Abstract
The ATR-Chk1 checkpoint pathway is activated by UV-induced DNA lesions and replication stress. Little was known about the spatio and temporal behaviour of the proteins involved, and we, therefore, examined the behaviour of the ATRIP-ATR and Rad9-Rad1-Hus1 putative DNA damage sensor complexes and the downstream effector kinase Chk1. We developed assays for the generation and validation of stable cell lines expressing GFP-fusion proteins. Photobleaching experiments in living cells expressing these fusions indicated that after UV-induced DNA damage, ATRIP associates more transiently with damaged chromatin than members of the Rad9-Rad1-Hus1 complex. Interestingly, ATRIP directly associated with locally induced UV damage, whereas Rad9 bound in a cooperative manner, which can be explained by the Rad17-dependent loading of Rad9 onto damaged chromatin. Although Chk1 dissociates from the chromatin upon UV damage, no change in the mobility of GFP-Chk1 was observed, supporting the notion that Chk1 is a highly dynamic protein.
1. Introduction
Eukaryotic cells are continuously threatened by DNA damage caused by environmental factors and intracellular metabolic processes. To protect themselves against these potential threats, cells have developed DNA damage checkpoint and repair mechanisms, which help to ensure transmission of an intact genome. Cell cycle checkpoints and DNA repair mechanisms together determine the ultimate faith of the cell after suffering DNA damage. Activation of the DNA damage checkpoint involves the activation of transducer kinases ATR/ATM and subsequently the effector kinases Chk1/Chk2 [1]. So-called mediator proteins, including Claspin and BRCA1, were additionally discovered, and function either in the recruitment of substrates to DNA lesions or as scaffolds on which protein complexes are assembled [2, 3]. In response to a variety of DNA damaging agents like UV light and replication stress, the ATR-mediated checkpoint pathway is activated. Biochemical data indicates that ATRIP, in complex with ATR, binds to RPA-coated single stranded DNA (ssDNA) [4]. Independently, the Rad17-RFC complex is also recruited to sites of damage. The Rad17-RFC protein complex facilitates the loading of the Rad9-Rad1-Hus1 (9-1-1 complex) sliding clamp onto the DNA [5–7]. Subsequently, TopBP1 is recruited to DNA lesions by binding to the Rad9 subunit of the 9-1-1 complex, thereby locating near the ATRIP-ATR heterodimer. Through an interaction with TopBP1, ATR becomes fully active, resulting in the activation of effector kinase Chk1 and subsequent checkpoint arrest [8–10].
Detection of DNA alterations after genotoxic stress is essential for the survival of cells and gaining more insight into the early events of the DNA damage response will give a better understanding of how DNA damage checkpoints function, how genome stability is achieved, and how cancer can develop. In recent years, biochemical work has provided invaluable insight into the requirements, substrates, and activities of proteins involved in the ATR-mediated checkpoint pathway [11–13]. Although the importance for protein localization after DNA damage induction has been reported for proteins involved in the response to double-stranded breaks (DSBs), not much is known about how the ATR-Chk1 pathway operates in living cells and moreover, how the spatio and temporal behaviour of proteins in this pathway influence the DNA damage response [14, 15]. We set out to study the behaviour of ATRIP-ATR and 9-1-1 DNA damage sensor complexes by creating cDNA constructs expressing GFP- (green fluorescent protein-) tagged proteins in human cells. The use of GFP-fusion proteins creates advantages over using standard immunofluorescence techniques as it avoids fixation methods and antibody artefacts. In addition, GFP-labelled proteins can be followed in time using live cell video microscopy. Furthermore, due to the spectral qualities of GFP and its variants it has become possible to perform quantitative fluorescent analysis [16–18]. The use of GFP-labelled proteins opens up a number of new possibilities in the DNA damage response field. First, the localization GFP-fusion proteins can be directly followed into DNA damage-induced nuclear foci. Second, the ability to measure in time makes it possible to establish an order of events occurring directly after DNA damage induction. Third, the existence of multiple spectral GFP-variants allows for the simultaneous detection of several fluorescently labelled proteins in a single cell [19–21]. Forth, live cell video microscopy in combination with GFP-photobleaching experiments can be applied to quantitatively determine changes in protein mobility in response to DNA damage. Collectively, these tools help to increase our understanding of cellular mechanisms involved in DNA damage response.
The accumulation of DNA damage response proteins at sites of damage, shown in cells as nuclear foci, is essential for downstream checkpoint events, although how, is not yet fully understood [22–24]. Many, if not most, proteins in the ATR-mediated checkpoint pathway are recruited to sites of damage into nuclear foci. As a result, a proportion of the total amount of free proteins becomes immobilized in these foci due to interactions with either the damaged chromatin or other proteins at the DNA lesion. To gain more understanding of the DNA damage-induced behaviour of a protein, its localization and mobility can be determined. The DNA damage-induced change in protein mobility has been studied for several proteins involved in the DNA damage response, mainly in DNA repair and in response to double stranded breaks (DSBs) [17, 25–27]. The studies performed on proteins involved in nucleotide excision repair (NER), for example, have been important in understanding this DNA repair process and helped explain some of the phenotypic characteristics seen in patients that harbour mutations in proteins in this pathway. This indicates the importance of these types of studies for research in cancer and related diseases associated with genome instability [28, 29].
A method was setup for the generation of stable cell lines expressing GFP-fusion proteins and different assays to validate and characterize GFP-expressing cell lines were developed. We analyzed the dynamic behaviour of multiple proteins involved in the ATR checkpoint pathway including all three components of the 9-1-1 complex, ATRIP, and of the downstream kinase Chk1. We compared the mobility changes of these proteins after UV-irradiation and together the results indicated a distinct dynamic behaviour of the studied proteins. Whereas effector kinase Chk1 was highly mobile and did not immobilize upon DNA damage, Rad9 and ATRIP were associated to sites of DNA damage. ATRIP directly bound to DNA lesions but in a more transient manner than Rad9, which displayed a relatively slow exchange with damaged chromatin. Together these data demonstrate the importance of spatio and temporal protein regulation in the cells response to DNA damage.
2. Materials and Methods
2.1. Cell Culture
U2OS, Hela, and 293T cells were grown using standard procedures. U2OS and Hela cells stably expressing eGFP-fusion proteins were grown in standard medium supplemented with 350–700 μg/mL of G418 (gentamicin).
2.2. Antibodies
Antibodies obtained from commercial sources were as follows: Orc2 (BD Pharmingen), Grb2 (BD Transduction), RPA (Ab2, Oncogene), Rad9 (Novus), γH2AX (Upstate), Chk1-pS317 (Cell Signaling), and Chk1 (Abcam). The following antibodies were obtained from Santa Cruz Biotechnology: CENP-F (H-260), Cyclin A (H-432), Cyclin B1 (GNS1), Ku86 (C-20), Chk1 (G-4), Rad1 (N-18), Rad9 (C-20), and ATR (N-19).
Rabbit polyclonal anti-Rad9, anti-Hus1, and anti-GFP were described before and a kind gift by R. Freire (Tenerife, Spain) [30, 31]. Rabbit polyclonal anti-ATRIP was a kind gift by P. M. Reaper (UK) [32].
2.3. Transfection
Plasmid DNA was transfected into cells using the calcium phosphate transfection method.
siRNA oligonucleotides (Dharmacon Research) were transfected into cells using Oligofectamine (Invitrogen) according to the manufacturer's instructions and as previously described [33]. Sequences of oligonucleotides were as follows:
Luc (CGUACGCGGAAUACUUCGAdTdT),
Chk1 (UCGUGAGCGUUUGUUGAACdTdT)
Rad9 (ACCACUAUAGGCAAUGAGGdTdT)
Rad9 3′UTR (CCAAGAACCUGAAGCCUGUUU/GAAUCCAGCUUUGACCUUUU).
2.4. Colony Survival Assays
Cells were counted and 1000 cells were seeded onto 60 mm diameter dishes whereafter left for 12 hours to attach. Cells were treated with different doses of UV (5, 10, 15, and 20 J/m2) and incubated for 7–14 days, after which the colonies were fixed, stained, and counted. All experiments were performed in triplicate.
