Abstract
PPARα is one of three members of the soluble nuclear receptor family called peroxisome proliferator-activated receptor (PPAR). It is a sensor for changes in levels of fatty acids and their derivatives that responds to ligand binding with PPAR target gene transcription, inasmuch as it can influence physiological homeostasis, including lipid and carbohydrate metabolism in various tissues. In this paper we summarize the involvement of PPARα in the metabolically active tissues liver and skeletal muscle and provide an overview of the risks and benefits of ligand activation of PPARα, with particular consideration to interspecies differences.
1. Introduction
Dietary fatty acids (FAs) are not only important for membrane structures and in signalling processes, but also have the ability to influence gene expression by binding to specific transcription factors [1]. One receptor family that acts as mediators to influence transcription according to nutritional state is the peroxisome proliferator-activated receptor (PPAR) family. There are three isoforms of PPAR receptors that have specific, but also overlapping target genes: α, β/δ, and γ [2–4]. Early on PPAR activity was thought to mainly influence lipid metabolism, inflammation, and glucose homeostasis. Later it became clear that PPARs also play a role in modulating the processes of cell proliferation and differentiation, apoptosis, and aging [5–8]. The receptors show a nuclear localization in the form of a heterodimer with the retinoid X receptor (RXR). A ligand activated PPARα-RXR heterodimer regulates the transcription of genes by binding to their peroxisome proliferator response elements (PPREs), a process called “transactivation” [9–11]. Besides, a mechanism based on “transrepression” has been described and is reviewed in [12]. The anti-inflammatory actions of PPARα ligands are mostly thought to be based on “transrepression” by the negative interference of PPARα with other transcription factor pathways [13, 14].
Here we focus on the first identified PPAR receptor, PPARα [15], and its activation in different tissues and physiological states in humans and mice. It is expressed at elevated levels in tissues with high metabolic rates, such as the liver, heart, skeletal muscle, kidney, and also in the intestine [12, 16]. Additionally, it is present in cells of the immune system (e.g., macrophages, monocytes, and lymphocytes) [17–19]. The receptor has a central role in fatty acid oxidation, lipid and lipoprotein metabolism, inflammatory responses, and oxidative stress. Its position in the centre of energy balance, lipid metabolism, and inflammation makes it an important factor in the development of obesity-related diseases, and therefore, presents a possible target to influence metabolic disorders. Ligands include saturated and unsaturated FA and their derivatives, hypolipidemic fibrates (ciprofibrate, clofibrate, fenofibrate, and gemfibrozil), and modified fatty acids (e.g., tetradecylthioacetic acid, TTA), as well as xenobiotics [20–22]. In particular during fasting, when free FAs are released into the blood, endogenous lipid-activation is of importance. The importance of PPARα in the cellular metabolic response to fasting was clearly shown in PPARα-null mice [23]. Whereas under normal conditions, these mice do not display a strong phenotype, the absence of PPARα causes lipid accumulation in liver and heart, hypoglycemia, hypothermia, ketonuria, and elevated free fatty acids during fasting ultimately leading to premature death [23]. In contrast, wildtype mice adapt to fasting by induction of hepatic and cardiac PPARα target genes that results in increased FA uptake and oxidation [24].
A great number of animal studies have demonstrated beneficial effects of specific PPAR activation in counteracting metabolic disorders. An increasing number of human studies supports the findings obtained in animal studies. When it comes to PPARα activation, however, it has become clear that not all results obtained in mice can be extrapolated to humans and caution is warranted in predicting tissue-specific effects.
This paper will focus on the tissues liver and skeletal muscle exploring tissue-specific effects of PPARα activation and stress the differences of human- and mouse-based studies.
