Abstract
We describe the potential of current Web 2.0 technologies to achieve data mashup in the health care and life sciences (HCLS) domains, and compare that potential to the nascent trend of performing semantic mashup. After providing an overview of Web 2.0, we demonstrate two scenarios of data mashup, facilitated by the following Web 2.0 tools and sites: Yahoo! Pipes, Dapper, Google Maps and GeoCommons. In the first scenario, we exploited Dapper and Yahoo! Pipes to implement a challenging data integration task in the context of DNA microarray research. In the second scenario, we exploited Yahoo! Pipes, Google Maps, and GeoCommons to create a geographic information system (GIS) interface that allows visualization and integration of diverse categories of public health data, including cancer incidence and pollution prevalence data. Based on these two scenarios, we discuss the strengths and weaknesses of these Web 2.0 mashup technologies. We then describe Semantic Web, the mainstream Web 3.0 technology that enables more powerful data integration over the Web. We discuss the areas of intersection of Web 2.0 and Semantic Web, and describe the potential benefits that can be brought to HCLS research by combining these two sets of technologies.
Keywords: Web 2.0, integration, mashup, Semantic Web, biomedical informatics, bioinformatics, life sciences, health care, public health
1. INTRODUCTION
Web 2.0 refers to a second generation of Internet-based services—such as social networking sites, wikis, communication tools, and folksonomies—that emphasize online collaboration and sharing among users (http://www.paulgraham.com/web20.html). If the first generation Web has revolutionized the way people access information on the Internet, Web 2.0 has revolutionized the way people communicate across the Internet. Web 2.0 has transformed the Web into an environment that provides richer user experiences by allowing for the combination of disparate information in a variety of data formats, the facilitation of interaction between multiple parties, and the collaboration and sharing of information. Web 2.0 consists of a variety of applications implemented using diverse technologies. In general, the variety of Web 2.0 applications can be classified as follows:
• Rich Internet applications
These applications behave very much like desktop applications, and are easy to install and easy to use. In particular, they provide a dynamic interface with interactive features like point-and-click/drag-and-drop. These interfaces are achieved with technologies such as Ajax (Asynchronous JavaScript and XML) (http://en.wikipedia.org/wiki/AJAX), and mini plug-in programs known variously as widgets, gadgets and snippets, which create a programming environment within the browser and allow the user to easily combine information and create a variety of graphical presentations. As a result of this progress, the gap between Web programming and desktop programming has been diminishing (http://blogs.adobe.com/shebanation/2007/02/desktop_application_programmin.html).
• Collaboration tools
These include asynchronous collaboration tools such as wikis and blogs, to which users do not need to be simultaneously connected at any given time to collaborate. This category also includes synchronous, real-time (or near real-time) collaboration enablers, such as leading-edge instant messaging tools.
• User-contributed content databases
These are large-scale environments—such as YouTube, a video posting Web site, and Flickr, a photo-sharing site—in which users share content in multimedia format.
• Integrative technologies enabling the Web as a platform
There are abundant services and data sources scattered over the Internet. While they may be accessed independently, it has been exceedingly challenging to integrate Web-based services to create novel functionality. Web 2.0 mashup offers a solution to this problem. Mashup tools like Yahoo! Pipes (http://pipes.yahoo.com/pipes/) offer a graphical workflow editor that allows the user to pipe Web resources together easily. Other tools like Dapper (http://www.dapper.net/) provide an easy way for users to extract (or scrape) Web contents displayed in heterogeneous formats and output the extracted contents in a standard format such as tab-delimited values and XML. Data visualization tools like Google Maps (http://maps.google.com/) and Google Earth (http://earth.google.com/) offer a GIS (Geographic Information System) interface for displaying and combining geographically related data. Despite their different functionalities, these tools may interoperate. For example, the output of Dapper may be fed into Yahoo! Pipes, and Yahoo! Pipes in turn can be linked to Google Map to process and display geographical data.
The rest of the paper is structured as follows. Section 2 gives an overview of data integration in health care and life sciences domains. Section 3 describes two scenarios demonstrating the use of a number of Web 2.0 tools/sites in achieving health care and life science data mashups. Section 4 discusses the strengths and weaknesses based on our experience with these Web 2.0 tools/sites. Section 5 introduces Web 3.0 with a main focus on Semantic Web and its potential application in health care and life sciences data mashup (semantic mashup). Section 6 discusses how Web 2.0 and Semantic Web can be combined to reap a greater benefit. Section 7 gives a conclusion. Finally, a glossary table is provided for defining/describing the terms related to Web 2.0/3.0 with examples.
2. HEALTH CARE AND LIFE SCIENCES DATA INTEGRATION
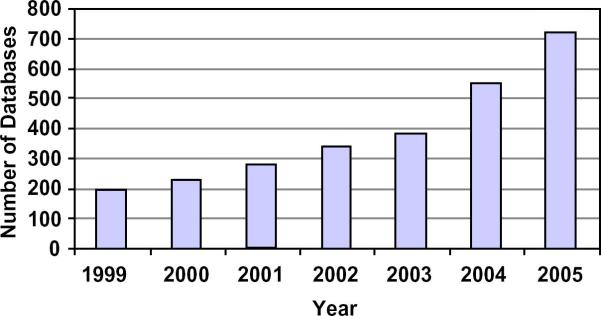
The popularity of the Web [1] and the success of the Human Genome Project (HGP) [2] have led to an abundance and diversity of biomedical data available via the Web. Figure 1 indicates the rate of growth in the number of Web-accessible biological databases that were published in the annual Database Issue of Nucleic Acids Research (NAR) between 1999 and 2005. These databases (which only represent a small portion of all biomedical databases in existence today) play an indispensable role in modern Health Care and Life Sciences (HCLS) research. They facilitate data mining and knowledge discovery [3]. The benefits for integrating these databases include the following:
HCLS data are more meaningful in context, while no single database supplies a complete context for a given HCLS research study.
New hypotheses are derived by generalizing across a multitude of examples from different databases.
Integration of related data enables validation and ensures consistency.
Figure 1.
Number of databases published in the NAR Database Issues between 1999 and 2005.
Via a Web browser, an HCLS researcher may easily access diverse information including DNA sequences, biochemical pathways, protein interactions, functional domains and annotations, gene expression data, disease information, and public health data. Integrating such data from diverse sources, however, remains challenging. Researchers wishing to analyze their own experimental data in combination with publicly available data face the cumbersome tasks of data preprocessing and cleaning [4], which includes scraping Web pages, converting file formats, reconciling incompatible schemas, and mapping between inconsistent naming systems. Even experienced programmers find such data integration tasks daunting and tedious.
A variety of approaches, including data warehousing [5, 6], database federation [6, 7], and Web services [8, 9], have been developed to facilitate data integration in the context of HCLS. One problem with these approaches is that they require their developers to have significant database/programming expertise. Moreover, these systems may not be able to anticipate or offer the flexibility needed by the end users (who may themselves not be well versed programmers). Furthermore, it is difficult if not impossible for these systems to keep up with the growth of Web data sources. There are very few such systems that allow the user to add new external data sources easily, especially ones that do not conform to standard data formats.
