Abstract
Multiple genetic disorders can be associated with excessive signalling by mutant G-protein-coupled receptors (GPCRs) that are either constitutively active or have lost sites where phosphorylation by GPCR kinases is necessary for desensitisation by cognate arrestins. Phosphorylation-independent arrestin1 can compensate for defects in phosphorylation of the GPCR rhodopsin in retinal rod cells, facilitating recovery, improving light responsiveness, and promoting photoreceptor survival. These proof-of-principle experiments show that, based on mechanistic understanding of the inner workings of a protein, one can modify its functional characteristics to generate custom-designed mutants that improve the balance of signalling in congenital and acquired disorders. Manipulations of arrestin elements responsible for scaffolding mitogen-activated protein kinase cascades and binding other signalling proteins involved in life-or-death decisions in the cell are likely to yield mutants that affect cell survival and proliferation in the desired direction. Although this approach is still in its infancy, targeted redesign of individual functions of many proteins offers a promise of a completely new therapeutic toolbox with huge potential.
Structural protein modules with specific functions, termed domains, are used and reused in evolution, with mixing and matching of various domains achieving combinations of functions. Successful elements, such as the kinase domain or immunoglobulin fold, are present in a surprising variety of proteins. The module composed of seven transmembrane α-helices (heptahelical domain; HD) is one of the greatest successes of evolutionary engineering: it constitutes the core of all G-protein-coupled receptors (GPCRs) – the most numerous protein family in the animal kingdom, with ~1000 members in most mammals (Refs 1, 2). Structurally, GPCRs are very diverse, ranging from the virtually HD-only light receptor rhodopsin (RHO), a prototype of the largest class A, to constitutively dimeric class C receptors, where each HD is fused to the Venus flytrap domain, or the enormous (>6000 amino acid long) ‘very large GPCR’ (GPR98), with a motley collection of domains attached to its HD (Ref. 3). Different GPCRs respond to incredibly diverse stimuli, from photons and small molecules to large proteins, metal ions, and extracellular protease activity (Refs 1, 3). Not surprisingly, GPCRs are targeted by about half of clinically used drugs (Ref. 2).
The key mechanisms of the activation of the HD core of GPCRs and signal transduction across the membrane appear to be fairly well conserved (Refs 4, 5). Receptor transition into an active conformation triggered by agonist binding involves the movement of the transmembrane helices relative to each other (Refs 6, 7), which significantly changes the cytoplasmic tip of the receptor. This conformational rearrangement dramatically increases its affinity for several intracellular signalling partners. Upon activation, the great majority of GPCRs bind heterotrimeric G proteins, catalysing the release of bound GDP and its replacement with GTP, the prevalent guanyl nucleotide in the cytoplasm. GTP-liganded Gα subunit loses its affinity for the Gβγ dimer, so that both parts of the G protein dissociate from the receptor and activate or inhibit various effectors. While it remains active, a single receptor can activate dozens of G protein molecules (Ref. 8), amplifying the signal to ensure high sensitivity.
In most cases, signalling is terminated by a two-step mechanism: active receptor is selectively phosphorylated by one or more GPCR kinases (GRKs), whereupon a member of the arrestin family of proteins binds with high affinity to the cytoplasmic tip of the active phosphoreceptor (Ref. 9), sterically precluding further G-protein interactions (Ref. 10) (Fig. 1a). Receptor-bound arrestin recruits GPCRs to coated pits for internalisation via its interactions with clathrin (Ref. 11) and the AP2 adaptor complex (Ref. 12). The arrestin–receptor complex often initiates another round of signalling via arrestin-mediated recruitment of mitogen-activated protein kinases (MAPKs) and other proteins (Refs 13, 14). Although the timing of action and relative contribution of individual members of arrestin and GRK families vary (Refs 15, 16), the general mechanism is conserved (Ref. 9). The ability of a single GPCR upon binding different ligands to assume distinct active conformations (Ref. 17) preferentially interacting with different G proteins and other signal transducers apparently underlies biased agonism (Refs 18, 19).
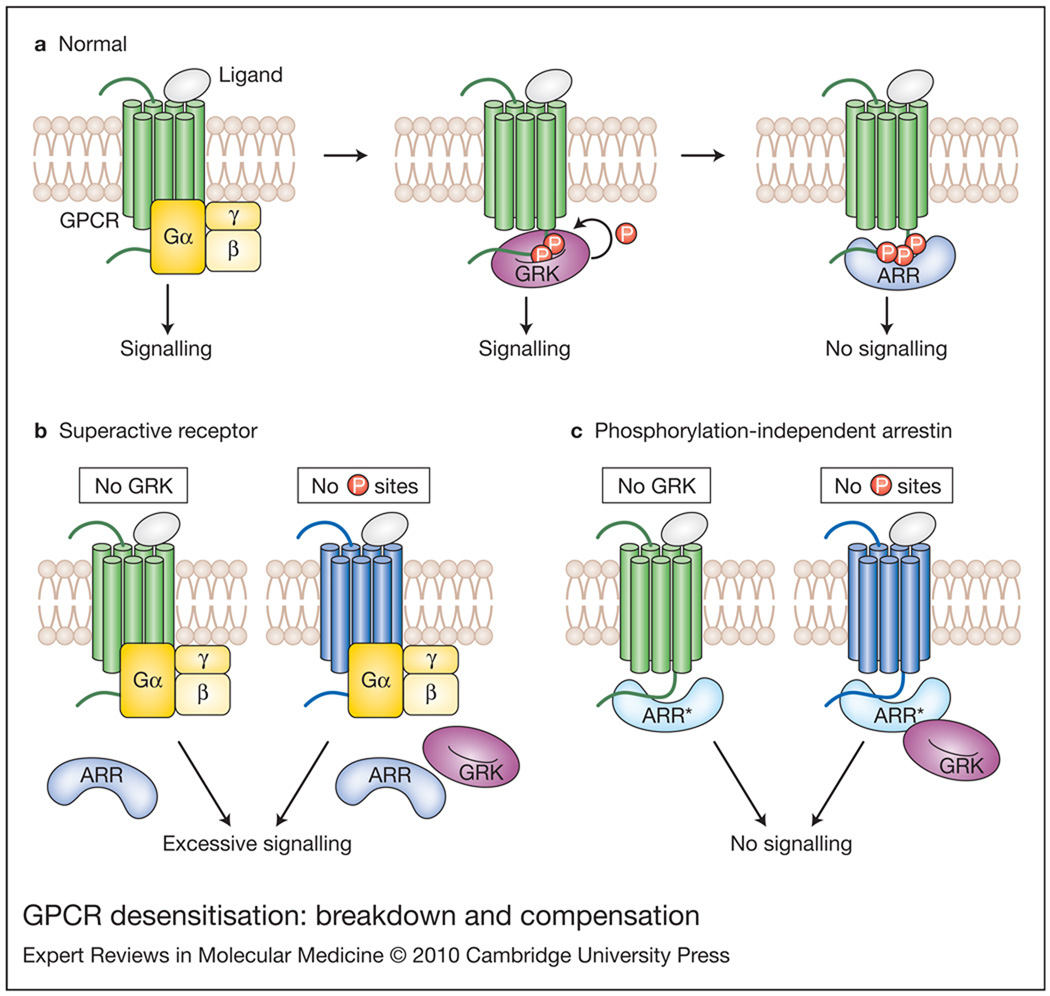
Figure 1. GPCR desensitisation: breakdown and compensation.
(a) Ligand-activated GPCRs bind heterotrimeric G proteins, catalysing exchange of GDP for GTP, and subsequent dissociation of Gα and Gβγ subunits (dissociation not shown), which regulate downstream signalling proteins. In normal situations, active GPCRs are phosphorylated by GPCR kinases (GRKs). Binding of wild-type arrestin (ARR; blue) to active phosphoreceptor stops the signalling in a timely fashion. (b) Normal GPCR (green) in the absence of GRK, or a mutant GPCR (dark blue) that does not have GRK phosphorylation sites, cannot be shut off by wild-type arrestins, which only bind phosphorylated GPCRs. In both cases, excessive receptor signalling results in pathology. (c) Expression of phosphorylation-independent arrestin (ARR*; lighter blue) compensates for the defects of receptor phosphorylation by binding unphosphorylated wild-type or mutant receptors, thereby reining in excessive signalling.
Diseases associated with excessive GPCR signalling
More than 30 human diseases have been directly linked to mutations in GPCRs (reviewed in Ref. 20). The majority of identified mutations are inactivating, and therefore recessive: disease phenotype develops only in individuals that are homozygous or compound heterozygous for the affected allele. However, activating mutations in different GPCRs underlie about a dozen disorders, ranging from dwarfism and hypo- and hyperthyroidism to several forms of cancer (Ref. 20). In some cases, constitutive activity of the receptor results in its excessive phosphorylation and association with arrestin. Constitutive desensitisation of a human vasopressin receptor mutant underlies nephrogenic diabetes insipidus (Ref. 21), whereas in the case of rhodopsin it results in night blindness or retinal degeneration (Ref. 22).
At the molecular level, receptor mutations are not the only cause of excessive GPCR signalling. For example, uncontrolled rhodopsin activity can be produced by activating mutations (Ref. 23), as well as desensitisation deficits due to absence of phosphorylation sites in mutant rhodopsin (Refs 24, 25, 26), and null mutations in rhodopsin kinase (Ref. 27) or arrestin (Ref. 28). All of these defects cause visual disorders ranging from night blindness to photoreceptor degeneration, which have been reproduced in genetically modified mice (Refs 29, 30, 31, 32). A growing body of evidence suggests that GPCR genes are protooncogenes (Ref. 33). Uncontrolled activation of wild-type M1, M3 and M5 muscarinic acetylcholine receptors (mAChRs; CHRM1, 3 and 5) by constant presence of an agonist (Ref. 34), ectopic expression of serotonin 5HT1C receptor (HTR2C) (Ref. 35), or expression of a constitutively active α1B-adrenergic receptor (ADRA1B) (Ref. 33) have all been shown to induce malignant transformation. The gene encoding the angiotensin receptor was first cloned as the mas oncogene (MAS1) (Ref. 36, 37) even before its pharmacological nature was established (Ref. 38). Signalling by wild-type GPCRs pushes the cell out of balance in most of these cases. Another example of excessive activity of a perfectly normal receptor in cells that apparently express all relevant GRKs and arrestins is supersensitivity of dopamine receptors associated with dopamine depletion in Parkinson disease or dyskinesia, developing upon prolonged treatment of parkinsonian patients with l-DOPA (Ref. 39).
One possible strategy to counteract excessive GPCR signalling is to boost the desensitization mechanisms that normally keep it in check. Increased expression of GRKs and/or arrestins is likely to decrease the signalling to normal levels where wild-type receptors are involved, but different approaches are needed in the case of mutant receptors that cannot be phosphorylated properly.
