Abstract
Many of the mitoses that produce pyramidal neurons in neocortex occur at the dorsolateral surface of the lateral ventricles in the embryo. RanBPM was found in a yeast two-hybrid screen to potentially interact with citron kinase (CITK), a protein shown previously to localize to the surface of the lateral ventricles and to be essential to neurogenic mitoses. Similar to its localization in epithelia, RanBPM protein is concentrated at the adherens junctions in developing neocortex. The biochemical interaction between CITK and RanBPM was confirmed in co-immunoprecipitation and protein overlay experiments. To test for a functional role of RanPBM in vivo, we used in utero RNAi. RanBPM RNAi decreased the polarization of CITK to the ventricular surface, increased the number of cells in mitosis and decreased the number of cells in cytokinesis. Finally, the effect of RanBPM knockdown on mitosis was reversed in embryos mutant for CITK. Together, these results indicate that RanBPM, potentially through interaction with CITK, plays a role in the progression of neocortical precursors through M-phase at the ventricular surface.
Keywords: citron kinase, RanBPM, midbody, mitosis, adherens junctions, neurogenesis
INTRODUCTION
Many mitoses and cytokineses in the embryonic neocortex are arranged along the very surface of the lateral ventricles. The full significance of this arrangement is not entirely clear, although there is accumulating evidence that adherens junctions, which also line the ventricular surface are critical regulators of neuronal output (Cappello et al., 2006; Chenn and Walsh, 2002). Citron kinase (CITK) is essential to neurogenic mitoses, and its disruption by mutation in rats (flathead mutants, Roberts et al., 2000; Sarkisian et al., 2002) and mice (Di Cunto et al., 2000) results in severe primary microcephaly, disrupted mitoses, failed cytokineses, and cell death in neuronal precursors throughout the developing central nervous system (Sarkisian et al., 2002; Di Cunto et al., 2000). In embryonic neocortex, CITK is polarized to the ventricular surface where it interacts with the product of the human microcephaly-related gene ASPM at cytokinesis furrows and midbodies (Sarkisian et al., 2002; Paramasivam et al., 2007). The mechanisms and protein interactions that polarize CITK to the ventricular surface have not been identified.
RanBPM was initially identified as a protein that interacts with Ran-GTPase (Nishitani et al., 2001; Nakamura et al., 1998). RanBPM localizes to the plasma membrane and adherens junctions of polarized epithelial cells, including bronchial epithelia, kidney tubules, and mammary glands (Denti et al., 2004). Although originally proposed as a novel regulator of microtubule polymerization (Nakamura et al., 1998), RanBPM was subsequently reported to interact both in vitro and in vivo with a wide range of transmembrane and intracellular proteins. RanBPM-interacting molecules include the neural adhesion molecule L1 (Cheng et al., 2005), the integrin LFA-1 (Denti et al., 2004), the plexin-A receptor for semaphorin 3A signaling (Togashi et al., 2006), the p75 neurotrophin receptor (Bai et al., 2003a), receptor tyrosine kinase MET (Wang et al., 2002), Cav3.1 T-type Ca2+ channel (Kim et al., 2008), and amyloid precursor protein/BACE1 (Lakshmana et al., 2009). These findings together are consistent with a role of RanBPM as a scaffolding protein that may serve to localize many proteins (Denti et al., 2004; Lakshmana et al., 2009).
In this study, we report that RanBPM is an interactor of CITK. In the developing rat neocortex, RanBPM co-localizes with junctional markers ZO-1 and β-catenin. Using in utero RNAi of RanBPM, we show that suppression of RanBPM expression increases the number of mitotic cells and decreases the number of cells entering cytokinesis at the surface of the neocortical ventricular zone. In the rat neuroepithelium, RanBPM expression is critical for the polarized localization of CITK during cell division. The junctional association of RanBPM does not however require CITK. Furthermore, the effect of RanBPM RNAi on the progression of mitosis is reversed in the CITK mutant (CITK fh/fh) embryos. Taken together, we propose that RanBPM acts to polarize CITK during mitosis to the VZ surface, and that this association facilitates the progression of mitosis in developing neuroepithelia.
MATERIALS AND METHODS
Animals
Wistar pregnant rats (Charles River Laboratories, Inc., Wilmington, MA) were used as the source of wildtype embryos. All CITK homozygous mutants (Citkfh/fh ; flathead rats), heterozygous and unaffected littermates were generated from a breeding colony maintained at the University of Connecticut. All animal care procedures were approved by the University of Connecticut IACUC.
Construction of bait yeast and yeast two-hybrid screen
A CITK bait was constructed from the first 1344 bp of 5’-region of rat CITK cDNA encoding amino acids 1–448. This sequence was inserted into the pGBKT7 vector (Clontech, Mountain View, CA) and transformed into the AH109 yeast host strain using the Yeastmakertm yeast transformation system 2 (Clontech). The Matchmakertm yeast two-hybrid system (Clontech) was used for screening the library. A pretransformed human fetal brain cDNA library in Y187 yeast strain (Clontech) was screened by yeast mating with another yeast strain, AH 109, transformed with the CITK bait construct. For mating, the bait yeast strain and the library yeast strain were mixed on 2X YPDA/Kan with gently swirling at 30 °C overnight. A lower stringency selection procedure (SD/-His/-Leu/-Trp) was used first to detect both strong and weak interactions followed by a higher stringency selection (SD/-Ade/-His/-Leu/-Trp/X-α-Gal). Final positive yeast plasmids were prepared using the Zymo yeast plasmid prep (Zymo research, Orange, CA) and transformed into the bacterial host cells, DH5α (Invitrogen, San Diego, CA). The individual plasmid inserts were sequenced, and sequences were analyzed using BLAST.
Expression plasmids
For co-immunoprecipitation, the following full-length plasmids were used: pCAG-myc-citron kinase, pcDNAI-RanBPM-flag, and pcDEB-T7-RanBPM (Nakamura et al., 1998). For the protein overlay assay, the following plasmids were generated from PCR-based cloning: pET-32a-NTCITK-HIS (amino acids 1–448), pGEX-5X-1-GST-RanBPM (amino acids 135–729), and pET-32a-MCPH-HIS (amino acids 1–480).
Co-immunoprecipitation of RanBPM and CITK
Cos7 cells and HEK 293T cells were grown to 90–95% confluency in Dulbecco’s Modified Eagle ’ s Medium (DMEM) containing 10 % FBS and 1% penicillin/streptomycin. Cells were co-transfected with citron kinase-myc (CITK-myc) and either RanBPM-flag or RanBPM-T7. Lysates were subjected to immunoprecipitation (IP) with rabbit polyclonal anti-myc antibody (Abcam, Cambridge, MA) and mouse monoclonal anti-T7 antibody (Novagen, Madison, WI) followed by immunoblotting with mouse monoclonal anti-RanBPM antibody and rabbit polyclonal anti-myc antibody, respectively. Lysates from rat forebrain at embryonic day12 (E12) and postnatal day 7 (P7) were subjected to IP with a rabbit polyclonal anti-citron antibody (CT295) followed by immunoblotting with a mouse monoclonal anti-RanBPM antibody for detecting interactions and another rabbit polyclonal anti-citron antibody (CT261) for detecting inputs.
Protein overlay assay of RanBPM and CITK
Different amounts of purified 5’-terminal region of CITK (NTCITK-HIS, 8 µg, 4 µg, 2 µg, 1 µg and 0.5 µg) were separated by SDS-PAGE and transferred to a PVDF membrane. The membranes were blocked by incubation in 5% nonfat dry milk in TBST for 2 hr. Purified RanBPM-GST and GST were diluted in TBST to 10 µg/ml, and incubated with membrane overnight at 4°C. The membranes were washed three times in TBST and subsequently incubated with rabbit polyclonal GST antibody for 3 hr. After washing three times with TBST, the membrane was incubated with a HRP-conjugated goat anti-rabbit antibody, and washed three times with TBST and then three times with TBS. Bound antibodies were detected with ECL™ (GE healthcare, Buckinghamshire, UK).
Electron microscopy
E12 and E15 wildtype rat brains were harvested, placed in fixative, and immediately dissected to expose the ventricular lumen. Fixative was composed of 2.5% paraformaldehyde, 2% glutaraldehyde, and 0.2% tannic acid in 0.15 M cacodylate buffer with 1.5 mM CaCl2 and 1.5 mM MgCl2 (pH7.4). After 1.5 hr tissues were washed in buffer, post-fixed for 1 hr in 1% OsO4 and 0.8% K3Fe(CN)6 in 0.15 M cacodylate buffer with 1.5 mM CaCl2 and 1.5 mM MgCl2 (pH7.4) and embedded in epoxy resin. Coronal sections from the dorsal medial part of the ventricular surface were cut using a diamond knife, collected onto copper mesh grids, heavy-metal stained with uranyl acetate and lead citrate and viewed on a FEI Tecnai G2 Biotwin electron microscope at 80kV. Midbodies were identified by the concentrated microtubules in cells undergoing cytokinesis (Bellairs and Bancroft, 1975).