2. PPARα in Liver
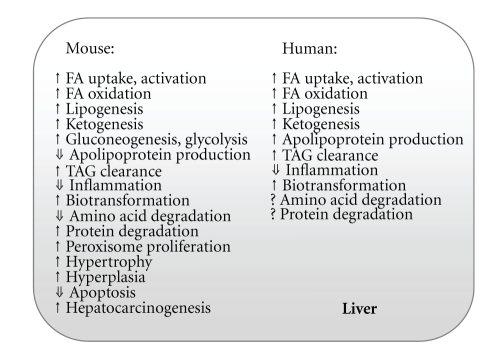
There are substantial differences between human and mouse target gene expression in terms of the effect of PPARα activation in the liver (Figure 1). Overall, the effect of activation by the PPARα agonist WY14643 is more prominent in mice than in humans [25]. In primary hepatocytes from mice and humans treated with WY14643, only a few target genes were affected similarly in the two species. However, both species share multiple changed gene ontology classes, including lipid metabolism. Individual PPARα regulation was observed for enzymes involved in biotransformation (chemical alterations of compounds in the body), as well as apolipoprotein and bile acid synthesis in human hepatocytes, and glucose homeostasis in mouse hepatocytes [25]. It was proposed earlier that the response might be dampened by quantitative differences of PPARα expression or different splice forms of PPARα. Indeed, there exist two splice variants of PPARα giving rise to an active and inactive receptor in humans [26]. To compare PPARα expression levels between human and mouse liver is, however, difficult due to daily variations [27] and differing reports have been published. Some reports show lower PPARα expression levels in human than in rodent liver [28–30], while another shows comparable expression levels between the two species [25].
Figure 1.
Examples of the multiple metabolic effects of PPARα activation in mouse or human liver. FA, fatty acid; TAG, triacylglycerol.
One of the main pathways involving PPARα regulation in mice and humans includes FA metabolism. In mice, PPARα activation is important for FA metabolism through the induction of genes coding for the fatty acid transporter CD36 [31] and the FA binding protein 1 (FABP1) that brings the FAs from the plasma membrane to the nucleus [32]. Another PPARα target gene is carnitine palmitoyl transferase 1 (Cpt1), that codes for a protein important for FA transport into mitochondria [25].Whereas CPT1 is localized to the outer membrane, CPT2, that is also regulated by PPARα, is found in the inner mitochondrial membrane. It converts acyl-carnitine to acyl-CoA and is strongly upregulated by PPARα agonists [33]. Most of the genes of FA metabolism are regulated by PPARα in both humans and mice, however Cd36 is an example of species-specific induction in mice [25].
Genes encoding for mitochondrial proteins of the β-oxidation pathway are induced by PPARα activation, such as acyl-CoA synthetase (Acs) coding for an enzyme responsible for activation of FA to their fatty acyl-CoA derivatives. Also genes of the short-, medium-, long- and very-long-chain acyl-CoA dehydrogenases (Acad -s, -m, -l, -vl) coding for proteins that catalyze the first step in FA oxidation in a chain length-specific manner, are under the control of PPARα. In addition, the expression of the gene encoding the enzyme acetyl-CoA acyltransferase 2 (ACAA2) involved in the final step of β-oxidation, is PPARα dependent. Furthermore, hepatic carnitine synthesis is enhanced by PPARα activation in mice [34, 35]. Carnitine is a conditionally essential nutrient that plays an important role in mitochondrial long-chain FA import for β-oxidation [36]. In PPARα-null mice, free carnitine levels were drastically suppressed in plasma and several tissues including liver, the primary site of carnitine biosynthesis. This was consistent with reduced hepatic expression of the genes involved in carnitine biosynthesis (Bbox1) and transport (Octn2) [37]. In an earlier study, Van Vlies and colleagues established a fasting-induced elevation of these genes that is PPARα-dependent [38]. Both studies point to an essential position for PPARα in carnitine metabolism in mice [37, 38]. No similar indications of PPARα-induced carnitine synthesis have been described in humans. However, pigs that also are a nonproliferative species and are considered similar to humans due to their metabolic features, show an increased carnitine production upon fasting [39]. It is therefore likely that also humans will prove to have a similar response.
