Abstract
Cortical microtubules (MTs) participate in the spatial control of cell expansion and division that is required for plant growth and morphogenesis. Well-ordered transverse cortical MTs promote cell elongation and restrict radial cell expansion. The molecular mechanism controlling their ordering is poorly understood. We report the first known signaling pathway that promotes the organization of cortical MTs into parallel arrays oriented perpendicular to the axis of cell elongation in plants. Well-ordered MTs locally restrict cell expansion to promote indentation formation in the jigsaw puzzle-shaped pavement cells of Arabidopsis leaves. Deleting ROP6, a Rho-family GTPase, randomized cortical MTs and released the localized restriction of cell expansion, whereas ROP6 overexpression enhanced MT ordering, turning the jigsaw puzzle appearance of cells into a cylindrical shape. ROP6 directly binds and activates MT-associated RIC1 to achieve the MT ordering. The ROP6-RIC1 pathway also affects MT ordering of hypocotyl cells, showing a broad role for this pathway in the spatial regulation of cell expansion.
Results and Discussion
Well-ordered cortical microtubules (MTs) are believed to restrict cell expansion to the direction perpendicular to their dominant orientation. Accordingly, transversely arranged MTs are important for cell elongation and plant growth along the apical-basal axis, whereas cells grow isotropically with cortical MTs arranged randomly. Several MT dynamic behaviors, including angle-dependent modification of sustained treadmilling, rotary movement, and polar co-alignment due to selective stabilization, have been suggested to regulate cortical MT ordering [1-4]. Furthermore, a number of MT-associated proteins (MAPs), e.g., RIC1, SPR1/SKU6, and SPR2, are known to affect the orientation of cortical MTs [5-7]. However, how these MT behaviors and MAPs are linked to developmental and environmental signals that control the ordering of cortical MTs remains mysterious.
We use the Arabidopsis leaf epidermal pavement cells (PCs) with their jigsaw puzzle appearance as a model system to investigate signaling mechanisms regulating the organization of the cytoskeleton in plants [5, 8-12]. In PCs, ordered cortical MTs are associated with indenting regions of PCs and are excluded from the protruding region that contains fine cortical actin microfilaments (MFs). We recently showed that ROP2 and ROP4, Arabidopsis members of the conserved Rho GTPase family [13-15], promote the protrusion by activating the localized actin accumulation and inhibiting MT organization by inactivating the microtubule-associated RIC1 protein in Arabidopsis PCs [5, 8]. In the indenting region, the ordering of cortical MTs is activated by RIC1, which belongs to the RIC family (ROP-interactive CRIB motif-containing proteins) of ROP effector proteins that interact with the active form of ROPs through the CRIB motif [5, 16]. In this report, we demonstrate that RIC1's function in promoting MT ordering is activated by another Rho family GTPase, ROP6.
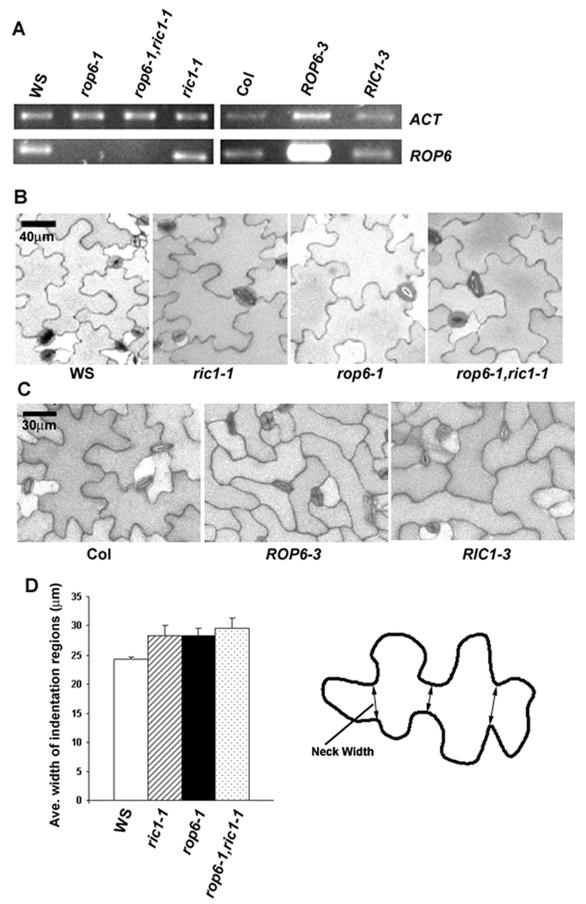