RanBPM RNA interference (RNAi) by in utero electroporation
Vector-based shRNA (short hairpin RNA) of RanBPM targeting either the 3’UTR region (shRNA1, 5’-GAG CTC ACA CTT ATA TTG T-3’; shRNA3, 5’- GTA GAC GGG CTC TGC TGA TCT GA-3’) or the coding region (shRNA2, 5’-GTG GAA GAC TAC CTA CAT T-3’; shRNA4, 5’-AGT GTC TGG GAC TGA TGG CTC GA-3’) and a 4 base-mutated shRNA of shRNA1 (shRNA1m4 or control shRNA, 5’- GAG CTG ACA CTA ATA ATC T-3’) were inserted into the mU6pro vector. Knockdown of RanBPM was examined by transfection into B104 cell line which was grown in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS and 1% penicillin/streptomycin. Two rounds of transfections were performed; with the second transfection performed three days after the initial one. Cell lysates were prepared five days after the initial transfection. RanBPM shRNAs and control shRNA constructs were electroporated with pCAGGS-eGFP into E13 rat embryonic neocortex in utero as described previously (Bai et al., 2003b). Briefly, untimed pregnant Wistar females in embryonic day 13 (E13, where the plug date was designated as E0) were anesthetized with Ketamine/Xylazine (100/10 mixture, 0.1 mg/g body weight, intraperitoneally (i.p.)), the uterine horns exposed, and 1–3 µl of plasmids with Fast Green (2 mg/ml; Sigma) was microinjected by pressure (picospritzer, General Valve corporation) through the uterus into the lateral ventricles of embryos by pulled glass capillaries (Drummond Scientific; 0.5 µg/µl for pCAGGS-eGFP and 1.5 µg/µl for RanBPM RNAi constructs). Electroporation was accomplished by discharging a 500-µF capacitor charged to 50–100 V with a sequencing power supply. The voltage pulse was discharged across a pair of copper alloy oval plates (1 X 0.5 cm) pinching the head of each embryo through the uterus.
Immunohistochemistry and antibodies
After in utero electroporation, brains were harvested in cold PBS and processed either by fixing in 4% paraformaldehyde in 0.1M phosphate buffer and sectioning at 70–75 µm on a vibratome in the coronal plane or by fixing in 0.5% paraformaldehyde, cryoprotecting, and sectioning on a cryostat at 7–20 µm. Sections were permeablized, blocked, and immunostained by standard methods.
The following primary antibodies and dilutions were used: a rabbit polyclonal anti-myc antibody (1:500, Abcam), a rabbit polyclonal anti-citron antibody (1:200, CT295), a mouse monoclonal anti-CITK antibody (1:200, BD Pharmingen), a mouse monoclonal anti-AuroraB antibody (1:200, BD Pharmingen), a rat monoclonal anti-ZO-1 antibody (1:300, Development Studies Hybridoma Bank at the University of Iowa), a rabbit polyclonal anti-β-catenin antibody (1:500, Sigma), a mouse monoclonal anti-nestin antibody (1:300, Millipore), a mouse monoclonal anti-phosphorylated-vimentin55 (1:500, MBL), a rabbit polyclonal anti-phospho-Histone H3 (1:500, Millipore), and a rabbit polyclonal anti-Ki67 antibody (1:500, Novocastra). The following secondary antibodies were used: a goat anti-rabbit IgG conjugated Alexa 488, a goat anti-rabbit IgG conjugated Alexa 568, a goat anti-mouse IgG conjugated Alexa 488, and a goat anti-mouse IgG conjugated Alexa 568 (1:200, Invitrogen). Alexa Fluor® 546 phalloidin (1:500, Invitrogen) was used for actin staining and TO-PRO®-3 iodide (1:3000, Invitrogen) was used for counter-staining.
Imaging and statistical analysis
All fluorescent images were acquired with a laser-scanning confocal microscope (Leica TCS SP2, laser lines at 488, 543, 633 nm excitation) using 20X PL APO N.A. 0.7, 40X PL APO N.A. 1.25 and 100X PL APO N.A. 1.40 oil immersion objectives. We used Adobe Photoshop CS3 to assemble images and ImageJ for the quantification used in Fig. 4. Quantification of data shown in Fig. 5 was done from confocal images of dorsomedial embryonic necortex tansfected with RanBPM shRNAs- or control shRNA. Double labelled cells (phosH3+, GFP+) were quantified from Z-series images of 5 µm total thickness collected at 1 µm steps. The number of phosH3+ GFP+ cells was determined for 200 µm of the dorsomedial ventricular surface and normalized to the number of total GFP+ in the same region (n=3 brains, 6 sections from each brain, Fig. 5C). We used two-sample Student’s t-tests to compare means of two independent groups, and ANOVA followed by Tukey-Kramer HSD and Wilcoxon/Kruskal-Wallis tests for comparisons of three or more groups. We considered values as significant when p<0.05. All data are presented as means. Error bars represent ± s.e.m.
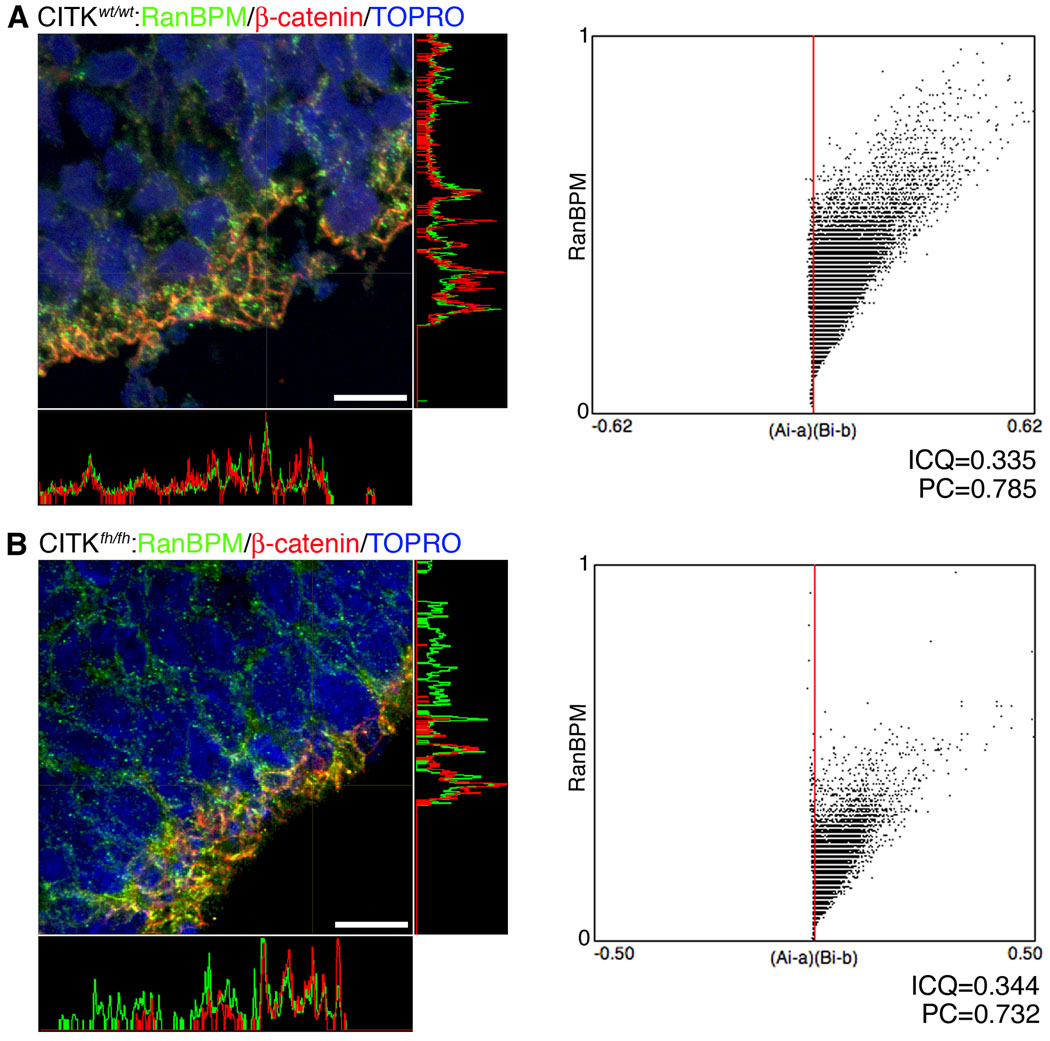
Figure 4. Citron kinase is not necessary for RanBPM expression at the adherens junctions of the VZ surface.
left panels, Endogenous RanBPM co-localizes with β-catenin at the VZ surface of wildtype (A) and CITKfh/fh mutant littermates (B). E17 brain sections from both were immunostained with anti-RanBPM antibody (green) and anti-β-catenin antibody (red). Co-localization of RanBPM and β-catenin is shown at exposed junctional complexes. Orthogonal projections confirming their co-localization show a very similar intensity pattern for the two proteins. Scale bars, 50 µm. right panels, Co-localization analysis with JACoP. Intensity correlation analysis (ICA) was performed for co-localization of RanBPM and β-catenin. A (CITKwt/wt) and B (CITKfh/fh) show intensity correlation plots of the images with the respective plots of the pixel intensities of RanBPM staining (y axis) against their (Ai-a)(Bi-b) values (Ai: individual green (RanBPM) pixel intensity, a: mean of green (RanBPM) pixel intensity, Bi: corresponding red (β-catenin) pixel intensity, and b: mean of red (β-catenin) pixel intensity). Both plots show different intensity co-localization, indicating that absence of CITK does not produce a change in RanBPM localization at the apical VZ junction. ICQ, Intensity correlation quotient value; PC, Pearson’s coefficient.
