Nucleotide supply, not local histone acetylation, sets replication origin usage in transcribed regions
Replication origin usage is not determined by local histone acetylation levels, but by the availability of nucleotides, which is modified following forced alterations of protein acetylation balance in the cell.
Keywords: CTP synthetase, molecular combing, replication dynamics, thymidylate synthase, trichostatin A
Abstract
In eukaryotes, only a fraction of replication origins fire at each S phase. Local histone acetylation was proposed to control firing efficiency of origins, but conflicting results were obtained. We report that local histone acetylation does not reflect origin efficiencies along the adenosine monophosphate deaminase 2 locus in mammalian fibroblasts. Reciprocally, modulation of origin efficiency does not affect acetylation. However, treatment with a deacetylase inhibitor changes the initiation pattern. We demonstrate that this treatment alters pyrimidine biosynthesis and decreases fork speed, which recruits latent origins. Our findings reconcile results that seemed inconsistent and reveal an unsuspected effect of deacetylase inhibitors on replication dynamics.
Introduction
In eukaryotic cells, initiation events should be adequately distributed throughout the chromosomes to ensure timely genome duplication. With the exception of Saccharomyces cerevisiae, this is achieved in the absence of specific sequence motifs marking the origins. In addition, only a fraction of the potential origins fires in each S phase. The molecular mechanisms that specify sequences to become origins and to commit a particular subset of origins to become active are only partly elucidated (Sclafani & Holzen, 2007). Chromatin structure, which provides more flexible heritable information than DNA sequence, is considered as a good candidate for controlling initiation. Indeed, inactivation of specific deacetylases increases the efficiency of latent origins in yeast and Drosophila (Pasero et al, 2002; Aggarwal & Calvi, 2004; Pappas et al, 2004). Similar results have been obtained in mammalian cells treated with trichostatin A (TSA), a deacetylase inhibitor (Kemp et al, 2005). Altogether, these results suggested that local histone hyperacetylation facilitates recruitment of pre-replicative complexes onto origins by enhancing DNA accessibility (Tabancay & Forsburg, 2006). In contrast to these results, obtained upon forced hyperacetylation, the study of some specific loci during differentiation and development in vertebrates did not show any correlation between the level of histone acetylation and origin usage (Prioleau et al, 2003; Dazy et al, 2006; Gregoire et al, 2006).
Previously, the relationship between acetylation and the relative efficiency of replication origins under physiological conditions has been explored mostly in a developmental context (Dazy et al, 2006; Gregoire et al, 2006), whereas the effects of TSA were examined using established mammalian cell lines (Kemp et al, 2005). In this study, we investigated the impact of the endogenous acetylation of histones and of TSA-induced hyperacetylation on origin activity in the same model of differentiated Chinese hamster cells. We focused on the AMPD2 (adenosine monophosphate deaminase 2) locus in two different fibroblast cell lines (474 and 422) amplified for this region (Debatisse et al, 1982; supplementary Fig S1 online). We have shown previously that a switch in origin usage occurs when cells face changes in growth conditions (Anglana et al, 2003; Courbet et al, 2008). For example, in cells from the cell line 474, replication forks travel at a speed of approximately 0.6 kb min−1, and initiation events occur at high density and are distributed evenly between the different origins of the locus (‘slow' replication dynamics). When these cells are supplemented with adenine and uridine (474AU) fork speed increases to 1.5 kb min−1, the density of initiation decreases and about 70% of the initiation events occur at an origin called oriGNAI3 (‘fast' replication dynamics). This model thus gives us the opportunity to study the endogenous levels of histone acetylation in correlation with origin efficiencies along the AMPD2 locus and, reciprocally, the impact of modulating origin usage on local histone acetylation.