We speculated that as a ROP effector protein RIC1 must be activated to promote MT organization by one of the 11 ROP GTPases in Arabidopsis. We anticipated that a loss of function mutant for a RIC1 activating ROP would mimic ric1 knockout mutants, and analyzed available Arabidopsis rop knockout mutants for PC-shape phenotype. Indeed, a ROP6 null mutant, rop6-1, resulting from a T-DNA insertion into the sixth exon, exhibited a phenotype similar to that of the ric1-1 knockout mutant. Lateral cell expansion in the indentation regions of PCs was significantly increased in rop6-1, which resulted in wider PC necks (Figure 1A, B and D; Figure S1A). To confirm that the wider neck region was due to the absence of ROP6, GFP-ROP6 driven by the ROP6 promoter was transformed into rop6-1. Several lines with a GFP-ROP6 transcript level similar to that of wild type ROP6 indeed showed the same PC shape as the wild type (Figure S2). Furthermore, ROP6 overexpression strongly inhibited lateral expansion of both lobes and indentations, as does RIC1 overexpression [5], which causes PCs to lose the intercalating jigsaw puzzle appearance (Figure 1C and Figure S3). Thus, ROP6 plays an important role in restricting lateral cell expansion, as does RIC1 [5]. PCs of the rop6-1 ric1-1 double mutant showed a phenotype identical to that of the ric1-1 or rop6-1 single mutant (Figure 1A,B and D, Figure S1A), which provides genetic evidence that ROP6 and RIC1 act in the same pathway.
Figure 1. Genetic analyses indicate that ROP6 and RIC1 act in the same pathway to promote the formation of narrow necks in pavement cells.
(A) RT-PCR analysis of ROP6 transcript levels in wild type (WS), rop6-1, rop6-1;ric1-1, ric1-1 (in WS background), wild type (Col), and ROP6 and RIC1 overexpressing lines (ROP6-3 and RIC1-3, in Col background).
(B) Pavement cell shapes of WS, rop6-1, ric1-1, and rop6-1;ric1-1 double mutants.
(C) Pavement cell shapes of Col, ROP6-3 and RIC1-3 mutants.
(D) Quantitative analysis of neck widths for WS, rop6-1, ric1-1, and rop6-1;ric1-1 double mutants. The differences in neck widths between WS and other mutants or double mutant line were significant (p<0.05, t-test), with no significant differences between the rop6-1;ric1-1 double mutant and rop6-1 or ric1-1 single mutant. In each line, a total of about 250 cells from 3 seedlings was measured. The carton on the right illustrates how the neck width was measured. All data are means ± standard deviation.
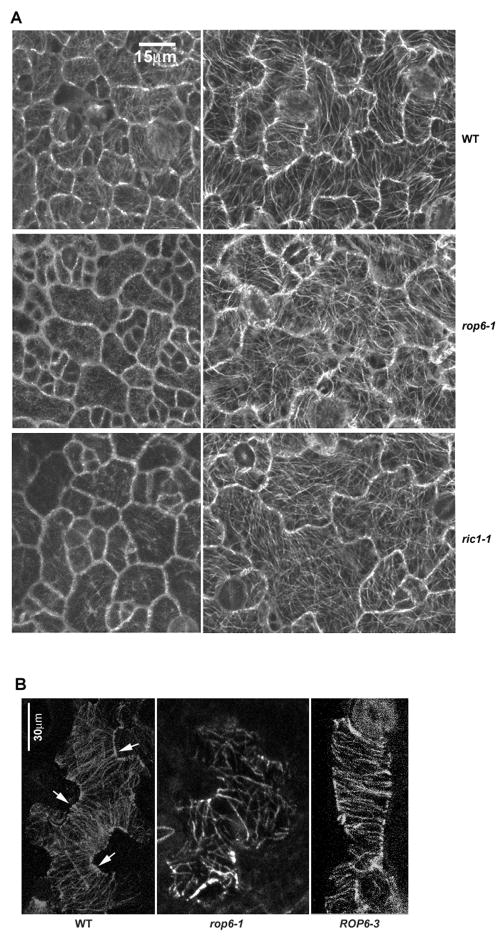
We next visualized MTs by GFP-tagged tubulin to investigate whether ROP6, like RIC1, affects the organization of cortical MTs (Figure 2A) [5, 8, 17]. Similar to young ric1-1 cells, young rop6-1 cells (stage I) contained fewer cortical MTs than the wild type (Figure 2A) [5]. Both rop6-1 and ric1-1 cells at stage II had more randomly oriented cortical MTs and fewer MT bundles than wild-type cells. Immunostaining with anti-tubulin antibody revealed a similar MT organization pattern in fixed rop6-1 PCs (Figure 2B). As in RIC1-overexpressing cells, ROP6-overexpressing PCs (ROP6-3) exhibited nearly all cortical MTs highly ordered in the orientation transverse to the axis of cell elongation (Figure 2B). These results also support the hypothesis that ROP6 and RIC1 function in the same pathway to promote the ordering of cortical MTs and to spatially control cell expansion.