Peroxisomal fatty acid oxidation is important for the partial oxidation of long, very long, and branched FAs. The first characterized PPARα target gene, acyl-CoA oxidase 1 (Acox1) encodes the rate-limiting enzyme of this process [40]. After ACOX1 has introduced a double bond to generate enoyl-CoA and H2O2, the bifunctional protein/enoyl-CoA hydratase (BIEN), that carries two enzymatic activities, performs the second step of β-oxidation resulting in 3-ketoacyl-CoA. 3-ketoacyl-CoA is then cleaved by acetyl-CoA acyltransferase 1 (ACAA1) to produce acetyl-CoA [41]. All the above-mentioned genes are under the regulation of PPARα in mice.
In addition to mitochondrial and peroxisomal β-oxidation, ω-hydroxylation occurs in smooth endoplasmic reticulum. In both mice and humans, this process is upregulated by the effect of PPARα on expression of cytochrome P450 4A11 (CYP4A11) [25, 42–44]. The hepatic cytochrome P450 4A11 catalyzes ω-hydroxylation of medium and long-chain FAs. Subsequently cytosolic dehydrogenases convert them to dicarboxylic acids, which can be further processed by peroxisomal β-oxidation. Human PPARα also is a transcriptional regulator of FA oxidation in the different organelles, but shows overlap with mice rather on the pathway than on the gene level [25]. To conclude, PPARα regulates enzymes important for uptake, traffic to final destination, activation, and oxidation of FAs in the three organelles mitochondria, peroxisomes, and microsomes in both mice and humans.
Paradoxically, at the same time as PPARα activation leads to an increase in FA oxidation, it also augments FA synthesis by affecting gene expression levels of several enzymes involved in lipogenesis. In mice, PPARα stimulates the conversion of malate into pyruvate to generate NADPH for lipogenesis by upregulating the expression of malic enzyme (ME1) [45]. Besides, the ∆5, ∆6, and ∆9 desaturases, rate-limiting enzymes in the synthesis of polyunsaturated FAs (PUFAs) from saturated FAs, are found in increased amounts after PPARα activation [46–48]. The induction of desaturases could help to ensure that there are always enough PUFAs for their diverse functions, including being effective PPARα agonists as proposed by others [46]. Likewise, PPARα activation in human hepatocytes induces the expression of several target genes involved in FA synthesis [25].
Other crucial processes requiring PPARα activation are lipoprotein synthesis and assembly. The impact of PPARα agonist on lipoprotein gene expression in humans or mice is distinct. The use of fibrates in humans leads to reduced plasma triacylglycerol (TAG) levels and increased high-density lipoprotein (HDL) cholesterol levels. In mice, plasma TAG as well as HDL levels are lowered. The liver, besides the intestine, determines the amount of HDL in plasma by regulating HDL synthesis and catabolism. The reason for the species-specific opposite effect of PPARα activation on HDL levels is probably increased production levels of apolipoprotein A-I (APOA1) and APOA2 in humans [49, 50] and suppressed (APOA1) or unchanged (APOA2) expression in mice [51]. These apolipoproteins are part of HDL cholesterol and are crucial for reverse cholesterol transport from peripheral cells to the liver, where excess cholesterol can be eliminated into the bile [52]. The liver is also the place where very low-density lipoprotein (VLDL) particles are assembled and then secreted into the plasma. The VLDL amount in peripheral cells is influenced by lipoprotein lipase (LPL). The hepatic expression of this hydrolase, which mediates VLDL triglyceride lipolysis, is upregulated by PPARα [53]. Moreover, its activity is stimulated by APOA5 and inhibited by APOC3. Activation of PPARα increases APOA5 [54–56] and decreases APOC3 [57] transcription, resulting in a plasma TAG lowering effect, thereby, together with increased HDL concentrations, reducing the risk for atherosclerosis in humans [58].