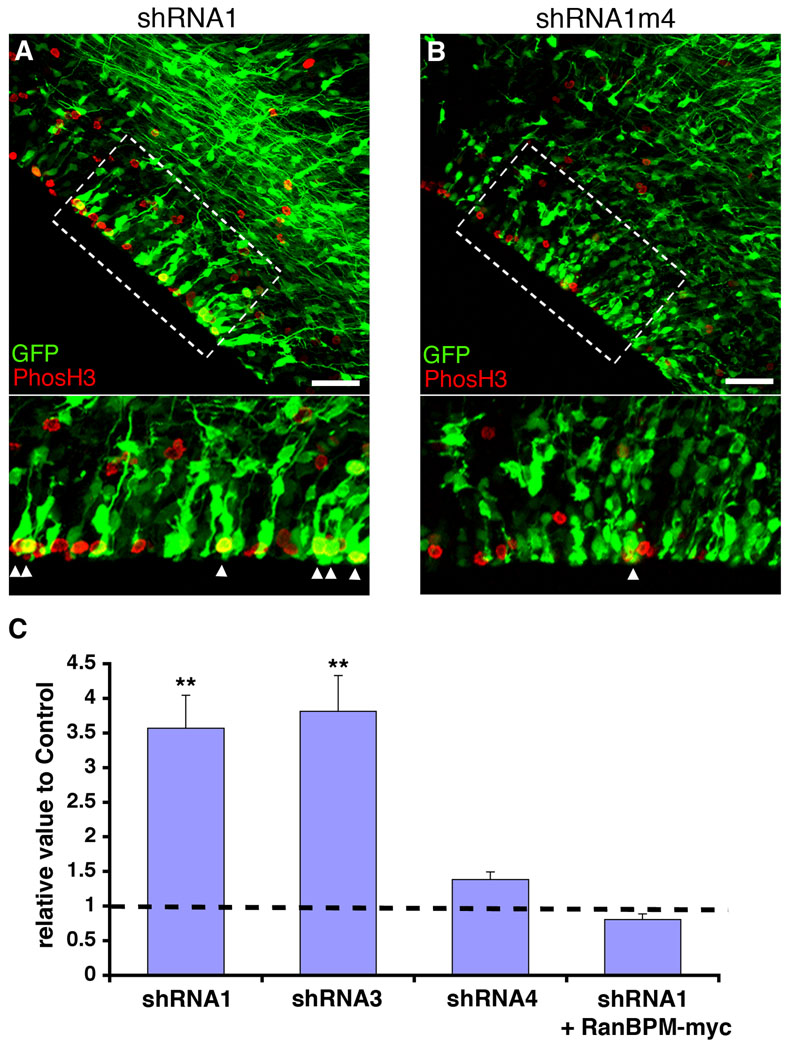
Figure 5. RNAi of RanBPM increases the number of cells in mitosis.
RanBPM-shRNA1 (shRNA1, A) and control shRNA1m4 (B) were co-transfected with GFP in E13 rat embryonic brains by in utero electroporation and analyzed with immunostaining 3 days after transfection. The boxed regions in the upper panels are shown at higher magnifications in the bottom panels (A and B). Note that larger numbers of both phosH3-positive and GFP-positive cells appear in RanBPM-shRNA1-transfected VZ than in control shRNA1m4-transfected VZ. Scale bars, 50 µm. C, The analysis of mitotic marker, phosH3, in RanBPM-shRNA1 and control shRNA1m4 treated VZ. Both phosH3-positive and GFP-positive cells at the VZ were counted and normalized to total GFP-positive cells at the VZ, and then normalized to control transfected condition (n=3 brains, 6 sections from each brain). The control value is shown as the black dotted line. Note that higher numbers of mitotic cells (phosH3+ GFP+/total GFP+ cells) are observed with shRNA1 and shRNA3 transfection than with shRNA4 transfection compared to control shRNA1m4 transfection. In contrast, by co-expression of RanBPM-myc with shRNA1 the number of mitotic cells was similar to that for cells transfected with GFP alone. **, p<0.01, determined by Student’s t-test. Error bars represent ± s.e.m.
RESULTS
RanBPM localizes to cell junctions at the ventricular zone surface
In an effort to identify novel interactors of CITK, we performed a yeast two-hybrid screen. A human fetal brain cDNA library was screened with a bait construct encoding the first 448 amino acids of CITK. One hundred clones were recovered in a first low stringency screen, and ten of those passed a subsequent higher stringency selection. Of these 10, sequences of 4 were not within known coding sequences and one matched a human chromosome 15 open reading frame (BC016725). Five clones contained plasmids with sequence matching coding sequence for RanBPM (GenBank Accession No. NM005493). Of the five isolated RanBPM clones, four matched sequence corresponding to amino acids 135–729, and one matched amino acids 132–729.
To determine the cellular expression of RanBPM within embryonic neocortex, we used a previously characterized monoclonal antibody raised against RanBPM (Denti et al., 2004). The antibody identified a single 90 kDa protein in lysates from rat E14 forebrain (Fig. 1A). We confirmed the specificity of the antibody for RanBPM with RNAi against RanBPM. The 90 kDa band was decreased in lysates from rat B104 cells transfected with shRNA vectors directed against RanBPM, RanBPM-shRNA1, but not by transfection of an shRNA with 4 bases mis-matched to RanBPM sequence, RanBPM-shRNA1m4 (Fig. 1B). Similarly, transfection of RanBPM-shRNA1 into rat neural progenitor cells maintained in culture decreased immuno-positivity for anti-RanBPM (Fig. 1C). In sections from E15 neocortex, RanBPM immuno-positivity was particularly intense at the VZ surface (Fig. 1D). Moreover, RanBPM immuno-positivity co-localized with proteins of the apical junction complex ZO-1 and β-catenin (Fig. 1E, F). The junctional pattern of RanBPM in the embryonic neocortex is consistent with the junctional localization previously described for RanBPM in adult lung, kidney and mammary cells (Denti et al., 2004).
Figure 1. RanBPM is localized to junctions at the VZ surface.
A, Immunoblot shows the expression of RanBPM in the E14 rat brain lysate. α-tubulin is used on the same blot as an internal control. B, Evaluation of RanBPM shRNA constructs by immunoblotting. RanBPM-shRNA1 (targeting 3’UTR region of RanBPM) is more effective than RanBPM shRNA2 (targeting coding region of RanBPM) in knocking down RanBPM expression in B104 cells. A four-base mutation construct of RanBPM-shRNA1 (RanBPM-shRNA1M4) is used as the control shRNA. α-tubulin is shown as control. C, A single confocal z section shows that RanBPM shRNA1 transfected dividing cells (left panels) decrease RanBPM on the membrane compared to the non-transfected cells (right panels). Arrowhead indicates high RanBPM expression on the cell membrane. Scale bar, 50 µm. D, RanBPM is polarized at the ventricular zone surface in E15 rat neocortex. Scale bar, 100 µm. E, Endogenous RanBPM and ZO-1 expression. E15 brain sections were immunostained with anti-RanBPM antibody and anti-ZO-1 antibody. RanBPM expression is shown as green and ZO-1 as red. F, Endogenous RanBPM (green) and β-catenin (red) expression in E15 rat brain. Scale bars, 10 µm. Insets show the dotted boxes with higher magnification. The orthogonal projected panels of single-z images confirm the co-localization of RanBPM and junctional proteins, ZO-1 and β-catenin.