Figure 2. ROP6 promotes the organization of cortical MTs into highly ordered parallel arrays in pavement cells.
(A) Cortical MTs in pavement cells in late stage I (left panel) and late stage II (right panel) from wild type (WT), rop6-1, and ricl-1. MTs were visualized by use of stably expressed tubulin-GFP [17]. MTs in rop6-1 and ric1-1 cells were less ordered and bundled than that in wild-type cells.
(B) Immunolocalization of cortical MTs by use of anti-tubulin antibody confirms rop6-1 cells with more randomly arranged cortical MTs than wild-type cells and that ROP6 overexpression (ROP6-3) promotes the formation of highly ordered transverse MTs aligned perpendicularly to the length of cells. Arrows indicate ordered transverse MTs in the neck regions of WT cells.
To further test this hypothesis, we analyzed the organization of fine cortical MFs in PCs altered in ROP6 expression, since we previously showed that the RIC1-MT pathway suppresses the accumulation of this MF population activated by the ROP2 pathway [5]. Transiently expressed GFP-mTalin was used to visualize the fine MFs in stage I and II PCs [5, 8]. Fluorescent aggregates of GFP-mTalin (indicated by arrowheads in Fig. S4) are usually associated with lobe regions of the cell cortex in WT cells [5, 8]. We found that this population of fine MFs was greatly reduced in 87% (n=31) of the ROP6-overexpressing cells (ROP6 OX-3 stage I and II cells), as we previously observed in RIC1-overexpressing cells [5]. In contrast, in rop6-1 cells (stage I and II), 81% of them (n=47) exhibited ectopic localization of GFP-mTalin aggregates in the indenting region of the cell cortex, similar to ric1-1 cells with 81% (n=23) of them exhibiting the ecotopic GFP-mTalin localization (Figure S4).
We then used fluorescence resonance energy transfer (FRET) analysis to determine whether ROP6 and RIC1 physically interact with each other in vivo. Strong FRET signals at the PM (or the cortical region closed to PM) were detected when CFP-RIC1 and YFP-ROP6 were co-expressed in rop6-1 ric1-1 double mutant cells (Figure 3A and B). Consistent with RIC1 being an effector of ROP6, CFP-RIC1 did not interact with an YFP-tagged inactive form of ROP6 (YFP-DN-rop6). The interaction between ROP6 and RIC1 was also confirmed by yeast two-hybrid assay (data not shown). To confirm the in vivo interaction between ROP6 and RIC1, we conducted co-immunoprecipitation using the transgenic line expressing GFP-ROP6 in rop6-1. As shown in Figure 3C, GFP-ROP6 immunoprecipitates (with anti-GFP antibody) indeed contained RIC1 protein (detected with anti-RIC1 antibody), but those of transgenic plants expressing GFP alone did not.
Figure 3. ROP6 physically interacts with RIC1 in vivo, is preferentially localized to the indenting region of the plasma membrane.