The removal of excess cholesterol from the body is via the bile, a fluid produced in the liver, stored in the gall bladder, and secreted into the small intestine. Cholesterol is eliminated either intact or as bile acids that are steroid acids made from cholesterol. In humans, the two main bile acids synthesized in the liver, are chenodeoxycholic acid (CDCA) and cholic acid (CA) [59, 60]. Due to their amphipathic character they aid in the small intestine for the digestion and absorption of dietary lipids. There is controversy in the literature regarding the regulation of the rate-limiting enzyme in hepatic bile acid synthesis, called cholesterol 7α-hydroxylase (CYP7A1). Some reports suggest a transcriptional upregulation of Cyp7a1 upon PPARα activation in mice [61, 62]. In particular, the upregulation of Cyp7a1 under fasting conditions and the downregulation of this enzyme in PPARα-null mice corroborate a PPARα regulatory involvement and suggest increased expression upon fasting-induced PPARα activation [62]. Other studies support a downregulation of this endoplasmic reticulum enzyme upon induction with PPARα agonists in both humans and rodents [63–67]. This could be a potential risk for gallstone formation, if in humans receiving treatment with fibrates, bile acid synthesis is decreased over a longer period of time by a hepatic decrease of CYP7A1 activity. On the other hand, gene expression of sterol 12α-hydroxylase (Cyp8b1), an enzyme involved in CA synthesis, is increased under fasting and also with ligand-induced PPARα-activation in both rodents and humans [62, 67, 68]. This protein of the cytochrome P450 family controls the balance between CA and CDCA levels. Upon Cyp8b1 induction, higher CA concentrations positively influence the bile acid composition by increasing cholesterol solubility.
Important under conditions of extended fasting is the process called ketogenesis. In mice and humans, the production of ketone bodies is under the control of PPARα that upregulates the gene expression of mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase (Hmgcs2), coding for the rate-limiting enzyme of ketogenesis [25, 69, 70]. Of particular importance in regulating ketogenesis, in addition to FA oxidation, TAG clearance, and de novo lipogenesis is the ‘hormone-like' fibroblast growth factor 21 (FGF21) [71–73]. Its hepatic expression is PPARα-dependent and is induced by fasting, a ketogenic diet, and WY14643 [25, 71, 74, 75]. FGF21 positively influences lipid and glucose metabolism, in addition to insulin sensitivity in animals [76].
Hepatic gluconeogenesis is also regulated during fasting, when the liver changes from glucose uptake and glycogen synthesis to glucose production. The chain of reactions converting glycerol, lactate, or glucogenic amino acids to glucose involves the two rate-limiting enzymes, phosphoenol-pyruvate carboxykinase (PEPCK) and pyruvate carboxylase (PYC). Of these two genes, only the promoter for Pepck was found to have a functional PPRE in mice [77]. The induction of other enzymes in this pathway is PPARα-dependent, such as glycerol-3-phosphate dehydrogenase (GPDH) and glycerol kinase (GK), as well as the aquaporins (AQP) 3 and 9 that act as liver glycerol import channels [78]. The observation that PPARα-null mice manifest lower fed and fasted glucose levels supports an involvement of PPARα in hepatic glucose production [77]. However, another report proposes as a reason for fasting hypoglycemia, the preferential channelling of glucose-6-phosphate to hepatic glycogen stores and shows unchanged glucose 6-phosphate synthesis in PPARα-null mice [79]. The pathway glycolysis/gluconeogenesis is specifically affected by PPARα activation in mice and shows no response in human primary hepatocytes [25].
The enzyme glyoxylate reductase/hydroxypyruvate reductase (GRHPR) is important in the channelling of carbons from the glyoxylate cycle into gluconeogenesis or into the urea cycle depending on the body energy demands. In mice, PPARα activation (e.g., in the fasted state) is crucial in inducing transcriptional activation of Grhpr, thereby favouring a conversion of hydroxypyruvate to D-glycerate, a substrate needed in glucose synthesis [80]. In humans however, GRHPR expression was shown to be PPARα-independent due to promoter reorganisation during primate evolution. Moreover, alanine:glyoxylate aminotransferase (AGT), an enzyme of the glyoxylate cycle with two enzymatic activities is positively regulated by PPARα [80]. Its transaminase activity leads to the production of glycine and hydroxypyruvate.