Results
Origin usage is not related to histone acetylation
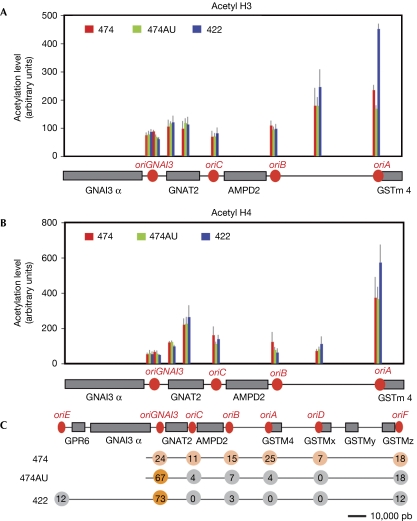
We first focused our study on a region encompassing oriGNAI3 and three other origins (oriA, B and C) in cells from the cell lines 474 and 474AU. The levels of histone acetylation were determined by chromatin immunoprecipitation (ChIP) experiments carried out with antibodies directed against acetylated histone H3 or H4 (Fig 1A,B). We observed that acetylation level of both histones varies along the locus but remains similar between cells from the cell lines 474 and 474AU, in which origin usage differs markedly. Furthermore, oriGNAI3 is nested in a region that is poorly acetylated even when used preferentially in fast conditions. By contrast, oriA presents a significantly high level of acetylation both in fast conditions when it is latent, and in slow conditions when it fires efficiently.
Figure 1.
Histone acetylation patterns do not correlate with origin efficiency. Chromatin immunoprecipitation experiments were performed with antibodies recognizing acetyl histone (A) H3 (06-599) or (B) H4 (06-598) and analysed by quantitative PCR using eight sets of primers (supplementary Tables S1 and S2 online). The acetylation level was normalized for each cell line to the median level along the locus (arbitrary units). Error bars represent s.e.m. values of three experiments. Grey boxes, genes; red circles, origins. (C) Schematic representation of origin efficiencies along the AMPD2 locus. Numbers represent origin efficiency in the indicated cell lines (data from the study by Anglana et al (2003) and circle colours symbolize this efficiency (orange, major origin; grey, minor origin; and tan, average efficiency). 474 versus 474AU and 474 versus 422 cells were compared for histone H3 and H4 acetylation profiles using Spearman and Wilcoxon's signed rank tests. In all cases, the correlation between mean values was significant (P<0.05) and the difference between medians of mean values was not significant, indicating that acetylation profiles are similar. Histone acetylation levels on oriGNAI3 were compared with those on oriA, oriB and oriC using the Mann–Whitney–Wilcoxon test. The levels of acetylation of both histones are significantly lower on oriGNAI3 (P=0.004 and P=5e6 for H3 and H4, respectively). Acetylation levels of oriA were similarly compared with those of oriGNAI3, oriB and oriC. The levels of acetylation of both histones are significantly higher on oriA (P=5e6 and P=6e6 for H3 and H4, respectively). See supplementary Table S3 online for details. AMPD2, adenosine monophosphate deaminase 2.
In cell line 474, the AMPD2 locus was rearranged during amplification (supplementary Fig S1 online). To determine whether amplification and/or this rearrangement affected the local acetylation profile, we studied the parental GMA32 cells and a second amplified line (cell line 422), in which no rearrangement occurred during amplification. Cells from cell line 422 show a replication pattern similar to that observed in cells from the cell line 474AU (Anglana et al, 2003). In GMA32 cells, forks travel at a speed of approximately 1.7 kb min−1 (data not shown). We did not study the replication dynamics along the AMPD2 locus by molecular combing because it remains a challenge to analyse single-copy regions using this technique. However, determination of leading-strand polarity has shown previously that oriGNAI3 is the major origin of the locus in non-amplified Chinese hamster cells (Svetlova et al, 2001). Thus cells from cell line 422 and non-amplified cells present ‘fast' replication dynamics. In both cells from cell line 422 (Fig 1A,B) and GMA32 cells (supplementary Fig S3 online), histone H3 and H4 acetylation profiles are the same as those observed in cells from cell line 474 (except for a high level of histone H3 acetylation in the oriA region in cells from cell line 422). These results indicate that neither the amplification process per se nor the rearrangement in cells from cell line 474 interfere with the acetylation status of the region and, correlatively, indicate that all the amplicons show similar profiles.