Localization of RanBPM and citron kinase at the VZ surface
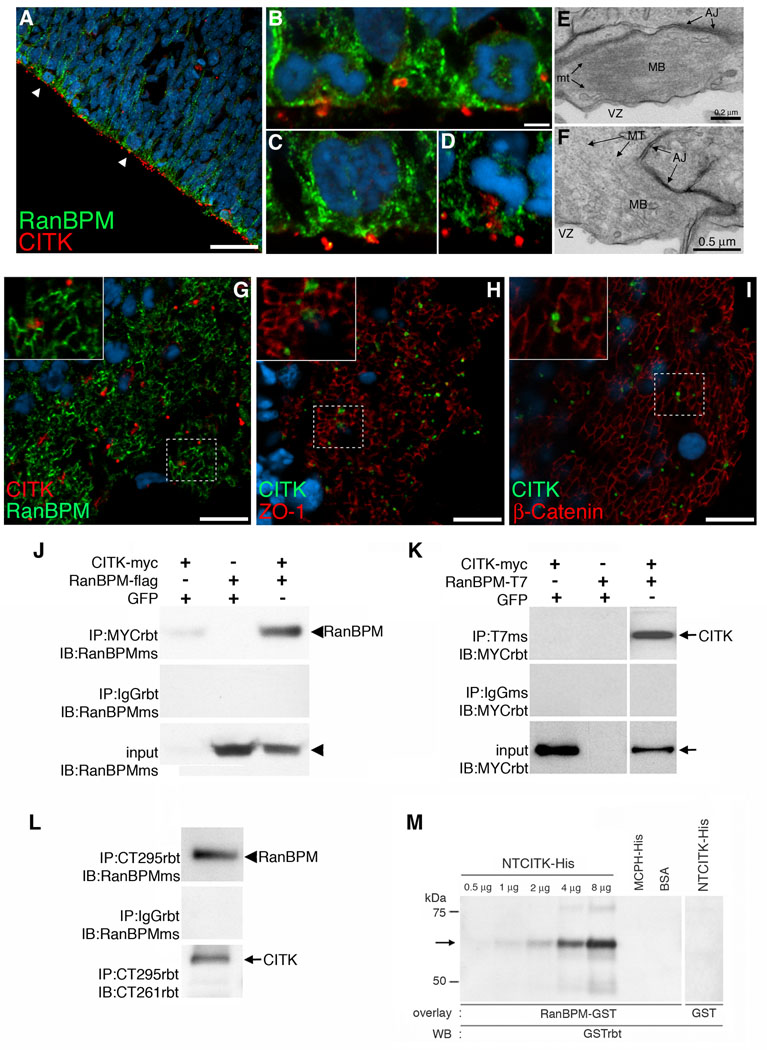
To determine the cellular compartments in which RanBPM and CITK potentially interact we performed double-label fluorescent immunohistochemistry. As previously reported, we found that CITK was highly polarized to the surface of the VZ in discrete points that correspond to cytokinesis furrows and midbodies of dividing precursors (Fig. 2A) (Sarkisian et al., 2002; Paramasivam 2007). In single confocal z sections, we found that RanBPM and CITK immuno-positivity overlapped at points along the VZ, but was not completely co-extensive with RanBPM immuno-positivity which extended beyond points of CITK positivity (Fig. 2B, C, D). Immunohistochemistry of the ventricular wall viewed en face showed that CITK immuno-positivity overlapped similarly with RanBPM, ZO1, and β-catenin (Fig. 2G, H, I). The adjacency of CITK immuno-positivity to junctional markers and RanBPM is consistent with previous electron microscopic analysis showing an adjacency between midbodies and adherens junction complex (AJC) (Bellairs and Bancroft, 1975). We similarly found in an EM analysis of E12 neocortex that midbodies (Fig. 2E, F) at the ventricular surface (VZ) make extensive junctional contacts (Fig. 2E, F). Together, these results indicate that a potential cellular site of interaction between CITK and RanBPM in embryonic neocortex is within the cellular compartment encompassing the AJC and cytokinesis furrow located at the surface of the ventricular zone.
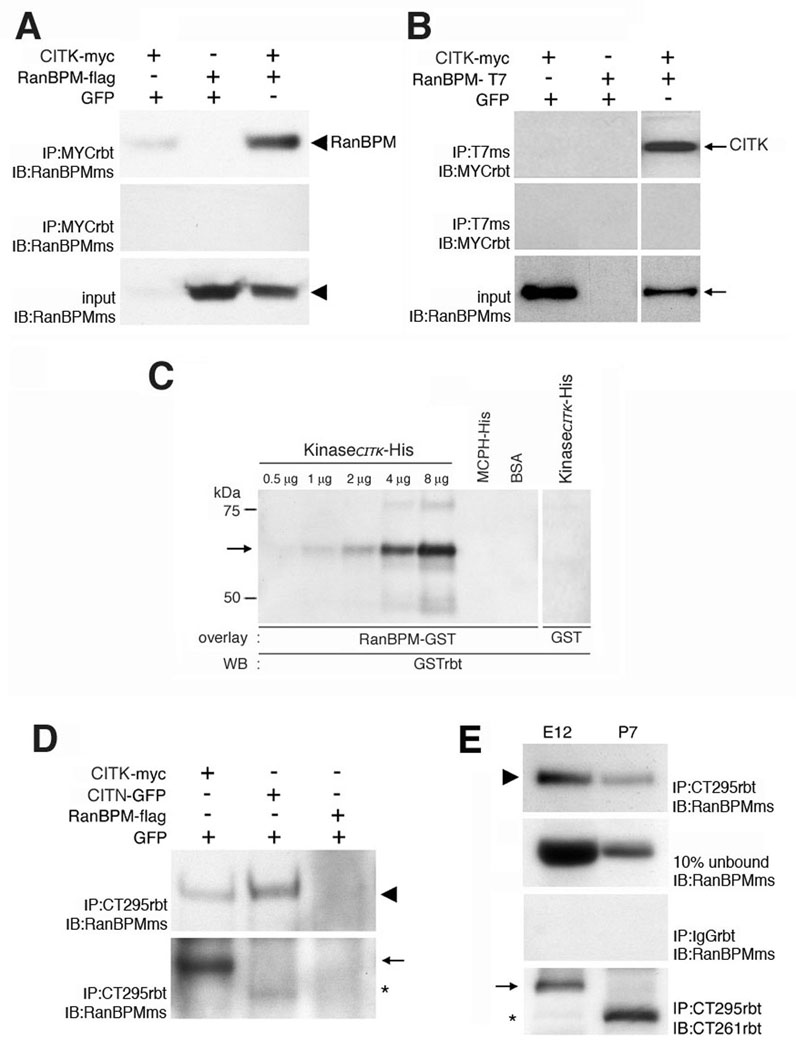
Figure 2. Adjacency of citron kinase and RanBPM at the VZ surface and biochemical confirmation of the CITK-RanBPM interaction.
A, Endogenous localization of Citron kinase (red) and RanBPM (green) is shown. E15 brain sections were immunostained with anti-Citron antibody and anti-RanBPM antibody. Arrows represent adjacent expression of both proteins in dividing cells at the VZ surface. Scale bar, 10 µm. Higher magnifications of labeled cells are shown in B–D (B:early telophase (left pair) and early anaphase (right pair), C:cytokinesis, D:metaphase). E–F, Transmission electron micrographs show that midbodies (MB) have prominent bundles of microtubules (MT). Note that the midbodies are nearly adjacent to adherens junctions (AJ). G–I, Citron kinase (red) localizes adjacent to RanBPM (G, green) similar to the pattern of localization between Citron kinase (green) and other junctional markers (red), ZO-1 (H) and β-Catenin (I). Scale bars, 10 µm. J, RanBPM-flag co-immunoprecipitates with CITK-myc in transfected Cos7 cells. Lysates of Cos7 cells transfected with CITK-myc were immunoprecipitated with either an anti-myc antibody (top panel) or a rabbit anti-IgG antibody (middle panel) followed by detection with an anti-RanBPM antibody. Endogenous RanBPM (90 kDa, arrowhead) in Cos7 cells co-immunoprecipitates with CITK-myc (lane 1); Lysates of Cos7 cells transfected with only RanBPM-flag were subjected to IP with an anti-myc antibody and detected with an anti-RanBPM antibody. Anti-myc antibodies did not immunoprecipitate RanBPM-flag (lane 2); Lysates of Cos7 cells co-transfected with CITK-myc and RanBPM-flag were immunoprecipitated with an anti-myc antibody followed by detection with an anti-RanBPM antibody. RanBPM-flag co-immunoprecipitates with CITK-myc, but not with rabbit IgG (lane 3). K, CITK-myc co-immunoprecipitates with RanBPM-T7 in transfected Cos7 cells. Lysates of Cos7 cells transfected with CITK-myc were immunoprecipitated with either an anti-T7 antibody (top panel) or a mouse anti-IgG antibody (middle panel) followed by detection with an anti-myc antibody. Anti-T7 antibodies did not immunoprecipitate CITK-myc (lane 1); Lysates of Cos7 cells transfected with RanBPM-T7 were subjected to IP with an anti-T7 antibody followed by detection with an anti-myc antibody. Anti-T7 antibodies did not co-immunoprecipitate CITK-myc (lane 2); Lysates of Cos7 cells co-transfected with CITK-myc and RanBPM-T7 were immunoprecipitated with an anti-T7 antibody followed by detection with an anti-myc antibody. CITK-myc (220 kDa, arrow) co-immunoprecipitates with RanBPM-T7, but not with a mouse IgG (lane 3). L, Interaction between endogenous RanBPM and CITK in rat developing brain. In E12 brain, anti-citron antibody (CT295) immunoprecipitated CITK and RanBPM co-immunoprecipitated with it (top panel). RanBPM does not co-immuoprecipitate with a rabbit IgG (second panel). The third panel shows that CT295 immunoprecipitates CITK in E12 brain lysate. Arrowhead:RanBPM, arrow:CITK. M, Protein overlay assay shows direct interaction between the N-terminal region of CITK (NTCITK, a.a.1–448) and RanBPM. NTCITK-His (65 kDa), MCPH-His (57 kDa), and BSA (67 kDa) were subjected to immunoblotting and overlayed with RanBPM-GST (left) and GST alone (right). The bound protein was probed with anti-GST antibodies and detected by chemiluminescence. RanBPM-GST bound to NTCITK-His, but to neither MCPH-His nor BSA. GST alone did not bind to NTCITK-His.
Biochemical interaction between RanBPM and citron kinase in vitro and in vivo
In order to test for a biochemical interaction between RanBPM and CITK, we performed co-immunoprecipitation assays in protein lysates prepared from co-transfected cell lines and from embryonic forebrain. In co-transfected cells, we tested for interaction between CITK and RanBPM in both directions. Myc-tagged CITK (CITK-myc) and flag-tagged RanBPM (RanBPM-flag) were co-transfected into Cos7 cells. Lysates were subjected to immunoprecipitation with a polyclonal rabbit anti-myc antibody followed by immunoblotting with a monoclonal mouse anti-RanBPM antibody. The results in figure 2J show that RanBPM co-immunoprecipitated with CITK-myc. To test the interaction in the opposite direction, CITK-myc and T7-tagged RanBPM (RanBPM-T7) were cotransfected into Cos7 cells. Lysates were immunoprecipitated with a monoclonal mouse anti-T7 antibody followed by immunoblotting with a polyclonal rabbit anti-myc antibody. The results showed that CITK-myc co-immunoprecipitated with RanBPM-T7 (Fig. 2K).