Molecular mechanics of arrestin proteins
The first arrestin was discovered in the visual system as a negative regulator of rhodopsin signalling. As arrestin nomenclature is rather confusing, we use systematic names in this article: arrestin1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin; SAG in HUGO Gene Nomeclature Committee database), arrestin2 (β-arrestin or β-arrestin1; ARRB1), arrestin3 (β-arrestin2; ARRB2), and arrestin4 (cone or χ-arrestin; ARR3). Arrestin1 was shown to bind active phosphorylated rhodopsin and suppress (arrest) further G-protein activation (Refs 40, 41). Subsequent cloning of additional family members (Refs 42, 43, 44, 45, 46) and elegant demonstration that arrestin subtypes preferentially regulate different GPCRs (Ref. 47) showed that the same mechanism is involved in the regulation of numerous receptors in this superfamily (reviewed in Refs 9, 48) (Fig. 1a).
Three families of cytoplasmic proteins preferentially bind active GPCRs: G proteins, GRKs and arrestins. In contrast to G proteins and GRKs, arrestins have no lipid modifications or other means of recruitment to the membrane except through receptor binding. Out of four functional forms of rhodopsin – inactive (Rh), active (Rh*), inactive phosphorylated (P-Rh), and active phosphorylated (P-Rh*) – arrestins bind with high affinity only one: P-Rh*. This raises an interesting question: how do these soluble proteins, diffusing in the cytoplasm and randomly encountering GPCRs in various functional states, specifically bind only P-Rh*? What is the molecular mechanism that translates the encounter with P-Rh* into high-affinity binding? The development of a direct binding assay with femtomolar sensitivity provided vital clues: arrestins were found to specifically bind to Rh* and inactive P-Rh, albeit with much lower affinity than to P-Rh* (Ref. 49). Importantly, a C-terminal truncation generates a mutant that still binds P-Rh* best, but shows greatly enhanced affinity for Rh* and inactive P-Rh, but not for Rh (Ref. 49). These data suggested that arrestin recognises receptor activation and phosphorylation states independently, but has internal restraints precluding high-affinity binding to anything except P-Rh*. GPCR activation involves a significant conformational rearrangement of the receptor, exposing elements that were inaccessible in the basal state (Refs 6, 7, 50). Arrestin must interact with these newly exposed elements to distinguish between active and inactive receptor. It also has to bind receptor-attached phosphates to discriminate between phosphorylated and unphosphorylated forms.
A model of sequential multisite binding (Ref. 51) explains arrestin selectivity for P-Rh*, as follows. Arrestin has separate sites interacting with elements in Rh* exposed by its activation and with receptor-attached phosphates. Individually these interactions are relatively weak, allowing arrestin to ‘probe’ the functional state of the receptor it encounters and dissociate quickly if it turns out to be an unpreferred one. Only when arrestin encounters P-Rh* are both of the primary sites simultaneously engaged. This releases internal constraints, allowing the arrestin molecule to undergo a conformational change of its own, which brings additional secondary binding sites into contact with the receptor, greatly increasing the energy of the interaction and hence the affinity (reviewed in Ref. 52). This model predicts that primary binding sites serve as sensors, and that the molecule works as a coincidence detector, swinging into action only when both sensors are activated simultaneously.
Obviously, a sensor in a protein can be artificially turned on by mutagenesis. One arginine in the first set of identified phosphate-binding residues behaved in this fashion: charge neutralisation generated an arrestin mutant with high binding to Rh* (Ref. 53). Subsequent demonstration that charge reversal increases Rh* binding even more, whereas Arg to Lys mutation preserving the charge does not, led to the idea that this arginine interacts with a negatively charged residue within arrestin, and that neutralisation of its charge either by receptor-attached phosphates or by mutagenesis breaks the salt bridge, thereby turning arrestin on (Refs 53, 54). Indeed, crystal structures of arrestin1 (Ref. 55), 2 (Refs 56, 57) and 4 (Ref. 58) revealed that between the two arrestin domains there is an arrangement of five charged residues that are largely solvent-excluded (unusual for a soluble protein), which was termed the polar core. Extensive mutagenesis proved that the salt bridge between an arginine and an aspartic acid in the polar core serves as the main phosphate sensor in all arrestins (Refs 59, 60, 61, 62, 63, 64, 65). Subsequent studies showed that the interaction between the β-strand I, α-helix I, and the arrestin C-tail serves as an auxiliary phosphate sensor and is also disrupted by receptor-attached phosphates (Refs 60, 66, 67, 68). Manipulation of this interaction yielded structurally distinct mutants with high affinity for Rh*. These gain-of-function mutants that bind either active form of their cognate receptors – P-Rh* and Rh* – with comparable affinity (Refs 61, 62, 63, 69) were termed phosphorylation-independent or enhanced arrestins (Ref. 65).
Phosphorylation-independent arrestins as tools for gene therapy
The strategy for gene therapy in the case of loss-of-function mutations is conceptually straightforward: expression of a missing functional protein is likely to solve the problem. With the exception of rhodopsin expressed in photoreceptors in enormous quantities, the levels of most GPCRs in other cells are relatively low, so that even a receptor mutant with severe folding defects is unlikely to cause additional trouble by overloading the cellular protein-degradation machinery consisting of proteasomes and lysosomes.
In contrast, dealing with gain-of-function mutations is much harder. These mutations are dominant, because normal protein encoded by the second allele cannot prevent excessive signalling. Traditionally, GPCR signalling is reduced by appropriate small-molecule antagonists (blockers), which are widely used for therapeutic purposes. Antagonists targeting normal GPCRs attenuate the amplitude of signalling, while leaving the regulation of the time course of signal transduction to cellular desensitisation mechanisms mediated by GRKs and arrestins. In the case of phosphorylation-deficient GPCR mutants this mechanism does not work, so an antagonist can decrease signal intensity, but cannot restore its normal time course, which is equally important biologically. Theoretically, mutant mRNA could be eliminated with an appropriately designed ribozyme. However, since even very low expression of constitutively active or desensitisation-deficient receptor is harmful (Ref. 30), the efficiency of this process must be extremely high. As mutant and normal mRNA encoded by the other allele could differ by as little as a single base (in the case of point and frameshift mutations), the required combination of very high efficiency and perfect selectivity is hardly realistic. Conceivably, one could deliver a ribozyme that indiscriminately destroys all endogenous mRNA for the receptor, along with the gene encoding normal protein in which the sequence is modified by silent mutations to make it resistant to this ribozyme. This places additional demands on the vector, which has to deliver two functional coding sequences instead of one. Thus, alternative strategies to dampen the signalling by gain-of-function receptor mutants are needed.
Logically, since excessive receptor signalling is at the root of the problem, increased activity of components of the desensitisation machinery could bring the system back into balance. For normal (nonmutant) GPCRs, GRK availability is rate-limiting, although extra arrestin further facilitates desensitisation in the presence of added GRK (Refs 70, 71). Obviously, where the mutation has eliminated or significantly reduced the number of GRK phosphorylation sites, an increase in the availability of wild-type GRKs and arrestins cannot achieve this goal (Fig. 1b). Theoretically, a small molecule that promotes the binding of wild-type arrestin to unphosphorylated mutant receptor in an activation-dependent manner could solve this problem. Indeed, two compounds that promote arrestin1 binding to light-activated unphosphorylated rhodopsin have been described: heparin (Ref. 72) and a synthetic phosphopeptide mimicking the fully phosphorylated rhodopsin C-terminus (Ref. 73). Although neither study directly addresses this possibility, both illustrate the same difficulties in adapting this approach for therapy. First, neither effective molecule is particularly small. Second, both carry multiple negative charges, which preclude crossing of the membrane to get inside the cell where they are expected to work. Third, both are effective at fairly high concentrations (high micromolar to millimolar). And last, but not least, both have bell-shaped dose–response curves: they promote arrestin binding to unphosphorylated Rh* only in a very narrow concentration range, and actually inhibit it above and below the optimum. While it could be possible to design smaller molecules enhancing arrestin binding to unphosphorylated receptor, a high concentration of negative charges is a must, as this molecule has to substitute for missing receptor-attached phosphates, activating the arrestin phosphate sensor. In fact, the great majority of clinically used drugs act on the extracellular side of the membrane, where the issue of membrane crossing does not arise. Still, some are hydrophobic enough to cross the membrane unaided or rely on transporters to get inside cells. Small molecules inhibiting or enhancing intracellular protein–protein interactions can and should be developed, although one has to keep in mind that this approach is not free of significant limitations. The ultimate solution might require the same type of delivery vehicle as gene transfer: one that targets specific tissues or cell types and delivers the cargo across the plasma membrane. Considering that drugs must be delivered repeatedly, whereas genes need to be delivered only once, the latter may turn out to be more feasible and economical.
Phosphorylation-independent arrestin mutants described above are promising tools for controlling excessive GPCR signalling (Fig. 1c). Their ability to blunt the signalling of normal GPCRs in the absence of receptor phosphorylation and that of phosphorylation-deficient mutants of rhodopsin (Ref. 63), μ- (Refs 74, 75) and δ-opioid (OPRM1, OPRD1) (Refs 61, 62), and β2-adrenergic (Refs 61, 62) (ADRB2) receptors has been convincingly demonstrated in vitro (Ref. 63), in exogenous expression systems (Refs 61, 62, 64, 74), and in primary cultures of GRK-deficient neurons (Ref. 75), paving the way for in vivo experiments. This compensational strategy was recently tested in the visual system of living animals (Ref. 65).
Testing the compensational approach in the visual system
Rod photoreceptors are neurons, in which the molecular machinery responsible for light-initiated signalling is localised in a specialised compartment, termed the outer segment, separated from the rest of the cell by a narrow cilium (Ref. 76). In a healthy retina, elongated outer segments of all photoreceptors are parallel and neatly arranged (Fig. 2c). Normal length of the outer segments is an indication of photoreceptor health; in visual disorders they often become shorter and visibly disorganised (Refs 29, 30, 31, 65). Photoreceptor cell bodies and nuclei make up the outer nuclear layer. The thickness of this layer is a good measure of the number of surviving photoreceptors; its thinning is a clear indication of disease-associated photoreceptor loss (Refs 29, 30, 31).
