i. Summary/Abstract
Due to the complexity of the mammalian central nervous system neuropeptidomic studies in mammals often yield very complicated mass spectra that make data analysis difficult. Careful sample preparation and extraction protocols must be employed in order to minimize spectral complexity and enable extraction of useful information on neuropeptides from a given sample. Controlling post-mortem protease activity is essential to simplifying mass spectra and to identifying low-abundance neuropeptides in tissue samples. Post-mortem microwave-irradiation coupled with cryostat dissection has proven to be effective in arresting protease activity to allow detection of endogenous neuropeptides instead of protein degradation products.
Keywords: Neuropeptide, Extraction, Microwave, Protease activity, Sample preparation, Mass spectrometry, HPLC, Peptide sequencing/identification
1. Introduction
Neuropeptides are small (5 to 100 amino acid residues) endogenous biomolecules that have the ability to act as neurotransmitters, neuromodulators, or neurohomones in the nervous system. These biomolecules play a role in many physiological functions including feeding, sleeping, learning, pain, anxiety, circadian rhythms, and memory (1,2). The biogenesis of neuropeptides occurs in the cell body of neurons. Here, pre-propeptides are synthesized in the rough endoplasmic reticulum (RER), secreted from the RER after the signal sequence is removed, and packaged into vesicles by the Golgi apparatus. Within these vesicles, propeptides are further processed and undergo post-translational modifications (i.e., glycosylation, C-terminal amidation, acetylation, phosphorylation, and disulfide bond formation) generating bioactive peptides (3).
The important biological role neuropeptides play has made these targets of many investigations. Many neuropeptides have been studied employing traditional techniques such as immunohistochemistry, radioimmunoassay, and Edman degradation. Although these techniques are valuable, mass spectrometry (MS) based techniques do not require a priori knowledge of peptide identity and allow for rapid elucidation of molecular species in a complex mixture. Mass spectrometry offers rapid and sensitive detection of ionizable species, but spectra can be complicated and low abundance target species (i.e., neuropeptides) can be masked due to salts, lipids, and surfactants (4). Thus, appropriate sample preparation methods that allow preferential identification of neuropeptides with minimal interference are often key to successful MS-based studies.
Important considerations must be taken during sample collection and preparation in order to obtain useful information in any neuropeptidomic study. The inhibition of active post-mortem proteases is one of the most important considerations a researcher must take into account. Once the animal is sacrificed and the tissue of interest harvested, proteases rapidly degrade larger proteins into smaller fragments that fall into the mass range of neuropeptides. These abundant protein fragments may suppress neuropeptide signal and make mass spectra interpretation very difficult. To minimize this spectra clouding from protein degradation, focused microwave-irradiation animal sacrifice (5-9), post-sacrifice microwave-irradiation of tissue (10), and cryostat dissection followed by a boiling extraction buffer (11) methods have all been used. Each of these techniques can effectively minimize the post-mortem protein degradation, but these techniques also possess their own drawbacks. Focused microwave-irradiation animal sacrifice, although an effective means to stop protease degradation, requires an expensive targeted-microwave emitting instrument and can introduce unnecessary stress on the animal that may affect neuropeptide expression. The use of a household microwave for post-mortem tissue fixation allows for the animal to be sacrificed by conventional methods, but can require a number of animals to develop a consistent protocol. Cryostat dissection followed by a boiling extraction buffer inhibits protease activity, but has a longer time gap between sacrifice and protease inhibition which allows some extracellular processing to occur.
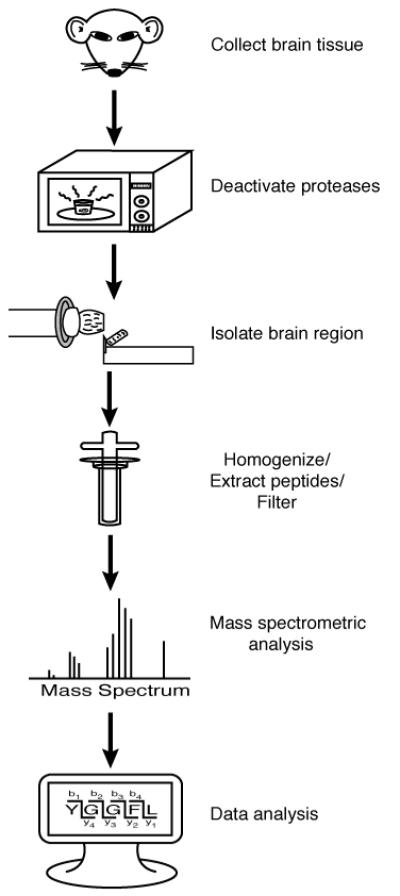
Presented in this chapter is a neuropeptidomic procedure that utilizes conventional microwave to inhibit proteases post-mortem, cryostat dissection to isolate specific tissue, acidified methanol to extract neuropeptides from tissue, and LC-QTOF-MS(/MS) to analyze the tissue extract. Figure 1 depicts a flowchart of the MS-based neuropeptide analysis procedure.