To address these problems, Web 2.0 mashups have emerged. A mashup is a Web application that combines multiple third-party services over the Web. Numerous mashup examples are available from www.programmableWeb.com. Most of the current mashups are for non-scientific use. The potential of data mashup in the HCLS domains has only recently been demonstrated by using Google Earth to geographically integrate and visualize different types of data, including epidemiological and public health data, to help track the global spread of avian influenza [10]. However, more HCLS use cases are needed to demonstrate the need and value of Web 2.0 mashups in the HCLS domains.
3. MASHUP SCENARIOS
We provide two scenarios that illustrate the use of several Web 2.0 mashup tools and sites to implement data integration in the HCLS domains. The first scenario, within a life sciences context, shows how to use Dapper and Yahoo! Pipes to integrate diverse data such as microarray measurements and gene annotation data. The second scenario, within a public health context, demonstrates how to geographically correlate cancer data with environmental data using Yahoo! Pipes, Google Maps, and GeoCommons (http://www.geocommons.com/).
3.1 Life Sciences Scenario
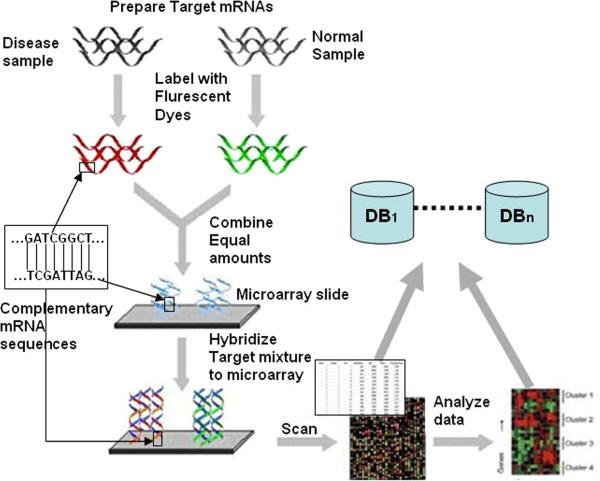
Figure 2 shows the workflow of a typical research study featuring the use of a spotted microarray, one kind of microarray technology. As shown in the figure, two biological samples (normal vs. disease), which consist of quantitatively distinct distributions of mRNA sequences, are labeled with fluorescent dyes. Sequences transcribed from the disease sample mRNA are labeled with the red fluorescent dye and sequences transcribed from the normal sample mRNA are labeled with the green fluorescent dye. Next, the two labeled samples are mixed in equal total amount, and that mixture is allowed to “hybridize” (bind) to the affixed reference sequences that have been deposited on the surface of a chemically-treated microscopic glass slide. Each spot on the slide contains many strands of the DNA sequence corresponding to one specific gene. A large number of spots, and therefore many gene sequences, may be featured on a given slide.
Figure 2.
A typical research workflow that involves the use of microarrays.
After hybridization is complete, the slide is scanned by a laser scanner that measures the amount of each dye at the scale of 5-10 μm pixels. Associated image processing software assembles the pixels into an image consisting of spots whose average pixel intensity values convey levels of gene expression. The color of a spot indicates how much the corresponding gene expresses in the disease sample relative to the normal sample. For example, a red or green spot means respectively that the gene is primarily expressed in the diseased or normal sample. If a spot is yellow, it means that the gene is equally expressed in both samples. If a spot is black, it means that the gene is not expressed or only meagerly expressed in both samples.
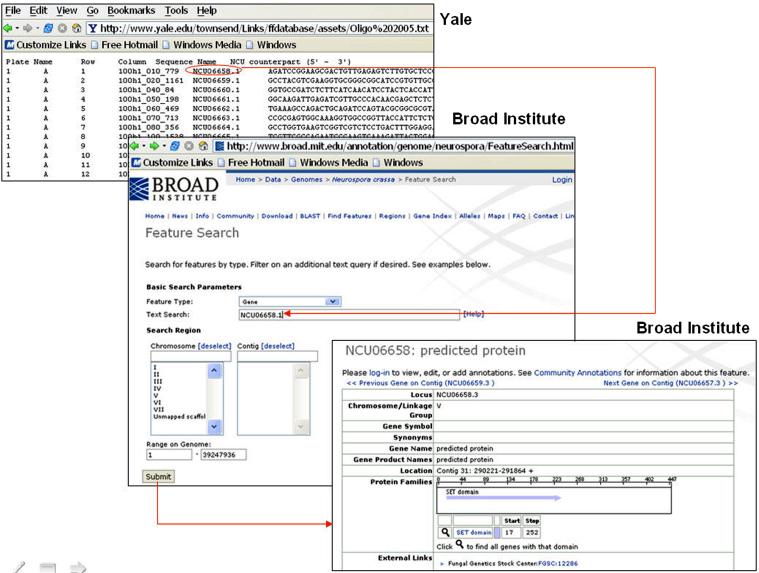
The imaging software processes the image data to produce a spreadsheet file of quantitative measurements of the image. This file, which contains rows corresponding to genes and columns corresponding to different types of measurements such as red intensities, green intensities and ratios, may be subjected to data analyses for statistical interpretation of the results. Such interpretation gains dramatically more meaning if the numerical output is integrated with known biological knowledge (e.g., gene annotation); yet such knowledge is frequently provided by diverse continuously-updated databases. In our scenario, we integrated data from two Web sites, one hosted at Yale University, and the other at the BROAD Institute [11]. The Yale site provides microarray data generated from microarray experiments studying the gene expression profiling of Neurospora crassa, a red bread mold. The data are presented in the form of a tab-delimited file, with the columns describing different properties of the spots of a microarray slide, including their locations, gene identifiers, and mRNA sequences. To find current information about each of the genes listed in this file, one may go to the BROAD Institute site to search for the gene annotation in its Neurospora crassa database. An example search and the corresponding search results are illustrated in Figure 3, where the gene identifier NCU06658.1 was used as the search term. The search result is a page containing assorted annotations of the gene, such as its name, chromosome number, and exact location in the genome.
Figure 3.
Microarray data and gene annotation provided by two sites.
Currently the most common way to perform this kind of data mashup is to write scripts (in languages such as Perl) to:
Parse the tab delimited file and extract the gene identifiers.
For each identifier, construct a URL that corresponds to the search result page of the gene, and retrieve the content of the page.
Parse the result page to extract the data fields of interest.
Merge the extracted data fields with the original tab-delimited file to produce the integrated dataset.
This traditional approach has a number of shortcomings:
Parsing HTML pages, especially those with potentially minor formatting discrepancies, is difficult and error-prone.
The scripts may not be easily updated when there are changes to the data sources.