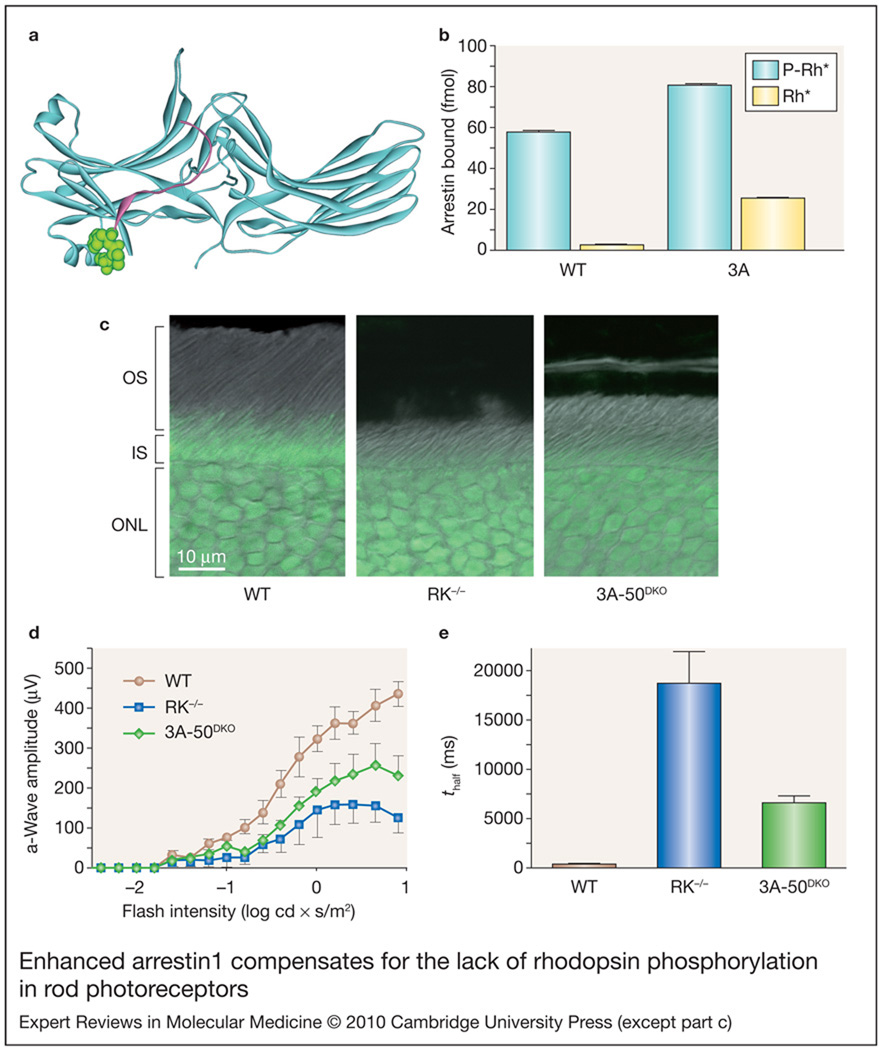
Figure 2. Enhanced arrestin1 compensates for the lack of rhodopsin phosphorylation in rod photoreceptors.
(a) Arrestin1 is an elongated two-domain molecule (crystal structure from Ref. 55, PDB 1CF1). Interdomain interactions and the anchoring of the C-tail (magenta) to the body of the molecule by bulky hydrophobic residues (side chains are shown in green) stabilise its basal (inactive) conformation. (b) Wild-type (WT) arrestin1 binds light-activated phosphorylated rhodopsin (P-Rh*) much better than active unphosphorylated rhodopsin (Rh*). Alanine substitution of the residues anchoring the C-tail (3A) yields an enhanced mutant with greatly increased affinity for Rh*. (c) Rhodopsin-containing outer segments (OSs) of healthy rod photoreceptors of WT animals are long and neatly organised. Despite normal expression of WT arrestin1, the loss of rhodopsin kinase (RK−/−) results in a dramatic shortening and dishevelled appearance of the outer segments. The expression of phosphorylation-independent arrestin1 in RK-deficient animals 3A-50RK−/−Arr1−/− [3A-50DKO (double knockout) on figure] increases the length of the outer segments, indicative of the improved overall health of the photoreceptors. (d) Photoreceptor response to light stimuli in vivo is measured by electroretinography. A typical electroretinogram begins with the negative a-wave, reflecting direct response of photoreceptor cells. The amplitude of a-waves robustly increases with flash intensity in WT animals (brown curve). It is greatly diminished in RK−/− mice expressing WTarrestin1 (blue curve). However, substitution with phosphorylation-independent arrestin1-3A in the double knockout line improves light sensitivity in the absence of rhodopsin phosphorylation (green curve). (e) The recovery of the photoreceptors is measured in a double-flash protocol. A desensitising flash is delivered first, and the response to the second flash delivered at variable time intervals after the desensitising flash is compared with the control response to the flash of the same intensity delivered without the preceding desensitising flash. The time of half recovery (thalf) is the interval between the two flashes allowing for half-maximum recovery of the response. Replacement of WT arrestin1 with arrestin1-3A facilitates the recovery about threefold. However, the recovery rate remains significantly slower than in WT animals, indicating that there is a lot of room for further improvement. The complete set of data was published in Ref. 65: part c is reproduced from Ref. 65, with permission from Elsevier (© 2009 Elsevier Ltd); parts b, d and e are compiled from data in Ref. 65. Abbreviations: IS, inner segment; ONL, outer nuclear layer.
Rhodopsin kinase (systematic name GRK1) knockout mice (RK−/−) (Ref. 31) display multiple deficits in the rod photoreceptors associated with excessive rhodopsin signalling. These animals express wild-type rhodopsin along with the normal complement of wild-type arrestin1, which cannot quench the signalling in the absence of rhodopsin phosphorylation (Fig. 2). Rod photoreceptors in vertebrates have extremely high light sensitivity (Ref. 77). To maintain it, rods are capable of shutting off rhodopsin signalling with rapid subsecond kinetics (Refs 32, 65, 78), which requires the concerted action of rhodopsin kinase phosphorylating rhodopsin at multiple sites (Refs 32, 79, 80) and arrestin1, which binds light-activated phosphorhodopsin (P-Rh*) with high affinity (Refs 51, 76). Very slow dissociation of arrestin1 from P-Rh* (Refs 51, 80) ensures high fidelity of the shut-off and rapid recovery (Refs 32, 79), whereas low arrestin1 affinity for light-activated unphosphorylated rhodopsin (Rh*) and a consequent much faster off-rate (Refs 51, 80) prevents reliable shut-off in the absence of phosphorylation and slows down rod recovery by two orders of magnitude (Refs 65, 81) (Fig. 2f,g). Therefore, even dim light completely desensitises rods of RK−/− mice due to prolonged rhodopsin signalling, causing night blindness (Ref. 31) similar to that observed in human patients with the same genetic defect (Ref. 27). Moreover, rods in RK−/− mice degenerate within a few months (Ref. 31). The fact that excessive signalling is at the root of all problems in RK−/− mice was convincingly demonstrated by dark-rearing of these animals, which effectively prevented photoreceptor degeneration (Ref. 31). Thus, RK−/− mice represent a perfect model to test the compensational potential of phosphorylation-independent arrestin mutants.
Arrestins are elongated two-domain molecules, with the C-tail anchored to the body of the N-domain (Fig. 2a) (Refs 55, 56, 57, 58). Receptor binding induces the release of the C-tail (Refs 67, 68, 82). The deletion of the arrestin C-tail (Refs 49, 83) or its detachment by alanine substitutions of three anchoring hydrophobic residues (3A mutation) (Ref. 66) facilitates arrestin transition into a high-affinity receptor-binding state, thereby generating mutants with greatly enhanced affinity for unphosphorylated active receptor (Fig. 2b). The functional capability of enhanced arrestin1-3A was directly compared with the wild-type arrestin1 in live mice (Fig. 2). To this end, the amplitude of the light response, the recovery kinetics, and photoreceptor survival in RK−/− mouse lines that have the normal complement of wild-type arrestin1 (RK−/−Arr1+/+) or express only the enhanced 3A mutant (3A-50RK−/−Arr1−/−) (Refs 65, 84) were compared. In phosphorylation-deficient animals, enhanced arrestin 1 in creases the length of the outer segments, improves rodsurvival, and improves the ability of rods to respond to repeated light stimulation, facilitating photoresponse recovery about threefold (Ref. 65) (Fig. 2).
These results clearly indicate that this novel compensational approach to the therapy of gain-of-function genetic disorders is feasible. However, the engineered single-step shut-off of Rh* by enhanced arrestin1-3A yields a time of half-recovery of ~6 s – that is, significantly slower than in wild-type animals (~0.4 s) (Ref. 65). Thus, additional re-engineering of the arrestin1 molecule involving the receptor-binding surface is necessary to achieve better compensation. Nonetheless, the fact that this strategy works in rod photoreceptors, equipped with the most sensitive GPCR-driven signalling system with the fastest shut-off on record, suggests that it can fairly efficiently rein in excessive signalling by other GPCRs that normally desensitise much more slowly. The difficulty of testing this idea in other disease models in vivo lies in the relative promiscuity of the two nonvisual arrestins, which bind hundreds of different GPCR subtypes (Ref. 52).
Can we construct arrestin mutants with high receptor specificity?
Rod photoreceptors are arguably the most specialised cells, expressing millimolar concentrations of rhodopsin and its cognate arrestin1 (Refs 76, 85, 86), which makes targeted compensation of rhodopsin shut-off defects by phosphorylation-independent arrestin1 fairly straightforward. In contrast, most neurons and other cells express 5–25 different GPCRs (Refs 87, 88) along with arrestin2 and arrestin3 (Refs 15, 89, 90, 91). Many activating mutations of nonvisual arrestins have been described (Refs 61, 62, 64, 69, 74, 75, 92, 93, 94, 95, 96), but the usability of their phosphorylation-independent versions for therapeutic purposes could be questionable, because both interact with multiple receptors (Refs 69, 92, 97). Thus, while an enhanced version of arrestin2/3 would likely dampen the signalling by an overactive GPCR mutant, it will also blunt the signalling by all the normal GPCRs in the same cell, likely producing unwanted side effects. Therefore, to achieve full therapeutic potential, phosphorylation-independent nonvisual arrestins may need to be rendered specific for the particular receptor they are meant to target. However, practical use of phosphorylation-independent nonvisual arrestins does not have to be entirely contingent on successful narrowing of their receptor specificity. Considering the devastating consequences of many activating GPCR mutations (Ref. 20) and the lack of viable therapeutic alternatives, existing mutants with broad receptor specificity, although imperfect, will be in many cases better than nothing. Selective expression in affected cell types can be achieved by targeted delivery and cell-type-specific promoters, both of which are being actively developed (for representative reviews see Refs 98, 99, 100, 101, 102). This will limit off-target effects to the cells where the overactive mutant receptor produces the most damage, so that the overall impact of the expression of enhanced arrestin will likely be positive.
Evolution made arrestin1 quite specific for rhodopsin, with only marginal affinity for nonvisual GPCRs (Refs 58, 92, 97), demonstrating that arrestin protein specific for a single receptor subtype can be designed. Although several research groups using a variety of methods identified a fairly extensive receptor-binding surface on arrestins (Refs 67, 92, 94, 103, 104, 105), much of it engages non-subtype-specific elements, such as receptor-attached phosphates (Refs 53, 54, 55, 58, 59, 60, 94), with only a few elements playing a key role in defining receptor preference (Ref. 106). Arrestin1 shows particularly low binding to the M2 mAChR, whereas arrestin2 binds this receptor fairly well (Refs 69, 92, 107). Relative specificity for rhodopsin and M2 mAChR of a series of arrestin1–arrestin2 chimeras with different parts of the molecule swapped between the two subtypes identified key elements whose exchange completely reversed receptor preference of these arrestins (Ref. 106). They turned out to be surprisingly small, localised on the concave sides of the two arrestin domains (Fig. 3). As a result of fairly high homology within the arrestin family (Ref. 108), only about a dozen residues in these regions are different, with even fewer nonconservative substitutions. Since these residues must be primarily responsible for receptor specificity, judicious manipulation of these amino acids by mutagenesis appears to be a natural approach to the construction of arrestin proteins with narrowed receptor specificity. Theoretically, random mutagenesis of even five positions yields 205, or >3000 000, combinations – that is, orders of magnitude more than can be tested by any assay within a reasonable time. However, the analysis of known arrestin sequences (Ref. 108) shows that in ~600 million years of arrestin evolution there were only a few different residues in each of the key positions (Fig. 3). Mammalian arrestins show even less variability, which brings the number of combinations to be tested down to a manageable ~30–240. This approach to designing arrestins with significantly enhanced receptor specificity looks promising, although it needs to be tested experimentally. Although mammalian arrestin2 and arrestin3 are generally regarded as promiscuous, and many GPCRs apparently bind both with the same or very similar affinity (Refs 92, 97), certain receptors demonstrate clear preference for arrestin3 (Refs 109, 110), whereas others prefer arrestin2 (Refs 111, 112). Even when a GPCR binds either nonvisual arrestin, its interaction with arrestin2 and 3 subtypes results in distinct trafficking patterns (Ref. 113) and signalling outcomes (Ref. 114). The construction of receptor-specific mutants can begin by increasing rudimentary receptor specificity already built into the two nonvisual subtypes. Obviously, engineered arrestins targeting particular receptors should be constructed on the basis of the native subtype they preferentially bind.
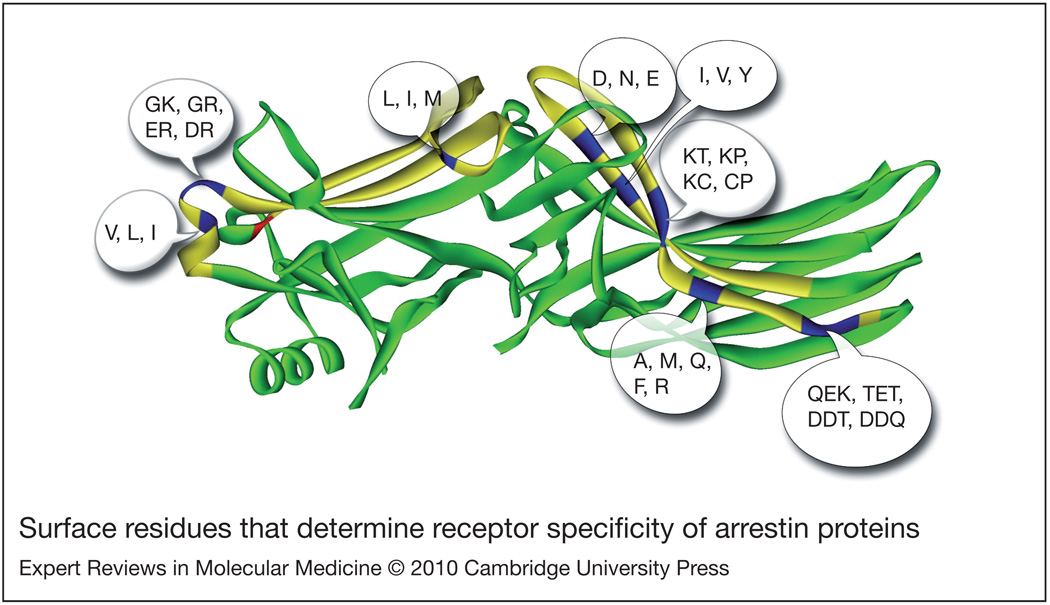
Figure 3. Surface residues that determine receptor specificity of arrestin proteins.
Swapping of two elements [shown in yellow on the crystal structure of arrestin2 (Ref. 56; PDB 1G4M)] localised on the concave sides of the two domains completely reverses receptor preference of arrestin1 and arrestin2 for rhodopsin and the M2 muscarinic acetylcholine receptor, respectively (Ref. 106). The positions of nonconservative substitutions in these elements are shown in blue, with alternative residues found in each position from Caenorhabditis elegans to mammals (Ref. 108) indicated in a single-letter code. The variability in the subset of mammalian arrestin proteins is significantly smaller. The position where visual arrestins with narrow receptor specificity carry a bulky hydrophobic residue that makes the N-domain more rigid is shown in red.
Manipulation of arrestin interactions with nonreceptor partners
In addition to GPCRs, arrestins bind dozens, if not hundreds, of surprisingly diverse proteins (Refs 13, 14). The view that only nonvisual arrestin2 and 3, and only in their receptor-bound conformation, engage these partners (Refs 13, 115) has been recently challenged by experimental evidence to the contrary (Refs 93, 95, 116, 117, 118, 119, 120). The emerging picture is that the three known conformational states of arrestins – free, receptor-bound, and microtubule-associated – play distinct roles in the regulation of cell signalling via direct interactions with numerous other proteins (Refs 13, 121). Just as modifications of receptor-binding elements generated arrestin mutants with unusual functional capabilities, re-engineering of parts mediating arrestin interactions with other partners is a viable strategy for construction of custom-designed arrestins with unique signalling properties that can be used for therapeutic purposes. However, the binding sites of very few non-GPCR partners have been elucidated with any degree of precision. Rare exceptions include clathrin (Refs 11, 122, 123), clathrin adaptor AP2 (Refs 12, 122), microtubules and unpolymerised tubulin αβ dimers (Refs 93, 118), cAMP phosphodiesterase PDE4D5 (Ref. 124), and calmodulin (Ref. 119).
Among arrestin binding partners, MAP kinases have attracted particular attention, as these proteins play a key role in life-or-death decisions in the cell. Arrestins facilitate the activation of ERK1/2 (MAPK3/MAPK1) (Ref. 125), which in simplistic terms are prosurvival and proproliferation kinases, as well as activation of JNK3 (MAPK10) (Ref. 126), which is largely antiproliferative and/or proapoptotic. Thus, judicious rechannelling of arrestin activity towards ERK1/2 would have therapeutic potential in neurodegenerative Alzheimer and Parkinson diseases and retinal degenerations, whereas shifting it towards JNK3 may counteract excessive proliferation in cancer. Originally, receptor-bound arrestin3 was shown to scaffold the ASK1–MKK4–JNK3 (MAP3K– MAP2K4–MAPK10) cascade (Ref. 126). Follow-up study by the same group found that in the absence of GPCR stimulation, arrestin3 promotes JNK3 activation in cells overexpressing ASK1 (Ref. 127), strongly implicating free arrestin3 in this process. Indeed, JNK3 was shown to bind free arrestin3 by three other research groups (Refs 95, 128, 129), and receptor-binding-deficient arrestin3 mutant ‘frozen’ in the basal conformation (Ref. 93) was found to promote JNK3 activation at least as well as wild-type arrestin3 (Ref. 117).
Two models of the organisation of a complex of arrestin and MAP kinases were proposed. The first, based on the original discoveries of arrestin-assisted signalling in ASK1–MKK4– JNK3 (Ref. 126) and c-Raf–MEK1–ERK1/2 (RAF1–MAP2K1–MAPK3/1) (Ref. 125) cascades, posits that upstream MAP3Ks (c-Raf or ASK1) bind to the N-domain and downstream MAPKs (ERK1/2 or JNK3) bind to the C-domain, whereas MAP2Ks (MEK1 or MKK4) do not bind arrestins directly, but interact with the complex via cognate MAP3K and MAPK (Refs 14, 130) (Fig. 4a). Recently, both MAP2Ks – MKK4 (Ref. 117) and MEK1 (Refs 117, 120) – were shown to bind arrestins directly. The two arrestin domains are independent folding units that can be expressed separately and retain certain functions (Refs 49, 51, 53, 92, 93). The demonstration that individual N- and C-domains bind JNK3, its upstream kinases ASK1 and MKK4, as well as ERK2, MEK1 and c-Raf, suggested a very different arrangement of the MAPK modules on the arrestin scaffold (Ref. 117). Recently, ASK1 interaction with sites in both arrestin domains were independently shown using peptide arrays (Ref. 131). The interaction of each kinase with both domains suggested an alternative model, where kinases are arranged side-by-side with their long axes parallel to the long axis of the arrestin, like three hotdogs on a single bun (Fig. 4b). Although several arrestin elements were recently implicated in the binding of MEK1 (Ref. 120) and components of the ASK1–MKK4– JNK3 cascade (Ref. 131), precise footprints of each kinase on arrestin remain to be elucidated.
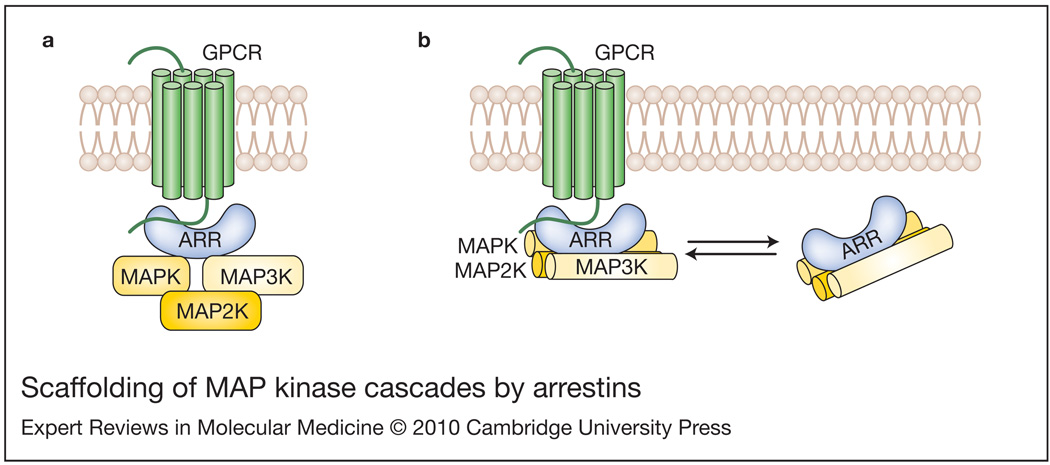
Figure 4. Scaffolding of MAP kinase cascades by arrestins.
(a) The original model of mitogen-activated protein kinase (MAPK) scaffolding by receptor-bound arrestin (Refs 125, 126, 127). The model posits that MAPK (ERK1/2 or JNK3) and MAPK kinase kinase (MAP3K; c-Raf-1 or ASK1) directly bind arrestin, whereas MAPK kinase (MAP2K; MEK1 or MKK4) is brought to the complex via interactions with MAPK and MAP3K. (b) An alternative model posits that all three kinases in the cascade directly interact with arrestins, and each kinase binds both arrestin domains (Ref. 117). Based on the ability of receptor-binding-deficient arrestin3 to facilitate ASK1-dependent JNK3 activation (Ref. 117), this model shows that both receptor-bound and free arrestins can scaffold MAPK cascades.
The same is true for the majority of arrestin binding partners, although in many cases we have some clues. First, any protein that binds the receptor–arrestin complex must interact with the elements that are not engaged by the receptor. Indeed, when the binding site of a protein on arrestin overlaps with the receptor-interaction surface, as was shown for microtubules (Refs 93, 118) and calmodulin (Ref. 119), as a result of higher affinity the receptor invariably outcompetes the other protein (Refs 84, 119). As the receptor-binding surface of arrestins is fairly well established, this narrows the field down to the sides of the molecule and the convex surfaces of the two domains (Refs 9, 121). Second, upon GPCR binding, arrestins undergo a significant conformational rearrangement (Refs 51, 96, 132, 133, 134), which involves the release of the C-tail and a movement of the two domains relative to each other (Refs 67, 68, 135). Recent comparison of wild-type, conformationally loose 3A mutants mimicking the receptor-bound state, and receptor-binding-deficient forms frozen in the basal state showed that arrestins in different conformations bind JNK3 with similar affinity (Refs 95, 116). The partners that show similar affinity for free and receptor-bound arrestin likely engage the elements equally exposed in both states. In contrast, proteins that preferentially bind receptor-associated arrestin [e.g. clathrin and AP2 (Refs 11, 122, 133), ubiquitin ligase AIP4 (ITCH) (Ref. 136)] must engage elements that become more exposed upon arrestin activation, whereas partners that prefer arrestin in the basal state [e.g. ubiquitin ligase MDM2 (Refs 95, 116)] either bind elements less exposed in the active conformation or require the exact spacing between the binding sites in the two domains that is only achieved in the basal conformation. Comparative studies of different arrestin subtypes provide additional information. Studies using arrestin2/3 shuttling between the nucleus and cytoplasm (Refs 95, 128, 129), as well as peptide arrays (Ref. 131), showed that arrestins 2 and 3 bind JNK3 equally well. Visual arrestins 1 (rod arrestin) and 4 (cone arrestin) were also shown to bind JNK3 (Refs 95, 116). Obviously, partners that bind all four vertebrate arrestins likely engage conserved residues, whereas arrestin-subtype-specific functions [e.g. JNK3 activation by arrestin3 (Refs 117, 126, 127)] must be mediated by residues absent in other arrestins. These considerations reduce the number of arrestin residues to be tested for their role in binding each partner, but there is still a lot of work to be done.
Although the task of creating arrestin mutants in which individual functions are disrupted or enhanced by targeted mutagenesis appears daunting, quite a few successes in this direction have already been achieved. Many of the relevant mutants were tested in cultured cells, and some even in live animals.
Components of the internalisation machinery: clathrin and AP2
In the process of identification of compact clathrin-and AP2-binding sites in the arrestin C-tail, several mutants were created with one or both of these sites disrupted (Refs 11, 12, 122). The expression of these mutants, which bind receptors normally but cannot recruit the receptor–arrestin complex to the coated pit, considerably reduced GPCR internalisation (Refs 11, 12, 122). Interestingly, expression of mutant arrestins with a disrupted phosphoinositide-binding site (Ref. 137) and the expression of the arrestin C-tail that carries clathrin- and AP2-interaction sites but does not bind receptors (Ref. 138) produces similar effects via different mechanisms: the former weakens the arrestin–receptor interaction, and the latter occupies arrestin-binding elements on clathrin and AP2 and outcompetes the arrestin–receptor complexes.
cAMP-specific phosphodiesterase PDE4D
Arrestins recruit two isoforms of cAMP-specific PDE4D to the activated β2-adrenergic receptor, thereby facilitating cAMP breakdown and reducing protein kinase A activity in the vicinity of the membrane (Ref. 139). Arrestin3 mutants in which the binding to PDE4D was disrupted by targeted mutagenesis bind the receptor normally, but do not recruit PDE4D and therefore, in contrast to wild-type arrestin3, do not affect PKA phosphorylation of the receptor (Ref. 124). Moreover, a recent in vivo study showed that arrestin3–PDE4D interaction in the amygdala is important for fear conditioning (Ref. 140). The authors found that the expression of a PDE4D–binding-deficient mutant does not rescue fear conditioning in arrestin3-knockout mice, whereas the expression of wild-type arrestin3 does (Ref. 140).
MEK1 and other MAP kinases
Arrestin2 is constitutively phosphorylated at the C-terminus by ERK2, and this phosphorylation reduces its affinity for clathrin, so that receptor internalisation requires dephosphorylation of arrestin2 (Ref. 141). An arrestin2 mutant deficient in MEK1 binding was recently constructed on the basis of identification of two key residues required for arrestin–MEK1 interaction (Ref. 120). In cells, this mutant is not phosphorylated by ERK2, demonstrating that arrestin2 is phosphorylated by ERK2 scaffolded and activated on it. As could be expected, the peptide disrupting arrestin2 association with MEK1 blocked receptor-activation-dependent ERK2 activation in cells, while promoting arrestin-dependent receptor internalisation (Ref. 120). Following recent identification of arrestin elements mediating the interactions with ASK1, MKK4 and JNK3 (Ref. 131), corresponding binding-deficient mutants need to be constructed and functionally tested.
Ubiquitination sites on arrestins
Activation of the β2-adrenergic receptor and arrestin binding were shown to induce the ubiquitination of both proteins, which plays distinct roles in receptor trafficking (Ref. 142). Arrestins and the receptor are ubiquitinated by two different E3 ligases – MDM2 (Ref. 142) and NEDD4 (Ref. 143), respectively – both of which are recruited to the complex via their interactions with arrestins (Refs 142, 143). Subsequent studies showed that MDM2 interacts with free arrestins (Ref. 129), and preferentially binds arrestins in their basal conformation (Refs 95, 116), suggesting that the receptor recruits a preformed arrestin–MDM2 complex; MDM2 then has a short time window to ubiquitinate receptor-bound arrestin, which assumes the active conformation, before MDM2 is released due to reduced affinity for this form (Ref. 95). In contrast, the activation of the chemokine receptor CXCR4 promotes arrestin2 interaction with E3 ubiquitin ligase AIP4 (Ref. 136). In this case, depletion of arrestin2 by small interfering RNA (siRNA) does not affect CXCR4 internalisation and initial ubiquitination, but prevents its sorting from endosomes to lysosomes and subsequent degradation (Ref. 136). Arrestins also recruit deubiquitinating enzymes USP33 and USP20, important for recycling and resensitisation to the β2-adrenergic receptor (Ref. 144).
Interestingly, particular lysines that are ubiquitinated in the same arrestin3 depend on the receptor it binds to: interaction with the angiotensin II type 1a receptor (AGTR1) promotes the modification of lysines 11 and 12, whereas binding to the vasopressin V2 receptor (AVPR2) directs ubiquitination to lysines 18, 107, 108, 207 and 296 (Ref. 145). Intriguingly, the elimination of individual sets of two (11 + 12) or five (18 + 107 + 108 + 207 + 296) lysines (by conservative Lys to Arg substitutions) selectively reduces arrestin3 affinity for the angiotensin or vasopressin receptor, respectively, and prevents arrestin-dependent ERK activation only by that receptor (Ref. 145). Thus, the two mutants described in this study selectively lost the ability to promote internalisation and postendocytic signalling by one specific GPCR, but not by another (Ref. 145). The effects of the same quintuple Lys to Arg arrestin3 mutant on M1 and M2 mAChRs were also receptor-specific: the mutation reduced stable arrestin–M2 association and M2 downregulation, while increasing downregulation of M1 (Ref. 146). Interestingly, the rate of M1 or M2 internalisation was not affected (Ref. 146). However, one should keep in mind that the M2 receptor belongs to the group of GPCRs that avidly bind and require arrestins for desensitisation, but normally internalise via arrestin-independent pathway(s) (Refs 69, 92, 107, 147, 148). An arrestin3 mutant where all 31 lysines that could be ubiquitinated were replaced with arginines was found to interact with AP2 and components of the ERK cascade normally, but displayed reduced affinity for the β2-adrenergic receptor and clathrin (Ref. 149). This mutant failed to enhance receptor internalisation and formation of stable signalosomes that retain activated ERK, whereas arrestin3–ubiquitin fusion showed the opposite phenotype (Ref. 149). Thus, manipulation of the ubiquitination sites on nonvisual arrestins changes their functional characteristics in a receptor-specific manner.
Interactions mediating arrestin oligomerisation
Arrestin1 self-associates, cooperatively forming dimers and tetramers (Refs 150, 151, 152). Interestingly, although arrestin1 invariably crystallises as a tetramer (Refs 55, 153), the biologically relevant solution tetramer was shown to be different from that observed in the crystal (Ref. 152), and considerable effort was required to elucidate its shape and the interaction interfaces between protomers (Ref. 154). Based on this information, two self-association-deficient arrestin1 mutants were constructed (Ref. 154), both of which retained normal ability to bind rhodopsin and microtubules. The receptor-binding surface of arrestin1 monomers within the tetramer and both possible dimers are shielded by sister subunits (Ref. 154), so it is not surprising that only monomeric arrestin1 can bind rhodopsin (Ref. 152). These data suggest that oligomers are a storage form, but it is unclear why arrestin1 robustly self-associates, whereas cone-specific arrestin4 does not (Ref. 155). The exact role of oligomerisation in the biology of arrestin1 remains to be elucidated, although the molecular tools necessary to establish it are ready (Ref. 154).
Arrestin2 forms dimers of different configuration in several crystal forms (Refs 56, 57). In some cases, the dimerisation interface appears to be significantly strengthened by inositol hexakisphosphate (InsP6) diffused into preformed crystals (Ref. 156). The ability of InsP6 to promote arrestin2 dimerisation was confirmed in solution (Refs 155, 156). In contrast, InsP6 reduces self-association of arrestin1 (Ref. 155), which could be interpreted as an indication that the shapes of arrestin1 and arrestin2 oligomers are different. Homo- and hetero-oligomers of both nonvisual arrestins were observed in overexpressing cells, suggesting that this process occurs in vivo (Ref. 157). Arrestin3 has a nuclear export signal (NES) in its C-terminus, whereas arrestin2 does not, so that arrestin3 is predominantly cytoplasmic, while arrestin2 is also present in the nucleus (Refs 95, 128, 129). Coexpression of arrestin3 suppresses nuclear localisation of arrestin2, suggesting that hetero-oligomerisation regulates subcellular localisation of arrestins (Ref. 157). Indeed, targeted disruption of arrestin2/3 oligomerisation increased nuclear localisation of arrestin2 without affecting its interactions with clathrin, AP2 or ERK2 (Ref. 156). Considering that arrestin2 plays a role in regulation of gene expression (Ref. 158), this may have important biological implications. Subsequent studies showed that disruption of arrestin3 oligomerisation by mutagenesis reduced its interactions with MDM2 and inhibited p53-dependent antiproliferative effects of arrestin3 (Ref. 159), as well as arrestin3 interaction with ERK1/2 (Ref. 160), identifying additional functions of arrestin3 oligomers. These clues suggest several possible biological functions of arrestin2/3 oligomerisation, although many mechanistic questions remain unanswered (see Ref. 161 for an excellent review).
Thus, a number of arrestin mutants with selectively disrupted individual functions have already been created and successfully used as research tools. Although many more kinds of designer arrestins are needed to explore and manipulate other facets of their function, some of the existing ones might well have therapeutic potential that needs to be explored.
Conclusions and future directions
Traditionally, medicine largely relies on small molecules that act as long as they are in the body. The main advantage of conventional drugs is the ease of delivery and straightforward ways of structure optimisation for maximum effect and acceptable pharmacokinetics. As far as killing invading bacteria or viruses with distinct biochemistry goes, where strong constant action is an advantage, small molecules are perfect therapeutic tools. However, problems arise when we need to suppress (or boost) the activity of endogenous enzymes and receptors. The key drawback is that enzyme inhibitors and receptor blockers affect a single process in the extremely complex network regardless of the functional state of the system. This single-mindedness of conventional therapies becomes a problem in many disorders, where hitting a single target too hard often results in drug-induced imbalances as bad as the disease itself. That is why therapeutic approaches are shifting towards types of drugs that act in a context-dependent manner: partial agonists, which increase or decrease signalling when the level of the endogenous agonist is low or high, respectively, and allosteric modulators, the effect of which is contingent on the presence of endogenous agonists (reviewed in Refs 162, 163). Proteins are the ultimate context-dependent signalling machines perfected over millions of years of rigorous evolutionary experimentation. Each function of every protein is controlled by sophisticated regulatory mechanisms that are already in place in the body and will likely prevent introduced protein from overdoing whatever we made it to do.
Mechanistic understanding of the inner workings of a protein paves the way to subtle manipulation of its individual functional characteristics for therapeutic purposes without destroying its biologically advantageous complexity and regulation. Recent findings show that re-engineering of arrestin proteins to create optimised versions with therapeutic potential is feasible (Ref. 65). Although this approach is still in its infancy, the ability of phosphorylation-independent arrestins to compensate for defects in receptor phosphorylation, as well as construction of other arrestin mutants with changed individual functional modalities, is very encouraging.
Potential benefits from the elucidation of molecular details of arrestin interactions with MAP kinases, ubiquitin ligases, and other signalling proteins are great enough to make this work worth doing: it will enable the engineering of mutants that channel the signalling to the pathways of choice. Arrestins designed to facilitate prosurvival signalling via ERK, MDM2 and/or other interaction partners could help the neurons affected by degenerative disorders to survive. By the same token, mutant arrestins channeling signalling to proapoptotic or antiproliferative pathways could be of interest in cancer, where rapid proliferation is the key problem. It is important to keep in mind that to achieve a therapeutic effect it is not necessary to kill malignant cells. Numerous precancerous cells emerge in the body every day, and our immune system disposes of them quite successfully. Only the ones that proliferate so fast that they outrun the immune response create problems. Thus, simply slowing down proliferation would likely make these cells vulnerable enough for the immune system to eliminate them.
Structurally, arrestins are inviting targets for creative protein engineering. One side of the molecule, where the concave surfaces of the two domains are localised, is responsible for receptor and microtubule binding. The opposite side carries the interaction sites for the majority of nonreceptor binding partners, which do not overlap with the receptor footprint. The third element, the detachable arrestin C-tail, carries the sites for the key components of the internalisation machinery of the coated pit: clathrin and AP2. Each of these elements can be manipulated without affecting the other two. Thus, necessary structural information will enable us to construct arrestins that connect the receptor of interest to the signalling pathway of our choosing. Although targeted re-engineering of all arrestin functions is still years away, several arrestin mutants with special functional characteristics have already been constructed and tested. Phosphorylation-independent mutants with high affinity for active unphosphorylated receptors were shown to normalise signalling in the absence of GPCR phosphorylation in vitro (Ref. 63), in model systems such as Xenopus oocytes (Refs 61, 62) or HEK293 cells (Ref. 64), in more challenging primary neuronal culture (Ref. 75), and even in rod photoreceptors of living mice (Ref. 65). We can also make arrestins that do not bind GPCRs very well but have increased affinity for microtubules instead (Ref. 93), which will be useful to direct specific signalling proteins to the cytoskeleton. The possibilities of further engineering are limited only by the available structural information.
Arrestins represent just four out of >20 000 proteins we have, and their versatility is unlikely to be unique. Virtually every protein consists of several structural elements, has multiple functions, and interacts with many binding partners. Thus, most proteins are amenable to subtle functional adjustments by targeted mutagenesis, as described here for arrestins. The difficulty of this approach is that a lot of precise structural and functional information about the protein of interest is necessary to make it feasible. In contrast, small molecules producing desired effects can often be identified by high-throughput screening of huge chemical libraries without the least knowledge of any molecular mechanisms. One only needs to know the desired target (e.g. a particular GPCR subtype or enzyme isoform) and proteins that should not be affected (e.g. highly homologous other GPCRs, or closely related other enzymes) to make a potential drug highly specific. After that, many well-established methods of therapeutic delivery of small molecules can be tested and the best one found. However, the very specificity so much sought after usually underlies the downside of small molecules: their single-mindedness makes it too easy to overdo whatever they are meant to do, causing side effects.
Intracellular signalling proteins, native or engineered, can be delivered only by gene transfer, which means that necessary methods are much more complicated and still need further development. This is well worth the effort: in contrast to commonly used single-purpose small molecules, which cannot be regulated in any way except via patient intake, proteins are sophisticated multifunctional machines, with their lifespan and every aspect of the function subject to complex regulation in the cell. Subtle changes in signalling pathways by an increase in expression of selected protein or by introducing its functionally modified version will remain under control of normal cellular mechanisms, minimising the chances of excesses. This approach will likely be developed first for congenital disorders, primarily those with molecular errors caused by gain-of-function mutations. However, when technical hurdles of targeting gene transfer to a particular tissue or cell type are overcome, one can envision many potential uses of these sophisticated molecular tools in a variety of disorders where normal cellular functions get out of hand. The most likely targets include diseases requiring adjustment of cell signalling, such as neurodegenerative disorders (caused by runaway neuronal death), cancer (runaway proliferation), and allergies and autoimmune disorders (runaway immune response).
Acknowledgments
Acknowledgements and funding
This work was supported in part by NIH grants EY011500, GM077561, GM081756 (V.V.G), and NS065868 (E.V.G). The authors thank Dr Robert J. Lefkowitz for critical reading of the manuscript, as well as the reviewers for constructive suggestions.
References
- 1.Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO Journal. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rompler H, et al. G protein-coupled time travel: evolutionary aspects of GPCR research. Molecular Interventions. 2007;7:17–25. doi: 10.1124/mi.7.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Fredriksson R, et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Molecular Pharmacology. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann KP, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends in Biochemical Sciences. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Hubbell WL, et al. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Advances in Protein Chemistry. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 7.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 8.Burns ME, Pugh ENJ. RGS9 concentration matters in rod phototransduction. Biophysical Journal. 2009;97:1538–1547. doi: 10.1016/j.bpj.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G protein-coupled receptors. Pharmacology & Therapeutics. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. This comprehensive review describes known molecular mechanisms of different aspects of arrestin functions. It focuses on arrestin interactions with GPCRs and other signalling proteins, highlighting mechanistic questions that remain unanswered.
- 10.Krupnick JG, Gurevich VV, Benovic JL. Mechanism of quenching of phototransduction: binding competition between arrestin and transducin for phosphorhodopsin. Journal of Biological Chemistry. 1997;272:18125–18131. doi: 10.1074/jbc.272.29.18125. [DOI] [PubMed] [Google Scholar]
- 11.Goodman OB, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 12.Laporte SA, et al. The interaction of beta-arrestin with the AP-2 adaptor is required for the clustering of beta 2-adrenergic receptor into clathrin-coated pits. Journal of Biological Chemistry. 2000;275:23120–23126. doi: 10.1074/jbc.M002581200. [DOI] [PubMed] [Google Scholar]
- 13. DeWire SM, et al. Beta-arrestins and cell signaling. Annual Review of Physiology. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. This comprehensive review describes different aspects of the recently discovered role of nonvisual arrestins in cell signalling, with particular emphasis on the ability of arrestins to bring elements of specific signaling pathways into close proximity and to recruit different proteins to activated phosphorylated GPCRs.
- 14.Kendall RT, Luttrell LM. Diversity in arrestin function. Cellular and Molecular Life Sciences. 2009;66:2953–2973. doi: 10.1007/s00018-009-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurevich VV, Gurevich EV. Rich tapestry of G protein-coupled receptor signaling and regulatory mechanisms. Molecular Pharmacology. 2008;74:312–316. doi: 10.1124/mol.108.049015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tobin AB, Butcher AJ, Kong KC. Location, location, location…site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends in Pharmacological Sciences. 2008;29:413–420. doi: 10.1016/j.tips.2008.05.006. This review describes GPCR phosphorylation as a dynamic regulatory process that occurs in a tissue-specific manner. The authors show how receptor phosphorylation offers a mechanism of regulating the outcome of GPCR signalling that can be tailored to achieve a specific physiological function.