Figure 1.
Flowchart of MS-based neuropeptide analysis.
2. Materials
2.1. Rat Dissection
Halothane, (Sigma; St. Louis, MO)
Cryoware cryogenic vials, (Nalgene; Rochester, NY)
Microwave, (General Electric, 1.5 kW)
Aluminum foil, (standard store purchased is suitable)
Ethanol
Dry ice
Cryostat, (Leica; Wetzlar, Germany)
Optimal cutting temperature compound (OCT), (Sakura; Torrance, CA)
Harris micro-punch, 3 mm, (Whatman; Clifton, NJ)
2.2. Neuropeptide Extraction
Methanol, purge and trap grade, (Fisher Scientific; Fair Lawn, NJ)
Glacial acetic acid
Water, double distilled by filtration system
Formic acid, Store in a 4 °C refrigerator.
10 kDa molecular weight cut-off tube, (Sartorius; Goettingen, Germany)
Handheld ground glass homogenizer, (Wheaten Science; Millville, NJ)
2.3. Mass Spectrometry Analysis
Quadrupole time-of-flight (QTOF) mass spectrometer (Waters; Milford, MA; QTOF MICRO) equipped with a nanoelectrospray ionization source (nESI). (see Note 1)
Ultra performance liquid chromatography (UPLC) (Waters; Milford, MA; nanoAcquity UPLC) (see Note 2)
Acetonitrile, HPLC grade, (Fisher Scientific; Fair Lawn, NJ)
Mobile phase A: 0.1% formic acid in double distilled water (v/v)
Mobile phase B: 0.1% formic acid in acetonitrile (v/v)
3. Methods
3.1. Rat Dissection
Rats are placed in a plastic cylinder, anesthetized with halothane, and then sacrificed by decapitation.
Immediately (< 60 s), the brain is removed from the rat and placed in a 15 mL cryogenic vial and immersed in warm water (see Note 3).
The cryogenic vial is then placed in the center of a microwave and heated for 7 seconds at full power. This allows the inside of the brain to rapidly reach a temperature of 80 °C (see Note 4, 5).
The cryojar is removed from the microwave, water is decanted off, and the brain is allowed to stand at room temperature for 1 to 3 minutes (see Note 6).
The brain is then snap-frozen in an ethanol/dry ice bath (see Note 7).
Place the snap-frozen brain on dry ice. Store at −80 °C.
Mount the frozen brain tissue on a chilled cryostat chuck (same temperature as the cryostat compartment, −15 °C to −20 °C) with OCT compound and allow one minute for the OCT compound to solidify. Add additional OCT compound to the base of the brain to provide additional support for cutting (see Note 8).
Section the brain into 300 μm slices by advancing objective without slicing until the desired width is achieved (see Note 9).
Use a tissue punch to isolate brain regions of interest. (In our case we used a 3mm tissue punch to collect hypothalamus and striatum regions of the rat brain.)
Place each tissue punch in a separate 1.5 mL microcentrifuge tube and add enough ice-cold acidified methanol (90% methanol, 9% glacial acetic acid, 1% doubly distilled water, DDW, v/v/v) to submerge the tissue (minimal volumes of acidified methanol are preferred). At this point you can store the tissue punches at −80 °C until extraction.
3.2. Neuropeptide Extraction
Place the frozen tissue directly into a 1 mL hand homogenizer containing 100 to 500 μL of ice-cold acidified methanol (see Note 10).
Immediately homogenize the tissue while keeping the temperature of the homogenate ice-cold (see Note 11, 12).
Place the homogenate in a 1.5 mL microcentrifuge tube and centrifuge the solution at 14000 g for 25 minutes at 4 °C (see Note 13).