Beyond the transcriptional activation of genes involved in lipid and glucose metabolism, the PPARα agonist WY14643 affects amino acid metabolism in rodents [81, 82]. The metabolic consequences include alterations in plasma amino acid levels. Whereas branched-chain amino acid amounts showed no change upon PPARα activation with WY14643, a significant increase in various glucogenic and some ketogenic amino acids was detected in rats [82]. Only one amino acid was lowered, namely arginine, a conditionally nonessential amino acid made in the urea cycle. mRNA levels of enzymes involved in the conversion of citrulline to arginine in the kidney are unknown, but hepatic levels of argininosuccinate synthetase (Ass) and argininosuccinate lyase (Asl) show a decrease [81, 82]. The exact mechanism of PPARα regulation of amino acid metabolism is unknown but certain genes involved in the regulation of amino acid degradation have also been shown to be negatively regulated, with the exclusion of Grhpr and arginase (Arg1) [81, 82]. The decreased amino acid degradation upon WY14643 treatment is accompanied by an increase in protein degradation. Some possible explanations for the observed amino acid mobilization upon PPARα induction are give in [82] and might be due to increased hepatic growth. The current findings are restricted to rodents and it is unclear at present if the situation is similar in humans that show no liver enlargement. One study points to a different situation in humans and describes increased plasma arginine levels after fenofibrate treatment of hypertriglyceridemic men [83]. The findings in rodents are limited to WY14643 treatment and it remains to be shown if they are of general character for PPARα ligands. The clofibrate-induced increased oxidation of branched-chain amino acids seems to be due to its direct inhibitory actions on branched-chain α-keto acid dehydrogenase kinase (BCKDK) that regulates the key enzyme of this process, and not due to effects mediated through PPARα activation [84].
Additionally, in mice, PPARα activation inhibits inflammatory gene expression by downregulation of acute phase proteins such as C-reactive protein (CRP), fibrinogen, and serum amyloid A (SAA) resulting in reduced hepatic inflammation and risk for cardiovascular disease and cancer [85]. Likewise in humans, there is a similar downregulation of plasma acute phase proteins after fenofibrate treatment [86]. Recently, it was demonstrated that the expression of the transcription factor CREBH that is exclusively found in the liver, is regulated by PPARα in both mice and humans [25]. It plays an important role in the activation of the acute inflammatory response and is also a regulator of hepatic gluconeogenesis [87, 88].
Described in mice is the reduced risk of liver damage by chemical-induced stress. Exposure to hepatotoxic agents like the environmental pollutant carbon tetrachloride (CCl4) induces reversible liver damage [89]. The underlying reason is a decreased resistance to oxidative stress that leads to lipid peroxidation, altered calcium homeostasis, and membrane damage. Stimulated mRNA expression of uncoupling protein 2 (Ucp2) by PPARα in rodents results in uncoupling of the proton gradient across the inner mitochondrial membrane and a downregulation of reactive oxygen species (ROS) induced by CCl4 metabolites [90, 91]. In addition, PPARα helps to protect from chemical-induced oxidative stress by upregulating genes of the chaperone family and of the proteasome, thereby influencing protein folding and degradation of harmed proteins in mice [92]. Furthermore, the observation that PPARα-null mice demonstrate decreased longevity, where stress response genes are of importance, and that PPARα expression decreases with age, suggests an involvement of PPARα in this process [7].
In rodents, long-term administration of PPARα leads to increased peroxisome proliferation, in addition to hepatic hypertrophy and hyperplasia that will ultimately result in liver tumors [93–98]. The carcinogenic response is based on enhanced cell replication that might increase the risk for DNA damage and altered oncogene and tumor suppressor gene expressions. Moreover, there is evidence for suppressed apoptosis in liver cells, a process important for the removal of damaged cells [99–102]. There is also a close relationship of PPARα-induced cancer formation with increased production of ROS due to peroxisome proliferation that might contribute to DNA damage [103].
Shah and colleagues have proposed changed hepatic microRNA (miR) expression via PPARα-regulation as the reason for liver cancer formation [104]. miRs are 21–23 nucleotide long sequences that are suggested to regulate the expression of up to 30% of all genes [105, 106]. