Abstract
Cell migration is essential in a number of processes, including wound healing, angiogenesis and cancer metastasis. Especially, invasion of cancer cells in the surrounding tissue is a crucial step that requires increased cell motility. Cell migration is a well-orchestrated process that involves the continuous formation and disassembly of matrix adhesions. Those structural anchor points interact with the extra-cellular matrix and also participate in adhesion-dependent signalling. Although these processes are essential for cancer metastasis, little is known about the molecular mechanisms that regulate adhesion dynamics during tumour cell migration. In this review, we provide an overview of recent advanced imaging strategies together with quantitative image analysis that can be implemented to understand the dynamics of matrix adhesions and its molecular components in relation to tumour cell migration. This dynamic cell imaging together with multiparametric image analysis will help in understanding the molecular mechanisms that define cancer cell migration.
Keywords: Cancer, Cell migration, Matrix adhesions, Dynamic imaging, Multiparametric analysis
Introduction
Cell migration, both single and collective, is a highly integrated multistep process that is essential in embryonic morphogenesis, tissue homeostasis and immune surveillance. While collective migration requires the movement of cohesive groups of cells [1], the single migrating cell is highly polarised with complex regulatory pathways that are spatiotemporally controlled [2]. Migration contributes to several important pathological processes, including cancer progression and metastasis formation. Metastasis, dissemination of malignant tumours to a distant organ, is the major cause of cancer mortality. Tumour cell motility is the hallmark of invasion and is an essential step in metastasis.
Cell migration can be seen as a cyclic process. The initial response of a cell to a chemotactic signal is to polarise and extend protrusions in the direction of movement. These protrusions are usually driven by actin polymerisation, and are stabilised by adhering to the extracellular matrix (ECM). These adhesions serve as traction sites for migration as the cell moves forward on top of them, and they are disassembled at the cell rear, allowing it to detach. This process depends on the cell type and environment. Matrix adhesion turnover is clearly visible in slow-moving cells such as fibroblasts or epithelial cells which show large protrusions, and is less visible in fast-moving cells such as neutrophils or cancer cells, which display small protrusions with a rapid turnover [2]. Interestingly, the movement of cell sheets shows some features of single-cell migration; however, the polarisation extends over the entire sheet [1]. In addition to typical matrix adhesions, typically referred to as focal adhesions, cells can form another variety of adhesive structures, namely podosomes and invadopodia, also termed podosome-type adhesions (PTAs) [3]. Those unique actin-rich adhesion structures are typically associated with sites of proteolytic degradation of the extracellular matrix components. This is thought to contribute to cellular invasiveness in physiological and pathological situations. Cell types that form podosomes include monocytic, endothelial and smooth muscle cells, whereas invadopodia are mostly observed in carcinoma cells [4–6]. In this review, we will focus on the study of matrix adhesions in relation to tumour cell migration and invasion; however, most of what we will discuss here is also relevant for the study of PTAs [7, 8].
Although matrix adhesion dynamics is very important for understanding cell migration behaviour, little is known about the molecular mechanisms that regulate adhesion dynamics and signalling during tumour cell migration and invasion. Advances in microscopic imaging techniques including spatiotemporally resolved imaging, fluorescent reporter reagents and multiparametric image analysis will contribute to a better insight into these processes. In this review, we will focus only on single cell migration and discuss the current and emerging imaging technologies that can be implemented to study adhesion dynamics and signalling in migrating cells. Tables 1 and 2 summarise all the existing techniques to study, respectively, protein dynamics and modes of tumour migration and can be used as guidelines. In parallel, we discuss the different multiparametric image analysis tools that can be applied after image acquisition to generate more detailed and reliable cellular and molecular measurements. Table 3 gives a non-exhaustive list of challenges and potential solutions for improving the ‘throughput’ of this systems microscopy approach. Finally, studying the fundamental mechanisms underlying cell migration and invasion using physiologically relevant experimental models together with dynamic imaging and multiparametric image analysis will help in discovering new interesting targets and therapeutics for inhibition of tumour invasion and consequently cancer metastasis formation.
Table 1.
Imaging techniques to study protein dynamics and interactions in adhesion
| Technology | Biology | What is next | References |
|---|---|---|---|
| Photoactivation |
Measure high resolution diffusion, trafficking and stability of protein Actin dynamics, protein movement, integrin receptor trafficking |
For in vivo imaging, to track in the long term photoconverted cells and study protein dynamics | [65–67] |
| FRAP or FLIP |
Measure t 1/2, k on, k off, diffusion coefficient but individual measurements Integrin, FAK, paxillin, zyxin, vinculin, and actin |
Comined FRAP and/or FLIP with TIRF and/or FRET | [70–81] |
| FRAP–FLIP |
Measure t 1/2, k on, k off, diffusion coefficient and protein mobility parameters. Include all bleached and unbleached matrix adhesions FAK, paxillin, vinculin, zyxin and actin |
Comined FRAP-FLIP with TIRF and/or FRET | Le Dévédec et al., unpublished data |
| FCS, ICS, RICS |
Determine rates of diffusion, degree of aggregation, number of fluorescent entities and flow velocities (mainly used in solution) α5-integrin, α-actinin, FAK, paxillin and actin |
In living cells to study protein distribution, dynamics and interactions at high time and spatial resolution | [87–91] |
| FSM |
Movement of structure, assembly dynamics, and subunit turnover Actin, integrin, FAK, talin and α-actinin |
[82–86] | |
| FRET |
Protein–protein interaction and protein activity Src, FAK, Rho GTPases, and matrix adhesion proteins |
Combined with FRAP and/or TIRF and in vivo | [93, 97–102, 106] |
Table 2.
Modelling distinct modes of tumour migration
| Environment | Models | Microscopy | Obtained information | References |
|---|---|---|---|---|
| In vitro/2D | Matrix coating | Wide-field, confocal | Insights into the organisation of molecular machineries underlying cell adhesion and migration | [24, 53, 120, 129, 159–163] |
| Patterned | ||||
| In vitro/3D | Matrigel | Wide-field, confocal, confocal reflection microscopy, SHG | Distinguish aspects of cell movement/invasion (collective/individual; mesenchymal/amoeboid). Visualise interactions cell. ECM (in particular, collagen fibers I) | [25, 109, 110, 112–114, 116–118, 121, 132, 159, 164] |
| On/in collagen gel | ||||
| In vivo | Zebrafish | Confocal, confocal reflection, Multiphoton (SHG and FLIM) | Aspects of cell movement in the primary environment. Visualise interaction between tumour cells and tumour environment (ECM, host cells and blood vessels). Visualise intravasation event when blood vessels are counterstained | [1, 49, 120, 123, 125–135, 165–171] |
| Mouse | ||||
| Rat |
Table 3.
Challenges and potential solutions to increase throughput of imaging techniques
| Challenge | Type | Potential solutions |
|---|---|---|
| Sample preparation | 2D | Development of predictive screening assays (→ use of primary or embryonic stem cells (ESCs) instead of easy to culture tumour cell-lines but genetically aberrant) |
| 3D | Development and validation of relevant 3D scaffolds (→ characterise ECM of patient tumour material) | |
| Improve 3D cell culture techniques for automated liquid handling robotics (→ collaboration between academia and pharmaceutical companies) | ||
| In vivo | Automated microinjection of tumour cells in ZF (→ automatic microinjector based on pattern recognition) | |
| Automated filling of the microwells plates with ZF preferably all similarly orientated (→ make use of adapted mould) | ||
| Automated image acquisition | 2D | Autofocus combined with z-scans for 3D imaging (→ image-based or reflection-based autofocusing) |
| Pre-optimisation of the acquisition settings (→ autoexposure algorithm to adjust integration time of detectors) | ||
| Automated object (cells of matrix adhesion) localisation (→ autoexposure algorithm to adjust integration time of detectors) | ||
| Intelligent microscope [158, 172] | ||
| 3D/in vivo | Higher throughput kinetic imaging microscopes suitable for automated 3D invasion studies (→ see commercially available kinetic imaging systems such as Incucyte or Cell-IQ [117]) | |
| Higher throughput kinetic imaging microscopes suitable for FRET, FRAP or FCS (→ in the future, intelligent microscope that recognises the object to be visualised) | ||
| Data handling | 2D | Storing terabytes of data (→ storage area network (SAN) which has multiterabyte to tens of terabytes capacity; commonly, data on the SAN are backed up on tape as well) |
| 3D/in vivo | ||
| Data management (→ development of databases retrieved [157, Table 2]) | ||
| Image analysis | 2D | Image segmentation (→ depending on imaging quality, choose between region-based, edge-based or region-growing method) |
| 3D/in vivo | ||
| Multiparametric image analysis (→ phenotypic profiling which involves computer vision methods) | ||
| Object tracking (→ high time resolution for imaging; adapted tracking algorithms for 3D imaging [173] and HTS data) | ||
| Data mining and modeling | 2D | Screening reproducibility and estimators (→ quality standards, e.g. coefficient of variations (CVs) and zscores should not exceed 5% and should be higher than 0.5, respectively) |
| 3D/in vivo | Significant behaviour changes detection | |
| Automated classification (→ supervised machine learning) | ||
| Development of computational models |
Adhesion proteins
Matrix adhesion complexes are cytoplasmic structures that have been originally identified by electron microscopy [9] or by interference reflection microscopy [10]. They are the closest site of contact between the cell and the underlying substratum. Integrins are the principal cell surface adhesion receptors mediating cell–matrix adhesions [11]. Integrins are heterodimeric receptors that directly bind extracellular matrix molecules and couple them to the actin cytoskeleton (for reviews, see [12, 13]). Integrin cytoplasmic domains form multi-molecular complexes with proteins involved in cell adhesion signalling and with adaptors that provide a connection to the cytoskeleton [11]. Approximately 150 proteins, which form the so-called ‘integrin adhesome’, have been retrieved to date to be part of the matrix adhesions including kinases, phosphatases and structural proteins (for review, see [14]). Even a larger number of over 600 proteins are defined to be involved in the spatiotemporal regulation of matrix adhesions (van Roosmalen et al., unpublished data). Upon attachment, integrins will cluster and promote local recruitment of structural proteins like vinculin, paxillin, talin, α-actinin and tensin, and signalling molecules including tyrosine kinases such as focal adhesion kinase (FAK), serine/threonine kinases and various adapter proteins. The molecular complexity of cell–matrix adhesions enables them to fulfill their dual role as modulators of both mechanical cell anchorage and transmembrane signalling [12]. Integrins are not the unique transmembrane receptors that have been described to control and regulate cell adhesion co-signalling; syndecans, discoidin domain receptors (DDR), CD44 and dystroglycan can also bind directly to the ECM, while growth factor receptors such as the epidermal growth factor receptor (EGFR) can crosstalk with the integrins to regulate the recruitment of cytoplasmic proteins to adhesion sites (for reviews, see [15, 16]). Syndecans as well as DDRs can bind to ECM molecules but most of all they synergise with integrins [17]. DDRs are a novel classs of receptor tyrosine kinases that bind to several collagens and stimulate matrix metalloproteinases (MMP) production. CD44, ICAM-1 and ICAM-2 and L-selectin are other adhesion receptors that are able to interact with ECM ligands as well as with ezrin, radixin, moesin (ERM) family proteins. Dystroglycan is a ubiquitously expressed heterodimeric adhesion receptor that can localise at matrix adhesions [18]. Dystroglycan in concert with integrins is involved in the spatial and temporal remodelling of adhesion numbers, types and distribution. It has emerged as a multifunctional adhesion platform with many interacting partners (for review, see [19]). One emerging area is the role of dystroglycan in cancer. Dystroglycan function appears to be disrupted in numerous epithelial-derived cancers [20]. A lot is known on the role of integrins in cell migration; in contrast, the mechanistic contribution of distinct adhesion systems such as syndecans, DDRs, CD44 and dystroglycan still remains incompletely understood. Advanced methods are necessary to systematically study the mechanism of regulation of these diverse adhesion systems.