It is difficult to reuse and share the scripts among different researchers. For instance, it is very common that when a graduate student or a postdoctoral fellow leaves a laboratory, the scripts written by him/her are not sufficiently documented for others to understand. In many cases other members of the laboratory resort to rewriting the scripts from scratch when the old ones fail to work due to changes at the data source side.
As we will discuss in Section 5, an ultimate solution to these problems involves standardizing data formats and adding semantic annotations, so that machines could process the data in a largely automated way. Yet before such semantically rich data are widely available, it is desirable to have some semi-automatic tools that facilitate data integration while minimizing the above issues. We have found that some Web 2.0 tools, such as Dapper and Yahoo! Pipes, serve this purpose well. Here we describe how such tools were used to perform the above data mashup task easily.
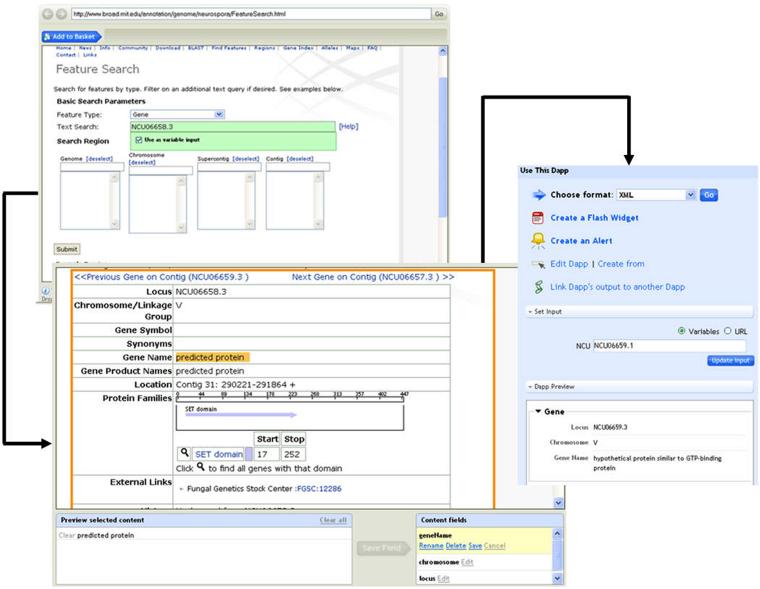
The parsing of HTML pages was handled by the Web tool Dapper. Use of the tool consisted of two phases: learning and applying. In the learning phase, Dapper took the search result pages of some genes as input (Figure 4, step 1), and asked the human trainer to mark on the screen the parts of the content that corresponded to the data fields of interest (Step 2, with the Gene Name field selected as an example). The gene identifier was set as a query parameter that would be changed dynamically for different genes (Step 1, green box). Using some machine learning algorithms, the back-end system of Dapper then learned the locations of the data fields in the HTML pages from the examples. The resulting product, called a “dapp”, was the data extraction proxy of the BROAD Institute site”. In the applying phase, when the dapp was presented a new gene identifier, it extracted the corresponding data values of the gene from the site and output them in standard XML format (Figure 4, step 3).
Figure 4.
A Dapper interface for querying and retrieving gene annotation.
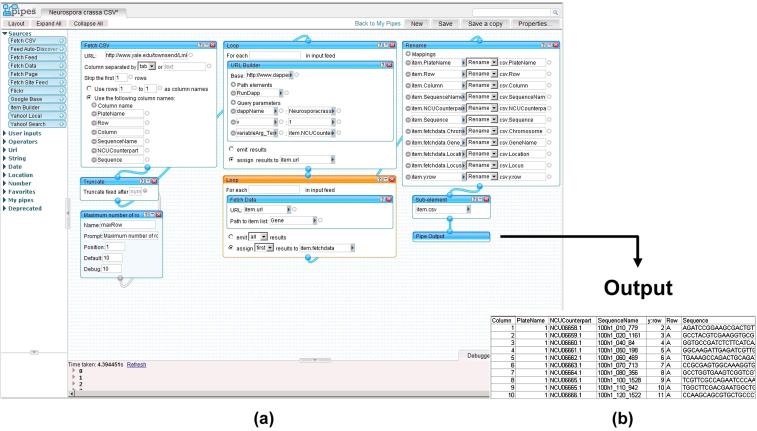
The dapp was then used as a data source to be integrated with the Yale tab-delimited file using Yahoo! Pipes. It is a tool that treats data as water flowing in pipes, and allows users to use different widgets to process their data, and connects the widgets like connecting pipes.
As shown in Figure 5, the Yahoo! Pipes tool has three panels: library, canvas, and debugger. The library panel lists categories of widgets that allow functions such as data fetching, filtering, and manipulation. The canvas panel allows the selected widgets to be placed, moved, and connected. The debugger panel is below the canvas panel, and it displays the output or error messages when the pipe is executed. The specific pipe used for our data mashup task is shown in the canvas panel. It starts with a “Fetch CSV” widget to fetch the tab-delimited data table from the Yale site. The output of the widget is piped to a “Truncate” widget for limiting the total number of rows in the result, which we set as 10 for demonstration. Then we used a “Loop” widget to iterate through each row to construct a URL to the dapp using the gene identifier, and another “Loop” widget to actually retrieve the content of the dapp output. Finally, all unwanted fields were filtered and the dataset was output as a comma-separated-value (CSV) file.
Figure 5.
(a) A Yahoo! pipe for mashup of microarray data and gene annotation and (b) integrated output.
The whole mashup process did not involve any coding. The user interfaces of the two tools were simple and intuitive enough for non-programmers to use. The difficult task of HTML parsing was handled by dedicated learning algorithms of Dapper, which, compared to most custom scripts, requires much less work by the user.
3.2 Public Health Scenario
Environmental health epidemiologists study the association between human diseases (e.g., cancer) and environmental factors. Such studies often require the integration of disparate data sources such as population census, air quality and environmental pollution release, and health care utilization data. These different data streams are typically produced by different agencies. Automated integration of data from these agencies is limited due to a variety of political and technological challenges. Web 2.0 mashups offer the potential for automating the integration of disparate health care data to enhance environmental health research. As an example, we demonstrate how to use Yahoo! Pipes and a Web 2.0 site called “GeoCommons” to geographically correlate cancer data with water pollution data in the United States.