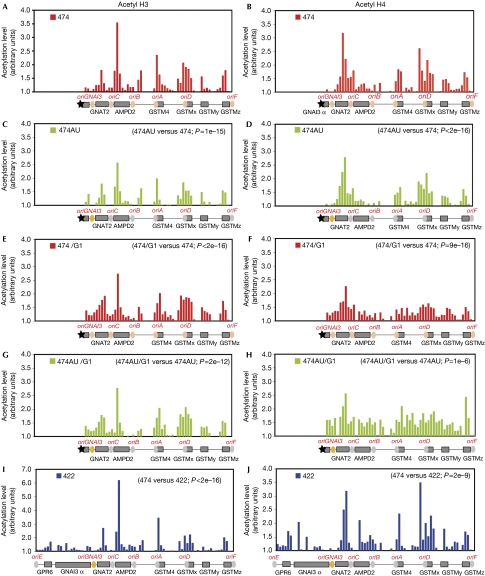
To get a broader view of histone H3 and H4 acetylation profiles at a higher resolution, we performed ChIP-on-chip experiments using DNA arrays covering the AMPD2 locus. In cells from cell lines 474, 474AU and 422, oriGNAI3, oriB and oriC are each nested in a region of low acetylation levels, whereas oriA and oriD are present in highly acetylated regions (Fig 2A–D,I,J). These results confirm and extend our previous observations and show that there is no correlation between local histone acetylation levels and origin efficiencies when exponentially growing cells were used for study (40% in G1 phase and 50% in S phase). As origin choice is set during the G1 phase, we also analysed the pattern of acetylation along the AMPD2 locus in cells from cell lines 474 and 474AU arrested in G1 by lovastatin (Fig 2E–H; Wu & Gilbert, 2000). Minor differences in histone acetylation profiles were observed between G1-arrested cells and growing cells; however, these differences are not statistically significant. In any case, we still do not find any correlation between origin efficiency and histone acetylation.
Figure 2.
Histone acetylation patterns in growing or G1-arrested cells correlate neither with localization nor with efficiency of origins. Chromatin immunoprecipitation experiments were performed with antibodies recognizing acetyl histone (A,C,E,G,I) H3 or (B,D,F,H,J) H4 and analysed by microarray hybridization. Results are expressed as the ratio of immunoprecipited DNA signal to the input signal (arbitrary units). Mean values of two experiments are reported. Circles symbolize: orange, major origin; grey, minor origin; tan, average efficiency. Black star: rearrangement breakpoint. Cells were compared for histone acetylation profiles by using the Spearman test. In all cases, the correlation between mean values was significant (P<0.05). Comparison and P-value results are indicated at the top right corner of each graph.
The possibility remained that acetylation on specific single positions sets the location or the relative efficiency of the origins. To verify this possibility, we also studied the level of acetylation on single residues of histone H3 (Lys 9, Lys 14, Lys 18, Lys 23, Lys 27 and Lys 56) and H4 (Lys 5, Lys 8, Lys 12, Lys 16 and Lys 91). For all these positions, we observed that profiles were similar in cells from cell lines 474, 474AU and 422 and origins were not associated with a high level of acetylation (supplementary Fig S2A,B online). Altogether our results strongly support the conclusion that acetylation levels of histones H3 and H4 are not sufficient to define the location or the efficiency of replication origins.