Adhesion types in 2D and 3D
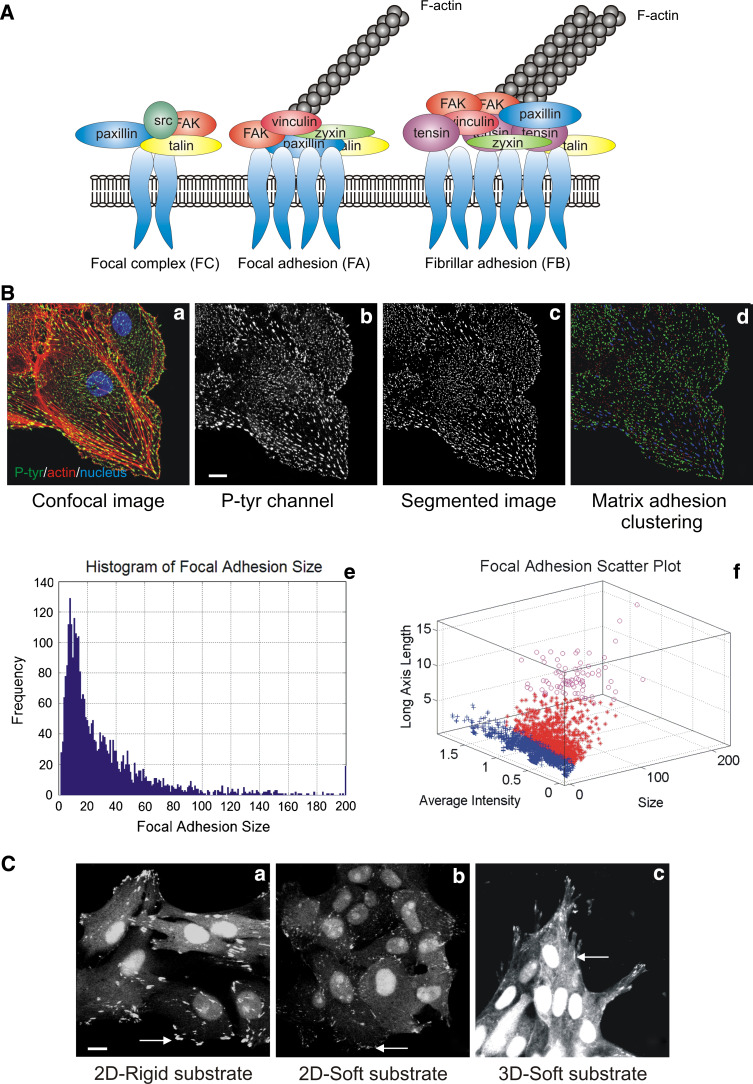
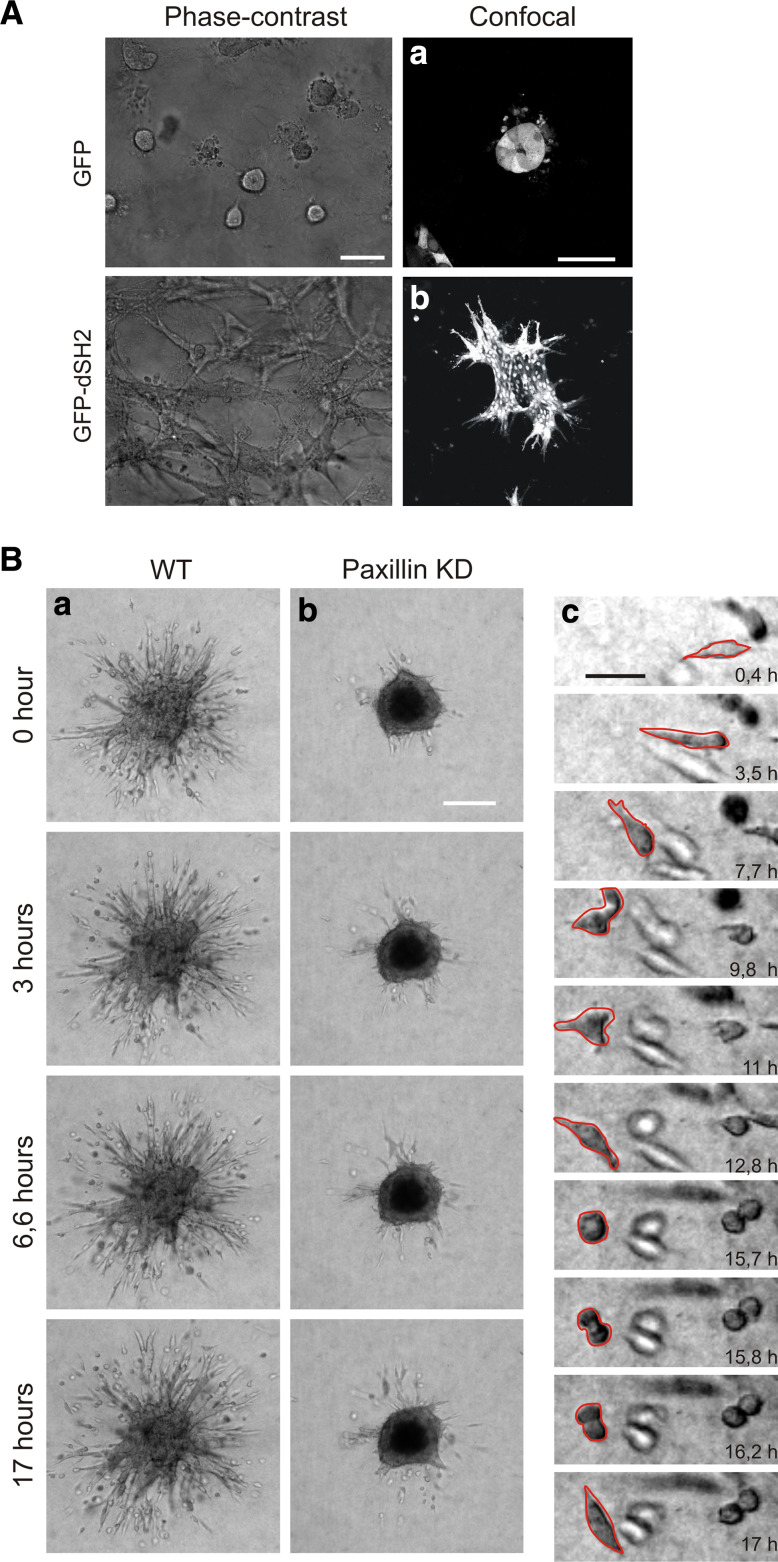
Matrix adhesions are diverse in molecular composition, size and shape. Their distribution over the whole cell body is also heterogeneous and depends on cell type and environment. Typically adherent cultured cells on two-dimensional substratum in vitro show a large number of matrix adhesions ranging in size from less than 1 to several μm2. Detailed analysis based on morphology, molecular composition and method of formation of mainly fibroblasts and epithelial cells allowed those adhesions to be classified in three different classes (Fig. 1a). The most common forms of integrin-mediated cell–matrix adhesions in cultured cells are focal adhesions (FA), fibrillar adhesions (FB) and focal complexes (FC) [12, 13]. Focal adhesions are oval structures, usually a few μm2 in area, and are associated with the termini of actin stress fibres. Fibrillar adhesions, which are derived from FAs, are elongated contact sites, associated with fibronectin fibrils. Focal complexes are small, dot-like adhesions that are mainly found at the cell edge and apparently nucleate FA formation (reviewed by [12]). Based on intensity-based segmentation, matrix adhesions can be analysed using fluorescence microscopy images obtained after immunofluorescence staining of cells for focal adhesion marker such as vinculin or paxillin. Indeed, the morphological and intensity parameters of adhesions can be quantified by analysing images following waterline image segmentation ([21] and Yan et al., unpublished data) (Fig. 1b). This segmentation defines adhesions as ‘‘objects’’ (Fig. 1b, part c), based on their fluorescence intensity and size range (Fig. 1b, part d). The image analysis provides various features such as size (Fig. 1b, part e), elongation, mean fluorescence-signal intensity and cell localisation with respect to the nucleus which can be used for further clustering (Fig. 1b, part f). Furthermore, matrix adhesions are heterogeneous in their molecular composition which can be studied using fluorescence ratio imaging. For example, FC do not contain zyxin whereas FB do not contain β3 integrin or phosphorylated FAK [22] but is highly enriched with tensin [13]. In general, quantitative multicolour fluorescence imaging can be used to unravel the molecular complexity of the various types of adhesions [23]. Matrix adhesion complexity also depends on the environmental stimuli. FAs are mechanosensors that sense the matrix physical properties whether it is rigid or soft and whether it is two- or three-dimensional [24]. Distinctive 3D matrix adhesions were described for the first time when cells were cultured in cell-derived 3D matrices: they appear like fibrillar adhesions also enriched in tensin but also rich in tyrosine phosphorylated proteins [25]. Epithelial cells stably expressing an alternative ‘reporter’ for protein tyrosine phophorylation GFP-dSH2 [26] show large peripheral and ventral focal adhesions on a rigid substrate, whereas on a soft substrate, the adhesions become smaller, dot-like and only localised in the periphery of the cell (Fig. 1c). In addition, the GFP-dSH2 construct localised in a fibrilla pattern alongside the elongated cell protrusions, in a 3D collagen matrix (Fig. 1c, part c). In conclusion, matrix adhesion complexity and diversity can be studied with static immunofluorescent images, but those sites are obviously very dynamic during cell migration in 2D and 3D environments.
Fig. 1.
Matrix adhesions diversity and composition. a Schematic view of the three classes of matrix adhesions found in adherent cells in vitro. b Image analysis of matrix adhesions. Confocal picture of epithelial cell stained for Hoechst (blue), P-Tyr (green) and F-actin (red) (a); scale bar 10 μm. Confocal picture of focal adhesions only (b). Matrix adhesions segmentation (c) and clustering according to size (d). Distribution of the matrix adhesions according to their size (e) and clustering according to matrix adhesions intensity and length (f). c Matrix adhesions differ in size and shape according to their environment: in 2D rigid versus soft and in 3D; scale bar 10 μm
Adhesion turnover during cell migration
Cell migration is an integrated process that requires the coordinated regulation of various structural and signalling molecules, including distinct kinases and phosphatases [27, 28]. Cell migration requires the establishment of cell polarity to create a leading edge and a trailing edge. The leading edge undergoes membrane protrusive activities driven by actin polymerisation that establish new matrix contacts, whereas at the trailing edge, cell adhesions are disassembled to promote retraction of the cell rear and forward cell movement. The rate of cell migration can be limited by the rate of rear retraction, and thus the dynamic formation and disassembly of cell–matrix adhesions are critical to cell migration [2, 28].
Formation of adhesions
The mechanism by which adhesions assemble in migrating cells is still under investigation. Some cells, particularly rapidly migrating ones such as leukocytes, have few visible integrin clusters, and thus very small submicroscopic adhesions are probably important for their migration. In other cells, small adhesions known as focal complexes can be observed at the leading edge. Formation of these adhesions depends on Rac- and Cdc42-GTPases, and these adhesions stabilise the lamellipodium by mediating attachment to the ECM, thereby contributing to efficient migration. However, cells with large integrin clusters (“focal adhesions”) are tightly adherent and are typically either non-migratory or move very slowly. The assembly of focal adhesions involves Rho-GTPase as well as myosin-induced contractility. During their formation, some protein components enter adhesions with similar kinetics, which suggests that they exist in preformed cytoplasmic complexes. However, other components enter adhesions with very distinct kinetics, which is consistent with a model in which a regulatory event initiates the serial addition of different proteins [27].
Adhesion disassembly at the front and the rear
Adhesion disassembly is observed both at the leading edge, where it accompanies the formation of new protrusions, and at the cell rear, where it promotes tail retraction. At the front of migrating cells, adhesions at the base of a protrusion disassemble as new adhesions form at the leading edge (for review, see [27]). However, some adhesions persist and mature into larger, more stable structures. Little is known about adhesion disassembly versus maturation; however, targeting of microtubules has been implicated as one factor that promotes adhesion disassembly (for review, see [29]). Both protein kinases and phosphatases also appear to be central to the regulation of adhesion turnover and stability [30]. For example, cells lacking the tyrosine kinases FAK or Src have more and larger adhesions and migrate poorly [31]. Adhesion turnover in migrating cells is also regulated by a complex of Rac-associated proteins [32, 33]. Also, the calpain family of intracellular proteases is implicated in regulating cell migration [28]. Calpain activity is tightly controlled in vivo as this intracellular protease irreversibly cleaves numerous signalling and structural proteins with widespread impacts on cell functioning and viability. The ubiquitous calpains can cleave a large number of adhesion complex components including talin, paxillin, vinculin, ezrin, alpha-actinin, FAK, E-cadherin, and the cytosolic tails of some of the beta-integrins [28, 34–36]. Calpain activity which relies on the removal of a protein or elimination of an enzymatic activity is a key regulator of matrix adhesion dynamics. For instance, the study of Franco and co-workers demonstrates that calpain-mediated proteolysis of talin is a rate-limiting step in adhesion disassembly [37]. In conlusion, functional studies that will sytematically determine the role of individual proteins involved in cell–matrix adhesions using dynamic imaging of the protein localisation, kinetics and interactions will shed a more complete light on the mechanisms of cell migration. RNAi (live) cell imaging-based screening approaches are key to such an analysis.
Cell migration/matrix adhesion and cancer
Metastasis is the most frequent cause of death for patients with cancer. Tumour cell motility is the hallmark of invasion and is an initial step in metastasis. In order to metastasise, cancer cells must first detach from the primary tumour, migrate, invade through tissues, and attach to a second site. Invasive carcinoma cells acquire a migratory phenotype associated with increased expression of several genes involved in cell motility such as matrix adhesion-associated genes. Many metastatic human carcinomas are characterised by the overexpression or constitutive activation of ErbB tyrosine kinase receptor (EGFR) family members involving activating mutations of the receptor kinases or an autocrine loop with EGF family ligands. High expression of EGFR and ErbB2, as well as another tyrosine kinase receptor c-Met, are associated with poor prognosis of breast cancer patients. In addition to stimulating cell differentiation and proliferation, EGF promotes tumour cell motility, invasion and metastasis [38–40]. Recent data provide evidence for a requirement for certain focal adhesion protein expression (e.g. integrin, FAK/Pyk2, paxillin, ILK, Ezrin) in metastatic dissemination. Integrins are known to regulate a diverse array of cellular functions crucial to the initiation, progression and metastasis of solid tumours. The importance of integrins in several cell types that affect tumour progression has made them an appealing target for cancer therapy [41]. Increased activity or overexpression of Src is a frequent occurence in many types of human cancer. Indeed, a prominent role of Src is played in invasion, epithelial-mesenchymal transition (EMT) and development of metastasis. Inhibitors of Src are promising drugs for cancer therapy (for reviews, see [42, 43]). Focal adhesion kinase (FAK), another well-studied non receptor tyrosine kinase, plays a key role in cancer progression. FAK expression and activity are enhanced in metastatic tumours of diverse origin [44–46]. FAK is considered as a promising gene candidate, and inhibitors of FAK are currently in clinical trial (for reviews, see [47, 48]). Not only activity of kinases but also activity of proteases such as calpain is enhanced upon transformation induced by the v-Src, v-Jun, v-Myc, k-Ras, and v-Fos oncoproteins. Elevated calpain activity commonly promotes focal adhesion remodelling, disruption of actin cytoskeleton, morphological transformation, and cell migration, although proteolysis of target substrates (such as focal adhesion kinase, talin, and spectrin) is differently specified by individual oncoproteins [34, 35]. Therefore, studying the motility mechanisms used by cancer cells would clarify some of the key events influencing metastasis in cancer. In addition, identification of the molecular pathways that play a role in cancer cell motility will provide new diagnostic approaches and targets for the treatment of metastatic cancer.