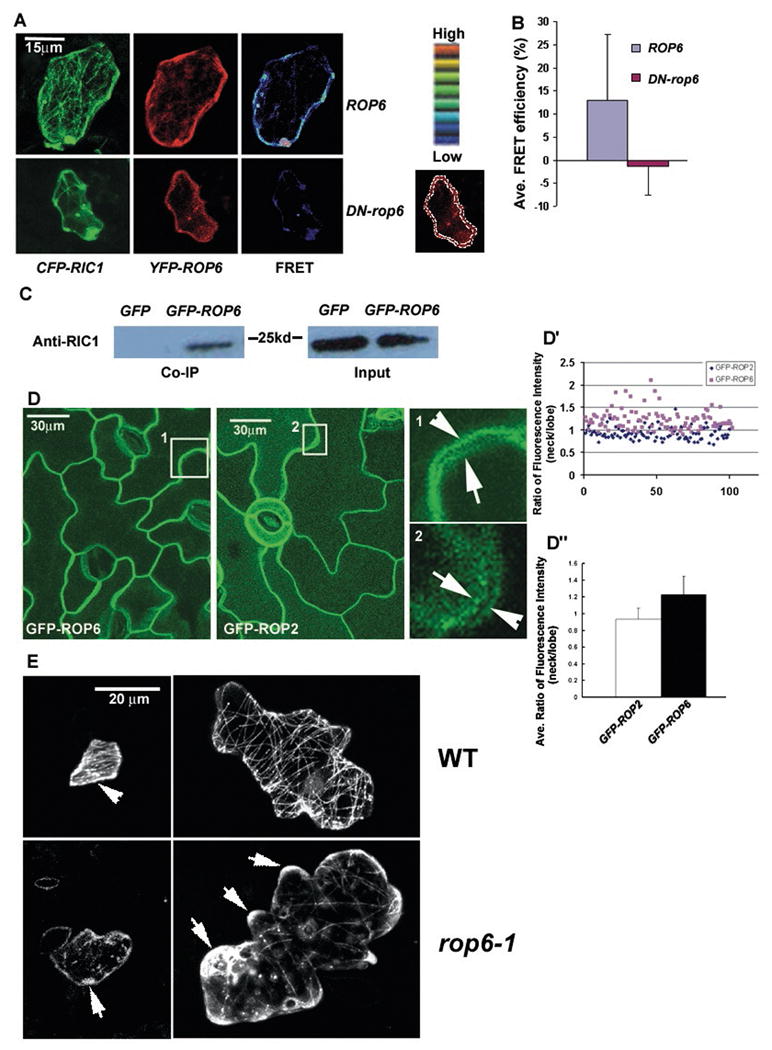
(A) Fluorescence resonance energy transfer (FRET) analysis was performed in pavement cells expressing CFP-RIC1 (pseudocolored green) and YFP-ROP6 or YFP-DN-rop6 (pseudocolored red). The pesudocolor scale was used to indicate the FRET signal intensity. The image under the pesudocolor scale indicates the region of the cell where the fluorescence intensity was measured.
(B) Quantitative analysis of FRET efficiency. See Methods for calculation of FRET efficiency. All data are means ± standard deviation.
(C) GFP-ROP6 co-immunoprecipitated with RIC1. Proteins isolated from a GFP-ROP6 transgenic line were immunoprecipitated with an anti-GFP antibody and analyzed by western blotting with an anti-RIC1 antibody. Anti-GFP immunoprecipitates from wild-type plants and plants expressing GFP alone did not contain RIC1 protein.
(D) –(D″) Subcellular localization of GFP-ROP6 and GFP-ROP2 and the quantitative analysis. (D) Pavement cells of transgenic lines expressing GFP-ROP6 or GFP-ROP2 were analyzed by confocal microscopy. Both GFP-ROP6 and GFP-ROP2 were localized to the plasma membrane (PM) (left and middle panel). Right panel shows magnified images taken from single optical sections near the mid-plane in boxes 1 and 2 of left and middle panel (projected from stacks of Z-series optical sectioning). Fluorescence intensity of GFP-ROP6 was higher in the indenting neck region (arrowhead) than in the complementary lobe tip (arrow) of the neighboring cell. In contrast, GFP-ROP2 shows preferential localization to the lobe tip (arrow). (D′) Scatter chart of the ratio of fluorescence intensity (indenting neck/neighboring lobe). Blue diamonds represent the ratio of fluorescence intensity in the indenting neck to that in the lobes in the GFP-ROP2 line and pink squares represent the ratio in cells expressing GFP-ROP6. (D″) The column graph shows the mean ratio of fluorescence intensity (indenting neck/neighboring lobe). The mean indentation-to-lobe ratio of fluorescence intensity in the GFP-ROP2 line was 0.93±0.13 [n=100], and the mean indentation-to-lobe ratio of fluorescence intensity in the GFP-ROP6 line was 1.22±0.22 [n=102]. The difference between GFP-ROP2 and GFP-ROP6 lines is significant (p<0.05, t-test). All data are means ± standard deviation.