TSA affects fork speed and origin hierarchy
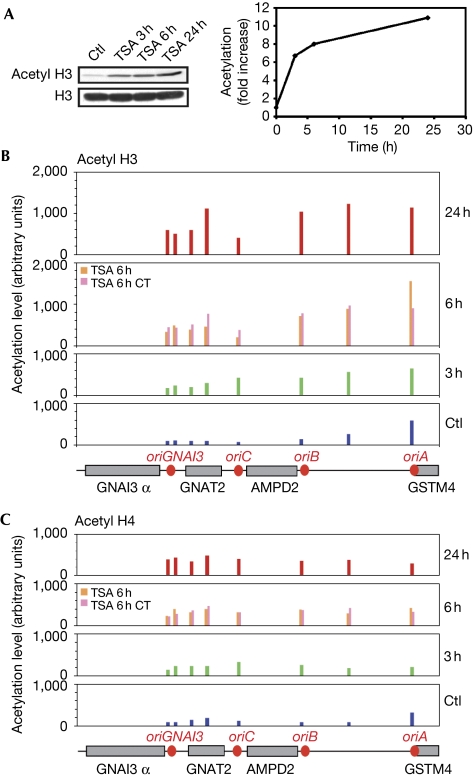
The above conclusion remains inconsistent with previous results showing that mammalian cells treated with TSA show changes in the pattern of origin usage (Kemp et al, 2005). We were thus interested in assaying the effect of this drug in our cell model. Treatment of cells from cell line 422 with TSA results in a progressive increase of bulk histone acetylation (Fig 3A). It also significantly raises histone H3 and H4 acetylation along the AMPD2 locus (Figs 3B,C). Analysis of the replication dynamics by molecular combing showed that the proportion of initiation events rises significantly in the bulk genome (Fig 4A). The distribution of these events along the AMPD2 locus was then studied by combining fluorescence in situ hybridization with detection of newly synthesized DNA (Anglana et al, 2003). We observed that TSA treatment results in a switch in origin usage along the locus (Fig 4B). Hence, for another locus and another mammalian species, our results fully confirm previous observations showing that TSA activates latent initiation sites (Kemp et al, 2005). However, as we did not detect specific histone hyperacetylation on origins mobilized on TSA treatment, this observation reinforces the aforementioned conclusion that origin efficiency is not governed by local acetylation patterns. We have reported previously that a reduction in fork speed triggers identical changes in the distribution of initiation events along the AMPD2 locus (Courbet et al, 2008). It was therefore of interest to determine whether TSA affects fork movement. As shown in Fig 4C, TSA does induce a progressive reduction of fork speed in cells from the cell line 422, which suggested that the changes in origin usage induced by TSA might be an indirect consequence of a slowing down of replication forks.
Figure 3.
Trichostatin A treatment induces global and locus-specific histone hyperacetylation. (A) Bulk histone H3 acetylation analysed by western blotting (left panel) with quantification of the signals (right panel). Similar results were obtained for histone H4 (data not shown). (B,C) Chromatin immunoprecipitation experiments were performed with antibodies recognizing acetyl histone (B) H3 or (C) H4 and analysed by quantitative PCR. Acetylation level was normalized for each condition to the median level along the locus at 0 h (Ctl). Mean values of two experiments are reported. Grey boxes, genes; red circles, origins. Control and TSA 24 h were compared for histone acetylation levels using the Wilcoxon's signed rank test. In all cases, the difference between medians of mean values was significant (P=0.008 and P=0.02 for H3 and H4, respectively) confirming that histone acetylation increases after TSA treatment. CT, cytidine and thymidine; TSA, trichostatin A.
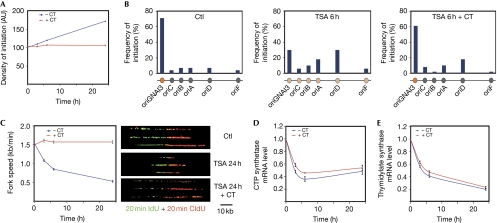
Figure 4.