Studying migration and adhesion dynamics in living cells
The above indicates a general requirement for advanced technologies to systematically study both cell adhesion dynamics and tumour cell migration. Recent advances in fluorescence probes and microscopy technologies have provided powerful approaches that present advantages over the traditional biochemical approaches. In particular, the newly developed recombinant fluorescent proteins (FPs) and genetically encoded biosensors are useful tools for imaging protein distribution, dynamics and interactions in live cells with high temporal and spatial resolutions enabling the elucidation of molecular mechanisms behind adhesion turnover responsible for the cell migration [49–54].
Phase contrast/Normaski imaging
The starting point for many investigations of cell movment is to characterise the behaviours of cells, by recording shape changes, direction and persistence of movement and the dynamics of subcellular membrane structures such as lamellipodia, filopodia, protrusions and membrane ruffling. This can be done with wide-field microscopy in either phase-contrast or Normarski Interference Contrast modes which is fast, not photo-toxic to cells and can be performed over a long period of time. Many software packages are now available that automatically track moving cells visualised in this way: e.g. CellTrack, ImageJ (plug-in MTrack2), Imaris, Image Pro and in-house-developed cell track systems [55, 56]. The tracking rates of those different softwares are very good, although application of fluorescence microscopy instead of phase contrast can improve the tracking efficiency [56]. Furthermore, due to high signal to noise ratio in phase-contrast or Normaski imaging, it remains difficult to analyse other cell features. Fluorescent imaging is an answer to that problem.
Fluorescent live cell imaging
Cell migration: wide-field
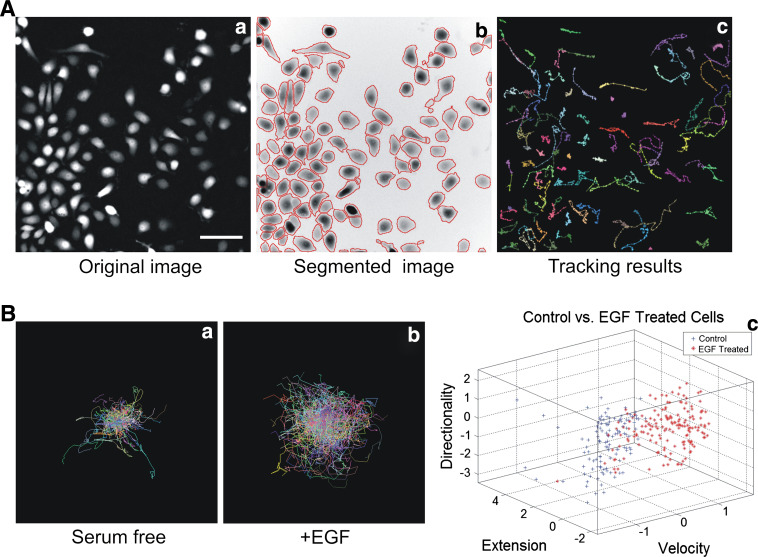
Wide-field fluorescent microscopes equipped with the appropriate excitation and emission filters and sensitive CCD camera are fast and sensitive enough to detect GFP stably expressing migrating cells. The dynamic imaging of 2D cell migration includes various assays such as directed cell migration (chemotaxis and woundhealing) and random cell migration [57]. The dynamic imaging of migrating cells which implies tracking of fast objects needs high temporal resolution. Indeed, to track fast-moving objects, the interval between two sequential frames should be short enough to ensure an overlap of the moving object between frame t and frame t + 1. Using cell-lines stably expressing GFP together with advanced image analysis tools allow the simultaneous detection of multiple cellular characteristics that define phenotypic response such as EGF response in rat mammary carcinoma cell-line (Fig. 2b). Application of advanced bioinformatics and statistical methods to multiparametric image data generates a non-biased phenotypic fingerprint that describes the effect of genetic or pharmacological manipulation (Fig. 2c). The extension of multiparametric image analysis to live cell studies can reveal further mechanistic insight (Yan K et al., unpublished data). Epi-fluorescent microscopy can also be used to visualise adhesion dynamics. The quantification of adhesion dynamics was first described by Webb and colleagues [58] using epi-fluorescence microscopy. Adhesion assembly and disassembly rate constants can be calculated by measuring the incorporation or loss of fluorescence (e.g. GFP-paxillin). Assembly will provide an increase in fluorescence intensity whereas disassembly will lead to a loss in intensity. Intensity values at various time points are plotted on a semi-logarithmic scale representing fluorescent intensity ratios over time. These ratios are calculated using the formula ln(I/I 0) for assembly and ln(I 0/I) for disassembly (where I0 is the initial fluorescent intensity and I is the fluorescent intensity at the indicated time). Rate constants can then be calculated from the slope of the resulting line of best fit. The study conducted by Webb et al. [58] revealed for the first time that fibroblasts with a knockout of FAK, Src and p130Cas decreased the disassembly rate constant of 1.2 × 10−1 min−1 in wild-type cells to between 6 × 10−3 and 9 × 10−3 min−1 in knockout cells.
Fig. 2.
Imaging and analysis of single cell migration. a Epifluorescent imaging and analysis of migrating MTLn3 cells ectopically expressing GFP. Epifluorescent pictures (a) are waterline based segmented (b) and cells are consequently tracked (c); scale bar 50 μm. b Individual cell tracks of MTLn3 stimulated (a) or not by EGF (b) and clustering analysis of both treatments based on directionality, extension and velocity (c)
Confocal laser scanning microscopy (CLSM)
While epi-fluorescence microscopy provides the high degree of time resolution which is required to visualise fast processes, confocal laser scanning microscopy (CLSM) offers several advantages over conventional optical microscopy, including controlled depth of field, the elimination of image degrading out-of-focus information, and the ability to collect serial optical sections from thick specimens. It provides high spatial resolution, but still many physiological processes and events take place faster than they can be captured by most CLSMs, which have image acquisition rates typically in the order of one frame per second. CLSMs using acousto-optical devices and a slit for scanning are faster than the galvanometer-driven point-scanning systems, and are more practical for physiological studies. These quicker designs combine good spatial resolution with good temporal resolution, which may be 30 frames per second at full screen resolution, or near video rate. The slower point-scanning microscope systems can achieve the best temporal resolution only by scanning a much reduced area on the specimen. If full spatial resolution is required, the frames must be collected less frequently, losing some temporal resolution. The confocal systems using disk-scanning or oscillating mirror-scanning methods are also capable of imaging rapid physiological or other transient events. Nowadays, new developments in confocal microscopy provide both high-speed and high-resolution imaging to capture intracellular biological processes. A resonant confocal scan head dramatically improves time, spatial and spectral resolutions and allows high-speed imaging up to 230 frames per second (512 × 64 pixels) which make it an ideal technique for bleaching and protein kinetic experiments (photo-activation and photo-bleaching, see below). A major drawback of confocal microscopy is photo-bleaching and photo-damage from the illuminating laser beam that can be cumulative over multiple scans. So the exposure to the beam should be kept to the minimum necessary to acquire the image. Another recent development is controlled light exposure microscopy (CLEM) which should be ideal for live imaging as it helps to reduce photo-bleaching and photo-toxicity; the two main limitations in live-cell confocal microscopy [59]. In CLEM illumination excitation light is reduced using two strategies. The first is based on the principle that if there is no signal, then no illumination is required (for example, when imaging the background). The second detects whether there is sufficient signal to acquire an image. If so, illumination is stopped.
Total internal reflection fluorescence (TIRF) microscopy
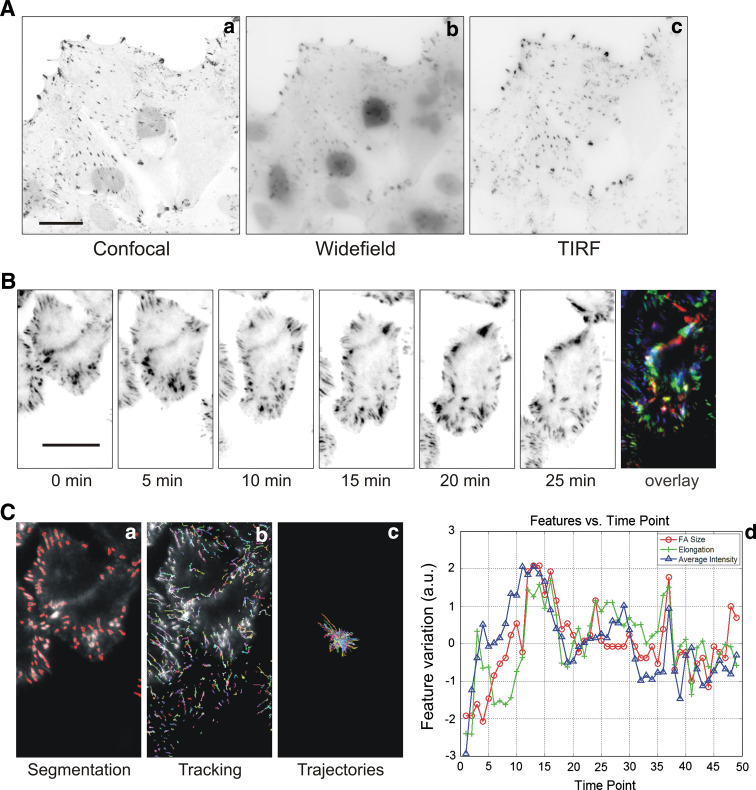
One key advance in the field of dynamic imaging of matrix adhesions is the recent commercial development of total internal reflection fluorescence (TIRF) microscopy. TIRF was first described almost 30 years ago (for review, see [60]): cell-substrate contacts of human skin fibroblasts, labelled with fluorescent lipids, were investigated by TIRF microscopy, which exploits the unique properties of an induced evanescent wave or field in a limited specimen region immediately adjacent to the interface between two media having different refractive indices. In practice, the most commonly utilised interface is the contact area between a specimen and a glass coverslip. In a typical experimental setup, fluorophores located in the vicinity of the glass–liquid or plastic–liquid surface can be excited by the evanescent field, provided they have potential electronic transitions at energies within or very near the wavelength bandwidth of the illuminating beam. Because of the exponential fall-off of evanescent field intensity, the excitation of fluorophores is restricted to a region that is typically less than 100 nm in thickness. By comparison, this optical section thickness is approximately one-tenth that produced by confocal fluorescence microscopy techniques. Because excitation of fluorophores in the bulk of the specimen is avoided, confining the secondary fluorescence emission to a very thin region, a much higher signal-to-noise ratio is achieved compared to conventional wide-field epifluorescence illumination. This enhanced signal level makes it possible to detect single-molecule fluorescence by the TIRF microscopy method. In Fig. 3a, we show the improvement of TIRF imaging above wide-field and confocal microscopy by eliminating out of focus background fluorescence. Another example of the application of high speed and resolution TIRF imaging of focal adhesion turnover is shown in Fig. 3b. The matrix adhesion turnover can be consequently quantified with multiparametric image analysis (Fig. 3c) (Yan K et al., unpublished data).
Fig. 3.
Imaging adhesions by confocal, wide-field and TIRF microscopy. a Z-scan series of the same renal epithelial LLC-PK1 cell overexpressing the reporter construct GFP-dSH2 performed with confocal (a), wide-field (b) and TIRF microscopy (c); scale bar 20 μm. Note the advantage of TIRF microscopy for visualising matrix adhesions. b Analysis of matrix adhesions dynamics with TIRF microscopy. Time lapse of a migrating MTLn3 cell expressing GFP-paxillin and overlay of the different frames to illustrate the focal adhesion turnover; scale bar 10 μm. Note the fast turnover of matrix adhesions in these cells. c Multiparametric analysis of matrix adhesion dynamics. a Matrix adhesion segmentation, b tracking of individual matrix adhesions, c plot of all individual matrix adhesion trajectories and lifetime, d example of possible plot of different features (FA size, elongation and intensity) of an individual matrix adhesion over the time. The different features are normalised so that the data distribution is scaled to 1 and the average of all features are shifted to zero. A FA size of −2 indicates that the FA size in this frame is smaller than its average size by 2 in the normalised feature space
Studying protein dynamics and interactions in adhesion
While imaging matrix adhesion proteins using fluorescent microscopy provides information about spatial localisation, it does not allow further measurements of protein modification that regulate the adhesion signalling. The goal of imaging matrix adhesions in migrating cells is not only to visualise their localisation but also to determine their function, the interaction of the different adhesome components, and the mechanisms that regulate adhesion formation and disassembly. To be able to quantify and model protein dynamics in adhesions, many quantitative measurements are needed: interaction between adhesion components, k on and k off of individual components and complexes, diffusion values, protein concentration in both the cytosol and the focal adhesions. The challenge is to develop methods for measuring these parameters during adhesion complex formation and disassembly in migrating cells and preferentially in a high-throughput manner (Table 1).
Photo-activation, Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photo-bleaching (FLIP)
Many fluorescent proteins (FPs) have been designed by mutagenesis to change their fluorescence intensity (photo-activation) or colour (photo-conversion) upon illumination by lights with specific wavelength, intensity and duration (for review, see [52]). Those FPs can be grouped into three classes: (1) irreversible changes in colour upon illumination which includes photo-activatible (PA)-GFP [61] that can be photo-activated with UV illumination and emits green fluorescence; (2) irreversible change in colour from green to red upon UV illumination which includes Kaede [62] and Dendra [63]; and (3) reversible change in intensity/colour upon UV illumination such as Dronpa [64] which is a very valuable marker for photo-conversion studies and fast cellular processes. Until now, mainly PA-GFP has been successfully used to investigate actin dynamics [65], protein adhesion movement [66] and trafficking of integrin receptors [67]. Those photo-switchable fluorophores are valuable probes than can be used in live cells to monitor with high resolution diffusion, trafficking and stability of protein targets.
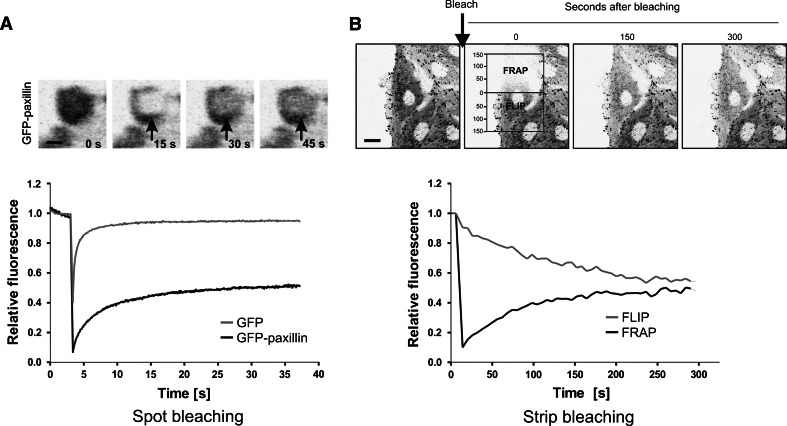
Fluorescence recovery after photo-bleaching is a commonly used technique to measure protein kinetics. In a typical FRAP experiment, a small defined region within a larger volume (for instance, cell nucleus or a focal adhesion) is briefly illuminated at high laser intensity (Fig. 4a) [68, 69]. Immediately after the bleach-pulse, the majority of the GFP-tagged proteins within the region have irreversibly lost their fluorescent properties: they are photo-bleached. Proteins from outside will then diffuse into the bleached region resulting in the increase of the fluorescence signal in the region until it reaches a defined signal intensity depending on the (transiently) immobile protein fraction. In summary, FRAP experiments provide information on protein mobility parameters including diffusion coefficient, immobile fraction and residence time. Several variants of FRAP have been developed including spot-FRAP, strip-FRAP, FLIP (Fluorescence loss in photo-bleaching), combined FLIP-FRAP. Spot-FRAP is based on photo-bleaching of a small spot within a focal adhesion (Fig. 4B), whereas in a strip-FRAP, a narrow strip spanning the cytoplasm in between focal adhesions is bleached. FRAP is widely and successfully used in adhesion biology. Several FRAP studies have addressed the dynamic properties of numerous adhesion properties [70–81].
Fig. 4.
Studying dynamics of matrix adhesion associated proteins by FRAP analysis. a Time lapse of a typical spot-bleaching experiment. A region of interest within a focal adhesion is defined, bleached with a high power laser intensity and subsequently followed over the time until fluorescence intensity reached a steady state. Fluorescence recovery over the time is plotted. Scale bar 1 μm. b Combined FLIP–FRAP experiment is performed over the whole cell which allows analysis of several adhesions in the same time. Average loss in fluorescence and recovery of fluorescence are plotted over the time. Scale bar 10 μm
In a FLIP experiment, the loss of fluorescence in a region or structure far from the bleached region is monitored. FRAP and FLIP can also be combined (FLIP–FRAP): two regions are monitored simultaneously after bleaching only one of them. We successfully developed a FLIP–FRAP bleaching procedure so that in one experiment we can analyse all the focal adhesions distributed over the whole cell body (Le Dévédec et al., unpublished data). Together with Monte Carlo simulation, we observed that FAK and paxillin had an equal diffusion rate but a differential residence time that is related to adhesion size and strength. With this technique, we are able to extract mobility parameters of adhesion proteins as well as a mapping of the protein kinetics according to focal adhesion size, type and localisation in the cell (Fig. 4b).
Fluorescence speckle microscopy (FSM)
Another fluorescent technique that detects protein dynamics, turnover and interaction is a method called fluorescent speckle microscopy (FSM) that uses a very low concentration of fluorescent subunits, conventional wide-field fluorescence light microscopy and digital imaging with a low-noise, cooled charged coupled device (CCD) camera [82]. In FSM, the fraction of fluorescently labelled molecules in the cell, relative to the level of endogenous unlabelled molecules, has to be very low (typically 0.5% or less). Labellled and unlabelled molecular subunits stochastically co-assemble into structures, giving a random and sparse distribution of fluorescent subunits with a ‘speckled’ appearance in high-resolution fluorescence images. The low level of fluorescent subunits reduces background fluorescence. Translation of the fluorescent speckle distribution indicates movement of structures whereas changes in speckle intensity and pattern reveal assembly dynamics and subunit turnover. Keys to successful FSM are the ability to image diffraction-limited regions (~0.25 mm) containing few (2–10) fluorophores, and the capacity to inhibit photo-bleaching which is only possible with sensitive imaging system that includes a low noise/high quantum efficiency camera. Extensive characterisation of actin dynamics using FSM has revealed two spatially, kinetically and kinematically distinct actin networks, with the local expansion of the lamella network being a source of persistent cell protrusion [83, 84]. There is also evidence that the actin network is dynamically coupled to adhesions [85]. A recent study using FSM measured the coupling of focal adhesion proteins to actin filament. Their FSM analysis of the dynamic interactions between matrix adhesion components and F-actin in living cells revealed that there is a hierarchy of motion from fast to slow, from actin-binding proteins to adhesion proteins within matrix adhesions and to integrins [86]. Those FSM-mediated molecular measurements provided considerable knowledge on the mechanism behind matrix adhesion dynamics.
Fluorescence correlation spectroscopy (FCS) and variants
Fluorescence correlation spectroscopy (FCS) analyses concentration fluctuations as a function of time to determine kinetic parameters, molecular associations and concentrations. This technique requires laser excitation of small focal volume and measure fluctuations in fluorescence intensity over many time intervals. Single or cross-correlation analysis (in the case of two different fluorophores) is applied in local areas across a time course to determine rates of diffusion, degree of aggregation, number of fluorescent entities and flow velocities. When two different fluorescent proteins are used, the cross-correlation function provides estimates of their fractional association and rates of co-transport. FCS is used most widely to study molecules in solution; only a few groups have applied FCS to analyse adhesion protein movements in intact living cells. A variant of FCS, image correlation spectroscopy (ICS), was implemented by Wiseman and co-workers [87] and allowed them to investigate the distribution, dynamics and interactions of α5-integrin, and α-actinin in the context of the formation and disassembly of adhesions during cell migration. Integrins are clustered throughout the cell and in nascent adhesions get 1.4 times more concentrated and 4.5 times more clustered and less mobile than in surrounding regions. Although FCS has a high temporal resolution (microseconds) but low spatial resolution, ICS has a low temporal resolution (seconds) but a high spatial resolution. A new analysis method, termed raster imaging correlation spectroscopy (RICS), can be applied on any confocal microscope [88] and bridges the timescales of FCS and ICS, as well as providing spatially resolved dynamic information such as the diffusion and binding of paxillin-GFP stably expressed in CHOK1 cells [89] and of FAK-GFP in MEFs [90]. Another variant of FCS, a general velocity-mapping technique termed spatio-temporal image correlation spectroscopy (STICS) has been described and provided new insights into the protein mobilities within the focal adhesions: while integrins were mostly immobile, paxillin and FAK immobile fractions were equal (74%), and actin was more rapidly diffusing (24%) [91]. Finally, a recent study that combined different fluorescence fluctuation approaches demonstrated that paxillin-GFP shows a heterogeneous dynamic within the cell [92]. In the cytoplasm, paxillin is uniformly distributed and diffuses freely as a monomer. Near adhesions, paxillin binds to protein partners and so its dynamics is reduced. These dynamic were different from assembling to disassembling adhesion regions, even within a single adhesion [92]. The implementation of complementary fluctuation methods will provide new data on the dynamics of protein adhesions during cell migration.
Fluorescence resonance energy transfer (FRET)
A powerful imaging method to study protein–protein interactions in living cells is fluorescence resonance energy transfer (FRET) [93, 94]. FRET is the non-radiative transfer of energy from a donor fluorophore in an excited state to a nearby acceptor fluorophore to allow energy transfer if within only 10 nm. Because this distance is in the range of protein sizes, FRET can also be used to study conformational changes of proteins tagged with a FRET donor and FRET acceptor. The most frequently used FRET methods are sensitised emission, ratio imaging, and acceptor photo-bleaching FRET [95] but the latter is not appropriate for studying rapid changes of protein interactions over time. The sensitised emission approach detects the emission of the acceptor fluorophore (often cyan fluorescent protein, CFP) while the donor fluorophore (often yellow fluorescent protein, YFP) is excited. Although still widely used, sensitised emission requires careful data processing [96] and due to signal-to-noise ratio can be poorly sensitive. Cross-talk and bleed through from one fluorophore to another makes the analysis highly dependent on control measurements of cells in which only one of the two fluorophores is present. An alternative approach to determine FRET is acceptor/donor ratio imaging (e.g. YFP/CFP) where both donor and acceptor emission are detected simultaneously when excited at the excitation wavelength of the donor. However, this method can be only applied when donor and acceptor are equally expressed in a cell system which is always the case when using FRET biosensors. In the study of adhesions, a few FRET biosensors have been designed to monitor in live cells the activity of a number of kinases, e.g. Src and FAK [97, 98] and GTPases, e.g. Rho, Rac and Cdc42 [99–102]. A fourth method to detect FRET is based on the reduced lifetime of excited donor molecules when they are in the proximity of acceptors [103, 104]. This technique is considerably more sensitive and accurate than intensity-based methods, but is slower and requires specific detector and often a pulsed light source such as a two-photon laser. Consequently, this has limited the application of FLIM in live cell studies. However, improvements in microscope design detector technology have reduced the time for data acquisition. Indeed, the group of P. French has developed a time domain optically sectioned FLIM microscope developed for high-speed live cell imaging. This single photon excited system utilises a spinning Nipkow disc microscope and can acquire fluorescence lifetime images of live cells at up to 10 frames per second, permitting high-speed FLIM of cell dynamics and protein interactions with potential for high throughput cell imaging and screening applications [105]. When correctly applied, FRET is a useful tool for investigating the molecular mechanisms that regulate integrin-mediated signalling in migrating cells [93, 97, 102, 106].
Optical imaging towards understanding tumour cell migration and intravasation
Of course, studying and understanding adhesion dynamics in cells migrating onto a rigid 2D substrate still does not reflect the in vivo situation. The critical comparison of 2D and 3D in vitro migration methods brought to light that there are differences in the observations made with these two systems. A combination of methods and environments is needed to investigate cell migration with regard to metastasis development. The biological relevance of focal adhesions was initially questioned, since equivalent structures to these prominent 2D adhesion structures were not easily observed in most tissues. However, focal adhesions have been found at point of high fluid shear stress in blood vessels [107] and in proximal tubular cell–ECM contact sites in kidney tissue [108]. Imaging migration and adhesions in 3D culture systems in vitro and in vivo are still in development. Despite the recent advances in dynamic imaging, a number of technical challenges remain to be overcome to allow functional and biochemical study of the tumour invasion mechanism in 3D ECM substrates and in vivo (Table 2).