Carefully transfer the supernatant to a clean microcentrifuge tube.
Re-extract neuropeptides from the tissue pellet by adding an additional aliquot of 100 to 500 μL of ice-cold acidified methanol, vortexing, and centrifuging for another 25 minutes.
Carefully transfer the supernatant to the microcentrifuge tube containing the first supernatant volume.
Filter the supernatants through a 10 kDa molecular weight cut off (MWCO) microcentrifuge tube at 14000 g for approximately 20 minutes at 4 °C. The flow-through volume contains the neuropeptides (see Note 14).
Vacuum dry the sample to dryness.
Resuspend the extracted neuropeptides in 20 to 25 μL of aqueous 0.1% formic acid.
Vortex to mix.
Centrifuge the re-suspended neuropeptides at 14000 g for 3 minutes.
Transfer the supernatant to a clean 0.6 mL microcentrifuge tube.
3.3. Mass Spectrometric Analysis
A Waters nanoAcquity UPLC system was used to deliver the specified volume of sample to a trap column (Waters Symmetry® C18, 180 μm × 20mm; Milford, MA) via a 5% flow of mobile phase B at a rate of 10 μL/min for 1.50 min. The flow rate was then switched to 200 nL/min, and the peptides were flushed onto the analytical column (Microtech C18, 75 μm x 15 cm; Vista, CA) and eluted via a mobile phase B: 5 – 95% linear gradient over 60 minutes into a nESI-QTOF mass spectrometer (see Note 15).
Mass spectra (MS) were collected for all eluted peptides and tandem mass spectra (MS2) were collected in data-dependent acquisition mode where MS2 survey scans were made when an eluted peptide had an ion count of 15 or greater.
3.4. Data analysis
Bioactive neuropeptides are oftentimes found to undergo extensive proteolytic cleavages or post-translational modifications, making it difficult to identify what protein precursor a neuropeptide originates from. Recently a binary logistic regression model (12) trained on mammalian prohormone cleavages has been developed that helps determine novel bioactive peptides from an organism’s genetic sequence information. Once optimized, this bioinformatics tool will minimize the time and effort required to analyze MS data and determine novel bioactive peptides from genetic sequence information. Currently database searching and de novo sequencing are the methods of choice for determining peptide identity.
The SwePep (13) database (http://www.swepep.org/) was developed to increase the throughput of identifying endogenous peptides in complex tissue samples analyzed by MS. This is a good place to begin when trying to identify neuropeptides in tissue extracts. Online databases like Mascot (Matrix Science; http://www.matrixscience.com/) and SEQUEST (Thermo Corp.; http://www.thermo.com/) are also powerful tools for identifying neuropeptides (14,15). In this case, Mascot was used to analyze QTOF data.
Use the software packaged with the mass spectrometer to process the LC-MS/MS data (in this case Micromass ProteinLynx 2.1).
Deisotope the data and convert the data into .pkl files.
Take the newly created .pkl files and search them against the Swiss-Prot database using the online version of Mascot.
Select “none” for the enzyme.
Set the peptide mass tolerance at 200 ppm and the MS/MS mass tolerance at 0.2 Da.
- Make three separate searches for variable modifications:
- Search 1: C-terminal amidation and N-terminal acetylation.
- Search 2: methionine oxidation and C-terminal amidation.
- Search 3: phosporylation of tyrosine, threonine, and serine and C-terminal amidation.
If peptides do not yield significant Mascot scores the sequences should be verified by de novo sequencing (see Note 16).
4. Notes
Other mass spectrometers capable of automated acquisition of tandem mass spectra may be used for peptide mass fingerprinting and de novo sequencing.
High performance liquid chromatography (HPLC) can be used in place of UPLC.
Protease degradation of proteins occurs immediately after death so the amount of time between sacrifice and protease inhibition should be as short as possible.
The microwave used does not have to be 1.5 kW. In fact, Che, et al. (10) demonstrated effective protease inhibition by placing a whole mouse head into a General Electric, 1.38 kW microwave for 5 to 13 seconds at full power. The only criteria for the microwave used is that it should have the ability to raise the brain temperature to > 80 °C within ten seconds (10). Reproducibility in heating between samples would also be benefited by placing the brain tissue in the same location of the microwave.
The brain tissue is easier to manipulate if it is allowed to dry for a few minutes directly after microwave-irradiation.