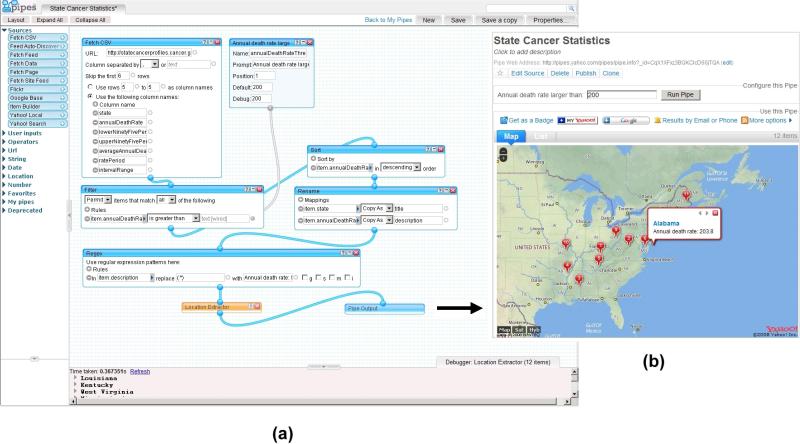
First, we identified a cancer profile dataset at the State Cancer Profiles Web site (http://statecancerprofiles.cancer.gov/map/map.noimage.php) developed by the National Cancer Institute (http://gis.cancer.gov/). This tabular dataset contains annual death rates for all types of cancers in different US states (the year of this data collection is 2004). We created a pipe as shown in Figure 6 (a) to fetch this cancer data table and applied a user-defined threshold against the annual death rates. The filtered output was fed to a “location extractor” widget that allows the states that have annual cancer death rates above the specified threshold to be displayed via Google Maps, as shown in Figure 6 (b). The map was then exported to a KML file (a standard XML format for Google Maps/Earth).
Figure 6.
(a) A Yahoo! pipe for filtering US state cancer profile data and (b) display the results using Google Maps.
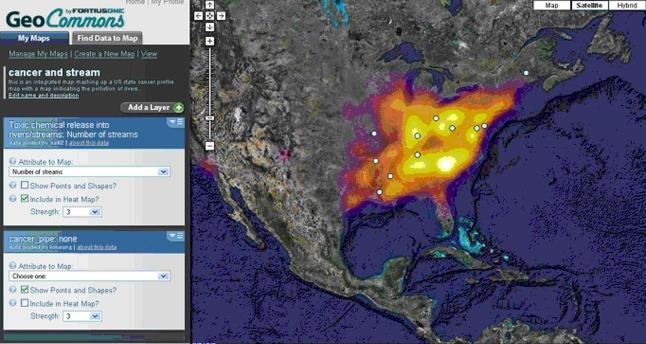
We uploaded the KML file to the GeoCommons Web site (http://www.geocommons.com). This site allows users to annotate and publish their uploaded maps as well as mashup the digital maps uploaded by other users. In this example, we found a “heat” map that details the number of polluted rivers/streams in the US. In a heat map, a brighter color corresponds to a higher number of polluted rivers/streams. Figure 7 shows a GeoCommons interface that allows the state cancer profile map to be superimposed with the water pollution map. We can see that most of the states with high cancer death rates are in the fire zone.
Figure 7.
A mashup of the state cancer profile map and water pollution map.
4. STRENGTHS AND WEAKNESSES
In this section, we discuss the general strengths and weaknesses of Web 2.0 mashup technologies based on our current experience in using them to integrate HCLS data. We have identified the following strengths.
• Applicability
The tools that we used in the mashup examples are useful for diverse areas of biomedical research. For example, Yahoo! Pipes supports a great variety of input and output data types that biomedical researchers need to deal with, from the most popular tab-delimited format to structured XML and semantically rich RDF. Common mashup tasks such as data integration by means of ID mapping can be performed without coding.
• Ease of use
As demonstrated by the mashup examples, tools like Dapper and Yahoo! Pipes provides an easy-to-use Web interface for extracting and integrating data from diverse sources. Extraction and integration with these intuitive tools is easier than writing code in a particular programming language (e.g., Perl) to parse and integrate data.
The tools in general have intuitive designs that require little learning time for beginners. New users are also greatly assisted by the active user community in solving their technical problems through reading or joining in related discussions at designated online message boards.
• Reusability and extensibility
Web 2.0 mashup tools like Yahoo! Pipes and Dapper are designed for sharing and reuse. For example, the Yahoo! Pipes site allows its users to describe and publish their pipes. Through its “Show off your Pipe” message board, users can comment and rank each other’s pipes. In addition, the shared pipes can be easily extended or modified by others to add new features. For instance, it is straightforward to take components from several publicly shared pipes to form a new, customized pipe.
• Interoperability
As shown in our examples, different Web 2.0 tools can be easily combined to enhance the mashup capability. For example, Yahoo! Pipes can be complemented by Dapper by allowing fetching of data in formats that are not supported by Yahoo! Pipes. In addition, Dapper provides an Application Programming Interface (API) that allows Web services for searching the dapps and software development toolkits (e.g., in Perl and Java) for accessing dapps programmatically.
• Active roles of users
Web 2.0 applications emphasize the active participation of users in reporting bugs, suggesting new functions, or even implementing new features through specific software development kits (SDK). These activities facilitate the improvement of applications much more rapidly than in traditional software engineering paradigms.
In spite of these strengths, we have experienced and would note several issues that arise in creating data mashups using the tools.
• Missing features and instability
Tools like Yahoo! Pipes and Dapper are relatively new, and are still under active development. Since many of these tools were initially designed for casual lightweight mashup tasks such as aggregating news feeds from a small number of Web sites, their designs did not incorporate a breadth of computational theory. For example, while Yahoo! Pipes provides operations commonly found in database query languages, such as selection and renaming, some other essential operations such as column selection (i.e., “projection” in database terms) and table-joining are currently either not supported or supported only in arcane ways. Many such features are needed in order for these tools to be widely adopted for daily research activities.
Additionally, these new tools still contain bugs. In particular, due to the heavy use of client-side scripting (e.g. JavaScript), these tools are especially prone to errors that arise from the many brands and versions of browsers that are in use today but not completely compatible. Moreover, as with any Web servers, a Web 2.0 site may become unreachable without prior warnings.
• Performance and scalability
Given the distributed nature of the Web and the limited speed of the network connections, mashing up large datasets from different sources can be very slow. We encountered this problem when attempting to integrate the whole microarray data table (consisting of tens of thousands of rows) with the corresponding annotation data. There was a timeout when we executed the pipe for the entire table. The largest number of rows that we were able to integrate successfully using our pipes was around 1500, and the task took about 1.5 minutes to run. In comparison, with all the datasets stored locally, integrating tens of thousands of rows should not take more than a few seconds using a customized script.
• Security
Most Web 2.0 sites do not have a strong security policy for their users. The users have to bear the security risks if they upload their data to these Web 2.0 sites. Although the user may choose not to publish their data to the public, he/she loses control of the data once the data are uploaded to a Web 2.0 server. The security is at the mercy of the person(s) in charge of the server security. Therefore, it is not recommended to use public Web 2.0 sites to share sensitive/confidential data.
• Flexibility
Although the Web 2.0 tools are found to be very useful in our two data mashup scenarios, by nature they are not as flexible as customized scripts. There are always some special cases that the standard widgets cannot handle properly. One solution, which is already adopted by Yahoo! Pipes, is to allow users to supply customized Web services as widgets. This is a promising approach in general, although standard Simple Object Protocol (SOAP) based Web services (http://xml.coverpages.org/soap.html) are still not yet supported.
• Quality of final output
Professional users are unlikely to switch to Web 2.0 tools until the aesthetic quality of the final graphical or tabular output matches the quality that may be achieved with local software.
