Abstract
TATA-binding protein (TBP) is a central component of the transcription apparatus and its association with promoters is dynamically regulated genome-wide. Recent work has shed new light on the functional specificity of Mot1 and NC2, two factors that control TBP distribution and activity. These studies underscore how regulation of TBP globally influences fundamental aspects of gene expression, including the balance of transcriptional output from different types of promoters, and the amplitude and timing of gene activation.
TATA-binding protein: linchpin of the RNA polymerase II preinitiation complex
Transcription is regulated by myriad mechanisms, many of which affect RNA polymerase II (Pol II) preinitiation complex (PIC) assembly. Although there is an increasing appreciation of the diversity and specialization in PIC organization [1], the general transcription factor (GTF) TATA-binding protein (TBP) has a widespread and crucial role in this process. TBP alone can support PIC formation on TATA-box-containing promoters, whereas TBP-associated factors (TAFs) are also typically required for TATA-less transcription [1]. How is TBP distributed among different types of promoters? What factors regulate its distribution and activity? Two new papers [2,3] employ complementary approaches to advance our understanding of how the activity of this crucial transcription apparatus component is regulated.
TBP is a saddle-shaped molecule, the binding of which to TATA-box DNA establishes an extensive hydrophobic protein–DNA interface of exceptional stability [4], thus, providing the protein–DNA platform required for assembly of a functional PIC. However, the extraordinarily long half-times for dissociation of TBP–DNA complexes measured in vitro (15 min to 1 hr or more; [5]) indicate that active mechanisms must be in place to ensure that TBP interacts with chromatin in vivo on time scales appropriate for rapid gene regulation. The problem is potentially acute because TBP binds with high affinity to a variety of DNA sequences [4] and, yet, numerous nonspecific sites would limit the available TBP pool. TBP binding to fortuitous sites within promoters could also inhibit PIC formation sterically.
Consistent with a role in initiating PIC assembly, many factors function by modulating TBP recruitment or activity. Although there are many interesting examples of transcriptional regulators that affect TBP function in gene-specific ways, here, I focus on modifier of transcription 1 (Mot1) and negative cofactor 2 (NC2), two regulators that have global effects on TBP activity. Mot1 is a member of the Snf2/Swi2 ATPase family that uses ATP hydrolysis to displace TBP from DNA [6]. In yeast, the robust catalytic activity of Mot1 is responsible for the highly dynamic behavior of virtually the entire TBP pool [7]. NC2 is a TBP-binding heterodimer with different activities. It was originally identified biochemically as a factor that blocks PIC assembly subsequent to TBP–DNA binding [8]. More recently, it was discovered that NC2 permits TBP relocalization along the DNA contour, an activity consistent with the architecture of the NC2–TBP–DNA complex, which resembles a clamp encircling DNA [9]. Individual NC2 subunits might also have separate functions, but this is not as well understood [3].