Depending on the microwave, method development may be needed in order to find a time and temperature setting that raises the rat brain to approximately 80 °C without altering the morphology of the brain.
Do not place the brain directly into the dry ice/ethanol bath to avoid tissue tear and preserve tissue integrity. Rather make a small envelope out of aluminum foil and place the brain within it. Then place the aluminum foil envelope containing the brain in the bath. Leave the envelope in the bath for approximately 3 minutes to ensure the inside of the brain completely freezes.
OCT compound can interfere with mass spectrometric analysis due to polymer contaminants. Thus, one should avoid applying it to tissue areas of interest for analysis. For example, if one wants to analyze striatum and hypothalamus punches mount the brain so that the cerebellum is in contact with the cryostat chuck and the OCT. Avoid getting OCT on the cryostat blade. If OCT happens to get on the blade either clean the blade thoroughly with 100% ethanol or replace the blade with an unused one.
Alternatively, a rat brain matrix (Zivic Instruments; Pittsburgh, PA) may be used to section the brain. If this technique is preferred it is recommended that you perform sectioning on dry ice so the brain does not thaw.
Minimal volume of acidified methanol is recommended. The key is to use a volume that allows for complete homogenization without over diluting the sample. For example, Dowell et al. (11) used an aliquot of 300 μL of ice cold acetic acid to homogenize a striatum punch that weighed 20 to 30 mg.
It is important to immediately homogenize the tissue in order to ensure complete deactivation of all protease activity (especially activity in the center of the tissue punch).
A microsonicator or electrical homogenizer may be substituted as long as you keep the temperature of the homogenate ice cold. Keep the temperature low by homogenizing the tissue in an ice bath.
A Pasteur pipet works well for transferring the homogenate.
Clean the MWCO tube with DDW and cold acidified methanol separately. Each species should be centrifuged at 14000 g for 3 minutes. Discard the resulting liquid. Repeat this cleaning twice in order to remove all glycerol from MWCO membrane.
Other mass spectrometric techniques can be used to detect neuropeptides in brain tissue samples. Offline fractionation of a tissue extract has been used to simplify complex biological samples (11). Matrix assisted laser desorption/ionization (MALDI) has also proven effective in identifying neuropeptides in mammalian biological samples (9, 11, 16, 17). In the case of MALDI, TOF, TOF/TOF and Fourier transform (FT) mass analyzers have successfully been used to identify neuropeptides. In addition to providing complementary ionization method, MALDI-based techniques also offer exciting capabilities to perform in situ direct tissue neuropeptide mapping or imaging (18-21).
A de novo sequence matching at least three consecutive amino acids of the presumed sequence was considered a positive identification in previous studies (11). It should be noted that it is often challenging to obtain complete fragmentation for de novo sequencing of neuropeptides. Various chemical derivatization methods have been employed to enhance fragmentation for improved sequence derivation (22, 23).
Acknowledgments
This work was supported in part by National Science Foundation CAREER Award (CHE-0449991), National Institutes of Health through grant 1R01DK071801, and an Alfred P. Sloan Research Fellowship (L.L.). R.M.S. acknowledges the National Institutes of Health Clinical Neuroengineering Training Program Grant 5T90DK070079. J.A.D. acknowledges an American Foundation for Pharmaceutical Education (AFPE) predoctoral fellowship.
Contributor Information
Robert M. Sturm, Department of Chemistry, University of Wisconsin-Madison, 777 Highland Avenue, Madison, WI 53705-2222, USA
James A. Dowell, School of Pharmacy, University of Wisconsin-Madison, 777 Highland Avenue, Madison, WI 53705-2222, USA
Lingjun Li, School of Pharmacy & Department of Chemistry, University of Wisconsin-Madison, 777 Highland Avenue, Madison, WI 53705-2222, USA.