(E) Transiently expressed GFP-RIC1 showed association with cortical MTs in wild-type (WT) cells. In WT, GFP-RIC1 was localized to both cortical MTs and other parts of the cell cortex (arrowhead) in young stage I cells, but mostly associated with cortical MTs in stage II or later. GFP-RIC1 was particularly enriched in indenting regions in these cells. In the rop6-1 line, GFP-RIC1 was sparsely associated with cortical MTs, but was highly enriched in the cortex of lobe regions (arrows) (this pattern was found in 71% (n=42) of rop6-1 cells examined).
If ROP6 acts in the RIC1 pathway to promote MT ordering, we would expect ROP6 to preferentially localize to the indenting regions of PCs. We examined ROP6 localization in the rop6-1 mutant expressing GFP-ROP6 driven by the ROP6 promoter. We expect GFP-ROP6 localization to reflect that of native ROP6, because GFP-ROP6 fully complements the rop6-1 knockout mutant (Figure S2). As shown in Figure 3D, GFP-ROP6 was localized exclusively to the plasma membrane (PM). Since the PM regions between neighboring cells are separated by a thin cell wall, it is difficult to optically separate these two membranes containing GFP-ROP6 fluorescence in the tangential side of the two contacting cells. However, the surfaces of leaf pavement cells are not completely flat, and thus we were able to identify an optical section with a separation between the membranes of two adjacent cells from Z serials of optical sections. This optical section was just above the top optical section with no observable spatial separation between the membranes of the two adjacent cells. We generally measured 2-3 regions per cell and a total 100 regions for each line. When we measured GFP-ROP6 signal in the indenting and protruding regions from the same cell, we found stronger GFP-ROP6 localization in the indenting regions than in the protruding lobes; the mean indentation-to-lobe ratio of fluorescence intensity was 1.24±0.28 (n=52). Preferential ROP6 localization to the indenting region was also found between the complementary indenting (arrowhead) and outgrowing (arrow) regions of neighboring cells (Figure 3D, box 1; Figure 3D′ and D″); the mean indentation-to-lobe ratio of fluorescence intensity was 1.22±0.22 (n=102). Conversely, GFP-ROP2 was preferentially localized to the lobe region [8] (see arrow in box 2, Figure 3D). The mean indentation-to-lobe ratio of GFP-ROP2 within the same cell was 0.91±0.1 (n=61). As shown in Figure 3D (box 2)-D″, the preferential localization of ROP2 to the outgrowing region was also observed between the complementary outgrowing and indenting regions of the neighboring cells; the mean indentation-to-lobe ratio of GFP-ROP2 was 0.93±0.13 (n=100). These results suggest that ROP2 and ROP6 are spatially regulated to coordinate the development of outgrowing and indenting regions, respectively. The opposing roles for these two ROPs are interesting, given that they belong to the same clade of the ROP family [13]. The distinct subcellular localization patterns, which may be determined by the hypervariable C-terminal region [13], may account for their distinct functions. In the outgrowing region, ROP2 promotes lobe formation by activating RIC4-dependent accumulation of fine MFs and inactivating RIC1 by sequestering it from cortical MTs [5], whereas in the indenting region ROP6 may activate RIC1 by promoting its association with cortical MTs.