2.5. Immunofluorescence
For immunostaining, cells were fixed in 2% paraformaldehyde containing 0.2% Triton X-100 for 20 minutes at RT and then permeabilized with 0.1% Triton X-100 for 5 minutes at RT. Samples were blocked in 1% FCS and immunostained with antibodies as indicated. For detection of GFP-tagged proteins in U2OS cells, living cells were studied or cells were fixed and permeabilized as above.
Images were made using a Cell Observer fluorescent microscope equipped with Axiovision software (Zeiss) or a Confocal Laser Scanning Microscope LSM 510 (Zeiss), equipped with a 488 nm Ar-laser and a 505–530 nm bandpass filter for green fluorescence. Red fluorescence was detected using a 543 nm laser and 560 nm longpass filter. For strip-FRAP experiments and time lapse imaging after UV laser induction, GFP-tagged proteins were detected in living cells using a Confocal Laser Scanning Microscope LSM 510 (Zeiss), equipped with a 488 nm Ar-laser and a 505–530 nm bandpass filter for green fluorescence.
2.6. Whole Cell Extracts and Cell Fractionation
For whole cell extracts, cells were washed in cold PBS, after which the cells were resuspended in Laemmli sample buffer (4% SDS, 20% glycerol, 120 mM Tris pH 6.8) and boiled for 5 minutes. Protein concentrations were determined using the Lowry protein assay. Chromatin fractionation was performed as previously described [34, 35].
2.7. Generation of DNA Damage and Photobleaching Techniques
UV-irradiation was performed using a 254 nm UV-C lamp (Philips) at 20 J/m2 and cells were processed 1 hour posttreatment. Cells were incubated with 5 μg/mL of aphidicolin (Sigma) for 20 hours prior to processing of samples. Cells were incubated with 200 nM camptothecin (Sigma) for 20 hours before fixation. IR (10 Gy) was induced using a 137Cs source or a linear accelerator (LINAC) and cells were processed 2 hours after treatment. Local UV laser induction was performed as previously described [36]. In short, cells were grown on a 25 mm quartz coverslip. A small area inside the nucleus was exposed for ~1 second to a 2 mW pulsed (7.8 kHz) diode laser emitting at 266 nm. Time-lapse images were made every minute immediately starting after local UV induction. The time course measurements were normalized to the plateau phase. Start of UV irradiation was defined as t = 0. Assembly curves were normalized to 1 and processed as previously described [26].
In the FRAP experiments, a strip spanning the nucleus was photobleached for 20 milliseconds using an Ar-laser (488 nm) at 100% laser intensity, which irreversibly bleaches all GFP molecules in that area. Subsequently, the redistribution of fluorescence in the strip was monitored by taking confocal images every 20 milliseconds for a total of 24 seconds at low laser intensity to avoid further bleaching. The fluorescence before bleaching (I t<0) was set to 1 and the intensity immediately after bleaching (I 0) was set to 0. The recovery of fluorescence was plotted against time.
In the FLIP experiments, half of the nucleus was continuously bleached using an Ar-laser (488 nm) at 50% laser intensity. Subsequently, the loss of fluorescence in the unbleached area was monitored taking confocal images every 10 seconds for a total of 300 seconds at low laser intensity. The fluorescence before bleaching (I t<0) was set to 1. The loss of fluorescence was plotted against time. FLIP-FRAP experiments were performed as previously described [33, 37].
3. Results
3.1. Generation of Cell Lines Stably Expressing GFP-Tagged Proteins
The limitations of conventional fixation methods were overcome by the introduction of GFP, the green fluorescent protein derived from Aequorea Victoria, a species of jellyfish [38, 39]. Through the cloning of the cDNA encoding GFP, it became possible to fluorescently label proteins and express them, thereby creating the opportunity for monitoring proteins in living cells. Different fluorescent versions of the GFP exist such as the red fluorescent (RFP), yellow fluorescent (YPF), and cyan fluorescent (CFP) protein, and many more spectral variants have been created since [40]. The expression of GFP and derivatives itself in mammalian cells is not harmful although the protein originates from a different species. Nonetheless, labelling of proteins with (fluorescent) epitopes can potentially influence the behaviour of the labelled protein. Validation and functional analysis of a GFP-fusion protein construct, therefore, is essential for the interpretation of experimentally obtained data.
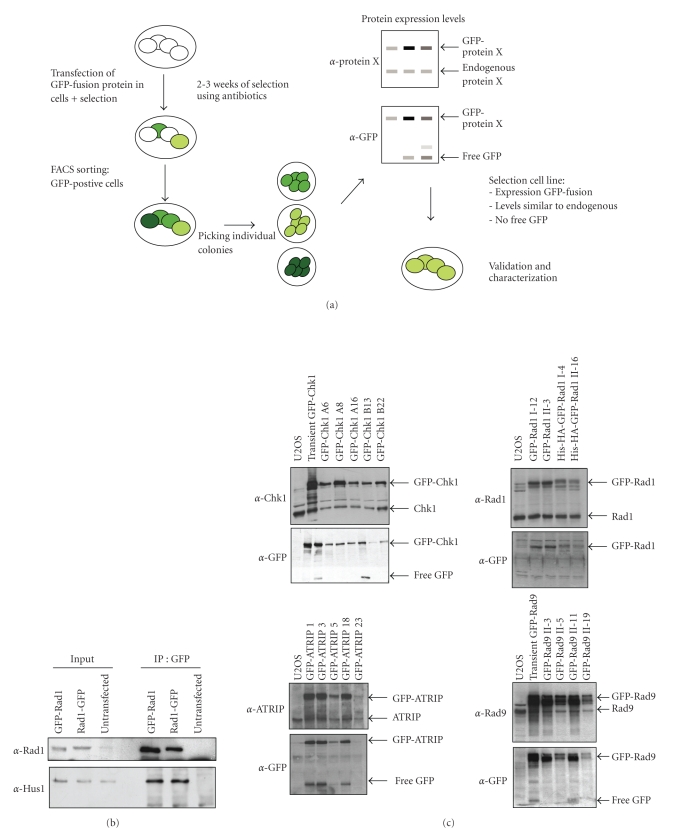
The amount of fluorescence signal in a cell population derived from a single cell clone is in principle identical in all cells and, therefore, comparing fluorescence intensities after different treatments becomes reliable. We setup a method for the creation, generation, and validation of GFP-fusion proteins for the purpose of live cell imaging. The approach is schematically depicted in Figure 1(a) and discussed below.
Figure 1.
Generation of stable cell lines expressing GFP-tagged proteins. (a) Stepwise protocol for stable expression of GFP-fusion proteins in cells. (b) 293T cells were transfected with GFP-Rad1, Rad1-GFP, or mock transfected, after which cells were lysed and immunoprecipitations were carried out using anti-GFP antibody. Western blot analysis of the immunoprecipitates using the indicated antibodies. (c) Protein expression analysis of different stable clones. Western blot analysis of whole cell extracts using the indicated antibodies.
3.1.1. Creation of GFP-Tagged Protein Constructs
After the discovery of GFP and its potential usefulness in biology, the protein itself became the subject of intensive research. Certain mutations in the GFP gene lead to different spectral variants and improvements in the protein characteristics, such as an increase in brightness (fluorescent intensity) and photo stability [41]. These qualities can be found in, for example, the GFP variant called “enhanced GFP” (eGFP). Vectors encoding for eGFP and fluorescent derivates are commercially available (Clontech) and have been published [40]. In the studies presented here, we made use of vectors containing eGFP or eGFP with the addition of two other, nonfluorescent epitopes, namely, an HA- and His-tag (eGFP will hereafter be referred to as GFP). The additional epitope creates the possibility to detect the GFP-fusion protein with antibodies directed against the HA- and/or His-tags. Additionally, these tags can be used for immunoprecipitation experiments to identify interaction partner proteins.
For proper cellular function a protein needs to be folded correctly upon its expression. Since GFP and the cDNA of interest encode for two proteins that are attached, proper protein folding might be influenced by sterical hindrance. As a possible solution to this problem, adding a small DNA sequence linker in between the sequence encoding for GFP and the cDNA this can be minimized [38, 42]. In addition, such a linker might help to restore the functionality of the protein, as a rigid tag might interfere with, for example, binding to (endogenous) partners.