Imaging adhesions and tumour invasion in 3D culture systems
Although studying the role of matrix adhesion in migrating cells in a 2D environment has provided a wealth of data, it is not surprising that there is an ongoing effort on setting up 3D invasion models adapted for dynamic live cell imaging that allow a more robust understanding of tumour cell invasion (for recent review, see [109]). A number of experimental methods incorporating different types of ECM substrates (Type I collagen, fibronectin, Matrigel, matrix polymers and tumour associated 3D matrices) have been developed to study 3D tumour invasion in vitro [110–115]. For instance, a recent patent application (US 20100056390) for a 3D cell culture system adapted for high-throughput drug screening has been accepted. Despite these recent advances in 3D substrates, it remains a technical challenge to visualise first the entire cell body of moving cells and second the matrix adhesions self. Several handicaps include background due to the matrix itself, signal to noise and the depth of the sample which limit the choice of objectives. Phase contrast imaging is a convenient imaging approach if signal to noise is not an issue for further data processing (Fig. 5a). It provides cellular and temporal resolution which is already enough to characterise the type of 3D invasion which can be depicted as either single (mesnchymal or amoeboid) or collective migration [1, 109, 110, 116]. Confocal microscopy is the most suitable imaging technique to collect fixed endpoints or time-lapse sequences of three-dimensional data of migrating cells and matrix adhesion activities [117–120]. Advanced bioinformatics is needed to further process the data and apply multiparametric image analysis to describe cell behaviour with several parameters [117, 121, 122]. In Table 3, we recapitulate the challenges and potential solutions to make the 3D culture system a versatile model for understanding tumour invasion mechanisms. Next to the dynamic imaging of cell migration, imaging of matrix adhesion turnover in a 3D environment is technically extremely challenging. In Fig. 5a, we show how confocal imaging allow high resolution imaging of the GFP-dSH2 reporter overexpressed in epithelial cells that have the ability to form tubulogenesis. Thanks to this reporter combined with fast confocal microscopy, we will be able to monitor tyrosine phosphorylation at adhesion sites when cells are migrating collectively through the collagen gels (Le Dévédec et al., unpublished data). Furthermore, by using our established 3D invasion assay for 4T1 mammary tumour cells together with GFP-tagged adhesion protein overexpression we are able to image adhesion dynamics in a 3D environment during tumour cell migration (Fig. 5b).
Fig. 5.
Imaging adhesion and cell migration in 3D culture system in vitro. a Phase contrast (scale bar 100 μm) and confocal pictures (scale bar 50 μm) of tubulogenesis assays conducted with LLC-PK1 cells overexpressing either GFP alone (a) or GFP-dSH2 (b) in Matrigel-collagen gels. b Time lapse series (of 17 h) of 4T1 mouse mammary carcinoma cells control (a) and paxillin knockdown (b) invading 3D collagen gels (made with the help of H. Truong). Scale bar 200 μm. c Detailed time lapse serie of one migrating 4T1 control cell. Scale bar 50 μm
Imaging migration and adhesions in vivo
Studying adhesion dynamics in migrating cells in an in vitro 3D matrix is already technically challenging. Obviously, it is even more challenging to do so within intact organisms. Suitable in vivo models for both cancer progression and high resolution imaging are necessary.
Zebrafish model for metastasis analysis
Recently, the zebrafish and its transparent embryos became a new model system to investigate tumour development, cancer cell invasion and metastasis formation [123–125]. In the transparent zebrafish embryos invasion, circulation of tumour cells in blood vessels, migration and micrometastasis formation can be followed in real-time. Moreover, a number of unique features make this animal model very attractive: zebrafish are inexpensive to maintain and breed in large numbers, develop rapidly ex vivo, and can be maintained in small volumes of water. Several independent studies have now shown that human melanoma cells and other cancer cell lines are able to induce neovascularisation when xenografted in the zebrafish [125–127]. The role of the small GTPase RhoC in tumour formation, angiogenesis and cell invasion was investigated in real-time in 1-month-old immunosuppressed zebrafish xenografted with the human breast cancer cell line MDA-435 [125]. This study achieved high-resolution imaging of the dynamic cell–vascular interface in transparent juvenile zebrafish. All these innovative studies established the use of the zebrafish xenotransplantation model for the analysis of cancer cell lines as we also did in our laboratory (Fig. 6). We screened a number of cancer cell-lines and established that very aggressive phenotype after injection in the yolk (Fig. 6a, part a), were able to outgrow and disseminate throughout the animal body (Fig. 6a, part b) (Ghotra et al., unpublished data). We also demonstrated that certain cell-types could trigger the angiogenesis process (Fig. 6a, parts c and d) (Ghotra et al., unpublished data). In a very recent study, it was shown that zebrafish embryos can even be used to directly transplant human tumour tissue and primary human tumour cells [128]. Zebrafish embryos thus provide a simple, fast and cost-effective method to test the metastatic behaviour of human cell-lines and primary tumours in an in vivo vertebrate animal model that also permits high throughput drug screening (see later section).
Fig. 6.
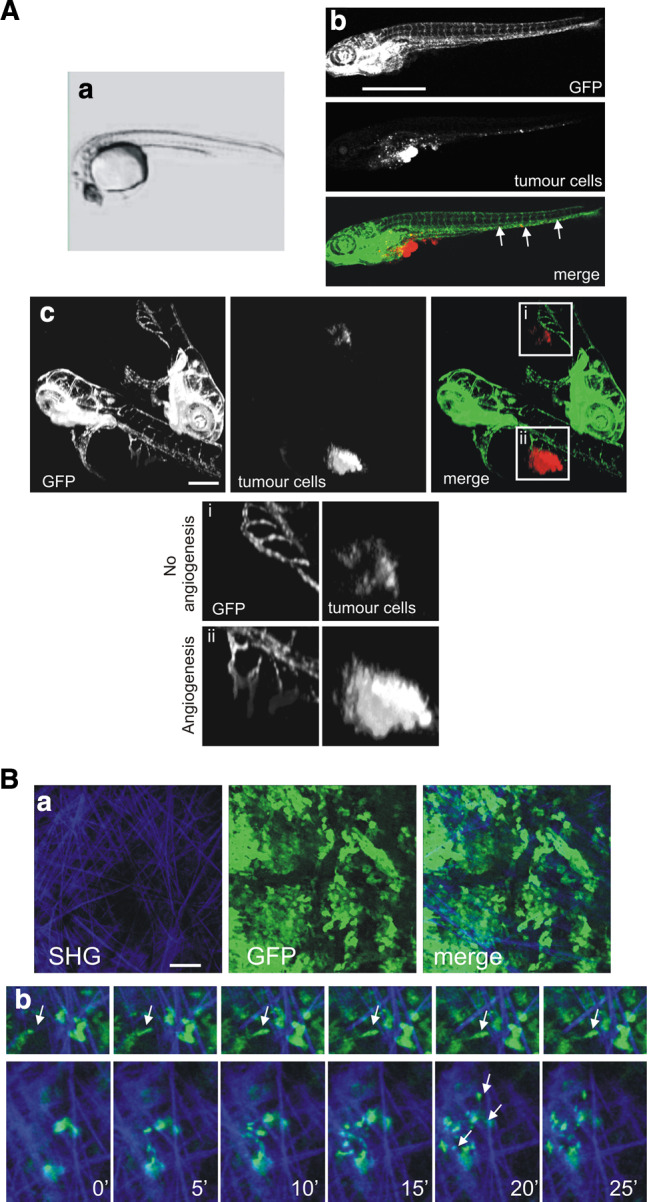
Imaging tumour cell migration in vivo. a Migration and cell mass formation of human tumour cells injected into the yolk sac of zebrafish embryos (Pictures obtained from V. Gothra, S. He, BE Snaar-Jagalska, and EHJ Danen). a Phase contrast overview picture of the yolk sac of zebrafish embryos. b An example of spreading of 4T1 breast tumour cells (red) in transgenic zebrafish embryos expressing GFP under an endothelial promotor. Cells invaded, migrated and formed distant micrometastases, which are indicated with arrows. Scale bar 1 mm. c Two examples of zebrafishes without angiogenesis (i) and with angiogenesis formed through the tumour cell mass formed (ii). Scale bar 200 μm. b Rat mammary carcinoma MTLn3 cells in orthotopic mammary tumours move show high motility in vivo with an amoeboid. a Multiphoton microscopy shows tumour mass (green) and extra cellular matrix visualised by second harmonic generation (blue). Scale bar 100 μm. b Time-lapse images of MTLn3 carcinoma cells as they extend protrusions along ECM fibres (arrowheads). Images shown are at 5-min intervals
Mouse metastasis model for imaging of tumour cell migration
The availability of multi-photon intravital microscopy has allowed researchers to visualise the dynamic behaviour of cancer cells in vivo [119, 120, 129, 130]. Multiphoton microscopy uses longer wavelengths (up to 1,200 nm) that are able to penetrate deeper into tissues and allow us to visualise more than 100 μm deep into the primary tumour. Multi-photon excitation also causes less photo-damage, permits good optical sectioning and 3D resolution [131] and non-invasive visualisation of the extra-cellular matrix thanks to the second harmonic generation phenomenon [132, 133]. In the past years, intravital imaging has been mainly used to follow up individual or group of cells fluorescently labelled within the primary tumour and study the interaction of moving cells with their microenvironment such as collagen matrix (Fig. 6b) [130, 134]. Recently, a new technical development, the mammary imaging window (MIW) has been shown to be advantageous for studying cell movement and adhesion with high resolution. The use of photo-convertible fluorophores such as Kaede or Dendra2 allows a precise monitoring of cellular movement in vivo no longer just over hours but over days [135], [136]. Despite all these technological advances, it was still not possible to visualise adhesions in migrating cells in vivo. Just recently, for the first time, a study on E-cadherin dynamics in living animals has been reported [137]. Photo-bleaching and photo-activation was used to compare the mobility of cell adhesion and plasma membrane probes in vitro and in tumours grown in mice and consequently demonstrate critical differences in molecular dynamics in vitro and in vivo.
Future directions and concluding remarks
FRAP/FRET, FRAP/TIRF and FRET/TIRF
An interesting alternative to the FRAP method is the combination of both FRAP and FRET that has already been applied in the nucleus [95, 138]. In the acceptor photo-bleaching FRET methodology, the bleaching of the acceptor results in an increase of the donor intensity. By applying FRAP on cells expressing adhesion proteins tagged with both FRET partners, e.g. CFP and YFP, and simultaneously recording in the bleached region the acceptor recovery and the redistribution of the increased donor signal, it is possible to compare the mobility of the interacting proteins (donor redistribution) relative to the mobility of the total pool of proteins (the YFP recovery as in a conventional FRAP experiment). Of course, it would be even more attractive to apply our previously described FLIP/FRAP methodology together with FRET to understand the complexity of both protein dynamics and interactions. Another very attractive combination of imaging technique is TIRF together with FRAP and/or FRET, a technique that is currently experiencing rapid growth in application [139–141]. This combination of imaging technologies will ensure an improved insight into adhesion protein dynamics and complexation in migrating cells since TIRF microscopy enormously enhances spatial resolution of fast-moving matrix adhesions.
High through put techniques (2D and 3D) for target identification
To explore the mechanisms underlying the regulation of cell migration by matrix adhesion dynamics, we described various qualitative and quantitative approaches that study adhesion dynamics and cell migration in a 2D and 3D environment. Such dynamic studies are of particular relevance to understand cancer cell motility. To provide a systematic analysis of genes that regulate cell migration or to study effect of potential drugs on tumour cell migration, high throughput image-based screening (HTS) is the most recent advance in imaging technology which has been made possible thanks to the availability of high-precision robotic liquid handling machinery, automated fluorescence microscopy and high-performance computing (for reviews, see [142] [143–146]). The capability of screening and segmenting diverse cellular populations combined with the possibility to detect protein–protein interactions or protein dynamics can offer a significant advantage in the fields of cellular proteomics and interactomics [147]. A high-throughput screening campaign includes the following: assay design (cell line, probes, kinetic, or end point), image acquisition (confocal vs wide-field, hardware specifications), image analysis (canned or custom algorithms), and data interpretation and image management (storage space, database, and visualisation). With the proper tools, HTS can be an invaluable tool to exploit the complexity of adhesion signalling pathways at a single cell level in a high-throughput manner (Fig. 7). The most recent image analysis packages provide capabilities that cover many microscopic applications suitable for HTS experiments, including the documentation of phenotypic readouts, the analysis of subcellular localisation and morphological changes, and the quantification of individual molecular modifications. Automated, cluster-based data analysis tools play an important role in analysing highly complex multiplexed HTS datasets into specific classes of phenotypic responses and assigning statistical significance to the changes in cellular processes that comprise the overall response [148]. However, first, not all microscopic applications are suitable for HTS experiments (e.g. FRAP, FCS, intravital imaging using mouse models), and second, kinetic HTS is still a technical and intellectual challenge (see Table 3 for detailed overview of the existing challenges and potential solutions).
Fig. 7.