We further tested whether ROP6 activates RIC1 to promote MT ordering by analyzing RIC1 localization in the rop6-1 knockout mutant. In wild-type young cells (stage I), GFP- or YFP-RIC1 was distributed in both PM and cortical MTs, and in stage II or later cells, RIC1 was mostly associated with MTs [5] (Figure 3E). GFP- or YFP-RIC1 was sparsely associated with cortical MTs and exhibited diffuse fluorescence (usually associated with lobes) in approximately 71% (n=42) of rop6-1 cells (Figure 3E). The ROP6-dependent MT association of RIC1 was further confirmed by RIC1 immunostaining. Similar to GFP-RIC1, in WT cells anti-RIC1 stained native RIC1 was localized as dots along cortical MTs [5], especially enriched in the indentation region, where thick transverse MT cables located, but was found enriched in the cortex of the lobe tip in rop6-1 PCs (Figure S5A and S5B). The loss of RIC1 association with cortical MTs was not due to absence of cortical MTs in rop6-1 cells, because cortical MTs, although less abundant and less organized, were clearly present in the rop6-1 PCs, as shown by immunostaining or tubulin-GFP (Figure 2B and S5A). These results indicate that on direct interaction with active ROP6, RIC1 associates with cortical MTs, where it promotes MT ordering. Taken together, these results show that ROP6 directly activates the association of RIC1 with cortical MTs and its function in MT ordering, constituting a signaling pathway leading to the organization of cortical MTs into transversely ordered parallel arrays in PCs.
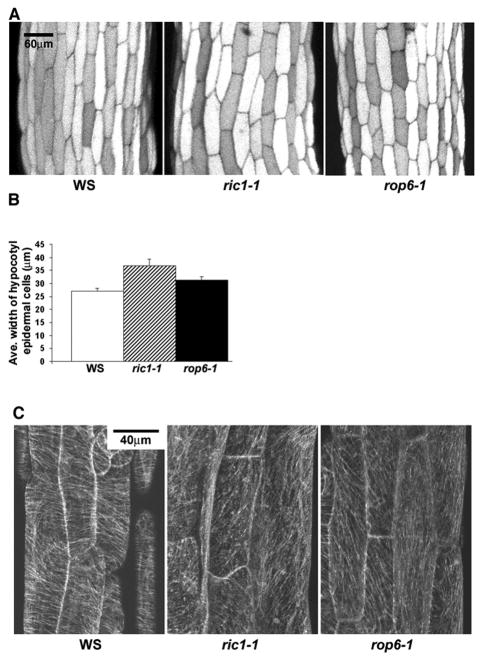
Transversely oriented MT arrangement is thought to allow anisotropic cell expansion preferentially in the direction perpendicular to the MTs, a process essential for plant development and morphogenesis. We next tested whether the ROP6-RIC1 pathway plays a role in the spatial regulation of this anisotropic cell expansion in hypocotyls. Knocking out ROP6 or RIC1 resulted in significantly wider cells (p<0.05) (Figure 4A and B; Figure S1B), whereas overexpression of ROP6 or RIC1 significantly reduced the width of hypocotyls cells, by 15% (30.3 μm ± 4.5, n=130) and 28% (25.6 μm ± 2.5, n=120), compared to wild type cells (35.4 μm ± 5.1, n=140), respectively. Furthermore, MT organization was affected in the hypocotyl epidermal cells of ric1-1 and rop6-1. Approximately 58% (n=36 from 4 seedlings) wild-type cells of 7-day-old seedlings displayed well-ordered transverse cortical MTs in parallel with each other and perpendicular to the direction of cell elongation (Figure 4C). In 36% wild-type cells, somewhat oblique (33%) or occasionally longitudinal (3%) but well-ordered MT arrays were observed. Only 6% wild type cells contained random oriented MTs. In most of the ric1-1 or rop6-1 cells, however, cortical MTs tended to be less bundled and oriented more obliquely and sometimes vertically compared to MTs in WT cells (Figure 4C). Approximately 39% ric1-1 cells (n=87 from 8 7-day-old seedlings) contained random cortical MT arrays. Well-ordered oblique or longitudinal MT arrays were found in In 32% or 26% of the ric1-1 cells, respectively. Transverse MTs were only found in 2% of the ric1-1 cells. Similarly, among rop6-1 cells (n=56 from 5 7-day-old seedlings, 41% of the cells had random oriented MTs, and well-ordered oblique or longitudinal MTs were found in 29% or 23%, respectively. Only 7% cells exhibited transverse MT arrays. The regulation of MT orientation involves rotary movements of cortical MT arrays [1], and thus ROP6 and RIC1 might participate in this regulation in hypocotyls. Taken together, our results clearly indicate that the ROP6-RIC1 pathway restricts radial or lateral cell expansion both in elongating cells and PCs with more complex spatial control by activating the ordering of cortical MT arrays.