5. HCLS 3.0
According to Spivacks (http://novaspivack.typepad.com/nova_spivacks_weblog/2006/11/web_30_versus_w.html), Web 3.0 refers to “a supposed third generation of Internet-based services — such as those using Semantic Web, microformats, natural language search, data-mining, machine learning, recommendation agents, and artificial intelligence technologies — that emphasize machine-facilitated understanding of information in order to provide a more productive and intuitive user experience.” Semantic Web (SW) technologies play a core role in this definition.
The World Wide Web Consortium (W3C) has launched the Semantic Web for Health Care and Life Sciences Interest Group (HCLSIG; http://www.w3.org/2001/sw/hcls/), which has been chartered to develop and support the use of SW technologies to improve collaboration, research and development, and innovation adoption in the HCLS domains [12]. One of the ongoing efforts involves converting a variety of HCLS data sources into the standard Semantic Web data formats endorsed by W3C: Resource Description Framework (RDF) (http://www.w3.org/RDF/) and Web Ontology Language (OWL) (http://www.w3.org/TR/owl-ref/) formats. While OWL is semantically more expressive than RDF (http://www.w3.org/TR/owl-ref/), OWL and RDF bear the same syntax. Datasets expressed in either format can be queried by the standard RDF query language — SPARQL (http://www.w3.org/TR/rdf-sparql-query/). For OWL datasets (ontologies), tools such as Pallet (http://www.mindswap.org/2003/pellet/), RacerPro (http://www.racersystems.com/), and Fact++ (http://owl.man.ac.uk/factplusplus/) can be used to perform OWL-based reasoning. At WWW 2007, a demonstration organized by the HCLSIG showed how to use SPARQL to query across a number of OWL ontologies in the Alzheimer’s disease research context. In addition, Semantic Web applications such as YeastHub [13], SWAN [14], and BioDash [15] have already emerged in the HCLS domains.
While Web 2.0 offers human-friendly tools for mashing up data, Semantic Web [16] better enables computers to help human users find and integrate information over the Internet, and to perform such activities in a more sophisticated way. As pointed out by Ankolekar et al. [17], Web 2.0 and Semantic Web are not two conflicting visions. They are, instead, complementary to each other. There is a potential benefit to mashing up Web 2.0 and Semantic Web in the context of HCLS. To implement the vision of Semantic Web, more datasets need to be converted into RDF/OWL formats. This conversion process may be facilitated by Web 2.0 tools that can be used to extract and aggregate non-SW content from numerous Web sites, producing data converted into RDF/OWL. Furthermore, Web 2.0 tools may be used to assist users to annotate a small amount of data. Such small annotated data sets may then be used as examples to train automatic annotation algorithms.
Currently, many Web 2.0 tools can process RSS feeds (which use a simple RDF structure). It would be desirable for these tools to be able to understand semantically richer formats like RDF Schema (RDFS) and OWL, thus supporting richer and possibly more intelligent integration. For example, SPARQL may be supported by future Web 2.0 tools for fetching, filtering, and aggregating RDF/OWL data sources. In addition, “RDF-attributes” or RDFa (http://www.w3.org/TR/xhtml-rdfa-primer/) has been proposed by W3C as an alternative to microformat for embedding ontological elements into existing HTML (more precisely XHTML) documents, mashing up human readability and machine readability. It would be logical for future Web 2.0 tools (e.g., Dapper) to recognize RDFa, even though RDFa parsing tools like GRDDL (Gleaning Resource Descriptions from Dialects of Languages) (http://www.w3.org/2004/01/rdxh/spec) are available.
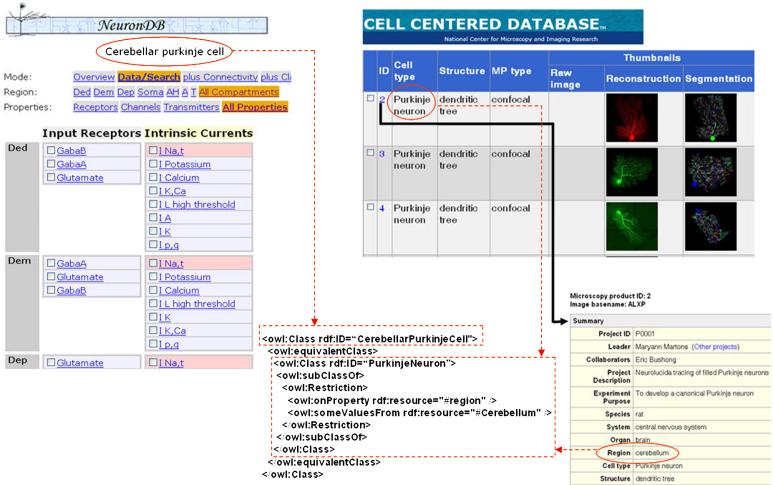
Figure 8 depicts an example demonstrating implementation of semantic mashup between existing Web pages using RDFa. On the left of Figure 8, a Web page of NeuronDB (http://senselab.med.yale.edu/neurondb/) shows the neuronal properties including receptors (e.g., GabaA and GabaB) and currents (e.g., I Potassium and I Calcium) located in different compartments (e.g., Dad, Dem, and Dep) of the “cerebellar purkinje cell”. On the right of Figure 8, there are 2 linked Web pages of the Cell Centered Database (CCDB) (http://ccdb.ucsd.edu/). The top Web page shows the different neuronal images for “purkinje neuron”, while the bottom page shows the detailed information about the “purkinje neuron”, including the region in the brain where the neuron is located. In this case, it is located in the “cerebellum”. Using RDFa, we can associate ontological fragments (in OWL format) with HTML elements. The OWL components (represented by dotted rectangles) corresponding to the circled HTML elements are shown in Figure 8. The semantic relationships are explicitly expressed using the OWL-DL syntax. For example, in CCDB, the class “PurkinjeNeuron” has a property named “region” whose value is “Cerebellum”. In addition, semantic mashup is achieved using the “equivalentClass” construct supported by OWL-DL. In this case, the NeuronDB class “CerebellarPukinjeCell” is equivalent to the CCDB class “PurkinjeNeuron” whose region property has the value “Cerebellum”.
Figure 8.
Semantic mashup between existing Web pages.
To take the concept of RDFa further, we may entertain the possibility of extending it to work for any XML format rather than limiting its domain to XHTML). One main benefit of such an extension is that existing visualization tools like Google Maps use XML as the input data format. Embedding ontologies in these XML formats would add a querying capability for ontology, while exploiting the visualization capability currently supported by existing tools. For example, if some geo-ontologies are integrated into Keyhole Markup Language (KML) (http://code.google.com/apis/kml/), geographic mashup by Google Maps/Earth may be performed in a fully semantic manner.