In order to determine whether endogenous CITK and RanBPM protein are in biochemical association in vivo we performed co-immunoprecipitation experiments using an antibody to CITK that is effective in immunoprecipitation of endogenous CITK (Zhang et al., 1999). The anti-citron antibody (CT295) immunoprecipitated CITK from E12 forebrain protein lysates (Fig. 2L). Moreover, RanBPM co-immunoprecipitated with CITK as evidenced by positivity for the 90 kDa band identified with the RanBPM monoclonal antibody (Fig. 2L). As a control, rabbit IgG was found to not immunoprecipitate CITK or RanBPM (Fig. 2L) from embryonic forebrain lysates. Next, we used a protein overlay assay to test whether CITK protein can directly bind to RanBPM protein (Fig. 2M). For this experiment, an affinity purified His-tagged 5’- terminal fragment of CITK (NTCITK-His), and two control proteins, His-tagged microcephalin (MCPH-His) and bovine serum albumin (BSA) were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Then purified GST-tagged RanBPM (RanBPM-GST) and GST only (GST) were incubated with the membranes. Bound proteins were then detected by immunoblotting with an anti-GST antibody. RanBPM-GST did not bind to MCPH or BSA indicating that RanBPM does not bind non-specifically to proteins immobilized on blots. Similarly, GST protein did not bind to CITK. In contrast, RanBPM-GST used as a probe bound to CITK in a dose dependent manner (Fig. 2L). Taken together, the interaction between CITK and RanBPM first identified in the yeast two-hybrid screen was confirmed by co-immunoprecipitation in cells and in tissue and by direct protein-protein interaction in vitro.
Knockdown of RanBPM prevents polarization of citron kinase
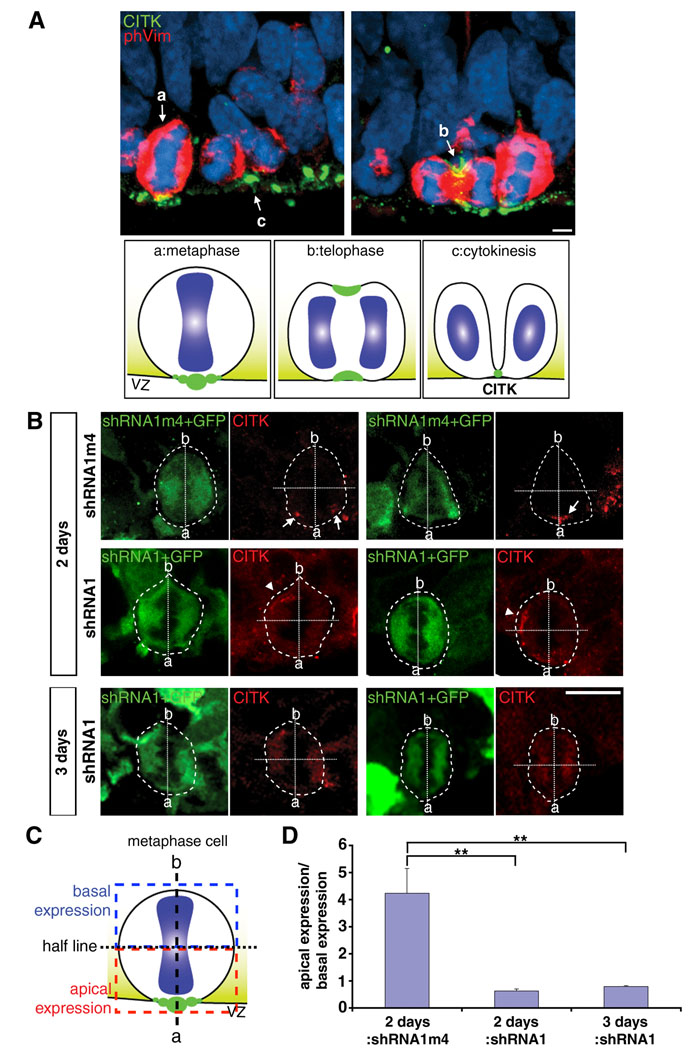
As RanBPM has been hypothesized to act as a scaffolding protein that serves to localize proteins to particular cellular compartments, we sought to determine whether it plays a role in localizing CITK. As shown above and in previous reports (Sarkisian et al., 2002; Paramasivam et al., 2007) CITK is highly polarized to the VZ surface. We further characterized the cellular localization pattern of CITK through M-phase, and found that CITK becomes polarized to the ventricular surface in metaphase. In metaphase cells, labeled by an anti-phospho-vimentin antibody, and identified further by condensed chromatin, CITK is concentrated and restricted to the apical/ventricular surface (Fig. 3A). The immunocytochemical pattern through mitosis suggests that through M-phase CITK polarizes to the VZ surface in metaphase and then extends around the circumference of the cell to form the cytokinesis furrow in telophase (Fig. 3A). As the furrow constricts CITK becomes increasingly concentrated to the midbody which remains anchored to the apical/ventricular surface (Fig. 3A).
Figure 3. Knockdown of RanBPM impedes apical polarization citron kinase.
A, CITK localization in cells at different stages of cell cycles in E15 rat brain. a, In metaphase cells, CITK aggregates apically towards the VZ surface. b, Then CITK dynamically moves to both the basal and apical sides of the cleavage furrow during telophase. c, In cytokinesis, CITK localizes to the midbody adjacent to the VZ surface. Scale bar, 5 µm. Diagrams depicting CITK localization (green) at the VZ surface during cell cycle. a, metaphase; b, telophase; c, cytokinesis. B, RanBPM-shRNA1 (shRNA1) results in mislocalization of CITK at the VZ surface 2 days and 3 days after transfection. Control (shRNA1m4) and shRNA1 were co-transfected with GFP in E13 rat embryonic brains by in utero electroporation. Phospho-vimentin (phVim, not shown) was used for labeling mitotic cells. RanBPM-shRNA1 disrupts the polarization of CITK (red, arrowheads) in metaphase cells (phVim positive) by 2 days after in utero electroporation and shows diffuse distribution of CITK 3 days after in utero electroporation, while control transfected cells maintained the polarization of CITK (arrows). Scale bars, 10 µm. The transfected metaphase cells are outlined, and ‘a’ stands for apical side and ‘b’ stands for basal side of mitotic cells. The dotted lines represent the apico-basal axis and perpendicular half lines. The graph in D shows the ratio of CITK expression in the apical half of metaphase cells relative to CITK expression in the basal half of metaphase cells as depicted in the schematic shown in C. The significance was tested by two-sample Student’s t-tests in two independent groups and by ANOVA in all three groups followed by post hoc Tukey-Kramer HSD and Wilcoxon/Kruskal-Wallis tests (**, p<0.01). Error bars represent ± s.e.m.
To test for a possible function of RanBPM in CITK polarization, RanBPM-shRNA1 constructs or control constructs (RanBPM-shRNA1m4) were co-transfected with a GFP expression plasmid (pCAGGS-eGFP) into E13 rat embryonic neocortex by in utero electroporation. The polarity of CITK immuno-positivity in phospho-vimentin positive cells in mitosis was then determined and compared between control and RNAi treated cells 2 and 3 days following transfection. For this analysis, single transfected cells in mitosis were identified, the apical and basal poles determined by the orientation of metaphase nuclei, and CITK immuno-positivity in the basal and apical poles quantified. Two days following RNAi treatment, there was CITK immuno-positivity in the basal half of mitotic cells, but not in control transfected cells (Fig. 3B). Three days after RNAi treatment, CITK immuno-positivity in RNAi treated cells was diffuse and remained un-polarized (Fig. 3B). These results indicate that RanBPM expression is required for polarization of CITK to the ventricular/apical surface in cells in mitosis.
We next examined whether RanBPM RNAi caused a change in the mitotic spindle orientation at the VZ surface in mitotic cells. As above, we selected shRNA-transfected cells that were in metaphase or anaphase for analysis. The angle of the metaphase plate plane relative to the ventricular surface was determined for 32 cells in each transfection condition. We found that the angle of 96.9% of the cells in each group was greater that 50° and that there were no significant differences in the angles between the shRNA1-treated cells (mean = 75.4°, s.e.m. ± 2.2) or the control-treated cells (mean = 78.8°, s.e.m. ± 2.1) (n=32 for each group, p > 0.1, determined by Student’s t-test and ANOVA). Thus, suppression of RanBPM expression in vivo affects the polarization of CITK without affecting mitotic spindle orientation.
Citron kinase is not necessary for RanBPM localization
To determine whether RanBPM localization to the adherens junction at the VZ surface is dependent upon CITK, we examined the co-localization of RanBPM with β-catenin in flathead mutant rats that are homozygous for a null allele of CITK (Sarkisian et al., 2002). Immunohistochemistry showed that the co-localization of RanBPM with β-catenin at the surface of the neocortical ventricular zone was not disrupted in CITKfh/fh mutant embryos (Fig. 4B). Orthogonal projections of images from wildtype (Fig. 4A) and CITKfh/fh mutants (Fig. 4B) showed that intense RanBPM immunoreactivity was present within the same pixels as positivity for β-catenin. The correlation of co-localization was 0.344 in CITKfh/fh mutants and 0.335 in wildtype, and the Pearson’s coefficient was 0.732 in CITKfh/fh mutants and 0.785 in non-mutants (by JACoP analysis; Bolte and Cordelières, 2006). Thus, RanBPM localization to the adherens junction is not dependent upon CITK.