Trichostatin-A-induced changes in replication dynamics of cells from cell line 422 are abrogated by exogenous precursors. (A) Relative frequency of initiation events at the genome-wide level as determined by the ratio of the number of initiation events relative to replication events (replication forks and initiations). Frequency of initiation was normalized to that of 0 h −CT. −CT and +CT, without and with cytidine and thymidine added in the medium. Values were normalized to that of 0 h. At least 100 DNA fibres were analysed for each time-point and culture condition. −CT and +CT conditions were compared by using a χ2-test. Densities of initiation are not significantly different between the two conditions at 0 h, whereas they are at 24 h (P=0.01). Similarly, densities of initiation are not significantly different between 0 h and 24 h in +CT conditions. (B) Localization of initiation events along the AMPD2 locus. At least 30 DNA fibres were analysed for each condition. Circles symbolize: orange, major origin; grey, minor origin; tan, average efficiency. Fisher's exact test indicated that initiation profiles are not significantly different between Ctl and TSA 6 h+CT, whereas they are different between Ctl and TSA 6 h (P=0.007). (C) Fork speed (left) and examples of combed DNA molecules labelled with IdU then with CldU (right). −CT and +CT conditions were compared using a Mann–Whitney–Wilcoxon test. Replication speeds are not significantly different between the two conditions at 0 h, whereas they are at 24 h (P<2e16). (D,E) CTP synthetase and TS messenger RNA levels. For each condition, levels were normalized to that of zero time point (arbitrary units). Errors bars represent standard error of the mean of two experiments. AMPD2, adenosine monophosphate deaminase 2; AU, arbitrary units; CldU, chlorodeoxyuridine; IdU, iododeoxyuridine; TS, thymidylate synthase.
TSA affects pyrimidine metabolism
A shortage of dNTPs is a well-known cause of fork slowing. We wondered whether the decrease in fork speed observed in the presence of TSA is a consequence of depletion in DNA precursors. Transcriptome analysis has shown previously that TSA treatment induces a change in the expression of 2–5% of human genes (Glaser et al, 2003). Among them, the expression of the genes coding for two enzymes involved in pyrimidine biosynthesis, CTP synthetase 1 and thymidylate synthase (TS), is downregulated. No other gene involved in nucleotide biosynthesis was observed to be affected. We tested the effect of TSA on these two genes in cells from cell line 422 (Fig 4D,E). We observed that their expression is reduced at least by a factor of two after 6 h of treatment and even further at 24 h in the case of TS. These observations were confirmed at the protein level (supplementary Fig S4A online). These effects of TSA are expected to reduce the supply of thymidine and possibly of cytidine derivatives in the cells. Quantification of dTTP and dCTP pools in cells from cell line 422 treated with TSA for 24 h showed a marked reduction—more than 80%—in the levels of both precursors (supplementary Fig S4B,C online, asynchronous cells). After 6 h of treatment, the dCTP pool was reduced twofold, whereas the dTTP pool was only slightly reduced. As dNTP pools are modulated during the cell cycle (Mathews, 2006), we enriched the population in S-phase cells (to 65%). In these conditions, dCTP and dTTP pools are reduced about threefold after 6 h in the presence of TSA. However, even though highly suggestive, these results do not formally prove that this reduction in pyrimidine pools is the cause of the changes observed in the replication dynamics after TSA treatment.
Exogenous nucleotide precursors block TSA effects
If a decrease in dTTP and dCTP pools was responsible for fork slowing, it should be possible to suppress the effects of TSA by adding cytidine and thymidine in the culture medium. Under these conditions, we observed that the decrease in fork speed induced by the drug is completely abrogated (Fig 4C), although the histone acetylation level along the locus is still high (Fig 3B,C) and the expression of the CTP synthetase and TS genes still reduced (Fig 4D,E). Noticeably, suppression of the effect of TSA on fork speed also suppresses its effects on the density of initiation events at the global level (Fig 4A) and on the hierarchy of the origins along the AMPD2 locus (Fig 4B), with oriGNAI3 remaining the major origin. These results establish that TSA reduces fork speed through a lowering of dCTP and dTTP pools and that the effects of TSA on the replication dynamics are an indirect consequence of pyrimidic deoxyribonucleotide starvation. The activation of latent origins after TSA treatment is a consequence of a slowing down of replication forks through the well-documented compensation mechanism (Courbet et al, 2008). As these effects are completely reversed by precursor addition, whereas local acetylation remains elevated, we conclude that TSA-induced hyperacetylation of histone or of other factors at the origins has no role in the modulation of the initiation pattern we observed.