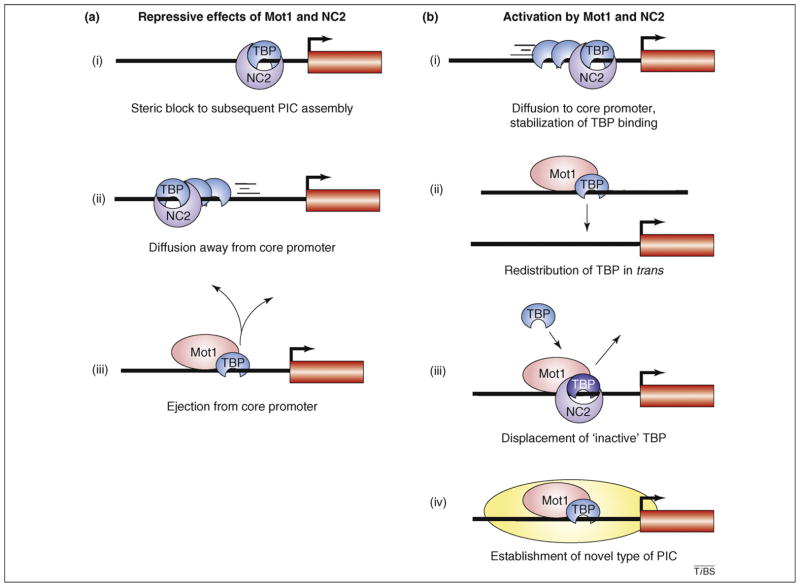
The biochemical activities of Mot1 and NC2 indicate roles as global transcriptional repressors (Figure 1a) and, indeed, genetic and genome-wide expression analyses in yeast support the notion that both factors can function in this way [10–14]. However, both factors also have global roles in gene activation that were unanticipated from biochemical analyses [11–16]. Two general models can explain how Mot1 and NC2 activate gene expression (Figure 1b). The first posits that the known biochemical activities of these factors can activate and repress transcription. Notably, Mot1-mediated TBP–DNA dissociation could ensure that a sufficient pool of free TBP is available for nucleating PIC formation; alternatively, removal of inactive, kinetically trapped forms of TBP might be essential to clear promoters for subsequent assembly of functional PICs [17,18]. NC2 might activate gene expression by transiently stabilizing weak TBP–DNA interactions [19] or by funneling TBP to core promoters by catalyzing diffusion along DNA. All of these ideas posit that Mot1 and NC2 form transcriptionally inactive complexes with TBP and that they foster gene activation indirectly by modulating TBP dynamic behavior. A second general model is that Mot1 and NC2 have novel activities in gene activation. Intriguingly, some evidence points to direct roles for Mot1 in gene activation [20,21], although such mechanisms have not been elucidated in molecular detail.
Figure 1.
General mechanisms of Mot1- and NC2-mediated gene regulation. The cartoons illustrate biochemical activities of Mot1 and NC2 and how they might contribute to gene regulation. (a) The thick black horizontal line represents intergenic DNA, the red box represents a gene and the arrow at the edge of the red box represents a transcription start site. The cartoons illustrate mechanisms by which NC2 (purple) and Mot1 (pink) might repress transcription. These include: (i) NC2-mediated inhibition of PIC formation; (ii) NC2-facilitated mobilization of TBP (blue) away from promoters; and (iii) Mot1-catalyzed dissociation of TBP from promoters. (b) The cartoons illustrate mechanisms by which NC2 and Mot1 might activate transcription, including NC2- or Mot1-mediated relocation of TBP from a non-promoter site to a core promoter in (i) cis or (ii) trans. (iii) Mot1-mediated removal of an inactive, kinetically-trapped TBP molecule (with or without NC2) would permit another molecule of TBP to bind, perhaps establishing an active complex. NC2 stabilizes TBP–DNA binding and, thus, might transiently stabilize TBP binding to weak DNA sites to facilitate PIC assembly. (iv) In yeast, Mot1 is a component of a transcriptionally active complex at promoters after stress (depicted in yellow) and has also been implicated in chromatin remodeling that is required for gene activation.
TBP regulatory circuit differentially affects different types of core promoters
Although the nature and distribution of TBP-binding sites is well-studied, most promoters lack TATA elements. Metazoan TATA-less promoters possess elements, such as the downstream promoter element (DPE), which specify PIC assembly and promoter activity. Hsu et al. [2] provide new insight into how the inherently ‘repressive’ NC2 and Mot1 factors contribute to transcriptional activation. The new work shows that TATA- and DPE-containing promoters in Drosophila melanogaster are differentially dependent on TBP, Mot1 and NC2. Consistent with expectation, TATA-dependent transcription relies strongly on TBP. In remarkable contrast, however, TBP depletion had little, if any, effect on DPE-dependent transcription [2]. Metazoans possess other TBP-related proteins (TRFs) and the results do not exclude the possibility that one of them, TRF2, participates directly in DPE-mediated transcription. Another possibility is that DPE-dependent transcription does not require any type of TBP-related protein. If so, DPE-dependent transcription in D. melanogaster is mechanistically distinct from TATA-less transcription in Saccharomyces cerevisiae, which is TBP dependent. In S. cerevisiae, NC2 and Mot1 repress TATA-containing promoters and activate TATA-less ones [12,18,22]. A similar trend was observed in D. melanogaster, although fewer genes have been analyzed to date and the effects of Mot1 and NC2 depletion on promoter activity are seemingly more nuanced [2]. Genome-wide analyses in D. melanogaster will be crucial for discerning large-scale patterns in promoter dependencies on TBP, Mot1 and NC2 to establish how the roles of these factors are similar and different in the two organisms. A key observation in the new work is that TBP overexpression inhibits DPE-dependent transcription [2]. This finding indicates that large-scale transcriptional output in D. melanogaster results from a balance between providing sufficient TBP to drive TATA-containing promoters and limiting TBP to prevent inhibition of DPE-driven transcription.