In addition to a neomycin resistance cassette for selection, the GFP vectors contain a CMV promoter that is compatible with expression in immortalized cells, such as the human osteosarcoma (U2OS) and epithelial cervical cancer (Hela) cell lines used in this study. As adding a tag might influence protein functionality, it is advisable to construct both N- and C-terminal labelled versions of the proteins. cDNA was cloned into the GFP-vectors via PCR and/or restriction enzyme cutting and religating by T4 DNA ligase, resulting in plasmids encoding an in-frame sequence of GFP and the cDNA of interest. After obtaining the right plasmid DNA the functionality of the GFP-fusion protein was tested by determining the interaction with its known binding partners. In this way a reasonable choice can be made to continue with either the N- or C-terminally tagged version of the protein, or both. After transient expression of the cDNA encoding the GFP-fusion protein and the known binding partner, cell extracts were prepared for immunoprecipitation using antibodies against GFP. The interaction with the known binding partners was determined by western blotting. The N- and C-terminally labelled proteins from the Rad9-Rad1-Hus1 complex were tested in this manner. Labelling of Rad1 at either site of the protein did not influence the interaction with endogenous Hus1 (Figure 1(b)). GFP-labelling of Rad9 at the C-terminus completely abrogated the interaction with its binding partners whereas Hus1 labelling at the N-terminus decreased the binding to its partners (data not shown). When binding partners of a GFP-fusion protein are not known or do not exist and/or antibodies are not available testing for functionality must be performed in a different manner. Later in this section, different assays that can be used to validate the function of the GFP-tagged protein in living cells are described.
3.1.2. Expression of GFP-Tagged Proteins in Human Cells
After obtaining cDNA encoding a GFP-fusion protein, cells were transfected to achieve random integration of the GFP-fusion construct. We used the well studied human U2OS and Hela cells for the expression of the GFP-fusion protein since these cells are well-suited for both live cell imaging and siRNA-mediated downregulation of proteins. We obtained a transient transfection efficiency of ~70% in both Hela and U2OS cells using the calcium-phosphate transfection method, whereafter we started with antibiotic selection. Importantly, multiple copies of the cDNA can be incorporated in the genome and the site (s) of integration additionally may create artefacts and influence cellular behaviour. In human cells this problem can only be tackled by using retroviral constructs that integrate only once. To minimize these possible artefacts we only selected individual clones expressing low amounts of GFP-fusion proteins.
3.1.3. Selection of GFP-Positive Cells Using an Antibiotic Selection Marker
To obtain cells that incorporated at least one copy of the GFP-fusion cDNA we used antibiotic selection through the neomycin-resistance cassette present in the GFP vector. To achieve stable integration of the cDNA in the genome of the transfected cells, cells were seeded in different densities and selected using 700 μg/mL G418 for at least two weeks, refreshing the medium with antibiotics every 3 days.
3.1.4. FACS Sorting of GFP-Positive Cells
G148 selection caused cell death of cells that did not integrate the cDNA construct in their genome. GFP-positive cells were sorted from the total population by FACS. Approximately 5%–10% of the cells became GFP-positive after two weeks of G418 selection. This small percentage is most likely due to an incorrect integration the GFP-fusion cDNA causing expression of the only part of the construct including the G418 resistance cassette. The sorted GFP-positive cells were replated in different dilutions so that single cells could grow into discrete colonies. Cells were kept in similar conditions of antibiotic selection as previously used and medium was refreshed at least twice a week.
3.1.5. Picking of Individual Cell Colonies
After the multiplication of a single cell into a colony of around 50–100 cells, the colonies were randomly picked under sterile conditions using an inverted light microscope. At least 24 colonies were picked for every cell line. The selected colonies were kept in individual wells and were cultured until completely confluent, whereafter split into two plates. One plate was frozen and the other one was used to prepare cell lysates for expression analysis.
3.1.6. Determining Protein Expression Levels of the GFP-Fusion
The cell lysates from individual clones were analyzed by SDS-PAGE and western blotting using antibodies specifically against the protein of interest and GFP. Expression levels of the endogenous compared to the GFP-fusion protein was determined as well as the amount of free GFP or the existence of degradation products.
3.1.7. Selecting Clones Expressing Relatively Low Amounts of GFP-Tagged Protein
Clones were selected on the basis of the following criteria: low expression levels of the GFP-fusion protein (not higher than that of the endogenous protein) and the absence of free GFP or other undefined products, based on the western blotting analysis. A number of 4 clones that meet these criteria were selected and thawed for further validation. Clones were cultured in medium containing 350 μg/mL G418 to maintain expression of the GFP-fusion protein.
With the method described above stable cell lines were obtained that express a GFP-fusion from the ATR-Chk1 pathway, namely, ATRIP, Rad17, Rad9, Rad1, Hus1, and Chk1 (Figure 1(c) and data not shown). We also generated control cell lines expressing GFP and H2B-GFP-Chk1.
3.1.8. Validation and Characterization of the Selected Stable Cell Lines
To validate the functionality of the GFP-fusion protein and characterize the stable cell lines we utilized a number of assays.
Immunoprecipitations (IP) of the GFP-fusion protein were performed to confirm the interaction of the GFP-fusion with its known endogenous protein partners. We published before that GFP-Rad9 interacts with its endogenous partners Rad1 and Hus1 [33]. A similar experiment was performed with GFP-ATRIP, which demonstrated that the GFP-fusion was able to bind endogenous ATR (Figure 2(a)).
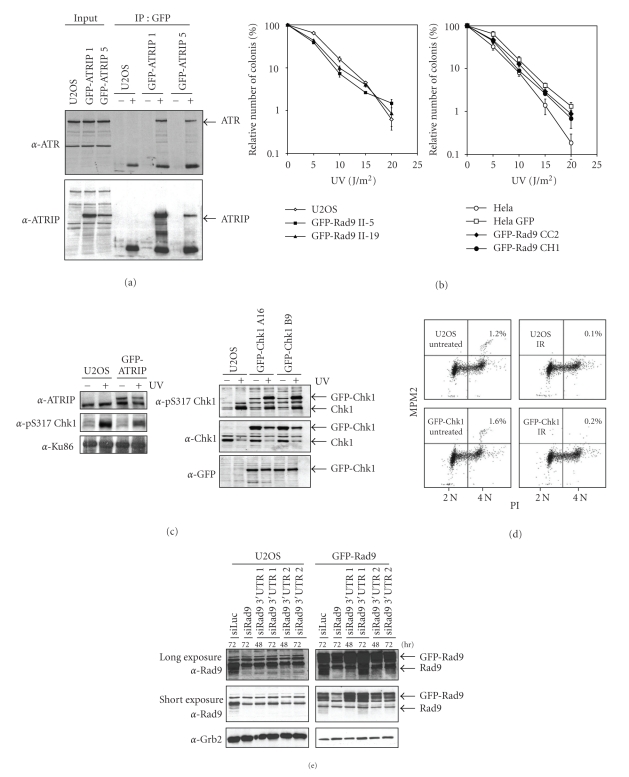
Figure 2.
Validation and functionality of stable cells expressing GFP-fusions. (a) Two independent U2OS clones expressing GFP-ATRIP were lysed after which immunoprecipitations were carried out using anti-GFP antibody. Western blot analysis of the immunoprecipitates using the indicated antibodies. (b) U2OS and Hela cells expressing GFP-Rad9 were seeded in low density and treated with different doses of UV. The number of surviving colonies after 10 days was counted. (c) Left panel: cells expressing GFP-ATRIP or control U2OS cells were left untreated or treated with UV. One hour later, cells were lysed and analyzed by western blotting with the indicated antibodies. Right panel: as left panel, but with two clones expressing GFP-Chk1. (d) Control U2OS cells or cells expressing GFP-Chk1 were left untreated or treated with 10 Gy IR. After 2 hours, cells were fixed and stained with PI and MPM2 and analyzed by FACS. The percentage of mitotic cells is indicated. (e) U2OS cells expressing GFP-Rad9 were transfected with siRNA oligos directed against Rad9 or the 3′UTR of Rad9 for the indicated time periods. Cell extracts were prepared and western blot analysis was performed using the indicated antibodies.
To check if stable expression of the GFP-fusion protein influences the cells response to DNA damage by functioning as a dominant negative, we determined the sensitivity of our cell lines to genotoxic stress. We compared the colony survival in response to UV light of the stable clones to the parental untransfected cell line. As shown in Figure 2(b), the expression of the GFP-Rad9 in both Hela and U2OS cells did not alter the sensitivity to UV damage, indicating that GFP-Rad9 does not function as a dominant negative. The expression of GFP alone in Hela cells also did not influence sensitivity to UV-irradiation.
The ATR-dependent checkpoint pathway is triggered in response to genotoxic stress and induces the ATR-mediated phosphorylation of Chk1, which results in a temporal cell cycle arrest [11]. To test if expression of the GFP-fusion proteins influences checkpoint activation we monitored the UV-induced phosphorylation of Chk1. Expression of GFP-ATRIP and GFP-Chk1 did not influence the levels of phosphorylated Chk1 (Figure 2(c)). GFP-Chk1 is phosphorylated in response to UV damage, indicating the GFP-tagged Chk1 behaves similarly to the endogenous one. To study the G2 phase arrest induced by DNA damage, we determined the mitotic index of cells expressing GFP-Chk1 and untransfected cells. A similar decrease in mitotic cells upon ionizing radiation (IR) was observed in GFP-Chk1 expressing and control cells, indicating that expression of GFP-Chk1 does not interfere with the DNA damage-induced G2 checkpoint (Figure 2(d)).
Although these assays demonstrate the absence of a dominant negative effect of the GFP-fusions, to definitively rule out problems due to GFP-tagging, complementation of a mutant or knock out cell line is required. Cells derived from Seckel syndrome patients that harbour a mutation in the ATR gene, leading to an abrogated expression of the ATR protein, are available [43], but most proteins in this pathway are essential for cell viability making a direct complementation impossible in human cells. In addition, knock outs of Rad9 and Hus1 in mouse embryonic stem cells are lethal, although the lethality of Hus1 disruption can be rescued by knocking out p21 [44–46]. Chicken DT40 cells can be successfully used for knocking out genes that are essential in mammalian cells, like Chk1, but are less suitable for the generation of stable GFP-fusion cell lines and live cell imaging, since these cells are genomically unstable and can only be cultured in suspension [47]. Another approach is siRNA-mediated downregulation of the endogenous protein and subsequent complementation by expression of the GFP-tagged protein. We favour this option since in this way we are able to study both the functionality of the GFP-fusion protein as well as its mobility in the same cell line.
To specifically downregulate the endogenous protein, different siRNA-mediated approaches are available. First, as the cDNA encoding for the GFP-fusion protein does not contain a 3′UTR, the use of siRNA oligos that specifically target this region of the protein of interest should result in knock down of the endogenous protein only. We applied this approach in cells expressing GFP-Rad9. Figure 2(e) demonstrates that 48 hours after transfection with siRNA oligo number 1, endogenous Rad9 is more efficiently downregulated than the GFP-labelled Rad9 protein. At 72 hours after transfection, the efficiency of downregulation became less, demonstrating that knock down is optimal 48 hours after transfection with siRNA oligo number 1. Transfection with siRNA oligo number 2 also lowered endogenous Rad9 levels more than the GFP-labelled Rad9 protein levels and the efficiency of downregulation was similar between 48 and 72 hours after transfection. In an alternative approach, a silent mutation is made in the cDNA of the GFP-tagged protein, thereby leaving the encoded protein unchanged. By using siRNA oligos that recognize the mRNA from the endogenous but not the GFP-fusion protein, the endogenous protein can be specifically downregulated, such that the functionality of the GFP-fusion protein can be assessed [33]. Collectively the performed validation assays indicate that the introduction and stable expression of GFP-fusions of Chk1, ATRIP, Rad9, Rad1, and Hus1 did not change the response of these cells to DNA damage. These GFP-fusion proteins can therefore be seen as functional equivalents of their endogenous counterparts.
3.2. Cellular Localization of GFP-Fusion Proteins in Response to DNA Damage
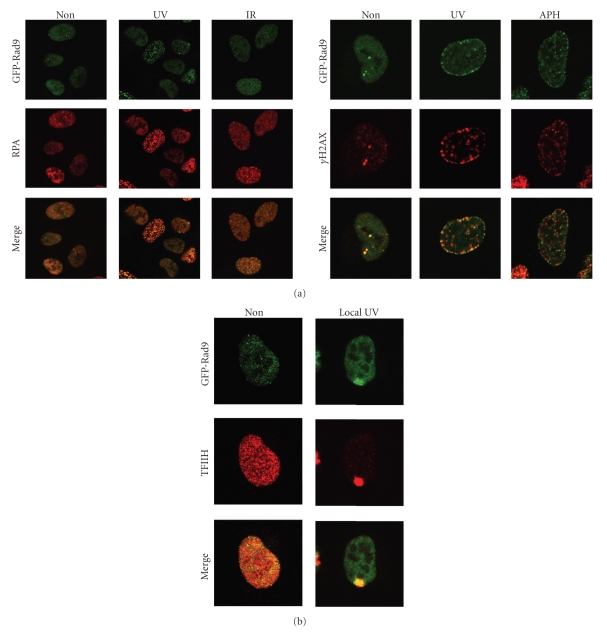
Numerous studies over the past years indicated that the accumulation of DNA damage response proteins at sites of damage is of crucial importance for downstream checkpoint events [14, 22, 24, 48, 49]. Although for many proteins involved in checkpoint regulation a (relatively small) fraction of the total pool of proteins is bound to chromatin in unperturbed cells, the majority of these proteins relocalize and accumulate at or near sites of DNA lesions to form so-called nuclear foci [50]. GFP-Rad9, for example, localizes into foci after the treatment with different DNA damaging agents (Figure 3(a)).
Figure 3.
DNA damage-induced foci formation of GFP-Rad9. (a) GFP-Rad9 expressing cells were left untreated or exposed to UV, IR, or aphidicolin. GFP-Rad9 was detected by direct fluorescence and RPA and γH2AX by immunofluorescence. (b) GFP-Rad9 expressing cells were treated with 40 J/m2 local UV-irradiation using an isopore filter. One hour later the cells were fixed and stained. GFP-Rad9 was detected by direct fluorescence and TFIIH by immunofluorescence.
To investigate the specific DNA substrate onto which a protein is recruited, different DNA damaging agents can be tested, as well as colocalization with other proteins involved in the DNA damage response. Biochemical experiments indicated that the Rad17-RFC and ATRIP-ATR protein complexes are recruited to DNA damage-induced RPA-coated ssDNA and this localization induces the activation of the downstream kinase Chk1 [4, 51, 52]. Rad9, a component of the 9-1-1 complex, is loaded onto the DNA by Rad17-RFC in response to DNA damage [5, 53]. We observed colocalization of GFP-Rad9 and RPA upon treatment of cells with both UV light and IR, suggesting that Rad9 is recruited to sites of RPA-coated ssDNA (Figure 3(a)). In response to DNA damage, histone H2AX is phosphorylated at Ser139 by ATM/ATR [54]. Although phosphorylation of H2AX (γH2AX) is commonly used to identify DSBs, γH2AX is also induced in response to UV light or treatment with other agents that do not directly induce DSBs, like the replication inhibitor aphidicolin (Figure 3(a)) [55]. We observed colocalization of GFP-Rad9 and γH2AX in all UV-induced foci whereas treatment of cells with aphidicolin or IR did not result in a complete colocalization of the two proteins (Figure 3(a) and data not shown). Close inspection demonstrated that foci induced by aphidicolin either contained both GFP-Rad9 and γH2AX or γH2AX only. Since the phosphorylation of H2AX is triggered in response to a wide variety of DNA lesions, we conclude that GFP-Rad9 is only recruited to a subset of aphidicolin-induced DNA lesions.
Not all proteins accumulate into foci, even though they are recruited to sites of DNA damage. Proteins involved in nucleotide excision repair, for example, do not form foci in response to genotoxic stress. The accumulation of these proteins onto damaged DNA can be visualized by applying local UV damage in the nucleus. By irradiating the cells while covered with an isopore filter, the UV light can only penetrate the pores and as a consequence, only a small part(s) of the nucleus contains damaged DNA [56, 57]. As shown in Figure 3(b), proteins involved in the ATR-mediated checkpoint pathway, such as Rad9, also accumulate onto local UV damage. This method requires fixation of cells and is therefore unsuitable for live cell imaging. The development of laser-induced DNA damage circumvented this problem [26, 36, 58]. Laser-induced DNA damage can be easily combined with live cell imaging and different types of lasers are commonly available [15, 59]. For example, recent studies make use of specific types of sources and lasers, specifically inducing subsets of DNA lesions such as CPDs and 6-4P induced by UV light, and DSBs [14, 36, 56, 60, 61]. Nonetheless caution is required, as often sources and lasers also produce other, unwanted, types of lesions, that may influence protein behaviour adversely [36, 62].
We did not observe the accumulation of the effector kinase Chk1 into nuclear foci in response to DNA damage nor onto locally applied UV-induced DNA damage using a filter (data not shown), in contrast to ATR, TopBP1, 9-1-1 members, and many other checkpoint mediator proteins. In this sense, Chk1 behaves similarly to Chk2, the effector kinase for the cellular response to DSBs [15]. In order to trigger a cell cycle arrest in response to DNA damage, these effector kinases phosphorylate a range of substrates that are present throughout the nucleus. Although both Chk1 and Chk2 do not associate to chromatin after DNA damage, Chk1 associates to chromatin in unperturbed cells and is released from the chromatin in response to DNA damage [35]. In the light of this, the absence of accumulated Chk1 at sites of DNA lesions was to be expected.
3.3. Quantitative GFP-Fusion Protein Dynamics
By measuring the change in mobility of a protein in response to DNA damage a number of parameters can be determined. First, the time the protein spends at the site of a DNA lesion and second, the fraction of the total amount of protein engaged in that process. Fluorescent Redistribution after Photobleaching (FRAP) is applied to study the mobility of GFP-fusion proteins in living cells. In a defined region (strip) of the nucleus, the fluorescence is irreversibly photobleached through brief illumination at high laser intensity (strip-FRAP). Redistribution or recovery of the total fluorescence in the bleached area is then measured in time, indicative of protein mobility [16, 63, 64].
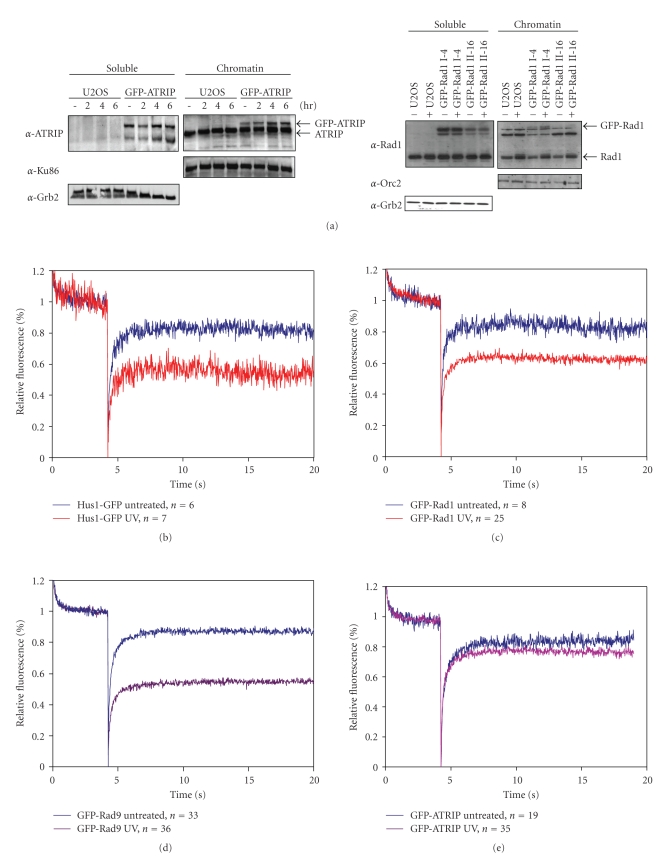
To gain further understanding of how the proteins in the ATR-pathway sense DNA lesions and subsequently respond to trigger the checkpoint-mediated cell cycle arrest, the dynamic behaviour of these proteins was studied. We demonstrated that GFP-Rad1, GFP-ATRIP, and GFP-Rad9 form foci and are recruited to chromatin in response to DNA damage, as shown by chromatin fractionation (Figure 4(a), data not shown) [21]. To address whether these proteins become immobilized at sites of DNA lesions, we determined the change in mobility of GFP-Rad1, GFP-Rad9, Hus1-GFP, and GFP-ATRIP upon UV-irradiation. Importantly, the total amount of GFP-labelled molecules could be different between cells and between GFP-fusion protein cell lines. These two discrepancies could potentially influence photobleaching results since relative immobilization is dependent on the total amount of protein available. To adjust for cell-to-cell differences in a population, we normalized the obtained measurements as described in Section 2. Furthermore, we used saturating amounts of UV-irradiation to be sure that all possible molecules would be engaged (data not shown). Although we did not measure the exact amount of GFP-fusion molecules per cell, western blotting analysis indicates that the different GFP-fusion proteins are expressed to a similar extent (Figure 1(b)).
Figure 4.
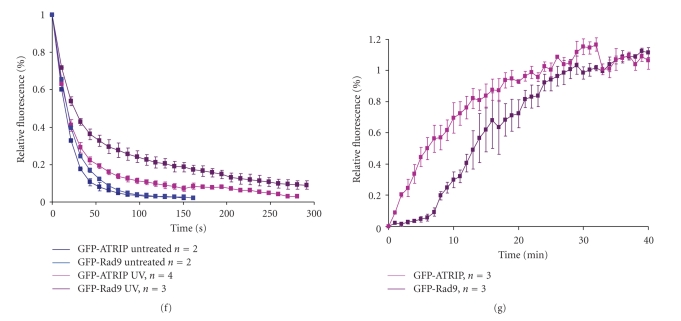
UV-induced changes in mobility of checkpoint proteins. (a) U2OS cells expressing GFP-ATRIP (left panel) and GFP-Rad1 (right panel) were left untreated or treated with UV. Two hours later, soluble and chromatin-bound proteins were isolated and analyzed by western blotting using the indicated antibodies. (b–e) Hus1-GFP (b), GFP-Rad1 (c), GFP-Rad9 (d), and GFP-ATRIP (e) expressing cells were left untreated or treated with UV and analyzed by strip-FRAP (see Section 2 for technical details). (f) U2OS cells expressing GFP-Rad9 or GFP-ATRIP were left untreated or treated with UV. After 1 hour cells were analyzed by FLIP (see Section 2 for technical details). Plotted is the loss of fluorescence during continuous bleaching. (g) U2OS cells expressing GFP-Rad9 or GFP-ATRIP were locally irradiated using a 266 nm UV laser. Plotted is the association of fluorescent signal at the locally damaged area.
In response to UV light, 20%–40% of the 9-1-1 complex proteins become immobilized (Figures 4(b)–4(e)). The GFP-Rad1 and Hus1-GFP FRAP curves show more variability compared to the GFP-Rad9 FRAP curve, which is probably due to the fluorescent variety of the GFP-Rad1 and Hus1-GFP cell lines and the amount of cells that were measured (Figures 4(b)–4(d)). Although the ATRIP-ATR complex is also recruited to sites of DNA damage, relatively less GFP-ATRIP is immobilized (5%–10%) in response to UV damage as compared to either of the proteins of the 9-1-1 complex (Figure 4(e)). These data demonstrate that the 9-1-1 and ATRIP-ATR complexes behave differently after DNA damage and suggest that ATRIP displays a higher turnover at sites of DNA lesions.
To further investigate the possible distinct behaviour between these two putative DNA damage sensor complexes Fluorescent Loss after photobleaching (FLIP) experiments were performed. During FLIP half of the nucleus is constantly bleached and the loss of fluorescence in the unbleached half of the nucleus is measured, representing the rate of dissociation. As shown in Figure 4(f), nondamaged cells expressing either GFP-Rad9 or GFP-ATRIP completely lost fluorescence 150 seconds after start of bleaching (k 1/2~15 seconds). After UV irradiation the dissociation rates were less fast. GFP-ATRIP completely lost fluorescence after 250 seconds (k 1/2 ~ 25 seconds) whereas GFP-Rad9 after more than 300 seconds (k 1/2~ 40 seconds). These FLIP experiments indicate that after DNA damage induction both ATRIP and Rad9 become less mobile, but the release (dissociation) of ATRIP from sites of damage is faster than Rad9. These data are in accordance to the FRAP experiments, that show relatively less immobilization of ATRIP upon UV irradiation as compared to the proteins of the 9-1-1 complex.
The rate at which proteins associate to sites of DNA lesions was addressed by measuring the accumulation of proteins upon local UV laser irradiation. Briefly, a small area inside the nucleus is irradiated with a 266 nm UV laser. GFP-Rad9 and GFP-ATRIP fluorescence in the locally damaged area was measured every 60 seconds until a plateau was reached. GFP-ATRIP quickly started accumulating at the damaged area and reaches a plateau ~20 minutes after damage induction (k 1/2~ 6 minutes) (Figure 4(g)). GFP-Rad9 accumulation in the first 8 minutes is relatively slow, but rises faster thereafter and reaches a plateau after ~30 minutes (k 1/2~ 12 minutes) (Figure 4(g)). These results indicate that ATRIP and Rad9 associate very differently to UV-damaged DNA and suggest that ATRIP is recruited directly to the sites of damage in an exponential manner, following Michaelis-Menten kinetics [65] whereas the accumulation kinetics of Rad9 follow a sigmoidal curve (non-Michaelis-Menten kinetics), representing cooperative binding [66]. Rad17-dependent loading of Rad9 onto the DNA might explain this cooperative binding of GFP-Rad9 after DNA damage induction [67], compared to ATRIP-ATR which binds directly to RPA-coated ssDNA [4].
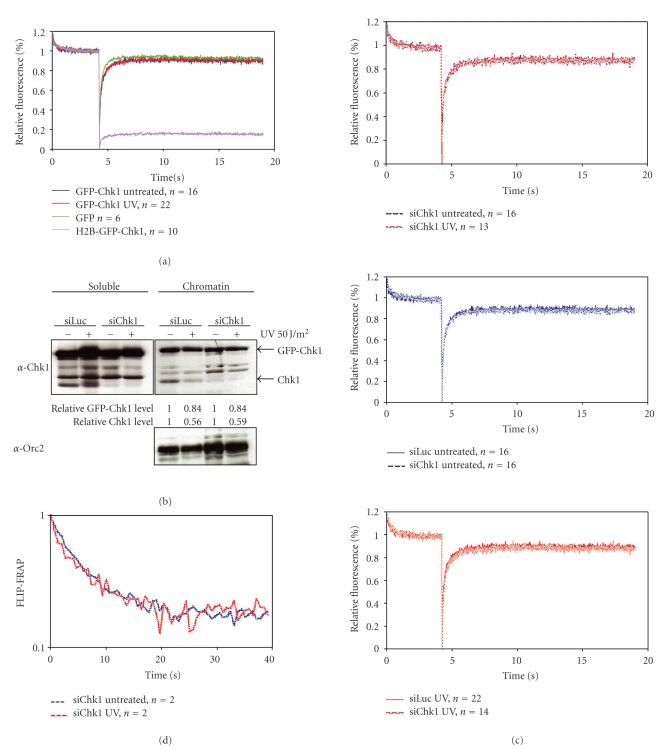
Chk1 is phosphorylated at or near the DNA lesion and thereafter released from the chromatin [35] and consequently, the amount of immobilized GFP-Chk1 measured by photobleaching experiments might become lower after DNA damage induction. To test this hypothesis, we performed strip-FRAP on cells expressing GFP-Chk1 treated with UV light. We compared the GFP-Chk1 mobility with that of free GFP and an artificially immobile form of Chk1, where GFP-Chk1 was fused to histone H2B (H2B-GFP-Chk1) [35]. Upon photobleaching GFP-Chk1 demonstrated a fast recovery of fluorescence both before and after UV damage (Figure 5(a)). The recovery was similar to that of free GFP, arguing against the existence of a stably bound GFP-Chk1 fraction. Since GFP-Chk1 was present in relatively high levels due to overexpression in addition to endogenous Chk1, we reasoned that the amount of Chk1 might be too high to observe any change in protein mobility upon DNA damage induction. Therefore, we lowered the total amount of Chk1 by RNA interference. Cells expressing GFP-Chk1 were transfected with either control luciferase or Chk1 siRNA oligos. Twenty four hours later cells were treated with UV light and chromatin fractions were prepared. As shown in Figure 5(b), both endogenous and GFP-labelled Chkl were released from chromatin upon DNA damage induction in control downregulated cells, although the GFP-tagged version less efficiently than endogenous Chk1 (endogenous 64%, GFP-labelled 16%). Transfection of Chk1 siRNA oligos resulted in downregulation of endogenous Chk1 and did not influence the release of Chk1 from chromatin. Although GFP-Chk1 is less efficiently released from the chromatin than endogenous Chk1, a small but considerable fraction of GFP-Chk1 (~15%) is released in response to DNA damage, which is comparable to the previous experiment without the depletion of endogenous Chk1.
Figure 5.
Mobility of GFP-Chk1 in response to DNA damage. (a) U2OS cells expressing GFP, H2B-GFP-Chk1, or GFP-Chk1 were left untreated or treated with UV and analyzed by strip-FRAP (see Section 2 for technical details). (b) Chromatin fractionation of GFP-Chk1 expressing cells transfected with siRNA oligos directed against luciferase or Chk1 for 24 hours before UV treatment and cell fractionation. Relative Chk1 levels as compared to untreated controls are indicated. Western blot analysis of the fractions was performed using the indicated antibodies. (c) GFP-Chk1 expressing cells were transfected with siRNA oligos against luciferase or Chk1 for 24 hours and left untreated or treated with UV. One hour later, cells were analyzed by strip-FRAP. (d) U2OS cells expressing GFP-Chk1 were transfected with Chk1 siRNA oligos for 24 hours and left untreated or treated with UV and subsequently analyzed by FLIP-FRAP (see Section 2 for technical details).
Next, we lowered the total amount of Chk1 in GFP-Chk1 expressing cells by siRNA-mediated downregulation. Subsequent strip-FRAP analysis indicated that nondamaged GFP-Chk1 expressing cells did not contain more immobile GFP-Chk1 molecules compared to UV-treated cells (Figure 5(c)). Downregulation of Chk1 did not influence the recovery of residual GFP-Chk1 fluorescence in untreated cells and accordingly it did not change GFP-Chk1 mobility after UV-treatment.
The strip-FRAP experiments demonstrated the absence of a stably bound Chk1 pool in these conditions. Nonetheless, these experiments were performed in a relatively short period of time (20 seconds). Since Chk1 released from the chromatin in response to UV-induced DNA damage could be a more gradual and thus time-dependent process, we additionally addressed GFP-Chk1 mobility by determining its residence time on DNA. Chk1 was downregulated in GFP-Chk1 expressing cells as previously described and subsequently the cells were irradiated with UV-light. Thereafter, half of the nucleus was bleached once and redistribution of GFP-Chk1 fluorescence was measured in the bleached and unbleached area over time. The time it takes to reach full fluorescent redistribution determines the proteins residence time. The results shown in Figure 5(d) indicated that GFP-Chk1 reached redistribution after 40 seconds, which is outside the timeframe of our original experiment. However, during the 40 seconds directly after bleaching we did not observe any difference in the mobility using this method. These results further indicate that Chk1 is a highly mobile protein and suggests that it only very transiently interacts with the chromatin.
4. Discussion
The initial response to DNA damage is of crucial importance for cell functioning and viability, since an improper DNA damage response can eventually lead to cancer and other human diseases [1, 68]. In response to UV light, the putative DNA sensors Rad17-RFC and ATRIP-ATR are independently recruited to DNA regions containing RPA-coated ssDNA [4, 52]. Rad17-RFC is responsible for the loading of the 9-1-1 complex onto the DNA [5]. These events are required for the subsequent activation of Chk1, which results in a cell cycle arrest [12]. To gain further insight into the events that take place after a genotoxic insult in living cells, we GFP-labelled several proteins of the ATR pathway and expressed these fusion proteins in human cells. We discussed how to generate and validate stable GFP-fusion cell lines.
The quantitative analysis of GFP-fusion proteins in living cells using video microscopy allowed us to determine the dynamic behaviour of different individual proteins of the pathway in response to DNA damage. With the use of photobleaching techniques (strip-FRAP, FLIP, FLIP-FRAP, local UV laser) we determined multiple parameters of the proteins, including the rate of accumulation onto locally induced UV laser damage, rate of diffusion, immobile fraction, and the average time of immobilization (residence time) before and after global UV-irradiation. These methods are useful tools to investigate the dynamics of proteins involved in the DNA damage response, as shown in this paper, since many of the involved proteins bind DNA or proteins present at the site of damage and become immobilized to a certain extent.
GFP-Rad9, GFP-Rad1, Hus1-GFP, and GFP-ATRIP accumulate into nuclear foci in response to DNA damage. However, a relatively larger fraction of the 9-1-1 complex proteins become immobilized in response to UV as compared to ATRIP. Biochemical experiments show that the 9-1-1 complex is loaded onto the 5′ ssDNA/dsDNA by Rad17-RFC, therefore, its association to DNA lesions might take some time and is the result of cooperative binding [51]. In contrast, ATRIP interacts directly with RPA-coated ssDNA, explaining its exponential binding behaviour [4]. The results obtained by FLIP suggest that a loaded 9-1-1 complex cannot easily dissociate from the DNA. This dissociation might be slower because the 9-1-1 complex forms a ring around the DNA. It, therefore, has to either slide off a DNA end or actively dissociate, which likely is a more time-consuming process [69]. In addition, the FRAP experiments show that 20%–40% of the 9-1-1 complex molecules become immobile whereas only 5%–10% of the ATRIP molecules is immobilized after DNA damage. These results also indicate a relatively more stable association of the 9-1-1 complex with damaged chromatin which is consistent with the FLIP results.
All together, the putative DNA damage sensors Rad9-Rad1-Hus1 and ATRIP-ATR show a different dynamic behaviour directly after induction and during the presence of UV-induced DNA damage. Nonetheless, both GFP-Rad9 and GFP-ATRIP molecules are exchanging rapidly in UV-induced foci since all the mobility measurements performed on both GFP-Rad9 and GFP-ATRIP DNA damage-induced-foci do not indicate a stable immobilized fraction, suggesting that these foci are not static but highly dynamic structures.
Recruitment of the ATRIP-ATR heterodimer to sites of DNA damage results in ATR kinase activation which leads to the phosphorylation and subsequent activation of the Chk1 kinase [70, 71]. To investigate the spatio and temporal behaviour of downstream effector Chk1 in living cells, we studied the dynamic behaviour of GFP-Chk1 in response to DNA damage. In vivo imaging experiments confirm the absence of a stable chromatin-bound Chk1 fraction. The data presented here signify that Chk1 is a very mobile protein both in the absence or presence of DNA damage. The observed chromatin binding of Chk1 that is observed in biochemical experiments is not indicative of a stably bound Chk1 chromatin fraction [35] (Figure 5(b)). This high nuclear mobility of Chk1 was hypothesized to ensure the access of additional Chk1 molecules to ATR and the transmission of the damage signal throughout the nucleus [15, 35, 72].
Measuring protein mobility is not always as straightforward as it seems. Problems that are commonly faced when performing these types of experiments are monitor bleaching, influx of proteins from the cytoplasm, and blinking. Monitor bleaching is caused by observing the cells before, during, and after photobleaching or time lapse experiments. By using low amounts of laser power this can be avoided, although with low fluorescent protein levels this could be problematic. Lowering the laser power could result in a low resolution, which in turn results in a large variability between measurements. Monitor bleaching can be corrected for by measuring the amount of bleaching in an unbleached area of the cell, but this only applies for freely diffusion proteins [63, 73]. Another potential problem is the presence of a cytoplasmic protein fraction that could influence nuclear measurements due through the influx of fluorescence from the cytoplasm. Before the start of the actual measurement, the cytoplasmic fraction can be bleached, thereby eliminating this potential influx. Blinking is the property of fluorescent molecules to be reactivated, which can also influence photobleaching results. By keeping experimental conditions similar in all conditions the influence of blinking is constant in all measurements and, therefore, insignificant. When comparing results obtained from different fluorescent variants, on the other hand, it needs to be taken into account that the blinking properties are different and, therefore, the amount of blinking needs to be determined for every variant and corrected for in the final analysis [63, 73].
To overcome the problem of GFP-fusion protein overexpression and the (competitive) presence of the endogenous counterpart, a GFP knock-in strategy using mice can be utilized [74] (Jeroen Essers, personal communication). This method places the GFP tag before the endogenous gene of interest in the mice genome, thereby being regulated by the endogenous promoter. As a result the expression levels of the GFP-fusion protein are identical to nontargeted protein in wild type cells, resulting in better quantitative measurements. The behaviour of proteins can additionally be studied in different tissues and cell types under physiological conditions [75]. Performing a similar approach for proteins involved in the ATR-Chk1 pathway may result in a more precise determination of the dynamic behaviour of this DNA damage checkpoint pathway. The ATR gene may act as a good candidate to start a GFP knock in approach, since it localizes to sites of damage and thereby becomes immobilized. Furthermore, ATR interacts with and phosphorylates a number of key target proteins.
Conclusively, we show that the GFP-labelling of ATRIP, Chk1, and the 9-1-1 complex proteins results in functional equivalents that can be studied by live cell video microscopy and quantitative fluorescence photobleaching. We compared the mobility changes of GFP-ATRIP, GFP-Chk1, GFP-Rad9, GFP-Rad1, and Hus1-GFP after UV-irradiation and collectively the results indicate a distinct dynamic behaviour between some of the studied proteins. Whereas effector kinase Chk1 is highly mobile and does not immobilize upon DNA damage, Rad9 and ATRIP stably associate to sites of DNA lesions. ATRIP directly binds to DNA lesions but more transiently than Rad9, which in contrast displays a more stable association with damaged chromatin. Together these data demonstrate the importance of spatio and temporal protein regulation in the DNA damage-induced ATR-Chk1 pathway.
Acknowledgments
The authors thank Raimundo Freire for sharing reagents and technical support and Philip Reaper for reagents. This work was supported by the Association for International Cancer Research (AICR 05-005), the Dutch Cancer Society (EMCR 2005-3412), and the Ramón y Cajal Program (Ministerio de Ciencia e Innovación) to V. A. J. Smits and the integrated project 512113 from the European Commission and the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NWO) to R. Kanaar.
References
- 1.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Reviews Cancer. 2003;3(3):155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 2.Huen MSY, Sy SMH, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nature Reviews Molecular Cell Biology. 2010;11(2):138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huen MSY, Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends in Biochemical Sciences. 2010;35(2):101–108. doi: 10.1016/j.tibs.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 5.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes and Development. 2002;16(2):198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294(5543):867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 7.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes and Development. 2001;15(21):2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. Journal of Biological Chemistry. 2007;282(38):28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 9.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K-I, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes and Development. 2007;21(12):1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majka J, Niedziela-Majka A, Burgers PJ. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Molecular Cell. 2006;24(6):891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou B-BS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 12.Shiotani B, Zou L. ATR signaling at a glance. Journal of Cell Science. 2009;122(3):301–304. doi: 10.1242/jcs.035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper JW, Elledge SJ. The DNA damage response: ten years after. Molecular Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Bekker-Jensen S, Lukas C, Kitagawa R, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. Journal of Cell Biology. 2006;173(2):195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nature Cell Biology. 2003;5(3):255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 16.Essers J, Houtsmuller AB, Kanaar R. Analysis of DNA recombination and repair proteins in living cells by photobleaching microscopy. Methods in Enzymology. 2006;408:463–485. doi: 10.1016/S0076-6879(06)08029-3. [DOI] [PubMed] [Google Scholar]
- 17.Essers J, Houtsmuller AB, Van Veelen L, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO Journal. 2002;21(8):2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Veelen LR, Cervelli T, Van De Rakt MW, Theil AF, Essers J, Kanaar R. Analysis of ionizing radiation-induced foci of DNA damage repair proteins. Mutation Research. 2005;574(1-2):22–33. doi: 10.1016/j.mrfmmm.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300(5616):87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 20.Easwaran HP, Leonhardt H, Cardoso MC. Cell cycle markers for live cell analyses. Cell Cycle. 2005;4(3):453–455. doi: 10.4161/cc.4.3.1525. [DOI] [PubMed] [Google Scholar]
- 21.Warmerdam DO, Freire R, Kanaar R, Smits VAJ. Cell cycle-dependent processing of DNA lesions controls localization of Rad9 to sites of genotoxic stress. Cell Cycle. 2009;8(11):1765–1774. doi: 10.4161/cc.8.11.8721. [DOI] [PubMed] [Google Scholar]
- 22.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Molecular Cell. 2008;30(3):267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair. 2004;3(8-9):997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320(5882):1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JHJ, Vermeulen W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 1999;284(5416):958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- 26.Zotter A, Luijsterburg MS, Warmerdam DO, et al. Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced DNA damage depends on functional TFIIH. Molecular and Cellular Biology. 2006;26(23):8868–8879. doi: 10.1128/MCB.00695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luijsterburg MS, Goedhart J, Moser J, et al. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. Journal of Cell Science. 2007;120(15):2706–2716. doi: 10.1242/jcs.008367. [DOI] [PubMed] [Google Scholar]
- 28.Giglia-Mari G, Coin F, Ranish JA, et al. A new, tenth subunit TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nature Genetics. 2004;36(7):714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 29.Giglia-Mari G, Miquel C, Theil AF, et al. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biology. 2006;4(6, article e156) doi: 10.1371/journal.pbio.0040156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toueille M, El-Andaloussi N, Fouin I, et al. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase β and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Research. 2004;32(11):3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rendtlew Danielson JM, Larsen DH, Schou KB, et al. HCLK2 is required for activity of the DNA damage response kinase ATR. Journal of Biological Chemistry. 2009;284(7):4140–4147. doi: 10.1074/jbc.M808174200. [DOI] [PubMed] [Google Scholar]
- 32.Jones GG, Reaper PM, Pettitt AR, Sherrington PD. The ATR-p53 pathway is suppressed in noncycling normal and malignant lymphocytes. Oncogene. 2004;23(10):1911–1921. doi: 10.1038/sj.onc.1207318. [DOI] [PubMed] [Google Scholar]
- 33.Medhurst AL, Warmerdam DO, Akerman I, et al. ATR and Rad17 collaborate in modulating Rad9 localisation at sites of DNA damage. Journal of Cell Science. 2008;121(23):3933–3940. doi: 10.1242/jcs.033688. [DOI] [PubMed] [Google Scholar]
- 34.Mendez J, Stillman B. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Molecular and Cellular Biology. 2000;20(22):8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smits VAJ, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Current Biology. 2006;16(2):150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 36.Dinant C, de Jager M, Essers J, et al. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. Journal of Cell Science. 2007;120(15):2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]
- 37.Essers J, Theil AF, Baldeyron C, et al. Nuclear dynamics of PCNA in DNA replication and repair. Molecular and Cellular Biology. 2005;25(21):9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsien RY. The green fluorescent protein. Annual Review of Biochemistry. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 39.Tsien RY, Miyawaki A. Seeing the machinery of live cells. Science. 1998;280(5371):1954–1955. doi: 10.1126/science.280.5371.1954. [DOI] [PubMed] [Google Scholar]
- 40.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312(5771):217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 41.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373(6516):663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 42.Prescott M, Nowakowski S, Nagley P, Devenish RJ. The length of polypeptide linker affects the stability of green fluorescent protein fusion proteins. Analytical Biochemistry. 1999;273(2):305–307. doi: 10.1006/abio.1999.4235. [DOI] [PubMed] [Google Scholar]
- 43.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nature Genetics. 2003;33(4):497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 44.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes and Development. 2000;14(15):1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss RS, Leder P, Vaziri C. Critical role for mouse Hus1 in an S-phase DNA damage cell cycle checkpoint. Molecular and Cellular Biology. 2003;23(3):791–803. doi: 10.1128/MCB.23.3.791-803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopkins KM, Auerbach W, Wang XY, et al. Deletion of mouse Rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Molecular and Cellular Biology. 2004;24(16):7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zachos G, Rainey MD, Gillespie DAF. Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Molecular and Cellular Biology. 2005;25(2):563–574. doi: 10.1128/MCB.25.2.563-574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes and Development. 2004;18(12):1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. Journal of Cell Biology. 2005;170(2):201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonner JA, Lawrence TS. Doxorubicin decreases the repair of radiation-induced DNA damage. International Journal of Radiation Biology. 1990;57(1):55–64. doi: 10.1080/09553009014550341. [DOI] [PubMed] [Google Scholar]
- 51.Majka J, Binz SK, Wold MS, Burgers PMJ. Replication protein a directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. Journal of Biological Chemistry. 2006;281(38):27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 52.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majka J, Burgers PM. Function of Rad17/Mec3/Ddc1 and its partial complexes in the DNA damage checkpoint. DNA Repair. 2005;4(10):1189–1194. doi: 10.1016/j.dnarep.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 54.Modesti M, Kanaar R. Dna repair: spot(light)s on chromatin. Current Biology. 2001;11(6):R229–R232. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- 55.Marteijn JA, Bekker-Jensen S, Mailand N, et al. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. The Journal of Cell Biology. 2009;186(6):835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moné MJ, Volker M, Nikaido O, et al. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Reports. 2001;2(11):1013–1017. doi: 10.1093/embo-reports/kve224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Volker M, Moné MJ, Karmakar P, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Molecular Cell. 2001;8(1):213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 58.Moné MJ, Bernas T, Dinant C, et al. In vivo dynamics of chromatin-associated complex formation in mammalian nucleotide excision repair. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(45):15933–15937. doi: 10.1073/pnas.0403664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mari P-O, Florea BI, Persengiev SP, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(49):18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aten JA, Stap J, Krawczyk PM, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303(5654):92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 61.Stap J, Krawczyk PM, Van Oven CH, et al. Induction of linear tracks of DNA double-strand breaks by α-particle irradiation of cells. Nature Methods. 2008;5(3):261–266. doi: 10.1038/nmeth.f.206. [DOI] [PubMed] [Google Scholar]
- 62.Williams ES, Stap J, Essers J, et al. DNA double-strand breaks are not sufficient to initiate recruitment of TRF2. Nature Genetics. 2007;39(6):696–698. doi: 10.1038/ng0607-696. [DOI] [PubMed] [Google Scholar]
- 63.Houtsmuller AB. Fluorescence recovery after photobleaching: application to nuclear proteins. Advances in Biochemical Engineering/Biotechnology. 2005;95:177–199. doi: 10.1007/b102214. [DOI] [PubMed] [Google Scholar]
- 64.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochemistry and Cell Biology. 2001;115(1):13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 65.Stroppolo ME, Falconi M, Caccuri AM, Desideri A. Superefficient enzymes. Cellular and Molecular Life Sciences. 2001;58(10):1451–1460. doi: 10.1007/PL00000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ricard J, Cornish-Bowden A. Co-operative and allosteric enzymes: 20 years on. European Journal of Biochemistry. 1987;166(2):255–272. doi: 10.1111/j.1432-1033.1987.tb13510.x. [DOI] [PubMed] [Google Scholar]
- 67.Zou L, Elledge SJ. Sensing and signaling DNA damage: roles of Rad17 and Rad9 complexes in the cellular response to DNA damage. Harvey Lectures. 2001;97:1–15. [PubMed] [Google Scholar]
- 68.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 69.Doré AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex—implications for clamp loading and regulation. Molecular Cell. 2009;34(6):735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 70.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes and Development. 2001;15(17):2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 71.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124(5):943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 72.Smits VAJ. Spreading the signal: dissociation of Chk1 from chromatin. Cell Cycle. 2006;5(10):1039–1043. doi: 10.4161/cc.5.10.2761. [DOI] [PubMed] [Google Scholar]
- 73.van Royen ME, Farla P, Mattern KA, Geverts B, Trapman J, Houtsmuller AB. Fluorescence recovery after photobleaching (FRAP) to study nuclear protein dynamics in living cells. Methods in Molecular Biology. 2009;464:363–385. doi: 10.1007/978-1-60327-461-6_20. [DOI] [PubMed] [Google Scholar]
- 74.Giglia-Mari G, Theil AF, Mari P-O, et al. Differentiation driven changes in the dynamic organization of basal transcription initiation. PLoS Biology. 2009;7(10, article e1000220) doi: 10.1371/journal.pbio.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan TLR, Essers J, Citterio E, et al. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Current Biology. 1999;9(6):325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]