCs and MΦ
RNA was extracted from total cell and supernatant samples using TRizol LS (Invitrogen) according to the manufacturer’s protocol at the indicated times post-inoculation. Samples were stored in RNAse free 2 ml tubes at −80°C.
Real time quantitative (q)RT-PCR assay
Extracted NV RNA was quantified by qRT-PCR as previously described (Guix et al., 2007). Briefly, a standard curve for absolute RNA quantification was generated in every assay by using RNA transcripts produced by in vitro transcription of a cDNA that contained ORF1- and ORF2-targeted regions (nt 4487 to 5671). Primers and probes used to amplify the ORF1 (nt 4641 to 4715) genomic region of NV have been previously described (Asanaka et al., 2005). Standard curves included five ten-fold dilutions and three replicate wells for each dilution. All samples were analyzed in at least triplicate wells. Reactions were performed using the TaqMan One-Step RT-PCR master mix reagent kit (Applied Biosystems) in an Applied Biosystems 7500 real-time PCR system. The thermal program consisted of 30 min at 48°C, followed by 10 min at 95°C, 45 cycles of 15 s at 95°C and 1 min at 60°C. For IFN-β mRNA expression analysis in PBMC-DCs treated with mock, γ-NV, NV, NV+ PB, LPS or Poly (I:C) after 10 hours, a comparative (CT) method or relative quantification (RQ) qRT-PCR analysis was performed using the IFN-β Taqman(R) Gene Expression Assay (Applied Biosystems). Normalization versus beta (β)-actin RNA was performed on these samples using the TaqMan β-actin control reagents (human) (Applied Biosystems).
Supplementary Material
Representative flow cytometric analysis showing phenotypic profiles of DCs obtained by the adherence method from the PBMC of a type O/Se+ individual (donor 369). PBMC-DCs stained with following antibodies: CD11c-FITC, CD103-PE, CX3CR1-Alexafluor 647, CD11b-APC and acquired on a CyAn ADP. FACS data were analyzed by using FlowJo software. (A) Forward scatter (FC) versus side scatter (SC) illustrating the morphology of PBMC-DCs by gating the FC-high and SC-high cells. (B) shows the frequency of CD11c+ DCs expressing CD11b. (C) Shows the frequency of CD11c+ DCs expressing CD103. (D) Shows the frequency of CD11c+ DCs expressing CX3CR1. The numbers in each panel represent the percentage of gated cells for the corresponding marker.
CD16+ monocytes (A) and CD14+ monocytes (B), from a type O/Se+ individual (donor 58), were isolated and treated with GM-CSF and IL-4 for at least 6 days to give rise immature CD14+ and CD16+ monocyte-derived DCs. Cells were harvested at day 0 and 8, and the expression of various cell surface markers was analyzed by flow cytometry. The Y axis of the histograms shows the number of positive cells. The percentage of positive cells and mean fluorescence intensity (within parentheses) are denoted. Green histograms represent the fluorescence of the isotype control.
Acknowledgments
We are grateful to Dr. Jacques LePendu for providing the primer sequences for the human FUT2 gene, and his approval to purchase the Custom TaqMan SNP Genotyping Assay for the allele 428 of the FUT2 gene from Applied Biosystems. This work was funded by the National Institutes of Health (P01 AI 57788, N01 AI 25465, and M01 RR-000188), and the Fulbright Scholar Program (FMECD2004/46139439).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asanaka M, Atmar RL, Ruvolo V, Crawford SE, Neill FH, Estes MK. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc Natl Acad Sci U S A. 2005;102:10327–10332. doi: 10.1073/pnas.0408529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. 2008;14:1553–1557. doi: 10.3201/eid1410.080117. PMC2609865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blutt SE, Conner ME. Rotavirus: to the gut and beyond! Curr Opin Gastroenterol. 2007;23:39–43. doi: 10.1097/MOG.0b013e328011829d. [DOI] [PubMed] [Google Scholar]
- Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, Stanley ER, Nussenzweig M, Lira SA, Randolph GJ, Merad M. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KO, George DW. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J Virol. 2007;81:12111–12118. doi: 10.1128/JVI.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology. 2006;353:463–473. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Changotra H, Jia Y, Moore TN, Liu G, Kahan SM, Sosnovtsev SV, Karst SM. Type I and type II interferons inhibit the translation of murine norovirus proteins. J Virol. 2009;83:5683–5692. doi: 10.1128/JVI.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham S, Souza M, Meulia T, Grimes S, Han MG, Saif LJ. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol. 2006;80:10372–10381. doi: 10.1128/JVI.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. Laboratory efforts to cultivate noroviruses. J Gen Virol. 2004;85:79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- Fankhauser RL, Noel JS, Monroe SS, Ando T, Glass RI. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J Infect Dis. 1998;178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- Gilpatrick SG, Schwab KJ, Estes MK, Atmar RL. Development of an immunomagnetic capture reverse transcription-PCR assay for the detection of Norwalk virus. J Virol Methods. 2000;90:69–78. doi: 10.1016/s0166-0934(00)00220-2. [DOI] [PubMed] [Google Scholar]