Discussion
By analogy with transcription, it was long proposed that local histone acetylation facilitates DNA accessibility for replication proteins and, consequently, increases origin firing. For example, a high level of histone H4 acetylation was observed at the human c-MYC, lamin B2 and β-globin origins (Kemp et al, 2005) and at developmentally regulated origins in Drosophila follicle cells (Aggarwal & Calvi, 2004). However, as all these origins lay close to active promoters, it complicates the evaluation of the relative contribution of histone acetylation to regulation of transcription and replication. In this study, we made use of the fact that only some origins of the AMPD2 locus co-localize with transcription promoters (supplementary Fig S1 online) to study the relationship between local histone acetylation and origin location or efficiency. We observed that the position of origins, whether major or minor, is not marked by a particularly high level of acetylation of histones H3 and H4. Furthermore, origin hierarchy in the locus can be changed without any effect on the profiles of histone acetylation. However, we observed that histone hyperacetylation induced by TSA treatment has no effect on origin hierarchy, provided the reduction in fork speed induced by the drug is compensated by a supply of exogenous pyrimidic precursors. Altogether our results establish that origin efficiency is unrelated to local histone acetylation, at least in euchromatic transcribed regions, such as the AMPD2 locus. Our results, obtained by analysis of mammalian fibroblasts, fully agree with previous studies carried out in a developmental context for several loci in different organisms (Prioleau et al, 2003; Dazy et al, 2006; Gregoire et al, 2006). Genome-wide analyses have also shown that more than half of active origins are localized neither in regions with a high level of histone H3 or H4 acetylation nor in decondensed chromatin regions (Cadoret et al, 2008; Karnani et al, 2010).
A recent study has shown that the activity of the histone acetyltransferase binding to ORC (HBO1) acetyltransferase is dispensable for the binding of the origin recognition complex to origins (Miotto & Struhl, 2010). Our results agree with this conclusion because we did not observe any evidence for a role of histone acetylation in setting origin location, which is determined by origin recognition complex binding. By contrast, HBO1 or general control nonderepressible 5 acetyltransferases were involved in the loading of the minichromosome maintenance complex during licensing (Iizuka et al, 2006), however, the authors did not assess an eventual impact on origin activity. In any case, our findings do not exclude that acetylation of factors other than histones might impinge on the replication dynamics.
As inactivation of some specific deacetylases also leads to an increased number of initiation events (Pasero et al, 2002; Aggarwal & Calvi, 2004; Pappas et al, 2004), and as CTP synthetase and TS expression is similarly reduced by other deacetylase inhibitors (Kemp et al, 2005), we infer that the effects we observed are not specific to TSA but result from hyperacetylation of histones or of other factors. In mammalian somatic cells, modulation of the acetylation balance could be a new pathway in the fine-tuning of replication in which CTP synthetase and TS are key molecular intermediates for the control of dNTP pools. This would imply that controlling dNTP pool level is not only dedicated to secure a continuous supply of precursors to the replication machinery but is used by the cell as a regulatory pathway to adjust replication dynamics. This would be the second example of such a regulation. It has been shown that, in yeast, modulation of dNTP pools is an effective way for controlling replication dynamics. In this organism, the DNA replication checkpoint is known to regulate the activity of ribonucleotide reductase (Zhao et al, 1998). In any case, our results reconcile previous findings that seemed inconsistent and establish an unsuspected pathway in the effects of TSA on replication dynamics via dNTP pool modulation.
Methods
Cell culture and drug treatment. Cell lines 422 and 474 were selected from GMA32 cells and propagated as described previously (Debatisse et al, 1982). For cells from the cell line 474AU, the medium was supplemented with 50 μM each of adenine and uridine. Cells were treated with TSA (Sigma, 200 ng ml−1) for 3, 6 or 24 h, as indicated. Cytidine and thymidine were added together with TSA at a final concentration of 10 μM. Arrest in G1 was obtained by lovastatin treatment (20 μM) for 22 h.