With regard to the cancer data mashup, we have encountered some cancer-related data that are tallied within geographic regions that exhibit different granularities. Some data may be collected at the city level, while other data may be collected at the county or state level. To support semantic mashup based on locations, one may define an ontology in which a city (e.g., North Haven) is located in a county (e.g., Greater New Haven), which is in turn located in a state (e.g., Connecticut). Given such an ontology, location-based inference may be performed when mashing up data.
The Semantic Web community has been working with data providers to convert their data into RDF/OWL ontologies. While the ultimate goal is to come up with heavy-weight (semantically rich) ontologies for supporting sophisticated machine reasoning, it may be worthwhile to also provide coarser ontologies that can be easily incorporated into future Web 2.0 tools. Currently these tools use tags and folksonomies to annotate and categorize content. A mashup of folksonomy and ontology merits exploration. For example, popular tags may evolve into standard terms. In addition, taxonomic or hierarchical relationships may be identified among existing tags. This bottom-up approach may allow social tagging to evolve into the development of standard ontologies. This evolution is reflected by the transformation of social wiki into semantic wiki. Instead of tagging wiki pages based on user-defined terms, semantic wiki tools such as ontowiki (http://ontowiki.net/Projects/OntoWiki) allow users to semantically (ontologically) annotate Web pages. The semantic mashup scenario depicted in Figure 8 can potentially be achieved using semantic wiki as well. In this case, OWL-formatted metadata will be generated for facilitating semantic data mashup.
HCLS represents flagship domains in which SW applications may be developed and shown to be successful (http://www.thestandard.com/internetnews/001301.php). One possible direction for future work may be to develop SW applications that would provide the infrastructure to support semantic mashup of HCLS data in a user-friendly and social-friendly fashion. We therefore envisage a transformation from Web 2.0 mashup to Web 3.0 semantic mashup, producing a better synergy between human and computer.
6. HCLS 2.0 + HCLS 3.0 = e-HCLS
e-Science describes science that is increasingly done through distributed global collaborations enabled by the Internet, using very large data collections, large-scale computing resources, and high performance visualization (http://e-science.ox.ac.uk/public/general/definitions.xml). It involves two components: semantic components and social components. e-HCLS is e-Science within the HCLS context. While the Semantic Web has the potential to play an important role in the semantic representation of e-Science, Web 2.0 has the potential to transform from the so-called “me-Science” (http://www.gridtoday.com/grid/963514.html), that is driven by an individual researcher or laboratory, into what we call “we-Science”, which is driven by community-based collaboration. The mashup scenarios described in our paper shed some light on the potential impact of social networking on HCLS.
Our public health data mashup scenario has demonstrated the benefit of sharing data (maps) in the community. Once the data are shared in a standard format (e.g., KML), visualization and integration may be readily achieved. While different groups have independently created different maps (e.g., cancer profiles and environmental pollution) to meet their own needs, new insights or knowledge can be derived when these maps are mashed up. This mashup is made possible by providing a global information commons like GeoCommons.
The microarray mashup scenario has illuminated the importance of data integration in data mining/analysis. Web 2.0 can potentially be used to create a social network that facilitates collaboration between microarray data providers and microarray data miners. In this case, via a microarray data commons (Web 2.0 site), data providers can publish their datasets, while data miners can publish their data analysis algorithms/programs. This way, not only can the data providers search for the appropriate tools for analyzing their datasets, but the data miners may also search for appropriate datasets for testing their analysis methods. They may furthermore make comments about their experience of using certain datasets/tools. Lastly, they can use the site to publish analysis results and to allow others to make comments about them. Currently, public microarray Web sites such as Gene Expression Omnibus (GEO) [18] and ArrayExpress [19] do not support this type of social networking.
A number of social networking sites/projects have emerged, which are tailored to the needs of different HCLS communities. For example, Alzforum (http://www.alzforum.org/) is a site that facilitates communication and collaboration within the Alzheimer’s Disease (AD) research community. It also allows its members to comment on AD research articles and publish such comments. Connotea (http://www.connotea.org/) is a free online reference management for all researchers, clinicians and scientists. myExperiment (http://myexperiment.org/) is a beta tool that allows scientists to contribute to a pool of scientific workflows, build communities and form relationships. In contrast to traditional peer-reviewed publication, Nature Precedings (http://precedings.nature.com/) is a site for researchers to share documents, including presentations, posters, white papers, technical papers, supplementary findings, and manuscripts. It provides a rapid way to disseminate emerging results and new theories, solicit opinions, and record the provenance of ideas. It would be interesting to see: i) how these sites would enable discovery, creativity and innovation, and ii) whether a larger social network can be formed if these social network sites are interoperable.
The Web 2.0/3.0 data mashup scenarios we have described are based on the assumption that the data are publicly accessible without the concern about security. However, this concern becomes real when mashing up sensitive healthcare data such as medical administrative data including hospital discharge data, claims data, medical records, and so on. The ability to integrate medical administrative data from different sources is crucial to outcome research [20]. The access to these medical administrative databases is restricted to approved researchers. In addition, it is often a requirement that manipulation, analysis, and transmission of such data need to be done in a secure manner. Developers have begun to explore how to provide a secure mechanism for mashing up sensitive data. For example, IBM has recently announced “SMASH”, which is a new technology for supporting secure data mashup (http://www.physorg.com/news124641823.html).
7. CONCLUSION
We have demonstrated the feasibility of using a variety of Web 2.0 mashup tools/sites including Dapper, Yahoo! Pipes, Google Maps, and GeoCommons to integrate complementary types of HCLS data provided by different sources in different formats. These tools may be used by people without programming experience to perform lightweight but useful data mashup over the Web. Despite their growing popularity in the civic domains, there is room for improvement of these tools to facilitate wider use in the scientific (e.g., HCLS) domains. Increased benefits will accrue if Web 2.0 is used to transition toward Web 3.0, such as Semantic Web, facilitating heavyweight semantic data mashup and social networking in the HCLS domains.
GLOSSARY.