- Glass PJ, White LJ, Ball JM, Leparc-Goffart I, Hardy ME, Estes MK. The Norwalk virus ORF 3 encodes a minor structural protein. J Virol. 2000;74:6581–6591. doi: 10.1128/jvi.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KY, Lew JF, Jiang X, Kapikian AZ, Estes MK. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J Clin Microbiol. 1993;31:2185–2191. doi: 10.1128/jcm.31.8.2185-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guix S, Asanaka M, Katayama K, Crawford SE, Neill FH, Atmar RL, Estes MK. Norwalk virus RNA is infectious in mammalian cells. J Virol. 2007;81:12238–12248. doi: 10.1128/JVI.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guix S, Estes MK. Caliciviridae and Astroviridae. In: Brasier AR, Garcia-Sastre A, Lemon SM, editors. Cellular Signaling and Innate Immune Responses to RNA Virus Infections. ASM Press; Washington D.C: 2009. pp. 389–402. [Google Scholar]
- Hanabuchi S, Watanabe N, Wang YH, Wang YH, Ito T, Shaw J, Cao W, Qin FX, Liu YJ. Human plasmacytoid predendritic cells activate NK cells through glucocorticoid-induced tumor necrosis factor receptor-ligand (GITRL) Blood. 2006;107:3617–3623. doi: 10.1182/blood-2005-08-3419. [DOI] [PubMed] [Google Scholar]
- Henderson KS. Murine norovirus, a recently discovered and highly prevalent viral agent of mice. Lab Anim (NY) 2008;37:314–320. doi: 10.1038/laban0708-314. [DOI] [PubMed] [Google Scholar]
- Herbst-Kralovetz M, Hjelm B, Lay M, Sarker S, Berta A, Atmar R, Arntzen C, Estes M, Nickerson C. Lack of Sucess in Culturing Noroviruses in 3-D Cell Culture Systems. 5th International Conference on Vaccines for Enteric Diseases; Malaga, Spain. 2009. [Google Scholar]
- Hutson AM, Airaud F, LePendu J, Estes MK, Atmar RL. Norwalk virus infection associates with secretor status genotyped from sera. J Med Virol. 2005;77:116–120. doi: 10.1002/jmv.20423. [DOI] [PubMed] [Google Scholar]
- Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis. 2002;185:1335–1337. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- Inden K, Kaneko J, Miyazato A, Yamamoto N, Mouri S, Shibuya Y, Nakamura K, Aoyagi T, Hatta M, Kunishima H, Hirakata Y, Itoh Y, Kaku M, Kawakami K. Toll-like receptor 4-dependent activation of myeloid dendritic cells by leukocidin of Staphylococcus aureus. Microbes Infect. 2009;11:245–253. doi: 10.1016/j.micinf.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Ito T, Liu YJ, Kadowaki N. Functional diversity and plasticity of human dendritic cell subsets. Int J Hematol. 2005;81:188–196. doi: 10.1532/IJH97.05012. [DOI] [PubMed] [Google Scholar]
- Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, Berg PL, Davidsson T, Powrie F, Johansson-Lindbom B, Agace WW. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med. 2008;205:2139–2149. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang M, Graham DY, Estes MK. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Katayama K, Hansman GS, Oka T, Ogawa S, Takeda N. Investigation of norovirus replication in a human cell line. Arch Virol. 2006;151:1291–1308. doi: 10.1007/s00705-005-0720-9. [DOI] [PubMed] [Google Scholar]
- Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, Stewart P, LePendu J, Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Marionneau S, Ruvoen N, Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology. 2002;122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa MC, Rodriguez LS, Franco MA, Angel J. Interaction of rotavirus with human peripheral blood mononuclear cells: plasmacytoid dendritic cells play a role in stimulating memory rotavirus specific T cells in vitro. Virology. 2007;366:174–184. doi: 10.1016/j.virol.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Mumphrey SM, Changotra H, Moore TN, Heimann-Nichols ER, Wobus CE, Reilly MJ, Moghadamfalahi M, Shukla D, Karst SM. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J Virol. 2007;81:3251–3263. doi: 10.1128/JVI.02096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess JH. Role of mucosal dendritic cells in inflammatory bowel disease. World J Gastroenterol. 2008;14:5138–5148. doi: 10.3748/wjg.14.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- Papafragkou E, Hewitt J, Park GW, Straub TM, Greeining G, Vinje J. Challenges of Culturing Human Norovirus in a 3-D Organoid Cell Culture Model. 109th General Meeting of the American Society for Microbiology; Philadelphia PA. 2009. [Google Scholar]