Figure 4. Both ROP6 and RIC1 spatially regulate cell expansion and the organization of transversely oriented parallel cortical microtubules (MTs) in hypocotyl cells.
(A) Hypocotyl epidermal cells in ric1-1 and rop6-1 lines are wider than wild-type cells.
(B) Quantitative analysis of mean cell widths in wild type (WT), ric1-1 and rop6-1 lines, with significant differences in widths between WT and ric1-1 or rop6-1 (P<0.05, t-test). The mean width of rop6-1 cells appeared slightly narrower than that of ric1-1 cells, but the difference was not significant (p>0.05, t-test). In each line, a total of more than 100 cells from 3 seedlings was measured from the region about 250 um to 750 μm away from the root-hypocotyl junction.
(C) Stably expressed GFP-tagged tubulin revealed the MT organization in hypocotyl cells. ric1-1 and rop6-1 were backcrossed with α-tubulin-GFP transgenic line 3 times to clean the background. Transverse MTs were dominant in cells from 7-day old wild-type (WT) seedlings. However, less bundled and thinner MTs tended to form oblique and vertical arrays in ric1-1 and rop6-1 mutant cells.
Our findings reveal the first known signaling pathway that promotes the organization of cortical MTs into parallel arrays oriented perpendicular to the axis of cell elongation in plants. The ROP6-RIC1 pathway acts to restrict cell expansion in the direction paralleled to the orientation of MT arrays. Overexpression of RIC1 or ROP6 turns less ordered cortical MTs into highly ordered transverse cortical MTs, consequently changing wavy pavement cells into elongated cells with straight outlines, which demonstrates that the ordering of cortical MTs that is activated by the ROP6-RIC1 pathway is clearly important for the spatial control of cell expansion. This is consistent with the hypothesis that cortical MTs guide the cellulose synthase and microfibril arrangements, which subsequently determine the spatial pattern of cell expansion [6, 18-25]. However, the rop6 or ric1 knockout mutants, though lacking transversely ordered cortical MTs, did not exhibit isotropic cell expansion, which typically results from severe disruption of cortical MTs induced by chemical treatments or mutations in genes required for the formation of cortical MTs. In agreement with the finding that the orientation of cellulose microfibrils does not necessarily correlate with well-ordered cortical MTs [26], this observation suggests that other population(s) of cortical MTs that are not ordered transversely and are ROP6-independent may also play an important role in the spatial control of cell expansion in the absence of the highly ordered cortical MTs.
Our FRET analysis suggests that RIC1 interacts with ROP6 at the PM, whereas RIC1 is found to be associated with cortical MTs as punctuates. How could RIC1 activation by ROP6 at the PM induce RIC1 association with cortical MTs as punctuates? At least two possible mechanisms can explain these observations: (1) A transient interaction of RIC1 with active ROP6 alters RIC1 conformation causing RIC1 to be translocated to MTs. (2) Alternatively, RIC1 association with ROP6 could induce RIC1 modification such as phosphorylation.
Our results demonstrate a surprising conservation of the signaling mechanism regulating the organization of MTs across plant and animal kingdoms, given how the plant MT nucleation and dynamics behaviors differ from those in animal cells [27, 28]. Rho-family GTPases promote MT organization by regulating the orientation of the MT organization center and MT stability directly through their effector proteins such as mDia proteins [5, 29]. An interesting future question is how the ROP6 effector protein RIC1 controls the organization of cortical MTs into transversely arranged parallel arrays.