| Terms | Description/Definition | Examples/URL’s |
|---|---|---|
| AJAX | It stands for “Asynchronous JavaScript and XML”, is a group of inter-related web development techniques used for creating interactive web applications. A primary characteristic is the increased responsiveness and interactivity of web pages achieved by exchanging small amounts of data with the server “behind the scenes” so that entire web pages do not have to be reloaded each time there is a need to fetch data from the server. This is intended to increase the web page’s interactivity, speed, functionality and usability. |
Google Maps (http://maps.google.com/) is an example of AJAX application. |
| Blog | It is a “Web journal” or “Web log”, which is a specialized Web service that allows an individual or group of individuals to share a running log of events and personal insights with online audiences. |
Life sciences blog (http://www.lsblog.org) Life Sciences Blog is an attempt to record anything that sounds interesting in the rapidly evolving universe of biosciences. It blogs about fields such as molecular biology, genetics, drug discovery, clinical trials, gene therapy, stem cell research, cancer research, cardiovascular diseases, diabetes and nanotechnology. |
| Folksonomy | It is also known as “social tagging”. It is the practice of collaboratively creating and managing tags to annotate and categorize content. In contrast to traditional subject indexing, metadata is not only generated by experts but also by creators and consumers of the content. Usually, freely chosen keywords are used instead of a controlled vocabulary. |
Del.icio.us (http://del.icio.us/) is a social bookmarking web service for storing, sharing and discovering web bookmarks. Users can tag each of their bookmarks with a number of freely chosen keywords. |
| Gadget/ Widget |
A Web gadget/widget is a mini-web application you can put in your web page, blog or social profile that can quickly and easily provide your visitors with, user specific information, extra functionality, and even a bit of fun and games. A gadget can be considered as a primitive widget. |
Google Gadgets (e.g., calculator, calendar and thermometer) are miniature objects that offer dynamic content that can be placed on a Web page. SnapShot (http://www.snap.com) is a widget that allows users to mouse-over links to get the most appropriate shot of content for that link. |
| Information commons |
An information commons provides access to information resources by a community of producers and consumers in an open access environment. |
An example is the recently launched Pathway Commons (http://www.pathwaycommons.org/) that serves as a central point of access to biological pathway information collected from public pathway databases. |
| Mashup | In the Web context, mashup is a Web application that combines data and/or functionality from more than one source. |
Geocommons (http://www.geocommons.com/) provides geo-mashup by providing a Web interface that allows users to select different maps and overlay them one on top of the other. |
| Ontology | In both computer science and information science, an ontology is a representation of concepts with a domain and the relationships between those concepts. It is a shared conceptualization of a domain. |
Gene ontology (http://www.geneontology.org) is a popularly used ontology in biomedical informatics. It provides a controlled vocabulary to describe gene and gene product attributes in any organism. It involves three categories of information, namely, biological processes, molecular functions, and cellular locations. |
| OWL | It stands for Web Ontology Language. It is a family of knowledge representation languages for encoding ontologies, and is endorsed by the World Wide Web Consortium. This family of languages includes: OWL-Lite OWL-DL, and OWL-Full. The semantics of OWL-Lite and OWL-DL are based on Description Logics, while OWL-Full uses a novel semantic model intended to provide compatibility with RDF Schema. |
Gene ontology is also available in OWL format (http://www.geneontology.org/GO.downloads.ontology.shtml). |
| RDF | It stands for Resource Description Framework. It represents a framework for representing information in the Web. It provides a graph data model. The underlying structure of any expression in RDF is a collection of triples (node-arc-node links), each consisting of a subject, a predicate, and object. Each subject, predicate, or object can be identified by a URI (Uniform Resource Identifier). |
Gene ontology is available in RDF format (http://www.geneontology.org/GO.downloads.ontology.shtml). |
| RDF Schema |
RDF schema provides constructs for defining the vocabularies (terms) users intend to use in RDF statements. These constructs include class, property, type, subClassOf, range, and domain. These constructs are expressed in RDF syntax. |
The following example illustrates an RDF schema defining four classes: DNASequence, Promoter, Protein, and TranscriptionFactor. Promoter is a subclass of DNASequence, whereas transcriptionFactor is a subclass of Protein. Bind is a property whose domain is TranscriptionFactor and whose range is Promoter. <DNASequence, type, Class> <Promoter,subClassOf,DNASequence> <Protein,type,Class> <TranscriptionFactor,subClassOf,Protein> <Bind, type,Property> <Bind, domain, TranscriptionFactor> <Bind, range, Promoter> |
| RDFa and GRDDL |
It stands for RDF attribute. It is a set of extensions to XHTML being proposed by W3C. RDFa uses attributes from XHTML’s meta and link elements, and generalizes them so that they are usable on all elements. This allows one to annotate XHTML markup with semantics. A simple mapping is defined so that RDF triples may be extracted. GRDDL is a markup format for “Gleaning Resource Descriptions from Dialects of Languages” such as RDFa. It is a W3C Recommendation, and enables users to get RDF out of XML and XHTML documents via XSLT. |
An illustrative of example of how to use RDFa and GRDDL in a digital library context is given via the following URL: http://wwwsop.inria.fr/acacia/personnel/Fabien.Gandon/tmp/grddl/rdfaprimer/PrimerRDFaSection.html. |
| Social networking |
It is a phenomenon defined by linking people to each other in some way. Users work together to rate news and are linked by rating choices or explicit identification of other members. Generally, social networks are used to allow or encourage various types of activity whether commercial, social or some combination of the two. |
Digg (http://www.digg.com/), which is a site for people to discover and share content from anywhere on the web, is an example of a social network (using social bookmarking). Digg has a tool called “Digg labs” that provides visualization of the social network beneath the surface of the Digg community’s activities. |
| SPARQL | It is an RDF query language (http://www.w3.org/TR/2006/WD-rdf-sparql-query-20061004/). It is standardized by the RDF Data Access Working Group (DAWG; http://www.w3.org/2001/sw/DataAccess/) of the World Wide Web Consortium. |
The following SPARQL query returns all neurons for each brain region within “Diencephalon”. PREFIX neurondb: <http://neuroweb.med.yale.edu/svn/trunk/ontology/senselab/neuron_ontology.owl#> SELECT ?brain_region_label ?neuron_label WHERE { ?neuron rdfs:label ?neuron_label. ?brain_region neurondb:has_part ?neuron; rdfs:label ?brain_region_label. neurondb:Diencephalon neurondb:has_part ?brain_region |
| Tag cloud | A tag cloud is a visual depiction of user- generated tags. The importance or popularity of a tag is shown with font size or color. For example, the bigger the font size, the more popular the tag. |
A tag cloud was created (http://en.wikipedia.org/wiki/Image:Web_2.0_Map.svg) to show the Web 2.0 related terms found in an article written by Tim O’Reilly summarizing his view of web 2.0 (http://www.oreillynet.com/lpt/a/6228). |
| Web 2.0 | As described in (http://www.oreillynet.com/pub/a/oreilly/tim/news/2005/09/30/what-is-web-20.html) , Web 2.0 includes the following key features: 1) User Centric and User Oriented; 2) Web Services, Web API’s; 3) Widgets, Gadgets, Mashup’s; 4) Blogs, Feeds, Wiki’s, Tagging; 5) Social networking; 6) Client rich technologies like AJAX |
Web sites like Flickr (http://www.flickr.com/), YouTube (http://www.youtube.com/), and MySpace (http://www.myspace.com/) possess Web 2.0 features. |
| Web 3.0 |
Web 3.0 is a term used to describe the future of the World Wide Web. Following the introduction of the phrase Web 2.0 as a description of the recent evolution of the Web, many people have used the term Web 3.0 to hypothesize about a future wave of Internet innovation. |
Semantic Web is a kind of Web 3.0 technology extending the Web such that the semantics of information and services on the Web is defined, making it possible for the Web to understand and satisfy the requests of people and machines to use the Web content. |
| Web service | A Web service is defined as a software system designed to support interoperable machine-to- machine interaction over a network. Web services are frequently just Web APIs that can be accessed over a network, such as the Internet, and executed on a remote system hosting the requested services. |
SOAP (Simple Object Access Protocol) (http://www.w3schools.com/SOAP/soap_intro.asp) is an XML-based protocol for accessing Web services over HTTP (HyperText Transfer Protocol). BioMoby (http://www.biomoby.org) is a registry for Bioinformatics (SOAP) Web Services. In contrast to SOAP, JSON (JavaScript Object Notation) is a non-XML, lightweight data-interchange format. It is easy for humans to read and write, while being machine readable. |
| Wiki | A wiki is a software program that allows users to collaboratively create, edit, link, and organize the content of a website, usually for reference material. Wikis are often used to create collaborative websites and to power community websites. A semantic wiki is a wiki that has an underlying model of the knowledge described in its pages. Semantic wikis allow the ability to capture or identify further information about the pages (metadata) and their relations. |
Besides the well-known Wikipedia, (http://www.wikiproteins.org/), Wikiprotein (http://www.wikiproteins.org/) is a new project that uses semantic wiki to facilitate community-based creation and curation of knowledge of proteins. |
| Workflow | The term is used in computer programming to capture and develop human to machine interaction. Workflow software aims to provide end users with an easier way to orchestrate or describe complex processing of data in a visual form, much like flow charts but without the need to understand computers or programming. |
Taverna (http://taverna.sourceforge.net/) is a client-based workflow editor that allows graphical connection and execution of Web Services without programming effort. It is designed for biologists/bioinformaticians to use. Yahoo! Pipes (http://pipes.yahoo.com/pipes/) is another example of a graphical workflow editor but it is in a Web 2.0 server environment and accepts JSON (not SOAP) Web services. |
8. ACKNOWLEDGMENTS
This work was supported in part by NSF grant BDI-0135442 to KC, and in part by NIH grant GM068087 to JPT, and in part by NIH grant T15LM007056 to MS. We gratefully acknowledge Mark Gerstein and Carole Goble for suggestions, comments, and views.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kei-Hoi Cheung, Center for Medical Informatics, Yale University.