Knockdown of RanBPM increases the number of precursors in mitosis
We next sought to determine the role of RanBPM in the cell cycle of precursor cells in vivo by decreasing the expression of RanBPM by in utero RNAi. Because it has been shown previously that loss of CITK function by mutation impairs mitosis (LoTurco et al., 2003) and cytokinesis (Sarkisian et al., 2002) we concentrated our analysis on effects during M-phase. For these experiments we electroporated shRNA vectors in vivo that were proven to knockdown expression levels of endogenous RanBPM in B104 neuroblastoma cells without affecting CITK expression (Supporting Figure 1). We then determined whether RNAi against RanBPM alters the number of cells in mitosis at the VZ surface by quantifying the number of transfected cells that are phospho-histone H3 (phosH3) positive at the VZ surface. Two effective RanBPM RNAi vectors, shRNA1 and shRNA3, caused a greater than 3 fold increase relative to control (shRNA1m4) transfected cells in the number of cells positive for phosH3 (Fig. 5). Because RanBPM immuno-positivity is membrane localized in vivo, this made it difficult to unambiguously determine co-labeling of transfected cells. Thus, we were unable to determine changes in RanBPM immuno-positivity in vivo following knockdown of RanBPM. To directly test for the specificity of the shRNA effect against RanBPM we performed add-back or rescue experiments in which we co-expressed RanBPM-myc along with shRNA1. In these rescue experiments the number of phosH3+ cells was similar to that for cells transfected with GFP alone and not targeted by RNAi (Fig. 5C). Together, these results indicate that the effect of shRNA1 on mitosis is specific to RanBPM knockdown, and that decreased RanBPM expression results in increases in the number of precursor cells in mitosis.
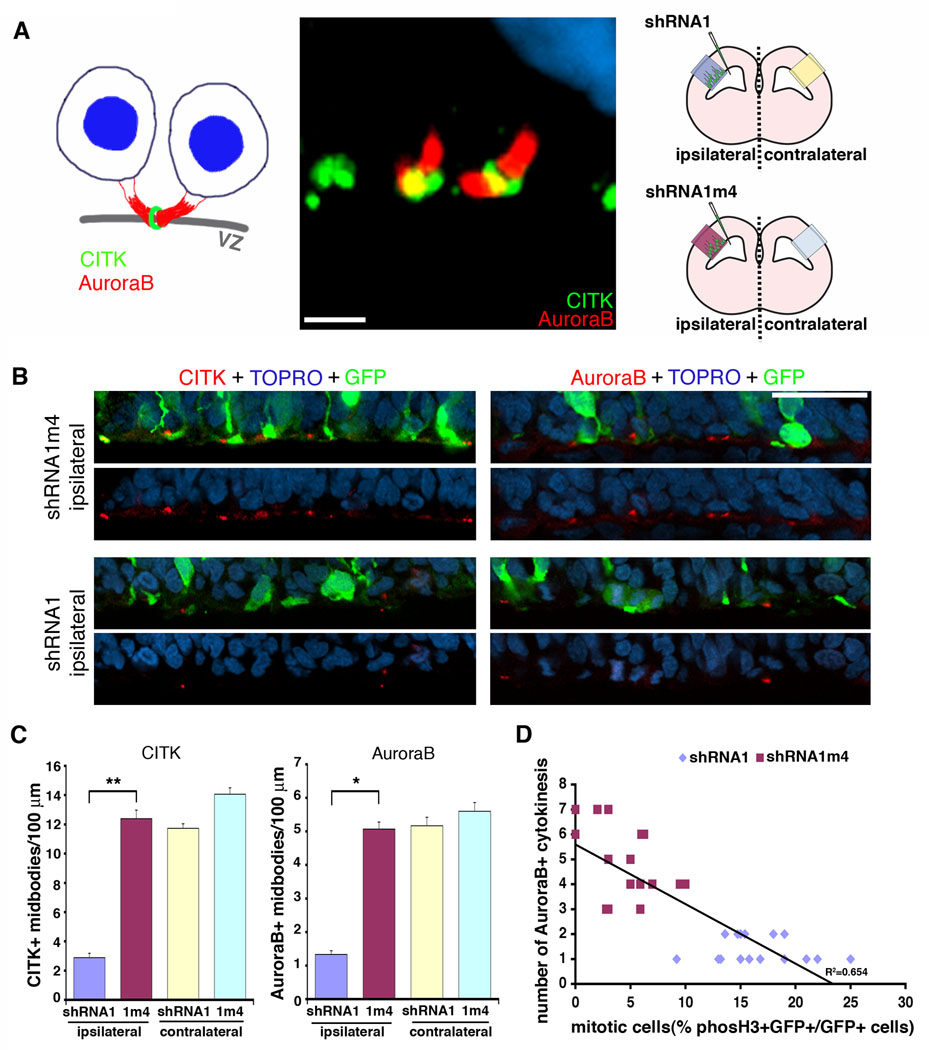
Knockdown of RanBPM decreases entrance into cyztokinesis
An increase in the number of cells in mitosis following RanBPM knockdown in vivo could be due to either an increase in the number of cells entering M-phase or a decrease in the number of cells exiting mitosis and progressing through cytokinesis. In order to distinguish between these two possibilities in vivo we assessed the progression through cytokinesis by quantifying the number of cytokinetic furrows and midbodies in the VZ following RanBPM RNAi. Antibodies to CITK and AuroraB label cytokinesis furrows and midbodies within the neocortical ventricular zone. Fig. 6A shows an example of CITK and AuroraB immunostaining of two distinct parts of the midbody at the VZ surface: CITK at the midbody ring and AuroraB at the flanking intercellular bridges. Following knockdown of RanBPM by shRNA1 in vivo there was a decrease in the number of midbodies as assessed by both CITK and AuroraB staining. Control (shRNA1m4) treated brains or contralateral VZs that received no transfection showed nearly 4-fold more midbodies than shRNA1 treated VZs (Fig. 6B, C). This decrease in cytokinesis, together with previous data indicating a large increase in mitotic cells (Fig. 5), suggests that RanBPM knockdown increases the number of cells in mitosis by slowing entry into cytokinesis. Indeed, the quantitative correlation between the fold mitotic cell increase and corresponding fold cytokinesis decrease (Fig. 6D) is consistent with an accumulation of cells in mitosis following RanBPM knockdown that is caused by a decrease in progression to cytokinesis.
Figure 6. RNAi of RanBPM decreases the number of midbodies at the ventricular zone surface.
A, Both the diagram (left) and confocal image (middle) show the markers, CITK and AuroraB, used to assess the RanBPM RNAi effects on cytokinesis. CITK (green) and AuroraB (red) label midbody structures in E15 brain. Citron kinase localizes to the midbody ring and AuroraB marks the flanking regions in U-shaped midbodies at the VZ surface. Scale bar, 5 µm. The diagram in right represents the regions that were analyzed in this experiment. B and C, RanBPM RNAi decreases the midbody formations at the VZ surface. RanBPM-shRNA1-transfected brains (bottom 2 panels) labeled with CITK (red, left panels) or AuroraB (red, right panels) display fewer midbodies than do control shRNA1m4-transfected brains (upper 2 panels). Scale bar, 50 µm. CITK-positive and AuroraB-positive midbodies were counted within 200 µm of the VZ surface. C, n=4 brains, 6 sections from each. *, p<0.01 and **, p<0.05, determined by Student’s t-test. Error bars represent ± s.e.m. D, The increase in mitotic cells by RanBPM-shRNA1 is correlated to the decreased numbers of cytokinesis showing the delay of M-phase progression.