5. References
- 1.Strand FL. Neuropeptides: general characteristics and neuropharmaceutical potential in treating CNS disorders. Prog Drug Res. 2003;100:1–37. doi: 10.1007/978-3-0348-8049-7_1. [DOI] [PubMed] [Google Scholar]
- 2.Clynen E, De Loof A, Schoofs L. The use of peptidomics in endocrine research. Gen Comp Endocrinol. 2003;132:1–9. doi: 10.1016/s0016-6480(03)00158-8. [DOI] [PubMed] [Google Scholar]
- 3.Strand FL. Models of neuropeptide action. New York Academy of Sciences; New York, N.Y.: 1994. [Google Scholar]
- 4.Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu. Rev. Anal. Chem. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 5.Theodorsson E, Stenfors C, Mathe AA. Microwave irradiation increases recovery of neuropeptides from brain tissues. Peptides. 1990;11:1191–1197. doi: 10.1016/0196-9781(90)90151-t. [DOI] [PubMed] [Google Scholar]
- 6.Mathe AA, Stenfors C, Brodin E, Theodorsson E. Neuropeptides in brain: effects of microwave irradiation and decapitation. Life Sci. 1990;46:287–293. doi: 10.1016/0024-3205(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 7.Nylander I, Stenfors C, Tan No K, Mathe AA, Terenius L. A comparison between microwave irradiation and decapitation: basal levels of dynorphin and enkephalin and the effect of chronic morphine treatment on dynorphin peptides. Neuropeptides. 1997;31:357–365. doi: 10.1016/s0143-4179(97)90072-x. [DOI] [PubMed] [Google Scholar]
- 8.Svensson M, Skold K, Svenningsson P, Andren PE. Peptidomics-based discovery of novel neuropeptides. J.Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 9.Parkin MC, Wei H, O’Callaghan JP, Kennedy RT. Sample-dependent effects on the neuropeptidome detected in rat brain tissue preparations by capillary liquid chromatography with tandem mass spectrometry. Anal. Chem. 2005;77:6331–6338. doi: 10.1021/ac050712d. [DOI] [PubMed] [Google Scholar]
- 10.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol. Cell. Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Dowell JA, Heyden WV, Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J. Proteome Res. 2006;5:3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 12.Amare A, Hummon AB, Southey BR, Zimmerman TA, Rodriquez-Zas SL, Sweedler JV. Bridging neuropeptidomics and genetics with bioinformatics: prediction of mammalian neuropeptide prohormone processing. J. Proteome Res. 2006;5:1162–1167. doi: 10.1021/pr0504541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falth M, Skold K, Norrman M, Svensson M, Fenyo D, Andren PE. SwePep, a database designed for endogenous peptides and mass spectrometry. Mol. Cell. Proteomics. 2006;5:998–1005. doi: 10.1074/mcp.M500401-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Ducret A, Van Oostveen I, Eng JK, Yates-III JR, Aebersold R. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 1998;7:706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatcher NG, Richmond TA, Rubakhin SS, Sweedler JV. Monitoring activity-dependent peptide release from the CNS using single-bead solid-phase extraction and MALDI TOF MS detection. Anal. Chem. 2005;77:1580–1587. doi: 10.1021/ac0487909. [DOI] [PubMed] [Google Scholar]
- 17.Che FY, Yan L, Li H, Mzhavia N, Devi LA, Fricker LD. Identification of peptides from brain and pituitary of Cpefat/Cpefat mice. Proc. Natl. Acad. Sci USA. 2001;98:9971–9976. doi: 10.1073/pnas.161542198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 19.Stoeckli M, Staab D, Staufenbiel M, Wiederhold KH, Signor L. Molecular imaging of amyloid beta peptides in mouse brain sections using mass spectrometry. Anal. Biochem. 2002;311:33–39. doi: 10.1016/s0003-2697(02)00386-x. [DOI] [PubMed] [Google Scholar]
- 20.DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barret-Wilt GA, Li L. Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J. Proteome Res. 2007;6:1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubakhin SS, Hatcher NG, Monroe EB, Heien ML, Sweedler JV. Mass spectrometric imaging of the nervous system. Curr. Pharm. Des. 2007;13:3325–3334. doi: 10.2174/138161207782360708. [DOI] [PubMed] [Google Scholar]
- 22.Fu Q, Li L. De novo sequencing of neuropeptides using reductive isotopic methylation and investigation of ESI QTOF MS/MS fragmentation pattern of neuropeptides with N-terminal dimethylation. Anal. Chem. 2005;77:7783–7795. doi: 10.1021/ac051324e. [DOI] [PubMed] [Google Scholar]
- 23.Ma M, Kutz-Naber KK, Li L. Methyl esterification assisted MALDI FTMS characterization of the orcokinin neuropeptide family. Anal. Chem. 2007;79:673–681. doi: 10.1021/ac061536r. [DOI] [PubMed] [Google Scholar]