RNA extraction and retro-transcription. Cells were collected and total RNA was extracted using the RNeasy midi kit (Qiagen). Retro-transcription was performed with 0.75 μg of RNA using the SuperScript III first-strand synthesis system for retro-transcription PCR (RT–PCR; Invitrogen). An aliquot of the reaction (1/250) was used for analysis by quantitative PCR.
ChIP. A detailed protocol is provided in the supplementary information online.
RT–PCR. A detailed protocol is provided in the supplementary information online.
Microarray hybridization. A detailed protocol is provided in the supplementary information online.
Western blots. Western blotting was performed using antibodies against histone H3 (Abcam, ab1791), acetyl histone H3 (Upstate Biotechnology, 06-599), thymidylate synthase (Abcam, ab7398) or proliferating cell nuclear antigen (Millipore, MAB424).
DNA combing and immunofluorescent detection. Combing and immunodetection were performed as described previously (Anglana et al, 2003; Labit et al, 2008).
Measurement of dNTP pools. A previously described enzymatic assay was used (Sherman & Fyfe, 1989).
Statistical analyses. A detailed description is provided in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank M. Schertzer for critical reading of the paper, Genomic Vision for making available the DNA combing technology and C. Chevalier for help with microarray spotting. S.G. and A.L. are supported by the Association pour la Recherche sur le Cancer. M.D. and team were supported by La Ligue Nationale contre le Cancer and the Agence Nationale de la Recherche.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN (2008) Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci USA 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M (2008) Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Dazy S, Gandrillon O, Hyrien O, Prioleau MN (2006) Broadening of DNA replication origin usage during metazoan cell differentiation. EMBO Rep 7: 806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Berry M, Buttin G (1982) Stepwise isolation and properties of unstable Chinese hamster cell variants that overproduce adenylate deaminase. Mol Cell Biol 2: 1346–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK (2003) Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2: 151–163 [PubMed] [Google Scholar]
- Gregoire D, Brodolin K, Mechali M (2006) HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary. EMBO Rep 7: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Matsui T, Takisawa H, Smith MM (2006) Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 26: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A (2010) Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Ghosh M, Liu G, Leffak M (2005) The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res 33: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H, Goldar A, Guilbaud G, Douarche C, Hyrien O, Marheineke K (2008) A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers. Biotechniques 45: 649–652, 654, 656–648 [DOI] [PubMed] [Google Scholar]
- Mathews CK (2006) DNA precursor metabolism and genomic stability. FASEB J 20: 1300–1314 [DOI] [PubMed] [Google Scholar]
- Miotto B, Struhl K (2010) HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas DL Jr, Frisch R, Weinreich M (2004) The NAD+-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev 18: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Gendron MC, Hyrien O (2003) Replication of the chicken β-globin locus: early-firing origins at the 5′ HS4 insulator and the ρ- and βA-globin genes show opposite epigenetic modifications. Mol Cell Biol 23: 3536–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM (2007) Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PA, Fyfe JA (1989) Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal Biochem 180: 222–226 [DOI] [PubMed] [Google Scholar]
- Svetlova EY, Razin SV, Debatisse M (2001) Mammalian recombination hot spot in a DNA loop anchorage region: a model for the study of common fragile sites. J Cell Biochem Suppl 36: 170–178 [DOI] [PubMed] [Google Scholar]
- Tabancay AP Jr, Forsburg SL (2006) Eukaryotic DNA replication in a chromatin context. Curr Top Dev Biol 76: 129–184 [DOI] [PubMed] [Google Scholar]
- Wu JR, Gilbert DM (2000) Lovastatin arrests CHO cells between the origin decision point and the restriction point. FEBS Lett 484: 108–112 [DOI] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.