- Parrino TA, Schreiber DS, Trier JS, Kapikian AZ, Blacklow NR. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N Engl J Med. 1977;297:86–89. doi: 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- Passlick B, Flieger D, Ziegler-Heitbrock HW. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood. 1989;74:2527–2534. [PubMed] [Google Scholar]
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. PMC2600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche PA, Soumelis V, Gorman DM, Clifford T, Liu M, Travis M, Zurawski SM, Johnston J, Liu YJ, Spits H, de Waal MR, Kastelein RA, Bazan JF. Human thymic stromal lymphopoietin preferentially stimulates myeloid cells. J Immunol. 2001b;167:336–343. doi: 10.4049/jimmunol.167.1.336. [DOI] [PubMed] [Google Scholar]
- Seidler S, Zimmermann HW, Bartneck M, Trautwein C, Tacke F. Age-dependent alterations of monocyte subsets and monocyte-related chemokine pathways in healthy adults. BMC Immunol. 2010;11:30. doi: 10.1186/1471-2172-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Ochsenbauer-Jambor C, Smythies LE. Intestinal macrophages: unique effector cells of the innate immune system. Immunol Rev. 2005;206:149–159. doi: 10.1111/j.0105-2896.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- Souza M, Azevedo MS, Jung K, Cheetham S, Saif LJ. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II.4-HS66 strain of human norovirus. J Virol. 2008;82:1777–1786. doi: 10.1128/JVI.01347-07. PMC2258707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub TM, Honer zu BK, Orosz-Coghlan P, Dohnalkova A, Mayer BK, Bartholomew RA, Valdez CO, Bruckner-Lea CJ, Gerba CP, Abbaszadegan M, Nickerson CA. In vitro cell culture infectivity assay for human noroviruses. Emerg Infect Dis. 2007;13:396–403. doi: 10.3201/eid1303.060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi S, Hashira S, Matsunaga T, Yoshida A, Shiota T, Tung PG, Khamrin P, Okitsu S, Mizuguchi M, Igarashi T, Ushijima H. Detection, genetic characterization, and quantification of norovirus RNA from sera of children with gastroenteritis. J Clin Virol. 2009;44:161–163. doi: 10.1016/j.jcv.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Taube S, Perry JW, Yetming K, Patel SP, Auble H, Shu L, Nawar HF, Lee CH, Connell TD, Shayman JA, Wobus CE. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol. 2009;83:4092–4101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, Kalchenko V, Geissmann F, Jung S. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007;204:171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt WD, Shakhar G, Jung S. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- White LJ, Ball JM, Hardy ME, Tanaka TN, Kitamoto N, Estes MK. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J Virol. 1996;70:6589–6597. doi: 10.1128/jvi.70.10.6589-6597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Thackray LB, Virgin HW. Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol. 2006;80:5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative flow cytometric analysis showing phenotypic profiles of DCs obtained by the adherence method from the PBMC of a type O/Se+ individual (donor 369). PBMC-DCs stained with following antibodies: CD11c-FITC, CD103-PE, CX3CR1-Alexafluor 647, CD11b-APC and acquired on a CyAn ADP. FACS data were analyzed by using FlowJo software. (A) Forward scatter (FC) versus side scatter (SC) illustrating the morphology of PBMC-DCs by gating the FC-high and SC-high cells. (B) shows the frequency of CD11c+ DCs expressing CD11b. (C) Shows the frequency of CD11c+ DCs expressing CD103. (D) Shows the frequency of CD11c+ DCs expressing CX3CR1. The numbers in each panel represent the percentage of gated cells for the corresponding marker.
CD16+ monocytes (A) and CD14+ monocytes (B), from a type O/Se+ individual (donor 58), were isolated and treated with GM-CSF and IL-4 for at least 6 days to give rise immature CD14+ and CD16+ monocyte-derived DCs. Cells were harvested at day 0 and 8, and the expression of various cell surface markers was analyzed by flow cytometry. The Y axis of the histograms shows the number of positive cells. The percentage of positive cells and mean fluorescence intensity (within parentheses) are denoted. Green histograms represent the fluorescence of the isotype control.