Inhibition of TATA-containing promoters by NC2 and Mot1 fits well with the biochemical data, but how might Mot1 and NC2 activate TATA-less transcription? The unanticipated effects of TBP on DPE-dependent transcription led Hsu et al. [2] to suggest a model in which both repression and activation mediated by Mot1 and NC2 stem from inhibitory effects on TBP function. Because the two types of promoters display a differential requirement for TBP, antagonism of TBP by Mot1 and NC2 results in two different transcriptional effects. The specific molecular explanation for how Mot1 and NC2 activate DPE promoters is unknown, but this study offers a simple concept for how factors that are fundamentally inhibitory toward TBP function induce reciprocal effects on gene expression depending on the nature of the core promoter.
Global landscape of GTF distribution
In yeast, TBP is associated with promoters approximately in proportion to transcription level [23]. Mot1 and NC2 are found at disproportionately high levels at target gene promoters, but they can nonetheless be detected at promoters (regardless of whether they possess a TATA box) in proportion to TBP [14,18,22]. Promoter localization of the NC2α subunit is correlated with gene activity in human cells as well [24]. These observations cast a seemingly different perspective on Mot1 and NC2 function than discussed earlier. Why do Mot1 and NC2 accrue at activated promoters? van Werven et al. [3] obtained yeast genome-wide chromatin immunoprecipitation (ChIP)-on-chip data to address this issue and provide additional insight into the interplay of these factors. A strength of the study is that the authors used oligonucleotide arrays to compare localization of seven different factors under two different growth conditions, providing a rich dataset. The results show striking co-localization of NC2, Mot1 and TBP, arguing that Mot1 and NC2 can function on the same promoters at the same time. Consistent with this idea, a discrete and essentially pure complex composed of NC2, Mot1, TBP and DNA was isolated from chromatin extracts. Because the complex lacks other GTFs or Pol II, this finding provides strong support for the idea that NC2 and Mot1 interact with a transcriptionally inactive form of TBP [17,18,20]. van Werven et al. [3] show that TBP and NC2 persist on a deactivated promoter in mot1 cells, extending prior observations [13,14,17,22] and solidifying the conclusion that Mot1 and NC2 limit transcription during the activated state. The results also implicate Mot1 in timely gene deactivation [3,17]. Thus, rather than functioning as an all-or-none-repressor, a function of Mot1 is to set the threshold of activated gene expression by antagonizing PIC formation.