A role of RanBPM in mitosis is dependent upon functional citron kinase
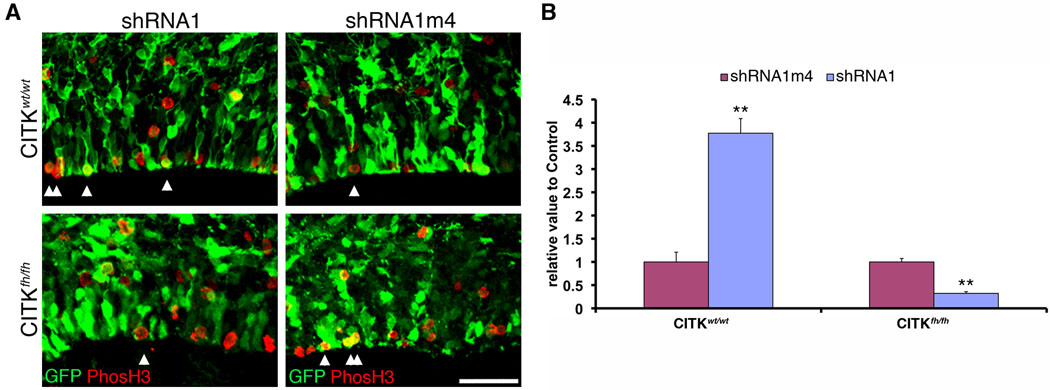
The change in localization of CITK in mitotic cells combined with the effects of RanBPM knockdown on mitosis is consistent with a model in which RanBPM is upstream of and necessary for recruiting CITK to the apical/ventricular surface during mitosis, and that this in turn is required for normal progression out of mitosis into cytokinesis. Two predictions from this working model are that 1) CITK mutants will also show an accumulation of cells in mitosis relative to non-mutants, and 2) RanBPM knockdown will not create an accumulation of cells in mitosis in flathead mutant embryos that lack CITK. Consistent with the first prediction we found that there was a two-fold increase in phosphoH3-positive cells in homozygous CITK mutants relative to wildtype littermates (Supporting Figure 2). To test the second prediction that RanBPM function in mitotic progression is dependent upon CITK function, we determined the effect of RanBPM knockdown on mitotic cells in flathead mutants (Fig. 7). E13 embryos (CITKwt/wt or CITKfh/fh) were co-transfected with either RanBPM-shRNA1 or control (RanBPM-shRNA1m4) together with pCAGGS-eGFP. Three days after surgery, embryos were harvested genotyped, and the relative number of phosH3-positive to transfected cells determined for shRNA1-treated and control neocortices. In contrast to the effect of RanBPM RNAi in wildtype animals where RanBPM knockdown caused a >3 fold increase in the number of mitotic cells, RanBPM knockdown in homozygous flathead mutants did not cause an accumulation of M-phase cells, but instead resulted in a decrease in mitotic cells (Fig. 7). Thus, the effect of RanBPM knockdown on mitosis accumulation is dependent upon functional CITK, and moreover, the reverse effect of RanBPM knockdown in the absence of CITK may reveal a CITK-independent role of RanBPM in cell cycle progression.
Figure 7. RanBPM function is dependent upon citron kinase.
A, RanBPM RNAi does not cause accumulation of mitotic cells in CITKfh/fh, as it does in CITKwt/wt. RanBPM-shRNA1 or control shRNA1m4 constructs were co-transfected with GFP into E13 embryonic CITK mutant brains by in utero electroporation. Double-immunopositive cells (GFP and phosH3) were quantified. Note that RanBPM-shRNA1 does not increase the numbers of mitotic cells, indicating that RanBPM RNAi function on mitosis through CITK. In addition, RanBPM may have a separate pathway by which it affects the VZ cell cycle. Arrowheads indicate both GFP- and phosH3-positive cells. Scale bar, 50 µm. Quantification is shown in B (n=3 brains, 4 sections from each brain). Two-way ANOVA was used to validate the significance between genotypes (p<0.01), RNAi and control (p<0.01), and their interaction (p<0.01).
DISCUSSION
In summary, we found that as in other epithelia (Denti et al., 2004) RanBPM localizes to the adherens junction complex in developing neocortex. We also report that RanBPM interacts physically with CITK in vivo and in vitro, and is required for normal progression through M-phase. Given this evidence, we propose that RanBPM links adherens junctions to cellular proteins necessary to progression through M-phase at the ventricular zone surface in developing necortex.
RanBPM has been reported to act as a scaffolding protein for a number of proteins in the developing nervous system. Recent studies showed that RanBPM is involved in amyloid β peptide synthesis by interacting with the lipoprotein receptor-related protein (LRP), β-secretase (BACE1), and amyloid precursor protein (APP) (Lakshmana et al., 2009). Plexin-A1 receptors also interact with RanBPM and the RanBPM/Plexin-A1 complex mediates Semaphorin3A signaling thus inhibiting axonal growth (Togashi et al., 2006). Our observations that RanBPM RNAi alters CITK localization (Fig. 3) adds to the functions of RanBPM in localizing developmentally important proteins.
RanBPM was originally identified as a Ran GTPase-binding protein in the microtubule-organizing center (Nakamura et al., 1998) implicating it in spindle formation during metaphase. Ectopic expression of RanBPM causes multiple microtubule organizing centers in dividing HeLa cells suggesting that RanBPM is involved in microtubule nucleation (Nakamura et al., 1998). Although these studies suggested to us the possibility that RanBPM could also play a role in spindle pole orientation in neocortical precursors, we were unable to observe such changes following RNAi.
RanBPM has also been previously linked to a possible function in mitosis by its phosphorylation by cell cycle kinases: cyclin-dependent kinase 11P46 (Mikolajczyk et al., 2003) and polo-like kinase 1 (Plk1) (Jang et al., 2004). Polo-like kinases are potent regulators of M-phase that phosphorylate substrate proteins on centrosomes, kinetochores, the mitotic spindle, and the midbody (Archambault and Glover, 2009). Disruption of Plk1 results in G2-delay and prometaphase arrests, and the dynamic release of Plk1 from early mitotic structures is crucial for mid- to late-stage mitotic events (Kishi et al., 2009). We hypothesize that RanBPM/Plk1 interaction may be upstream of CITK as CITK is particularly important for terminal stages of the cell cycle in neural precursor cells.
Citron kinase is a Rho activated kinase and we initially hypothesized that interactors with the kinase domain would identify potential substrates of CITK. We attempted to demonstrate that CITK phosphorylates RanPBM in heterologous expression systems and with cell free assays using purified CITK and RanBPM. However, while we were consistently able to observe phosphorylation of a control substrate, histone H1, we failed to detect phosphorylation of RanBPM by CITK (Supporting Figure 3). Thus, whether or not the kinase function of CITK is essential to its role in neurogenic cell division remains unknown.
RNAi experiments in the CITK mutants showed that RanBPM knockdown has additional effects on the cell cycle that do not depend upon CITK. In fact, knockdown of RanBPM had an opposite effect in the mutant relative to that in wildtype embryos. A general working model to explain this result is that RanBPM interacts with two pathways: one through CITK that facilitates progression through mitosis and into cytokinesis, and another not through CITK that normally limits the number of cells that are in mitosis at the VZ surface. Further assessment of phases of the cell cycle altered after RanBPM RNAi in CITK mutants may help elucidate these CITK independent roles further. One possibility is that by inhibiting minibrain-related kinase (Mirk)/Dyrk1B activity (Zou et al., 2003) RanBPM could stabilize p27Kip1, a cyclin-dependent kinase inhibitor (Deng et al., 2004). Enhanced levels of p27Kip1 could in turn reduce the number of cells in the cell cycle (Li et al., 2009) in the CITK mutant following knockdown of RanBPM. RanBPM may therefore play a dual role in both limiting and facilitating neurogenic cell divisions.
Supplementary Material
A, Evaluation of RanBPM-shRNA constructs by immunoblotting. RanBPM-shRNA1 (targeting 3’UTR region of RanBPM) is more effective than RanBPM-shRNA2 (targeting coding region of RanBPM) in knocking down RanBPM expression in B104 cells. A four-base mutated construct of RanBPM-shRNA1 (RanBPM-shRNA1m4) is used as the control shRNA. α-tubulin is shown as control. Note that CITK expression is not affected by RanBPM-shRNA1. B, RanBPM RNAi does not decrease CITK or β-catenin expression in cultured progenitor cells. C, RanBPM-shRNA3 decreases RanBPM, while RanBPM-shRNA4 does not.
CITKfh/fh have more mitotic progenitors within their VZ and SVZ than do CITKwt/wt littermates. E17 CITKfh/fh and wildtype littermate brain sections were stained with anti-phosphohistone H3 (phosH3, green) and anti-nestin (red) antibodies. Double-positive cells were counted within 400 µm2 of the VZ and SVZ. Scale bars, 50 µm. Quantification is shown in graph (n=3 brains, 6 sections from each brain). **, p<0.01 from Student’s t-test.
Purified KCITK-HIS was incubated with either purified RanBPM-GST and GST (A) or RanBPM-GST and Histone H1 (B) in the kinase reaction buffer, which contains γ-P32 labeled ATP. The products were analyzed by 12% acrylamide gel followed by autoradiography (left in A and B) and coomassie staining (right in A and B). Purified GST (A) and Histone H1 (B) are controls. In A, neither RanBPM-GST (asterisk) nor GST (double asterisks) are phosphorylated by KCITK-HIS, demonstrating that neither of them are a substrate for KCITK-HIS. In B, phosphorylation of Histone H1, KCITK substrate, is not affected by RanBPM-GST. left, lane 1, KCITK-HIS+ RanBPM-GST 5 µg; lane 2, KCITK+RanBPM 50 µg; lane 3, KCITK+RanBPM 100 µg; lane 4, KCITK+RanBPM 200 µg; lane 5, KCITK+GST 50 µg; lane 6, KCITK+GST 100 µg. right, lanes are same as left, coomassie stained gel. B, KCITK-HIS phosphorylates Histone H1. left, lane 1, KCITK+H1; lane 2, KCITK+H1+RanBPM 5 µg; lane 3, CITK+H1+RanBPM 50 µg; lane 4, CITK+H1+RanBPM 100 µg; lane 5, CITK+H1+RanBPM 200 µg. right, lanes are same, coomassie stained gel.
ACKNOWLEDGMENTS
We thank Dr. Mary Kennedy and Leslie Schenker for anti-citron antibodies (CT295 and CT261), Dr. Pascal Madaule for pCAG-myc-CITK, and Dr. Jeff Talbot for pcDEB-T7-RanBPM. We thank Dr. Marie Cantino, Steve Daniels and Jim Romanow for their technical assistance in the University of Connecticut electron microscopy laboratory. This work has been supported by NIH grant MH056524 to JJL.