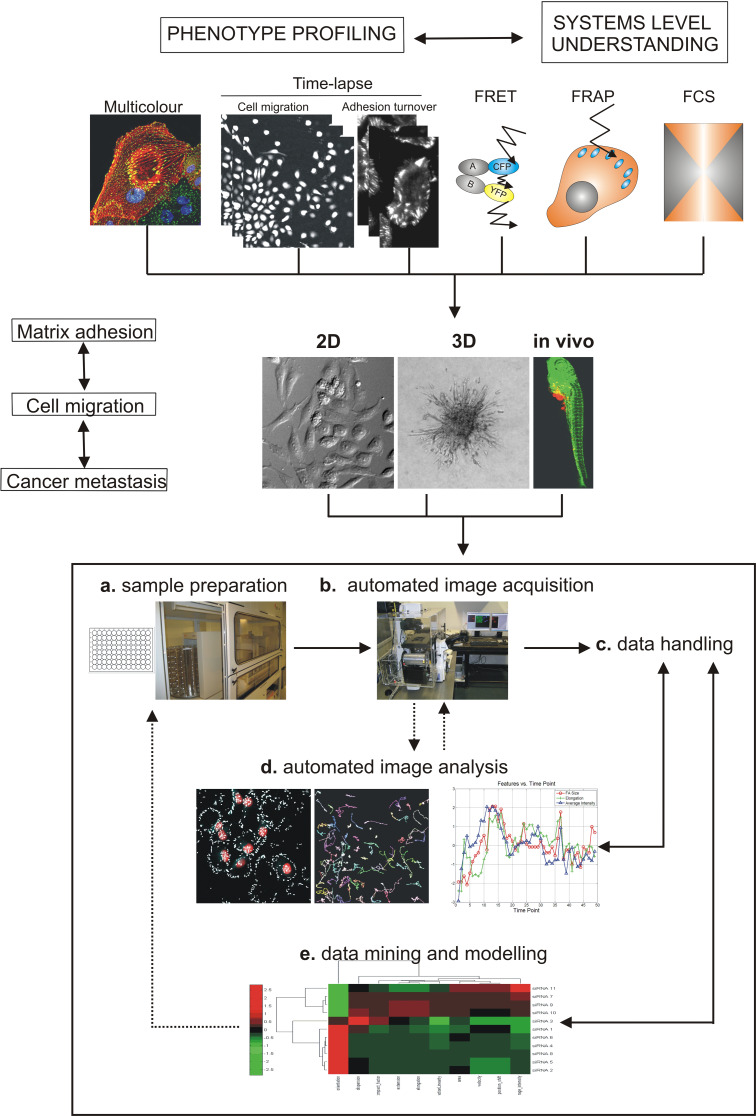
Steps for high content systems microscopy approach to understand cancer metastasis. Overview of imaging techniques that allow the phenotypic profiling of proteomics and cellomics (fixed multicolour and time-lapse) and the understanding at systems level (FRET, FRAP and FCS). To enhance our understanding of cancer metastasis, the high-throughput fluorescence microscopy should be applied in 2D, 3D and finally in vivo. a Sample preparation including cell transfection, exposure or immunostaining is nowadays conducted in multi-well dishes using robotics. b Automated image acquisition of fixed or living cells is done using automated microscopes (see Table 1, Available High Content Screening (HCS) instrumentation in [157]). Images can be acquired using different fluorescence microscopy techniques (e.g. fixed multicolour, time-lapse, FRET, FRAP, FCS. c Image data storage requires specialised software and hardware for data handling. d Automated image analysis which needs to be adapted or developed for each assay and is currently a challenge in HTS field. e Another big challenge in the field is the data mining and modelling which requires different disciplines such as statistics and bioinformatics (see Table 2 and available HCS informatics tools in [157]) (adapted from [142])
High-throughput fluorescence microscopy using fixed assays
The Geiger laboratory published two different screens which provide helpful methodologies and data on cell adhesion and migration: the first used high resolution microscopy to profile the effect of a library of natural extract on cell adhesion [149], while the second used a modified phagokinetics tracks with MCF7 and identified novel pro-migratory, cancer-associated genes [150]. Very recently, a third screen using high-throughput, high-resolution, microscopy-based assay together with human kinases, phosphatases and adhesome libraries was also performed and it provided a model for the molecular hierarchy of FA formation [151]. Another very elegant study used the traditional wound-healing assay with MCF-10A breast epithelial and screened siRNAs targeting 1,081 human genes encoding phosphatases, kinases and proteins predicted to influence cell migration and adhesion [152]. Extensive validation of all the hits yielded 66 high confidence genes that, when downregulated, either accelerated or impaired migration; 42 of these high confidence genes were not previously associated with motility or adhesion [152]. Although, the results of these screens are very promising and provide new data on cell migration, the analysis was performed with fixed samples.
High-throughput time-lapse fluorescence microscopy
Time-resolved live imaging would provide the highest content for assays on complex dynamic processes such as cell migration and invasion. Indeed, despite the recent described advances, a number of technical and analytical challenges remain to be solved to allow functional genomics screening together with dynamic imaging in a 2D and 3D environment (see Table 3). The major technical challenge is the need for (ultra-) fast image acquisition as well as to obtain ‘‘high content’’ information about the migratory behaviour of many cells, including dynamic features such as migration velocity. Only fast microscopes with high sensitive CCD camera and adapted bioinformatics can fulfill those requirements. In our laboratory, we are setting up a screen based on dynamic imaging of fast-moving cells to obtain cell behaviour measurements. Post-image acquisition, we also fix the plate and stain for focal adhesion markers so that we can correlate changes in cell motility with altered focal adhesion morphometry. Very recently, for the first time, a large study provided time-resolved profiles of RNAi-induced loss-of-function phenotypes resulting from siRNAs targeting the entire genome [153]. Here, a high-throughput phenotypic screening platform combining potent gene silencing by RNA interference, 2-day time-lapse microscopy and computational image processing was established. This was applied for a genome-wide phenotypic profiling of mitototic events for each of the approximately 21,000 human protein-coding genes. Cellular phenotypes were scored quantitatively by computational image processing, which allowed the identification of hundreds of genes involved in diverse biological functions including cell division, migration and survival. Such a systems microscopy approach will also be applicable in profiling tumor cell migration and invasion. The next step is the development of high-throughput methods that make use of functionally tagged transgenes in live cells to analyse protein–protein interactions and post-translational modifications. Their subcellular localisation and dynamics are needed to go from phenotypic profiling of the human genome to a systems-level understanding of the functional principles of the encoded protein machinery of human cells[143]. Indeed, in the future, it might be possible to analyse living cells using high-throughput approaches to determine intermolecular interactions using FRET, or protein dynamics using FRAP or FCS. These techniques are extremely powerfull in single-cell experiments since they provide quantitative data on the dynamics of biochemical reactions in living cells. However, except for FRET, none of these techniques have been successfully applied to systematic studies using HTS. As already mentioned, French reported an automated optically sectioning fluorescence lifetime imaging (FLIM) multiwell plate reader for high content analysis (HCA) in drug discovery and accelerated research in cell biology [105, 154]. Finally, the ultimate goal would be to perform those high-throughput screenings using time-lapse imaging in a relevant 3D culture environment and, even more challenging, in the zebrafish.
Perspectives and challenges to drug discovery and clinical treatment of cancer metastas formation
The above systems microscopy approach integrates different imaging techniques and relevant models for all stages of target-based drug discovery. This involves the study of cells, including target discovery, drug screening in cell-based assays, early safety evaluation, mode-of-action studies and in vivo studies to monitor cell fate. A prerequisite for reaching conclusions with low failure rates in drug discovery is the improvement of throughput that provides reliable data. The level of complexity comprising both changes in cellular morphology and macromolecular subcellular localisation and expression level makes HTS ideal among current approaches used for drug screening [155, 156]. Automated microscopy platforms that run in medium- or high-throughput mode to screen thousands of cDNAs, siRNAs, aptamers, proteins and antibodies using endpoints 2D migration and 3D invasion would benefit target discovery and validation [122, 148]. For the lead identification and optimisation, endpoint and time-lapse microscopy combined with multiparametric kinetic analysis incorporating both cell invasion assays and assessment of target activity with FAST/FLIM would be the next step [117]. However, as previously mentioned, kinetic HTS is still a technical and intellectual challenge which requires adapted fluorescent labelling techniques, automated ‘intelligent’ microscopes and well-developed software platforms [157, 158] (Table 3). In the later stages of the drug discovery trajectory, intravital imaging of tumour invasion in various relevant in vivo models can be used to evaluate the response of the drug in development and assess its activity when combined with FRET/FLIM. In the future, the combination of other microscopy techniques such as FRAP or FCS will help to obtain additional mechanistic information so that the drug development process can be more efficient. Nonetheless, a number of significant challenges remain related to the use of more biology- and disease-relevant predictive cell systems and the development of biosensors and bioreporters to measure and manipulate cellular events. Finally, the identification of relevant biomarkers specific to distinct invasion mechanisms will facilitate the development of clinical and preclinical studies and help administrate appropriate anti-invasive therapies to cancer patients [117].
Concluding remarks
Here, we presented a systems microscopy approach which integrates different imaging techniques developed over the past years to study adhesion signalling in relation to tumor cell migration and cancer metastasis [145]. In Table 1, we summarised the different techniques to study protein dynamics and interactions in matrix adhesions. In Table 2, an overview of the techniques used to model the various modes of tumour migration was given. In Table 3, we recapitulated the challenges and potential solutions to increase the throughput of imaging and multiparametric analysis. Understanding the movement of single cells and cells in tissue requires the analysis of complex processes under normal and perturbed conditions. Ideally, one would like to quantitatively determine movement and shape changes of cells and correlate these with the spatio-temporal dynamics of the cytoskeletal elements and the extra- and intracellular signalling pathways controlling these behaviours in isolated cells as well as in cells in tissue. Imaging together with multiparametric analysis and data modelling is the most appropriate method to reach that ultimate goal. Optimisation of current techniques and systems that combine diverse techniques will improve both our spatial and temporal resolutions of the role of matrix adhesions in migrating cells. Also, more and more bioinformatics tools will be generated for rapid and detailed image analysis and data processing. The additional knowledge obtained will hopefully provide insights into the molecular mechanisms behind tumour cell migration and help in developing new anticancer therapies.
Acknowledgments
The authors would like to thank Leo Price for technical support for the 3D invasion assays. This work was financially supported by grants from the Dutch Cancer Society (UL 2007-3860), the EU FP7 Health Program Metafight (Grant agreement no.201862) and ZF Cancer (Grant agreement no.201862), and the Netherlands Organization for Scientific Research (902-21-229 and 911-02-022).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Destaing O, Saltel F, Geminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi H, Pixley F, Condeelis J. Invadopodia and podosomes in tumor invasion. Eur J Cell Biol. 2006;85:213–218. doi: 10.1016/j.ejcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Baldassarre M, Ayala I, Beznoussenko G, Giacchetti G, Machesky LM, Luini A, Buccione R. Actin dynamics at sites of extracellular matrix degradation. Eur J Cell Biol. 2006;85:1217–1231. doi: 10.1016/j.ejcb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Chan KT, Cortesio CL, Huttenlocher A. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J Cell Biol. 2009;185:357–370. doi: 10.1083/jcb.200809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath JP, Dunn GA. Cell to substratum contacts of chick fibroblasts and their relation to the microfilament system. A correlated interference-reflexion and high-voltage electron-microscope study. J Cell Sci. 1978;29:197–212. doi: 10.1242/jcs.29.1.197. [DOI] [PubMed] [Google Scholar]
- 10.Izzard CS, Lochner LR. Cell-to-substrate contacts in living fibroblasts: an interference reflexion study with an evaluation of the technique. J Cell Sci. 1976;21:129–159. doi: 10.1242/jcs.21.1.129. [DOI] [PubMed] [Google Scholar]
- 11.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 12.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 13.Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 14.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huveneers S, Danen EH. Adhesion signaling—crosstalk between integrins, Src and Rho. J Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt S, Friedl P. Interstitial cell migration: integrin-dependent and alternative adhesion mechanisms. Cell Tissue Res. 2010;339:83–92. doi: 10.1007/s00441-009-0892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shintani Y, Fukumoto Y, Chaika N, Svoboda R, Wheelock MJ, Johnson KR. Collagen I-mediated up-regulation of N-cadherin requires cooperative signals from integrins and discoidin domain receptor 1. J Cell Biol. 2008;180:1277–1289. doi: 10.1083/jcb.200708137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson O, Moore CJ, Hussain SA, Kleino I, Peckham M, Hohenester E, Ayscough KR, Saksela K, Winder SJ. Modulation of cell spreading and cell-substrate adhesion dynamics by dystroglycan. J Cell Sci. 2010;123:118–127. doi: 10.1242/jcs.047902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore CJ, Winder SJ. Dystroglycan versatility in cell adhesion: a tale of multiple motifs. Cell Commun Signal. 2010;8:3. doi: 10.1186/1478-811X-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cross SS, Lippitt J, Mitchell A, Hollingsbury F, Balasubramanian SP, Reed MW, Eaton C, Catto JW, Hamdy F, Winder SJ. Expression of beta-dystroglycan is reduced or absent in many human carcinomas. Histopathology. 2008;53:561–566. doi: 10.1111/j.1365-2559.2008.03157.x. [DOI] [PubMed] [Google Scholar]