Concluding remarks and future perspectives
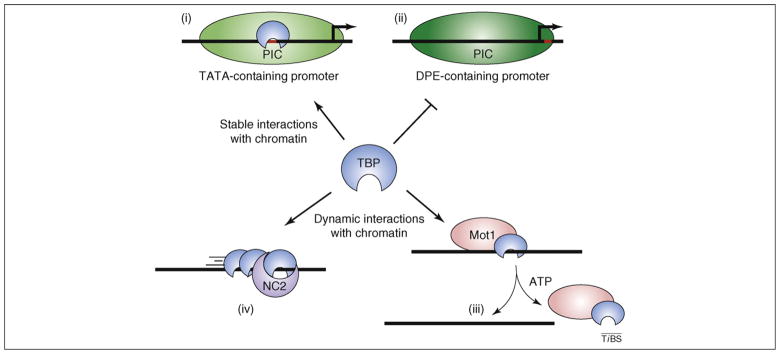
The effects of Mot1 and NC2 on gene expression are diverse and, yet, the studies highlighted here reinforce the notion that many observations – including some aspects of gene activation – can be explained by the repressive properties of these factors toward TBP. Important questions remain about the regulation and function of TBP dynamics in vivo (Figure 2). What is the dynamic behavior of TBP at particular chromatin sites? Do TBP dynamics regulate transcription start site utilization? Fundamental questions remain about the structure and function of PICs formed on TATA-less promoters, including the role of TBP. Do Mot1 and NC2 have direct roles in DPE-mediated transcription? At yeast TATA-less promoters, evidence for direct roles for TBP, Mot1 and NC2 in activation is strong, however, little is known biochemically about TATA-less transcription in yeast, much less about how Mot1 and NC2 might participate as constituents of functional PICs. Indeed, a proteomic analysis hints that the complexity of interactions involved in regulatory control by these factors is greater than imagined [25]. Recent work provides an excellent foundation for exploration of these issues.
Figure 2.
Interplay of forces governing TBP activity globally. The horizontal black lines represent DNA and the green ovals represent transcription preinitiation complexes (PICs) formed on each of two different types of promoters as indicated. The different shades of green indicate that PICs formed on these two types of promoters have different constituents. TBP transcriptional activity genome-wide is a consequence of a balance between stable interactions with chromatin [assembly into PICs (i) and regulatory factors responsible for its dynamic behavior (iii) and (iv)]. TBP activity level can influence the balance between (i) TATA-containing and (ii) TATA-less transcription. Color code: TBP, blue; Mot1, pink; NC2, purple.
Acknowledgments
This work was supported by National Institute of Health grant GM55763 to D.T.A. I am grateful to Dan Engel, Rebekka Sprouse, Jim McNally and members of the Auble Laboratory for comments on the manuscript.
References
- 1.Muller F, et al. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 2.Hsu JY, et al. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Werven FJ, et al. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008;22:2359–2369. doi: 10.1101/gad.1682308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patikoglou GA, et al. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprouse RO, et al. Function and structural organization of Mot1 bound to a natural target promoter. J Biol Chem. 2008;283:24935–24948. doi: 10.1074/jbc.M803749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auble DT, et al. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 7.Sprouse RO, et al. Regulation of TATA binding protein dynamics in living yeast cells. Proc Natl Acad Sci U S A. 2008;105:13304–13308. doi: 10.1073/pnas.0801901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meisterernst M, Roeder RG. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 9.Schluesche P, et al. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol. 2007;14:1196–1201. doi: 10.1038/nsmb1328. [DOI] [PubMed] [Google Scholar]
- 10.Davis JL, et al. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasgupta A, et al. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci U S A. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisberg JV, et al. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisberg JV, et al. Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:8122–8134. doi: 10.1128/MCB.22.23.8122-8134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madison JM, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TBP to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collart MA. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta A, et al. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huisinga KL, Pugh BF. A TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol. 2007;8:R46. doi: 10.1186/gb-2007-8-4-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cang Y, Prelich G. Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP) Proc Natl Acad Sci U S A. 2002;99:12727–12732. doi: 10.1073/pnas.202236699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisberg JV, Struhl K. Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Mol Cell. 2004;14:479–489. doi: 10.1016/j.molcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Topalidou I, et al. Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J. 2004;23:1943–1948. doi: 10.1038/sj.emboj.7600199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc Natl Acad Sci U S A. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Iyer VR. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol Cell Biol. 2004;24:8104–8112. doi: 10.1128/MCB.24.18.8104-8112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albert TK, et al. Global distribution of negative cofactor 2 subunit-α on human promoters. Proc Natl Acad Sci U S A. 2007;104:10000–10005. doi: 10.1073/pnas.0703490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnett DR, et al. A proteomic analysis of yeast Mot1p protein-protein associations: insights into mechanism. Mol Cell Proteomics. 2008;7:2090–2106. doi: 10.1074/mcp.M800221-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]