REFERENCES
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Bai D, Chen H, Huang BR. RanBPM is a novel binding protein for p75NTR. Biochem Biophys Res Commun. 2003a;309:552–557. doi: 10.1016/j.bbrc.2003.08.033. [DOI] [PubMed] [Google Scholar]
- Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat. Neurosci. 2003b;6:1277–1283. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- Bauer CR, Epstein AM, Sweeney SJ, Zarnescu DC, Bosco G. Genetic and systems level analysis of Drosophila sticky/citron kinase and dFmr1 mutants reveals common regulation of genetic networks. BMC Syst Biol. 2008;2:101. doi: 10.1186/1752-0509-2-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellairs R, Bancroft M. Midbodies and beaded threads. Am J Anat. 1975;143:393–398. doi: 10.1002/aja.1001430310. [DOI] [PubMed] [Google Scholar]
- Berto G, Camera P, Fusco C, Imarisio S, Ambrogio C, Chiarle R, Silengo L, Di Cunto F. The Down syndrome critical region protein TTC3 inhibits neuronal differentiation via RhoA and Citron kinase. J Cell Sci. 2007;120:1859–1867. doi: 10.1242/jcs.000703. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Camera P, Schubert V, Pellegrino M, Berto G, Vercelli A, Muzzi P, Hirsch E, Altruda F, Dotti CG, Di Cunto F. The RhoA-associated protein Citron-N controls dendritic spine maintenance by interacting with spine-associated Golgi compartments. EMBO Rep. 2008;9:384–392. doi: 10.1038/embor.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Bräuninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, Brakebusch C, Götz M. The Rho- GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Cheng L, Lemmon S, Lemmon V. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J Neurochem. 2005;94:1102–1110. doi: 10.1111/j.1471-4159.2005.03254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Deng X, Mercer SE, Shah S, Ewton DZ, Friedman E. The cyclin-dependent kinase inhibitor p27Kip1 is stabilized in G(0) by Mirk/dyrk1B kinase. J Biol Chem. 2004;279:22498–22504. doi: 10.1074/jbc.M400479200. [DOI] [PubMed] [Google Scholar]
- Denti S, Sirri A, Cheli A, Rogge L, Innamorati G, Putignano S, Fabbri M, Pardi R, Bianchi E. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem. 2004;279:13027–13034. doi: 10.1074/jbc.M313515200. [DOI] [PubMed] [Google Scholar]
- Di Cunto F, Imarisio S, Hirsch E, Broccoli V, Bulfone A, Migheli A, Atzori C, Turco E, Triolo R, Dotto GP, Silengo L, Altruda F. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–127. doi: 10.1016/s0896-6273(00)00090-8. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds JW, Ruffett TL. Cell proliferation in the neural tube: an electron microscopic and golgi analysis in the mouse cerebral vesicle. Z Zellforsch Mikrosk Anat. 1971;115:226–264. doi: 10.1007/BF00391127. [DOI] [PubMed] [Google Scholar]
- Jang YJ, Ji JH, Ahn JH, Hoe KL, Won M, Im DS, Chae SK, Song S, Yoo HS. Polo-box motif targets a centrosome regulator, RanGTPase. Biochem Biophys Res Commun. 2004;325:257–264. doi: 10.1016/j.bbrc.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Kishi K, van Vugt MA, Okamoto K, Hayashi Y, Yaffe MB. Functional dynamics of Polo-like kinase 1 at the centrosome. Mol Cell Biol. 2009;29:3134–3150. doi: 10.1128/MCB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmana MK, Yoon IS, Chen E, Bianchi E, Koo EH, Kang DE. Novel role of RanBP9 in BACE1 processing of amyloid precursor protein and amyloid beta peptide generation. J Biol Chem. 2009;284:11863–11872. doi: 10.1074/jbc.M807345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tang X, Jablonska B, Aguirre A, Gallo V, Luskin MB. p27(KIP1) regulates neurogenesis in the rostral migratory stream and olfactory bulb of the postnatal mouse. J Neurosci. 2009;29:2902–2914. doi: 10.1523/JNEUROSCI.4051-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk M, Shi J, Vaillancourt RR, Sachs NA, Nelson M. The cyclin-dependent kinase 11(p46) isoform interacts with RanBPM. Biochem Biophys Res Commun. 2003;310:14–18. doi: 10.1016/j.bbrc.2003.08.116. [DOI] [PubMed] [Google Scholar]
- Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Masuda H, Horii J, Kuma K, Yokoyama N, Ohba T, Nishitani H, Miyata T, Tanaka M, Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Hirose E, Uchimura Y, Nakamura M, Umeda M, Nishii K, Mori N, Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- Paramasivam M, Chang YJ, LoTurco JJ. ASPM and citron kinase co-localize to the midbody ring during cytokinesis. Cell Cycle. 2007;6:1605–1612. doi: 10.4161/cc.6.13.4356. [DOI] [PubMed] [Google Scholar]
- Roberts MR, Bittman K, Li WW, French R, Mitchell B, LoTurco JJ, D'Mello SR. The flathead mutation causes CNS-specific developmental abnormalities and apoptosis. J Neurosci. 2000;20:2295–2306. doi: 10.1523/JNEUROSCI.20-06-02295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian MR, Li W, Di Cunto F, D'Mello SR, LoTurco JJ. Citron-kinase, a protein essential to cytokinesis in neuronal progenitors, is deleted in the flathead mutant rat. J Neurosci. 2002;22:RC217. doi: 10.1523/JNEUROSCI.22-08-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebahn A, Rose M, Enz R. RanBPM is expressed in synaptic layers of the mammalian retina and binds to metabotropic glutamate receptors. FEBS Lett. 2008;582:2453–2457. doi: 10.1016/j.febslet.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Togashi H, Schmidt EF, Strittmatter SM. RanBPM contributes to Semaphorin3A signaling through plexin-A receptors. J Neurosci. 2006;26:4961–4969. doi: 10.1523/JNEUROSCI.0704-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Li Z, Messing EM, Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J Biol Chem. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Totsukawa G, Yamakita Y, Sasaki Y, Madaule P, Ishizaki T, Narumiya S, Matsumura F. Citron kinase, a Rho-dependent kinase, induces diphosphorylation of regulatory light chain of myosin II. Mol Biol Cell. 2003;14:1745–1756. doi: 10.1091/mbc.E02-07-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Vazquez L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. G protein control of microtubule assembly. Annu Rev Cell Dev Biol. 2004;20:867–894. doi: 10.1146/annurev.cellbio.20.012103.094648. [DOI] [PubMed] [Google Scholar]
- Zou Y, Lim S, Lee K, Deng X, Friedman E. Serine/threonine kinase Mirk/Dyrk1B is an inhibitor of epithelial cell migration and is negatively regulated by the Met adaptor Ran-binding protein M. J Biol Chem. 2003;278:49573–49581. doi: 10.1074/jbc.M307556200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, Evaluation of RanBPM-shRNA constructs by immunoblotting. RanBPM-shRNA1 (targeting 3’UTR region of RanBPM) is more effective than RanBPM-shRNA2 (targeting coding region of RanBPM) in knocking down RanBPM expression in B104 cells. A four-base mutated construct of RanBPM-shRNA1 (RanBPM-shRNA1m4) is used as the control shRNA. α-tubulin is shown as control. Note that CITK expression is not affected by RanBPM-shRNA1. B, RanBPM RNAi does not decrease CITK or β-catenin expression in cultured progenitor cells. C, RanBPM-shRNA3 decreases RanBPM, while RanBPM-shRNA4 does not.
CITKfh/fh have more mitotic progenitors within their VZ and SVZ than do CITKwt/wt littermates. E17 CITKfh/fh and wildtype littermate brain sections were stained with anti-phosphohistone H3 (phosH3, green) and anti-nestin (red) antibodies. Double-positive cells were counted within 400 µm2 of the VZ and SVZ. Scale bars, 50 µm. Quantification is shown in graph (n=3 brains, 6 sections from each brain). **, p<0.01 from Student’s t-test.
Purified KCITK-HIS was incubated with either purified RanBPM-GST and GST (A) or RanBPM-GST and Histone H1 (B) in the kinase reaction buffer, which contains γ-P32 labeled ATP. The products were analyzed by 12% acrylamide gel followed by autoradiography (left in A and B) and coomassie staining (right in A and B). Purified GST (A) and Histone H1 (B) are controls. In A, neither RanBPM-GST (asterisk) nor GST (double asterisks) are phosphorylated by KCITK-HIS, demonstrating that neither of them are a substrate for KCITK-HIS. In B, phosphorylation of Histone H1, KCITK substrate, is not affected by RanBPM-GST. left, lane 1, KCITK-HIS+ RanBPM-GST 5 µg; lane 2, KCITK+RanBPM 50 µg; lane 3, KCITK+RanBPM 100 µg; lane 4, KCITK+RanBPM 200 µg; lane 5, KCITK+GST 50 µg; lane 6, KCITK+GST 100 µg. right, lanes are same as left, coomassie stained gel. B, KCITK-HIS phosphorylates Histone H1. left, lane 1, KCITK+H1; lane 2, KCITK+H1+RanBPM 5 µg; lane 3, CITK+H1+RanBPM 50 µg; lane 4, CITK+H1+RanBPM 100 µg; lane 5, CITK+H1+RanBPM 200 µg. right, lanes are same, coomassie stained gel.