- 21.Zamir E, Katz BZ, Aota S, Yamada KM, Geiger B, Kam Z. Molecular diversity of cell-matrix adhesions. J Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- 22.Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem Soc Trans. 2004;32:416–420. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- 23.Zamir E, Geiger B, Kam Z (2008) Quantitative multicolor compositional imaging resolves molecular domains in cell-matrix adhesions. PLoS ONE. doi:10.1371/journal.pone.0001901 [DOI] [PMC free article] [PubMed]
- 24.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 25.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 26.Kirchner J, Kam Z, Tzur G, Bershadsky AD, Geiger B. Live-cell monitoring of tyrosine phosphorylation in focal adhesions following microtubule disruption. J Cell Sci. 2003;116:975–986. doi: 10.1242/jcs.00284. [DOI] [PubMed] [Google Scholar]
- 27.Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells–over and over and over again. Nat Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 28.Carragher NO, Frame MC. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol. 2004;14:241–249. doi: 10.1016/j.tcb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Small JV, Kaverina I. Microtubules meet substrate adhesions to arrange cell polarity. Curr Opin Cell Biol. 2003;15:40–47. doi: 10.1016/S0955-0674(02)00008-X. [DOI] [PubMed] [Google Scholar]
- 30.Larsen M, Tremblay ML, Yamada KM. Phosphatases in cell-matrix adhesion and migration. Nat Rev Mol Cell Biol. 2003;4:700–711. doi: 10.1038/nrm1199. [DOI] [PubMed] [Google Scholar]
- 31.Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Tomar A, Schlaepfer DD. Focal adhesion kinase: switching between GAPs and GEFs in the regulation of cell motility. Curr Opin Cell Biol. 2009;21:676–683. doi: 10.1016/j.ceb.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner CE, West KA, Brown MC. Paxillin-ARF GAP signaling and the cytoskeleton. Curr Opin Cell Biol. 2001;13:593–599. doi: 10.1016/S0955-0674(00)00256-8. [DOI] [PubMed] [Google Scholar]
- 34.Carragher NO. Calpain inhibition: a therapeutic strategy targeting multiple disease states. Curr Pharm Des. 2006;12:615–638. doi: 10.2174/138161206775474314. [DOI] [PubMed] [Google Scholar]
- 35.Carragher NO, Fonseca BD, Frame MC. Calpain activity is generally elevated during transformation but has oncogene-specific biological functions. Neoplasia. 2004;6:53–73. doi: 10.1016/s1476-5586(04)80053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carragher NO, Frame MC. Calpain: a role in cell transformation and migration. Int J Biochem Cell Biol. 2002;34:1539–1543. doi: 10.1016/S1357-2725(02)00069-9. [DOI] [PubMed] [Google Scholar]
- 37.Franco SJ, Rodgers MA, Perrin BJ, Han J, Bennin DA, Critchley DR, Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- 38.Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai KL, Zhang ZY, Sahai E, Condeelis J, Segall JE. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66:192–197. doi: 10.1158/0008-5472.CAN-05-1242. [DOI] [PubMed] [Google Scholar]
- 39.Le Devedec SE, van Roosmalen W, Maria N, Grimbergen M, Pont C, Lalai R, van de WB. An improved model to study tumor cell autonomous metastasis programs using MTLn3 cells and the Rag2(−/−) gammac (−/−) mouse. Clin Exp Metastasis. 2009;26:673–684. doi: 10.1007/s10585-009-9267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi K, Ito F. EGF receptor in relation to tumor development: molecular basis of responsiveness of cancer cells to EGFR-targeting tyrosine kinase inhibitors. FEBS J. 2010;277:316–326. doi: 10.1111/j.1742-4658.2009.07450.x. [DOI] [PubMed] [Google Scholar]
- 41.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 43.Guarino M. Src signaling in cancer invasion. J Cell Physiol. 2010;223:14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 44.Golubovskaya VM, Kweh FA, Cance WG. Focal adhesion kinase and cancer. Histol Histopathol. 2009;24:503–510. doi: 10.14670/HH-24.503. [DOI] [PubMed] [Google Scholar]
- 45.Owens LV, Xu L, Craven RJ, Dent GA, Weiner TM, Kornberg L, Liu ET, Cance WG. Overexpression of the focal adhesion kinase (p125FAK) in invasive human tumors. Cancer Res. 1995;55:2752–2755. [PubMed] [Google Scholar]
- 46.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 47.van Nimwegen MJ, van de WB. Focal adhesion kinase: a potential target in cancer therapy. Biochem Pharmacol. 2007;73:597–609. doi: 10.1016/j.bcp.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 48.Yafeng M, van de Water B, Le Dévédec SE. Focal Adhesion Kinase inhibition as a potential strategy for anticancer therapies. Drugs Future. 2009;34:477–484. [Google Scholar]
- 49.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 50.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 51.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Shyy JY, Chien S. Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annu Rev Biomed Eng. 2008;10:1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 53.Dormann D, Weijer CJ. Imaging of cell migration. EMBO J. 2006;25:3480–3493. doi: 10.1038/sj.emboj.7601227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 55.Sun YN, Lin CH, Kuo CC, Ho CL, Lin CJ. Live cell tracking based on cellular state recognition from microscopic images. J Microsc. 2009;235:94–105. doi: 10.1111/j.1365-2818.2009.03186.x. [DOI] [PubMed] [Google Scholar]
- 56.Hand AJ, Sun T, Barber DC, Hose DR, MacNeil S. Automated tracking of migrating cells in phase-contrast video microscopy sequences using image registration. J Microsc. 2009;234:62–79. doi: 10.1111/j.1365-2818.2009.03144.x. [DOI] [PubMed] [Google Scholar]
- 57.Entschladen F, Drell TL, Lang K, Masur K, Palm D, Bastian P, Niggemann B, Zaenker KS. Analysis methods of human cell migration. Exp Cell Res. 2005;307:418–426. doi: 10.1016/j.yexcr.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 58.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 59.Hoebe RA, Van Oven CH, Gadella TW, Jr, Dhonukshe PB, Van Noorden CJ, Manders EM. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat Biotechnol. 2007;25:249–253. doi: 10.1038/nbt1278. [DOI] [PubMed] [Google Scholar]
- 60.Axelrod D, Thompson NL, Burghardt TP. Total internal inflection fluorescent microscopy. J Microsc. 1983;129:19–28. doi: 10.1111/j.1365-2818.1983.tb04158.x. [DOI] [PubMed] [Google Scholar]
- 61.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 62.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 64.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 65.Osborn EA, Rabodzey A, Dewey CF, Jr, Hartwig JH. Endothelial actin cytoskeleton remodeling during mechanostimulation with fluid shear stress. Am J Physiol Cell Physiol. 2006;290:C444–C452. doi: 10.1152/ajpcell.00218.2005. [DOI] [PubMed] [Google Scholar]
- 66.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 67.Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson KI, McCaffrey MW, Ozanne BW, Norman JC. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell. 2007;13:496–510. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 68.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 69.van Royen ME, Farla P, Mattern KA, Geverts B, Trapman J, Houtsmuller AB. Fluorescence recovery after photobleaching (FRAP) to study nuclear protein dynamics in living cells. Methods Mol Biol. 2009;464:363–385. doi: 10.1007/978-1-60327-461-6_20. [DOI] [PubMed] [Google Scholar]
- 70.Ballestrem C, Hinz B, Imhof BA, Wehrle-Haller B. Marching at the front and dragging behind: differential alphaVbeta3-integrin turnover regulates focal adhesion behavior. J Cell Biol. 2001;155:1319–1332. doi: 10.1083/jcb.200107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hamadi A, Bouali M, Dontenwill M, Stoeckel H, Takeda K, Ronde P. Regulation of focal adhesion dynamics and disassembly by phosphorylation of FAK at tyrosine 397. J Cell Sci. 2005;118:4415–4425. doi: 10.1242/jcs.02565. [DOI] [PubMed] [Google Scholar]
- 72.Cluzel C, Saltel F, Lussi J, Paulhe F, Imhof BA, Wehrle-Haller B. The mechanisms and dynamics of (alpha)v(beta)3 integrin clustering in living cells. J Cell Biol. 2005;171:383–392. doi: 10.1083/jcb.200503017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geuijen CA, Sonnenberg A. Dynamics of the alpha6beta4 integrin in keratinocytes. Mol Biol Cell. 2002;13:3845–3858. doi: 10.1091/mbc.02-01-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lele TP, Pendse J, Kumar S, Salanga M, Karavitis J, Ingber DE. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol. 2006;207:187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- 75.Edlund M, Lotano MA, Otey CA. Dynamics of alpha-actinin in focal adhesions and stress fibers visualized with alpha-actinin-green fluorescent protein. Cell Motil Cytoskeleton. 2001;48:190–200. doi: 10.1002/1097-0169(200103)48:3<190::AID-CM1008>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 76.von Wichert G, Haimovich B, Feng GS, Sheetz MP. Force-dependent integrin-cytoskeleton linkage formation requires downregulation of focal complex dynamics by Shp2. EMBO J. 2003;22:5023–5035. doi: 10.1093/emboj/cdg492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lele TP, Thodeti CK, Pendse J, Ingber DE. Investigating complexity of protein-protein interactions in focal adhesions. Biochem Biophys Res Commun. 2008;369:929–934. doi: 10.1016/j.bbrc.2008.02.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wolfenson H, Lubelski A, Regev T, Klafter J, Henis, YI, Geiger B (2009) A role for the juxtamembrane cytoplasm in the molecular dynamics of focal adhesions. PLoS ONE. doi:10.1371/journal.pone.0004304 [DOI] [PMC free article] [PubMed]
- 79.Fraley TS, Pereira CB, Tran TC, Singleton C, Greenwood JA. Phosphoinositide binding regulates alpha-actinin dynamics: mechanism for modulating cytoskeletal remodeling. J Biol Chem. 2005;280:15479–15482. doi: 10.1074/jbc.M500631200. [DOI] [PubMed] [Google Scholar]
- 80.Chandrasekar I, Stradal TE, Holt MR, Entschladen F, Jockusch BM, Ziegler WH. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J Cell Sci. 2005;118:1461–1472. doi: 10.1242/jcs.01734. [DOI] [PubMed] [Google Scholar]
- 81.Goetz JG, Joshi B, Lajoie P, Strugnell SS, Scudamore T, Kojic LD, Nabi IR. Concerted regulation of focal adhesion dynamics by galectin-3 and tyrosine-phosphorylated caveolin-1. J Cell Biol. 2008;180:1261–1275. doi: 10.1083/jcb.200709019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/S0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 83.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 84.Ponti A, Matov A, Adams M, Gupton S, Waterman-Storer CM, Danuser G. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative Fluorescent Speckle Microscopy. Biophys J. 2005;89:3456–3469. doi: 10.1529/biophysj.104.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 86.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 87.Wiseman PW, Brown CM, Webb DJ, Hebert B, Johnson NL, Squier JA, Ellisman MH, Horwitz AF. Spatial mapping of integrin interactions and dynamics during cell migration by image correlation microscopy. J Cell Sci. 2004;117:5521–5534. doi: 10.1242/jcs.01416. [DOI] [PubMed] [Google Scholar]
- 88.Brown CM, Dalal RB, Hebert B, Digman MA, Horwitz AR, Gratton E. Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J Microsc. 2008;229:78–91. doi: 10.1111/j.1365-2818.2007.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Digman MA, Gratton E. Analysis of diffusion and binding in cells using the RICS approach. Microsc Res Tech. 2009;72:323–332. doi: 10.1002/jemt.20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brown CM, Hebert B, Kolin DL, Zareno J, Whitmore L, Horwitz AR, Wiseman PW. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J Cell Sci. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 92.Digman MA, Brown CM, Horwitz AR, Mantulin WW, Gratton E. Paxillin dynamics measured during adhesion assembly and disassembly by correlation spectroscopy. Biophys J. 2008;94:2819–2831. doi: 10.1529/biophysj.107.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ballestrem C, Erez N, Kirchner J, Kam Z, Bershadsky A, Geiger B. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J Cell Sci. 2006;119:866–875. doi: 10.1242/jcs.02794. [DOI] [PubMed] [Google Scholar]
- 94.Parsons M, Vojnovic B, Ameer-Beg S. Imaging protein-protein interactions in cell motility using fluorescence resonance energy transfer (FRET) Biochem Soc Trans. 2004;32:431–433. doi: 10.1042/BST0320431. [DOI] [PubMed] [Google Scholar]
- 95.van Royen ME, Dinant C, Farla P, Trapman J, Houtsmuller AB. FRAP and FRET methods to study nuclear receptors in living cells. Methods Mol Biol. 2009;505:69–96. doi: 10.1007/978-1-60327-575-0_5. [DOI] [PubMed] [Google Scholar]
- 96.van Rheenen J, Langeslag M, Jalink K. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys J. 2004;86:2517–2529. doi: 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cai X, Lietha D, Ceccarelli DF, Karginov AV, Rajfur Z, Jacobson K, Hahn KM, Eck MJ, Schaller MD. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol. 2008;28:201–214. doi: 10.1128/MCB.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Chien S. Analysis of integrin signaling by fluorescence resonance energy transfer. Methods Enzymol. 2007;426:177–201. doi: 10.1016/S0076-6879(07)26009-4. [DOI] [PubMed] [Google Scholar]
- 99.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakamura T, Kurokawa K, Kiyokawa E, Matsuda M. Analysis of the spatiotemporal activation of rho GTPases using Raichu probes. Methods Enzymol. 2006;406:315–332. doi: 10.1016/S0076-6879(06)06023-X. [DOI] [PubMed] [Google Scholar]
- 101.Pertz O, Hahn KM. Designing biosensors for Rho family proteins–deciphering the dynamics of Rho family GTPase activation in living cells. J Cell Sci. 2004;117:1313–1318. doi: 10.1242/jcs.01117. [DOI] [PubMed] [Google Scholar]