Kevin Y. Yip, Department of Computer Science, Yale University
Jeffrey P. Townsend, Department of Ecology & Evolutionary Biology, Yale University
Matthew Scotch, Center for Medical Informatics, Yale University.
9. REFERENCES
- [1].Berners-Lee T, Cailliau R, Luotonen A, Nielsen HF, Secret A. The World-Wide Web. ACM Communications. 1994;37(3):76–82. [Google Scholar]
- [2].Cantor CR. Orchestrating the Human Genome Project. Science. 1990;248:49–51. doi: 10.1126/science.2181666. [DOI] [PubMed] [Google Scholar]
- [3].Fayyad U, Uthurusamy R. Data mining and knowledge discovery in databases. Communications of the ACM. 1996;39(11):24–26. [Google Scholar]
- [4].Fayyad U, Piatetsky-Shapiro G, Smyth P, Uthurusamy R. Advances in knowledge and data mining. American Association for Artificial Intelligence; Menlo Park, CA: 1996. pp. 1–34. [Google Scholar]
- [5].Lee TJ, Pouliot Y, Wagner V, Gupta P, Stringer-Calvert DW, Tenenbaum JD, Karp PD. BioWarehouse: a bioinformatics database warehouse toolkit. Bioinformatics. 2006;7:170. doi: 10.1186/1471-2105-7-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shah SP, Huang Y, Xu T, Yuen MM, Ling J, Ouellette BF. Atlas - a data warehouse for integrative bioinformatics. BMC Bioinformatics. 2005;6:34. doi: 10.1186/1471-2105-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haas LM, Schwarz PM, Kodali P, Kotlar E, Rice JE, Swope WC. DiscoveryLink: A system for integrated access to life sciences data sources. IBM Systems Journal. 2001;40(2):489–511. [Google Scholar]
- [8].Stevens R, Robinson A, Goble C. myGrid: personalised bioinformatics on the information grid. Bioinformatics. 2003;19(Suppl. 1):I302–I304. doi: 10.1093/bioinformatics/btg1041. [DOI] [PubMed] [Google Scholar]
- [9].Wilkinson M, Links M. BioMoby: An open source biological web services proposal. Brief Bioinform. 2002;3(4):331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]
- [10].Butler D. Mashups mix data in global service. Nature. 2006;439:6–7. doi: 10.1038/439006a. [DOI] [PubMed] [Google Scholar]
- [11].Dunlap JC, Borkovich KA, Henn MR, GE GET, Sachs MS, Glass NL, McCluskey K, Plamann M, Galagan JE, Birren BW, Weiss RL, Townsend JP, Loros JJ, Nelson MA, Lambreghts R, Colot HV, Park G, Collopy P, Ringelberg C, Crew C, Litvinkova L, DeCaprio D, Hood HM, Curilla S, Shi M, Crawford M, Koerhsen M, Montgomery P, Larson L, Pearson M, Kasuga T, Tian C, Basturkmen M, Altamirano L, Xu J. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Advances in Genetics. 2007;57:49–69. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen H, Doherty D, Forsberg K, Yong G, Kashyap V, Kinoshita J, Luciano J, Marshall MS, Ogbuji C, Rees J, Stephens S, Wong G, Wu E, Zaccagnini D, Hongsermeier T, Neumann E, Herman I, Cheung K. Advancing translational research with the Semantic Web. BMC Bioinformatics. 2007;8(Suppl. 3):S2. doi: 10.1186/1471-2105-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cheung K-H, Yip KY, Smith A, deKnikker R, Masiar A, Gerstein M. YeastHub: a semantic web use case for integrating data in the life sciences domain. Bioinformatics. 2005;21(suppl_1):i85–96. doi: 10.1093/bioinformatics/bti1026. [DOI] [PubMed] [Google Scholar]
- [14].Gao Y, Kinoshita J, Wu E, Miller E, Lee R, Seaborne A, Cayzer S, Clark T. SWAN: A Distributed Knowledge Infrastructure for Alzheimer Disease Research. Journal of Web Semantics. 2006;4(3) [Google Scholar]
- [15].Neurmann EK, Quan D. Pacific Symposium on Biocomputing. World Scientific Publishing Co.; Maui, Haiwaii: 2006. Biodash: a semantic web dashboard for drug development. [PubMed] [Google Scholar]
- [16].Berners-Lee T, Hendler J, Lassila O. The semantic web. Scientific American. 2001;284(5):34–43. [Google Scholar]
- [17].Ankolekar A, Krotzsch M, Tran T, Vrandecic D. WWW 2007. ACM; Banff, Alberta, Canada: 2007. Mashing up Web 2.0 and the Semantic Web. [Google Scholar]
- [18].Edgar R, Domrachev M, Lash A. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara G, Oezcimen A, Rocca-Serra P, Sansone S. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Research. 2003;31(1) doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lurie N. Administrative data and outcomes research. Medical Care. 1999;28(10):867–869. doi: 10.1097/00005650-199010000-00001. [DOI] [PubMed] [Google Scholar]