Supplementary Material
Acknowledgments
We thank David Ehrhardt and members of the Yang laboratory for stimulating discussions during this work, Takashi Hashimoto for generous gift of the tubulin-GFP line, and the Ohio State University Arabidopsis Stock Center for providing the T-DNA insertion population. This work is supported by funding from National Institute of General Medical Science (grant #1R01GM081451-01 to ZY) and National Natural Science Foundation of China (grant # 90817105 to YF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chan J, Calder G, Fox S, Lloyd C. Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat Cell Biol. 2007;9:171–175. doi: 10.1038/ncb1533. [DOI] [PubMed] [Google Scholar]
- 2.Dixit R, Chang E, Cyr R. Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol Biol Cell. 2006;17:1298–1305. doi: 10.1091/mbc.E05-09-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixit R, Cyr R. Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell. 2004;16:3274–3284. doi: 10.1105/tpc.104.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 5.Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell. 2005;120:687–700. doi: 10.1016/j.cell.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 6.Paradez A, Wright A, Ehrhardt DW. Microtubule cortical array organization and plant cell morphogenesis. Curr Opin Plant Biol. 2006;9:571–578. doi: 10.1016/j.pbi.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Sedbrook JC. MAPs in plant cells: delineating microtubule growth dynamics and organization. Curr Opin Plant Biol. 2004;7:632–640. doi: 10.1016/j.pbi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Fu Y, Li H, Yang Z. The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell. 2002;14:777–794. doi: 10.1105/tpc.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu JL, Jilk R, Marks MD, Szymanski DB. The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell. 2002;14:101–118. doi: 10.1105/tpc.010346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith LG. Cytoskeletal control of plant cell shape: getting the fine points. Curr Opin Plant Biol. 2003;6:63–73. doi: 10.1016/s1369-5266(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 11.Wasteneys GO, Galway ME. Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol. 2003;54:691–722. doi: 10.1146/annurev.arplant.54.031902.134818. [DOI] [PubMed] [Google Scholar]
- 12.Lavy M, Bloch D, Hazak O, Gutman I, Poraty L, Sorek N, Sternberg H, Yalovsky S. A Novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol. 2007;17:947–952. doi: 10.1016/j.cub.2007.04.038. [DOI] [PubMed] [Google Scholar]
- 13.Zheng ZL, Yang Z. The Rop GTPase: an emerging signaling switch in plants. Plant Mol Biol. 2000;44:1–9. doi: 10.1023/a:1006402628948. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z. Small GTPases: versatile signaling switches in plants. Plant Cell. 2002;14 Suppl:S375–388. doi: 10.1105/tpc.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131:1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G, Gu Y, Li S, Yang Z. A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell. 2001;13:2841–2856. doi: 10.1105/tpc.010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda K, Matsuyama T, Hashimoto T. Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma. 1999;206:201–206. [Google Scholar]
- 18.Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- 19.Smith LG, Oppenheimer DG. Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol. 2005;21:271–295. doi: 10.1146/annurev.cellbio.21.122303.114901. [DOI] [PubMed] [Google Scholar]
- 20.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 21.Korolev AV, Buschmann H, Doonan JH, Lloyd CW. AtMAP70-5, a divergent member of the MAP70 family of microtubule-associated proteins, is required for anisotropic cell growth in Arabidopsis. J Cell Sci. 2007;120:2241–2247. doi: 10.1242/jcs.007393. [DOI] [PubMed] [Google Scholar]
- 22.Ishida T, Kaneko Y, Iwano M, Hashimoto T. Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007;104:8544–8549. doi: 10.1073/pnas.0701224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto T, Kato T. Cortical control of plant microtubules. Curr Opin Plant Biol. 2006;9:5–11. doi: 10.1016/j.pbi.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Ehrhardt DW, Shaw SL. Microtubule dynamics and organization in the plant cortical array. Annu Rev Plant Biol. 2006;57:859–875. doi: 10.1146/annurev.arplant.57.032905.105329. [DOI] [PubMed] [Google Scholar]
- 25.Abe T, Thitamadee S, Hashimoto T. Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol. 2004;45:211–220. doi: 10.1093/pcp/pch026. [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto K, H R, Williamson RE, Wasteneys GO. Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell. 2003;15:1414–1429. doi: 10.1105/tpc.011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 28.Murata T, Hasebe M. Microtubule-dependent microtubule nucleation in plant cells. J Plant Res. 2007;120:73–78. doi: 10.1007/s10265-006-0054-z. [DOI] [PubMed] [Google Scholar]
- 29.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.