- 17.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends in Pharmacological Sciences. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Kenakin T. Functional selectivity through protean and biased agonism: who steers the ship? Molecular Pharmacology. 2007;72:1393–1401. doi: 10.1124/mol.107.040352. [DOI] [PubMed] [Google Scholar]
- 19.Drake MT, et al. beta-arrestin-biased agonism at the beta2-adrenergic receptor. Journal of Biological Chemistry. 2008;283:5669–5676. doi: 10.1074/jbc.M708118200. [DOI] [PubMed] [Google Scholar]
- 20.Scho¨neberg T, et al. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacology & Therapeutics. 2004;104:173–206. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Barak LS, et al. Constitutive arrestin-mediated desensitization of a human vasopressin receptor mutant associated with nephrogenic diabetes insipidus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:93–98. doi: 10.1073/pnas.011303698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rim J, Oprian DD. Constitutive activation of opsin: interaction of mutants with rhodopsin kinase and arrestin. Biochemistry. 1995;34:11938–11945. doi: 10.1021/bi00037a035. [DOI] [PubMed] [Google Scholar]
- 23.Rao VR, Cohen GB, Oprian DD. Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature. 1994;367:639–642. doi: 10.1038/367639a0. [DOI] [PubMed] [Google Scholar]
- 24.Apfelstedt-Sylla E, et al. Ocular findings in a family with autosomal dominant retinitis pigmentosa and a frameshift mutation altering the carboxyl terminal sequence of rhodopsin. British Journal of Ophthalmology. 1993;77:495–501. doi: 10.1136/bjo.77.8.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim RY, et al. Dominant retinitis pigmentosa associated with two rhodopsin gene mutations. Leu-40-Arg and an insertion disrupting the 5′-splice junction of exon 5. Archives of Ophthalmology. 1993;111:1518–1524. doi: 10.1001/archopht.1993.01090110084030. [DOI] [PubMed] [Google Scholar]
- 26.Restagno G, et al. A large deletion at the 3′ end of the rhodopsin gene in an Italian family with a diffuse form of autosomal dominant retinitis pigmentosa. Human Molecular Genetics. 1993;2:207–208. doi: 10.1093/hmg/2.2.207. [DOI] [PubMed] [Google Scholar]
- 27.Cideciyan AV, et al. Null mutation in the rhodopsin kinase gene slows recovery kinetics of rod and cone phototransduction in man. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:328–333. doi: 10.1073/pnas.95.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs S, et al. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nature Genetics. 1995;10:360–362. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, et al. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–509. doi: 10.1038/39068. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, et al. Mechanisms of rhodopsin inactivation in vivo as revealed by a COOsteopathic Hospitals-terminal truncation mutant. Science. 1995;267:374–377. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- 31.Chen CK, et al. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez A, et al. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 33.Allen LF, et al. G-protein-coupled receptor genes as protooncogenes: constitutively activating mutation of the alpha 1B-adrenergic receptor enhances mitogenesis and tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:11354–11358. doi: 10.1073/pnas.88.24.11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutkind JS, et al. Muscarinic acetylcholine receptor subtypes as agonist-dependent oncogenes. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:4703–4707. doi: 10.1073/pnas.88.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julius D, et al. Ectopic expression of the serotonin 1c receptor and the triggering of malignant transformation. Science. 1989;244:1057–1062. doi: 10.1126/science.2727693. [DOI] [PubMed] [Google Scholar]
- 36.Young D, et al. Isolation and characterization of a new cellular oncogene encoding a protein with multiple potential transmembrane domains. Cell. 1986;45:711–719. doi: 10.1016/0092-8674(86)90785-3. [DOI] [PubMed] [Google Scholar]
- 37.Young D, et al. Characterization of the rat mas oncogene and its high-level expression in the hippocampus and cerebral cortex of rat brain. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:5339–5342. doi: 10.1073/pnas.85.14.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson TR, et al. The mas oncogene encodes an angiotensin receptor. Nature. 1988;335:437–440. doi: 10.1038/335437a0. [DOI] [PubMed] [Google Scholar]
- 39.Guigoni C, et al. Pathogenesis of levodopa-induced dyskinesia: focus on D1 and D3 dopamine receptors. Parkinsonism & Related Disorders. 2005;11:S25–S29. doi: 10.1016/j.parkreldis.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Letters. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 41.Pfister C, et al. Retinal Santigen identified as the 48K protein regulating light-dependent phosphodiesterase in rods. Science. 1985;228:891–893. doi: 10.1126/science.2988124. [DOI] [PubMed] [Google Scholar]
- 42.Lohse MJ, et al. beta-Arrestin:a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 43.Attramadal H, et al. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. Journal of Biological Chemistry. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 44.Sterne-Marr R, et al. Polypeptide variants of beta-arrestin and arrestin3. Journal of Biological Chemistry. 1993;268:15640–15648. [PubMed] [Google Scholar]
- 45.Craft CM, Whitmore DH, Wiechmann AF. Cone arrestin identified by targeting expression of a functional family. Journal of Biological Chemistry. 1994;269:4613–4619. [PubMed] [Google Scholar]
- 46.Murakami A, et al. X-arrestin: a new retinal arrestin mapping to the X chromosome. FEBS Letters. 1993;334:203–209. doi: 10.1016/0014-5793(93)81712-9. [DOI] [PubMed] [Google Scholar]
- 47.Lohse MJ, et al. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. Journal of Biological Chemistry. 1992;267:8558–8564. [PubMed] [Google Scholar]
- 48.Carman CV, Benovic JL. G-protein-coupled receptors: turn-ons and turn-offs. Current Opinion in Neurobiology. 1998;8:335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 49.Gurevich VV, Benovic JL. Cell-free expression of visual arrestin. Truncation mutagenesis identifies multiple domains involved in rhodopsin interaction. Journal of Biological Chemistry. 1992;267:21919–21923. [PubMed] [Google Scholar]
- 50.Li J, et al. Structure of bovine rhodopsin in a trigonal crystal form. Journal of Molecular Biology. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 51.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin: Sequential multisite binding ensures strict selectivity towards light-activated phosphorylated rhodopsin. Journal of Biological Chemistry. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 52.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends in Pharmacological Sciences. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin: diverse functional roles of positively charged residues within the phosphorylation-recignition region of arrestin. Journal of Biological Chemistry. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 54.Gurevich VV, Benovic JL. Mechanism of phosphorylation-recognition by visual arrestin and the transition of arrestin into a high affinity binding state. Molecular Pharmacology. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 55.Hirsch JA, et al. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 56.Han M, et al. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 57.Milano SK, et al. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- 58.Sutton RB, et al. Crystal structure of cone arrestin at 2.3Å: evolution of receptor specificity. Journal of Molecular Biology. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 59.Vishnivetskiy SA, et al. How does arrestin respond to the phosphorylated state of rhodopsin? Journal of Biological Chemistry. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 60.Vishnivetskiy SA, et al. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. Journal of Biological Chemistry. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 61.Kovoor A, et al. Targeted construction of phosphorylation-independent b-arrestin mutants with constitutive activity in cells. Journal of Biological Chemistry. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 62.Celver J, et al. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. Journal of Biological Chemistry. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 63.Gray-Keller MP, et al. Arrestin with a single amino acid sustitution quenches light-activated rhodopsin in a phosphorylation-independent fashion. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- 64.Pan L, Gurevich EV, Gurevich VV. The nature of the arrestin x receptor complex determines the ultimate fate of the internalized receptor. Journal of Biological Chemistry. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- 65. Song X, et al. Enhanced arrestin facilitates recovery and protects rod photoreceptors deficient in rhodopsin phosphorylation. Current Biology. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. This is the first study where specifically redesigned arrestin protein was used in living animals to compensate for a genetic defect. Using the visual system as a model, this work demonstrates both the potential of the compensational approach to gene therapy and the difficulties that must be overcome to achieve this goal.
- 66.Gurevich VV. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. Journal of Biological Chemistry. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 67.Hanson SM, et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vishnivetskiy SA, et al. The role of arrestin alpha-helix I in receptor binding. Journal of Molecular Biology. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gurevich VV, et al. Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. Journal of Biological Chemistry. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- 70.Krasel C, et al. Beta-arrestin binding to the beta2-adrenergic receptor requires both receptor phosphorylation and receptor activation. Journal of Biological Chemistry. 2005;280:9528–9535. doi: 10.1074/jbc.M413078200. [DOI] [PubMed] [Google Scholar]
- 71.Moore CA, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annual Review of Physiology. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- 72.Gurevich VV, et al. Visual arrestin binding to rhodopsin: Intramolecular interaction between the basic N-terminus and acidic C-terminus of arrestin may regulate binding selectivity. Journal of Biological Chemistry. 1994;269:8721–8727. [PubMed] [Google Scholar]
- 73.Puig J, et al. Synthetic phosphopeptide from rhodopsin sequence induces retinal arrestin binding to photoactivated unphosphorylated rhodopsin. FEBS Letters. 1995;362:185–188. doi: 10.1016/0014-5793(95)00225-x. [DOI] [PubMed] [Google Scholar]
- 74.Celver J, et al. Threonine 180 is requred for G protein-coupled receptor kinase 3 and b-arrestin mediated desensitization of the m-opioid receptor in Xenopus oocytes. Journal of Biological Chemistry. 2001;276:4894–4900. doi: 10.1074/jbc.M007437200. [DOI] [PubMed] [Google Scholar]
- 75.Macey TA, Lowe JD, Chavkin C. Mu opioid receptor activation of ERK1/2 is GRK3 and arrestin dependent in striatal neurons. Journal of Biological Chemistry. 2006;281:34515–34524. doi: 10.1074/jbc.M604278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gurevich VV, et al. How rod arrestin achieved perfection: regulation of its availability and binding selectivity. In: Fliesler SJ, Kisselev O, editors. Signal Transduction in the Retina. Boca Raton, FL, USA: CRC Press; 2007. pp. 55–88. [Google Scholar]
- 77.Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. Journal of Physiology. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- 78.Krispel CM, et al. RGSexpression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 79.Doan T, et al. Multiple phosphorylation sites confer reproducibility of the rod’s single-photon responses. Science. 2006;313:530–533. doi: 10.1126/science.1126612. [DOI] [PubMed] [Google Scholar]
- 80.Vishnivetskiy SA, et al. Regulation of arrestin binding by rhodopsin phosphorylation level. Journal of Biological Chemistry. 2007;282:32075–32083. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burns ME, et al. Deactivation of phosphorylated and nonphosphorylated rhodopsin by arrestin splice variants. Journal of Neuroscience. 2006;26:1036–1044. doi: 10.1523/JNEUROSCI.3301-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Palczewski K, et al. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. Journal of Biological Chemistry. 1991;266:18649–18654. [PubMed] [Google Scholar]
- 83.Pulvermuller A, et al. Functional differences in the interaction of arrestin and its splice variant, p44, with rhodopsin. Biochemistry. 1997;36:9253–9260. doi: 10.1021/bi970772g. [DOI] [PubMed] [Google Scholar]
- 84.Nair KS, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanson SM, et al. Eachrhodopsin molecule binds its own arrestin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strissel KJ, et al. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. Journal of Neuroscience. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Penn RB, et al. Arrestin specificity for G protein-coupled receptors in human airway smooth muscle. Journal of Biological Chemistry. 2001;276:32648–32656. doi: 10.1074/jbc.M104143200. [DOI] [PubMed] [Google Scholar]
- 88.Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends in Neurosciences. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gurevich EV, Benovic JL, Gurevich VV. Arrestin2 and arrestin3 are differentially expressed in the rat brain during postnatal development. Neuroscience. 2002;109:421–436. doi: 10.1016/s0306-4522(01)00511-5. [DOI] [PubMed] [Google Scholar]
- 90.Gurevich EV, Benovic JL, Gurevich VV. Arrestin2 expression selectively increases during neural differentiation. Journal of Neurochemistry. 2004;91:1404–1416. doi: 10.1111/j.1471-4159.2004.02830.x. [DOI] [PubMed] [Google Scholar]
- 91.Luo J, Busillo JM, Benovic JL. M3 muscarinic acetylcholine receptor-mediated signaling is regulated by distinct mechanisms. Molecular Pharmacology. 2008;74:338–347. doi: 10.1124/mol.107.044750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gurevich VV, et al. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. Journal of Biological Chemistry. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 93.Hanson SM, et al. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. Journal of Molecular Biology. 2007;368:375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. Journal of Biological Chemistry. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song X, et al. Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. Journal of Biological Chemistry. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carter JM, et al. Conformational differences between arrestin2 and pre-activated mutants as revealed by hydrogen exchange mass spectrometry. Journal of Molecular Biology. 2005;351:865–878. doi: 10.1016/j.jmb.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 97.Barak LS, et al. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. Journal of Biological Chemistry. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 98.Miller AM, Dean DA. Tissue-specific and transcription factor-mediated nuclear entry of DNA. Advanced Drug Delivery Reviews. 2009;61:603–613. doi: 10.1016/j.addr.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 99.Beck C, et al. Tissue-specific targeting for cardiovascular gene transfer. Potential vectors and future challenges. Current Gene Therapy. 2004;4:457–467. doi: 10.2174/1566523043346138. [DOI] [PubMed] [Google Scholar]
- 100.Sadeghi H, Hitt MM. Transcriptionally targeted adenovirus vectors. Current Gene Therapy. 2005;5:411–427. doi: 10.2174/1566523054546189. [DOI] [PubMed] [Google Scholar]
- 101.Lundberg C, et al. Applications of lentiviral vectors for biology and gene therapy of neurological disorders. Current Gene Therapy. 2008;8:461–473. doi: 10.2174/156652308786847996. [DOI] [PubMed] [Google Scholar]
- 102.Baum C, et al. Retrovirus vectors: toward the plentivirus? Molecular Therapy. 2006;13:1050–1063. doi: 10.1016/j.ymthe.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 103.Ohguro H, et al. Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Science. 1994:2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pulvermuller A, et al. Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. Journal of Biological Chemistry. 2000;275:37679–37685. doi: 10.1074/jbc.M006776200. [DOI] [PubMed] [Google Scholar]
- 105.Dinculescu A, et al. Insertional mutagenesis and immunochemical analysis of visual arrestin interaction with rhodopsin. Journal of Biological Chemistry. 2002;277:11703–11708. doi: 10.1074/jbc.M111833200. [DOI] [PubMed] [Google Scholar]
- 106.Vishnivetskiy SA, et al. Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. Journal of Biological Chemistry. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 107.Gurevich VV, et al. Binding of wild type and chimeric arrestins to the m2 muscarinic cholinergic receptor. Journal of Biological Chemistry. 1993;268:16879–16882. [PubMed] [Google Scholar]
- 108.Gurevich EV, Gurevich VV. Arrestins are ubiquitous regulators of cellular signaling pathways. Genome Biology. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oakley RH, et al. Differential affinities of visual arrestin, barrestin1, and barrestin2 for G protein-coupled receptors delineate two major classes of receptors. Journal of Biological Chemistry. 2000;275:17201–17210. doi: 10.1074/jbc.M910348199. [DOI] [PubMed] [Google Scholar]
- 110.DeGraff JL, Gurevich VV, Benovic JL. The third intracellular loop of alpha2-adrenergic receptors determines subtype specificity of arrestin interaction. Journal of Biological Chemistry. 2002;277:43247–43252. doi: 10.1074/jbc.M207495200. [DOI] [PubMed] [Google Scholar]
- 111.Macey TA, Gurevich VV, Neve KA. Preferential Interaction between the dopamine D2 receptor and Arrestin2 in neostriatal neurons. Molecular Pharmacology. 2004;66:1635–1642. doi: 10.1124/mol.104.001495. [DOI] [PubMed] [Google Scholar]
- 112.Dale LB, et al. Agonist-stimulated and tonic internalization of metabotropic glutamate receptor 1A in human embryonic kidney 293 cells: agonist-stimulated endocytosis is beta-arrestin1 isoform-specific. Molecular Pharmacology. 2001;60:1243–1253. doi: 10.1124/mol.60.6.1243. [DOI] [PubMed] [Google Scholar]
- 113.Mundell SJ, et al. Arrestin isoforms dictate differential kinetics of A2B adenosine receptor trafficking. Biochemistry. 2000;39:12828–12836. doi: 10.1021/bi0010928. [DOI] [PubMed] [Google Scholar]
- 114.Ahn S, et al. Reciprocal regulation of angiotensin receptor-activated extracellular signal-regulated kinases by beta-arrestins 1 and 2. Journal of Biological Chemistry. 2004;279:7807–7811. doi: 10.1074/jbc.C300443200. [DOI] [PubMed] [Google Scholar]
- 115.Gurevich VV, Gurevich EV. The new face of active receptor bound arrestin attracts new partners. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 116.Song X, Gurevich EV, Gurevich VV. Cone arrestin binding to JNK3 and Mdm2: conformational preference and localization of interaction sites. Journal of Neurochemistry. 2007;103:1053–1062. doi: 10.1111/j.1471-4159.2007.04842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Song X, et al. How does arrestin assemble MAP kinases into a signaling complex? Journal of Biological Chemistry. 2009;284:685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hanson SM, et al. Visual arrestin binding to microtubules involves a distinct conformational change. Journal of Biological Chemistry. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu N, et al. Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. Journal of Molecular Biology. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meng D, et al. MEK1 binds directly to betaarrestin1, influencing both its phosphorylation by ERK and the timing of its isoprenaline-stimulated internalization. Journal of Biological Chemistry. 2009;284:11425–11435. doi: 10.1074/jbc.M806395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gurevich VV, Gurevich EV, Cleghorn WM. Arrestins as multi-functional signaling adaptors. Handbook of Experimental Pharmacology. 2008;186:15–37. doi: 10.1007/978-3-540-72843-6_2. [DOI] [PubMed] [Google Scholar]
- 122.Kim YM, Benovic JL. Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. Journal of Biological Chemistry. 2002;277:30760–30768. doi: 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- 123.Kang DS, et al. Structure of an arrestin2/clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. Journal of Biological Chemistry. 2009;284:29860–29872. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baillie GS, et al. Mapping binding sites for the PDE4D5 cAMP-specific phosphodiesterase to the N- and C-domains of beta-arrestin using spot-immobilized peptide arrays. Biochemical Journal. 2007;404:71–80. doi: 10.1042/BJ20070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luttrell LM, et al. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McDonald PH, et al. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 127.Miller WE, et al. Identification of a motif in the carboxyl terminus of beta -arrestin2 responsible for activation of JNK3. Journal of Biological Chemistry. 2001;276:27770–27777. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- 128.Scott MG, et al. Differential nucleocytoplasmic shuttling of beta-arrestins. Characterization of a leucine-rich nuclear export signal in beta-arrestin2. Journal of Biological Chemistry. 2002;277:37693–37701. doi: 10.1074/jbc.M207552200. [DOI] [PubMed] [Google Scholar]
- 129.Wang P, et al. Subcellular localization of beta-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus. Journal of Biological Chemistry. 2003;278:11648–11653. doi: 10.1074/jbc.M208109200. [DOI] [PubMed] [Google Scholar]
- 130.Miller WE, Lefkowitz RJ. Expanding roles for beta-arrestins as scaffolds and adapters in GPCR signaling and trafficking. Current Opinion in Cell Biology. 2001;13:139–145. doi: 10.1016/s0955-0674(00)00190-3. [DOI] [PubMed] [Google Scholar]
- 131.Li X, et al. A scanning peptide array approach uncovers association sites within the JNK/betaarrestin signalling complex. FEBS Letters. 2009;583:3310–3316. doi: 10.1016/j.febslet.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 132.Schleicher A, Kuhn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 133.Xiao K, et al. Activation-dependent conformational changes in {beta}-arrestin 2. Journal of Biological Chemistry. 2004;279:55744–55753. doi: 10.1074/jbc.M409785200. [DOI] [PubMed] [Google Scholar]
- 134.Nobles KN, et al. The active conformation of beta-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of beta-arrestins1 and -2. Journal of Biological Chemistry. 2007;282:21370–21381. doi: 10.1074/jbc.M611483200. [DOI] [PubMed] [Google Scholar]
- 135.Vishnivetskiy SA, et al. Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. Journal of Biological Chemistry. 2002;277:43961–43967. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 136.Bhandari D, et al. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. Journal of Biological Chemistry. 2007;282:36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- 137.Gaidarov I, Keen JH. Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. Journal of Cell Biology. 1999;146:755–764. doi: 10.1083/jcb.146.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krupnick JG, et al. Modulation of the arrestin-clathrin interaction in cells. Characterization of beta-arrestin dominant-negative mutants. Journal of Biological Chemistry. 1997;272:32507–32512. doi: 10.1074/jbc.272.51.32507. [DOI] [PubMed] [Google Scholar]
- 139.Perry SJ, et al. Targeting of cyclic AMP degradation to beta 2-adrenergic receptors by beta-arrestins. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 140.Li Y, et al. Regulation of amygdalar PKA by beta-arrestin-2/phosphodiesterase-4 complex is critical for fear conditioning. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21918–21923. doi: 10.1073/pnas.0906941106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin FT, et al. Feedback regulation of beta-arrestin1 function by extracellular signal-regulated kinases. Journal of Biological Chemistry. 1999;274:15971–15974. doi: 10.1074/jbc.274.23.15971. [DOI] [PubMed] [Google Scholar]
- 142.Shenoy SK, et al. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 143.Shenoy SK, et al. Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. Journal of Biological Chemistry. 2008;283:22166–22176. doi: 10.1074/jbc.M709668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Berthouze M, et al. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO Journal. 2009;28:1684–1696. doi: 10.1038/emboj.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shenoy SK, Lefkowitz RJ. Receptor-specific ubiquitination of beta-arrestin directs assembly and targeting of seven-transmembrane receptor signalosomes. Journal of Biological Chemistry. 2005;280:15315–15324. doi: 10.1074/jbc.M412418200. [DOI] [PubMed] [Google Scholar]
- 146.Mosser VA, et al. Differential role of beta-arrestin ubiquitination in agonist-promoted down-regulation of M1 vs M2 muscarinic acetylcholine receptors. Journal of Molecular Signaling. 2008;3:20. doi: 10.1186/1750-2187-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pals-Rylaarsdam R, et al. Internalization of the m2 muscarinic acetylcholine receptor: arrestin-independent and -dependent pathways. Journal of Biological Chemistry. 1997;272:23682–23689. doi: 10.1074/jbc.272.38.23682. [DOI] [PubMed] [Google Scholar]
- 148.Lee KB, et al. Arrestin binding to the M2 muscarinic acetylcholine receptor is precluded by an inhibitory element in the third intracellular loop of the receptor. Journal of Biological Chemistry. 2000;275:9284–9289. doi: 10.1074/jbc.275.13.9284. [DOI] [PubMed] [Google Scholar]
- 149.Shenoy SK, et al. Ubiquitination of beta-arrestin links seven-transmembrane receptor endocytosis and ERK activation. Journal of Biological Chemistry. 2007;282 doi: 10.1074/jbc.M700852200. 29549-29546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schubert C, et al. Visual arrestin activity may be regulated by self-association. Journal of Biological Chemistry. 1999;274:21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- 151.Imamoto Y, et al. Concentration-dependent tetramerization of bovine visual arrestin. Biophysical Journal. 2003;85:1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hanson SM, et al. Structure and function of the visual arrestin oligomer. EMBO Journal. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Granzin J, et al. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 154.Hanson SM, et al. A model for the solution structure of the rod arrestin tetramer. Structure. 2008;16:924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hanson SM, et al. Opposing effects of inositol hexakisphosphate on rod arrestin and arrestin2 self-association. Biochemistry. 2008;47:1070–1075. doi: 10.1021/bi7021359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Milano SK, et al. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. Journal of Biological Chemistry. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- 157.Storez H, et al. Homo- and hetero-oligomerization of beta-arrestins in living cells. Journal of Biological Chemistry. 2005;280:40210–40215. doi: 10.1074/jbc.M508001200. [DOI] [PubMed] [Google Scholar]
- 158.Kang J, et al. A nuclear function of beta-arrestin1 in GPCR signaling: regulation of histone acetylation and gene transcription. Cell. 2005;123:833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 159.Boularan C, et al. beta-arrestin 2 oligomerization controls the Mdm2-dependent inhibition of p53. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18061–18066. doi: 10.1073/pnas.0705550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xu TR, et al. Mutations of beta-arrestin 2 that limit self-association also interfere with interactions with the beta2-adrenoceptor and the ERK1/2 MAPKs: implications for beta2-adrenoceptor signalling via the ERK1/2 MAPKs. Biochemical Journal. 2008;413:51–60. doi: 10.1042/BJ20080685. [DOI] [PubMed] [Google Scholar]
- 161.DeFea KA. Beta-arrestin multimers: does a crowd help or hinder function? Biochemical Journal. 2008;413:e1–e3. doi: 10.1042/BJ20081009. [DOI] [PubMed] [Google Scholar]
- 162.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nature Reviews Drug Discovery. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kenakin TP. 7TM receptor allostery: putting numbers to shapeshifting proteins. Trends in Pharmacological Sciences. 2009;30:460–469. doi: 10.1016/j.tips.2009.06.007. [DOI] [PubMed] [Google Scholar]