- 102.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 103.Parsons M, Messent AJ, Humphries JD, Deakin NO, Humphries MJ. Quantification of integrin receptor agonism by fluorescence lifetime imaging. J Cell Sci. 2008;121:265–271. doi: 10.1242/jcs.018440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bastiaens PI, Squire A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999;9:48–52. doi: 10.1016/S0962-8924(98)01410-X. [DOI] [PubMed] [Google Scholar]
- 105.Grant DM, McGinty J, McGhee EJ, Bunney TD, Owen DM, Talbot CB, Zhang W, Kumar S, Munro I, Lanigan PM, Kennedy GT, Dunsby C, Magee AI, Courtney P, Katan M, Neil MA, French PM. High speed optically sectioned fluorescence lifetime imaging permits study of live cell signaling events. Opt Express. 2007;15:15656–15673. doi: 10.1364/OE.15.015656. [DOI] [PubMed] [Google Scholar]
- 106.Fan Y, Gong Y, Ghosh PK, Graham LM, Fox PL. Spatial coordination of actin polymerization and ILK-Akt2 activity during endothelial cell migration. Dev Cell. 2009;16:661–674. doi: 10.1016/j.devcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Romer LH, Birukov KG, Garcia JG. Focal adhesions: paradigm for a signaling nexus. Circ Res. 2006;98:606–616. doi: 10.1161/01.RES.0000207408.31270.db. [DOI] [PubMed] [Google Scholar]
- 108.Alderliesten M, de Graauw M, Oldenampsen J, Qin Y, Pont C, van Buren L, van de WB. Extracellular signal-regulated kinase activation during renal ischemia/reperfusion mediates focal adhesion dissolution and renal injury. Am J Pathol. 2007;171:452–462. doi: 10.2353/ajpath.2007.060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 111.Yates C, Shepard CR, Papworth G, Dash A, Beer SD, Tannenbaum S, Griffith L, Wells A. Novel three-dimensional organotypic liver bioreactor to directly visualize early events in metastatic progression. Adv Cancer Res. 2007;97:225–246. doi: 10.1016/S0065-230X(06)97010-9. [DOI] [PubMed] [Google Scholar]
- 112.Kenny HA, Krausz T, Yamada SD, Lengyel E. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int J Cancer. 2007;121:1463–1472. doi: 10.1002/ijc.22874. [DOI] [PubMed] [Google Scholar]
- 113.Abhyankar VV, Toepke MW, Cortesio CL, Lokuta MA, Huttenlocher A, Beebe DJ. A platform for assessing chemotactic migration within a spatiotemporally defined 3D microenvironment. Lab Chip. 2008;8:1507–1515. doi: 10.1039/b803533d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Benton G, George J, Kleinman HK, Arnaoutova IP. Advancing science and technology via 3D culture on basement membrane matrix. J Cell Physiol. 2009;221:18–25. doi: 10.1002/jcp.21832. [DOI] [PubMed] [Google Scholar]
- 115.Zaman MH, Trapani LM, Siemeski A, Mackellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hooper S, Marshall JF, Sahai E. Tumor cell migration in three dimensions. Methods Enzymol. 2006;406:625–643. doi: 10.1016/S0076-6879(06)06049-6. [DOI] [PubMed] [Google Scholar]
- 117.Carragher NO. Profiling distinct mechanisms of tumour invasion for drug discovery: imaging adhesion, signalling and matrix turnover. Clin Exp Metastasis. 2009;26:381–397. doi: 10.1007/s10585-008-9222-y. [DOI] [PubMed] [Google Scholar]
- 118.Gobel W, Kampa BM, Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat Methods. 2007;4:73–79. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- 119.Wolf K, Friedl P. Functional imaging of pericellular proteolysis in cancer cell invasion. Biochimie. 2005;87:315–320. doi: 10.1016/j.biochi.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 120.Friedl P. Dynamic imaging of cellular interactions with extracellular matrix. Histochem Cell Biol. 2004;122:183–190. doi: 10.1007/s00418-004-0682-0. [DOI] [PubMed] [Google Scholar]
- 121.Nystrom ML, Thomas GJ, Stone M, Mackenzie IC, Hart IR, Marshall JF. Development of a quantitative method to analyse tumour cell invasion in organotypic culture. J Pathol. 2005;205:468–475. doi: 10.1002/path.1716. [DOI] [PubMed] [Google Scholar]
- 122.Adanja I, Debeir O, Megalizzi V, Kiss R, Warzee N, Decaestecker C. Automated tracking of unmarked cells migrating in three-dimensional matrices applied to anti-cancer drug screening. Exp Cell Res. 2010;316:181–193. doi: 10.1016/j.yexcr.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 123.Goessling W, North TE, Zon LI. New waves of discovery: modeling cancer in zebrafish. J Clin Oncol. 2007;25:2473–2479. doi: 10.1200/JCO.2006.08.9821. [DOI] [PubMed] [Google Scholar]
- 124.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, Look AT. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 125.Stoletov K, Montel V, Lester RD, Gonias SL, Klemke R. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc Natl Acad Sci USA. 2007;104:17406–17411. doi: 10.1073/pnas.0703446104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haldi M, Ton C, Seng WL, McGrath P. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis. 2006;9:139–151. doi: 10.1007/s10456-006-9040-2. [DOI] [PubMed] [Google Scholar]
- 127.Nicoli S, Presta M. The zebrafish/tumor xenograft angiogenesis assay. Nat Protoc. 2007;2:2918–2923. doi: 10.1038/nprot.2007.412. [DOI] [PubMed] [Google Scholar]
- 128.Marques IJ, Weiss FU, Vlecken DH, Nitsche C, Bakkers J, Lagendijk AK, Partecke LI, Heidecke CD, Lerch MM, Bagowski CP. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer. 2009;9:128. doi: 10.1186/1471-2407-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sahai E. Illuminating the metastatic process. Nat Rev Cancer. 2007;7:737–749. doi: 10.1038/nrc2229. [DOI] [PubMed] [Google Scholar]
- 130.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3:921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 131.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 132.Campagnola PJ, Loew LM. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat Biotechnol. 2003;21:1356–1360. doi: 10.1038/nbt894. [DOI] [PubMed] [Google Scholar]
- 133.Kedrin D, Wyckoff J, Sahai E, Condeelis J, Segall JE (2007) Imaging tumor cell movement in vivo. Curr Protoc Cell Biol 19.7.1–19.7.17 [DOI] [PubMed]
- 134.Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia. 2007;12:143–152. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]
- 135.Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gligorijevic B, Kedrin D, Segall JE, Condeelis J, van Rheenen J (2009) Dendra2 photoswitching through the Mammary Imaging Window. J Vis Exp. doi:10.3791/1278 [DOI] [PMC free article] [PubMed]
- 137.Serrels A, Timpson P, Canel M, Schwarz JP, Carragher NO, Frame MC, Brunton VG, Anderson KI. Real-time study of E-cadherin and membrane dynamics in living animals: implications for disease modeling and drug development. Cancer Res. 2009;69:2714–2719. doi: 10.1158/0008-5472.CAN-08-4308. [DOI] [PubMed] [Google Scholar]
- 138.van Royen ME, Cunha SM, Brink MC, Mattern KA, Nigg AL, Dubbink HJ, Verschure PJ, Trapman J, Houtsmuller AB. Compartmentalization of androgen receptor protein-protein interactions in living cells. J Cell Biol. 2007;177:63–72. doi: 10.1083/jcb.200609178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bal M, Zaika O, Martin P, Shapiro MS. Calmodulin binding to M-type K+ channels assayed by TIRF/FRET in living cells. J Physiol. 2008;586:2307–2320. doi: 10.1113/jphysiol.2008.152777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang L, Bittner MA, Axelrod D, Holz RW. The structural and functional implications of linked SNARE motifs in SNAP25. Mol Biol Cell. 2008;19:3944–3955. doi: 10.1091/mbc.E08-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Raveh A, Riven I, Reuveny E. The use of FRET microscopy to elucidate steady state channel conformational rearrangements and G protein interaction with the GIRK channels. Methods Mol Biol. 2008;491:199–212. doi: 10.1007/978-1-59745-526-8_16. [DOI] [PubMed] [Google Scholar]
- 142.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 143.Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 144.Conrad C, Gerlich DW. Automated microscopy for high-content RNAi screening. J Cell Biol. 2010;188:453–461. doi: 10.1083/jcb.200910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Megason SG, Fraser SE. Imaging in systems biology. Cell. 2007;130:784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 146.Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 147.Esposito A, Dohm CP, Bahr M, Wouters FS. Unsupervised fluorescence lifetime imaging microscopy for high content and high throughput screening. Mol Cell Proteomics. 2007;6:1446–1454. doi: 10.1074/mcp.T700006-MCP200. [DOI] [PubMed] [Google Scholar]
- 148.Denner P, Schmalowsky J, Prechtl S. High-content analysis in preclinical drug discovery. Comb Chem High Throughput Screen. 2008;11:216–230. doi: 10.2174/138620708783877780. [DOI] [PubMed] [Google Scholar]
- 149.Paran Y, Ilan M, Kashman Y, Goldstein S, Liron Y, Geiger B, Kam Z. High-throughput screening of cellular features using high-resolution light-microscopy; application for profiling drug effects on cell adhesion. J Struct Biol. 2007;158:233–243. doi: 10.1016/j.jsb.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 150.Naffar-Abu-Amara S, Shay T, Galun M, Cohen N, Isakoff SJ, Kam Z, Geiger B (2008) Identification of novel pro-migratory, cancer-associated genes using quantitative, microscopy-based screening. PLoS ONE. doi:10.1371/journal.pone.0001457 [DOI] [PMC free article] [PubMed]
- 151.Winograd-Katz SE, Itzkovitz S, Kam Z, Geiger B. Multiparametric analysis of focal adhesion formation by RNAi-mediated gene knockdown. J Cell Biol. 2009;186:423–436. doi: 10.1083/jcb.200901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, Brugge JS. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 153.Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters JM, Hyman AA, Durbin R, Pepperkok R, Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Talbot CB, McGinty J, Grant DM, McGhee EJ, Owen DM, Zhang W, Bunney TD, Munro I, Isherwood B, Eagle R, Hargreaves A, Dunsby C, Neil MA, French PM. High speed unsupervised fluorescence lifetime imaging confocal multiwell plate reader for high content analysis. J Biophotonics. 2008;1:514–521. doi: 10.1002/jbio.200810054. [DOI] [PubMed] [Google Scholar]
- 155.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–453. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 156.Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nat Rev Drug Discov. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 157.Zanella F, Lorens JB, Link W. High content screening: seeing is believing. Trends Biotechnol. 2010;28:237–245. doi: 10.1016/j.tibtech.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 158.King RD, Rowland J, Oliver SG, Young M, Aubrey W, Byrne E, Liakata M, Markham M, Pir P, Soldatova LN, Sparkes A, Whelan KE, Clare A. The automation of science. Science. 2009;324:85–89. doi: 10.1126/science.1165620. [DOI] [PubMed] [Google Scholar]
- 159.Bershadsky AD, Ballestrem C, Carramusa L, Zilberman Y, Gilquin B, Khochbin S, Alexandrova AY, Verkhovsky AB, Shemesh T, Kozlov MM. Assembly and mechanosensory function of focal adhesions: experiments and models. Eur J Cell Biol. 2005;85:165–173. doi: 10.1016/j.ejcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 160.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/S0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 161.Roy P, Rajfur Z, Pomorski P, Jacobson K. Microscope-based techniques to study cell adhesion and migration. Nat Cell Biol. 2002;4:E91–E96. doi: 10.1038/ncb0402-e91. [DOI] [PubMed] [Google Scholar]
- 162.Webb DJ, Brown CM, Horwitz AF. Illuminating adhesion complexes in migrating cells: moving toward a bright future. Curr Opin Cell Biol. 2003;15:614–620. doi: 10.1016/S0955-0674(03)00105-4. [DOI] [PubMed] [Google Scholar]
- 163.Wouters FS, Verveer PJ, Bastiaens PI. Imaging biochemistry inside cells. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/S0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- 164.Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 165.Peter M, Ameer-Beg SM. Imaging molecular interactions by multiphoton FLIM. Biol Cell. 2004;96:231–236. doi: 10.1016/j.biolcel.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 166.Condeelis JS, Wyckoff J, Segall JE. Imaging of cancer invasion and metastasis using green fluorescent protein. Eur J Cancer. 2000;36:1671–1680. doi: 10.1016/S0959-8049(00)00155-6. [DOI] [PubMed] [Google Scholar]
- 167.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 168.Hoffman RM, Yang M. Subcellular imaging in the live mouse. Nat Protoc. 2006;1:775–782. doi: 10.1038/nprot.2006.109. [DOI] [PubMed] [Google Scholar]
- 169.Pinner S, Sahai E. Imaging amoeboid cancer cell motility in vivo. J Microsc. 2008;231:441–445. doi: 10.1111/j.1365-2818.2008.02056.x. [DOI] [PubMed] [Google Scholar]
- 170.Hoffman RM. Imaging cancer dynamics in vivo at the tumor and cellular level with fluorescent proteins. Clin Exp Metastasis. 2009;26:345–355. doi: 10.1007/s10585-008-9205-z. [DOI] [PubMed] [Google Scholar]
- 171.Provenzano PP, Eliceiri KW, Keely PJ. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clin Exp Metastasis. 2009;26:357–370. doi: 10.1007/s10585-008-9204-0. [DOI] [PubMed] [Google Scholar]
- 172.King RD, Rowland J, Oliver SG, Young M, Aubrey W, Byrne E, Liakata M, Markham M, Pir P, Soldatova LN, Sparkes A, Whelan KE, Clare A. Make way for robot scientists. Science. 2009;325:945. doi: 10.1126/science.325_945a. [DOI] [PubMed] [Google Scholar]
- 173.Rabut G, Ellenberg J. Automatic real-time three-dimensional cell tracking by fluorescence microscopy. J Microsc. 2004;216:131–137. doi: 10.1111/j.0022-2720.2004.01404.x. [DOI] [PubMed] [Google Scholar]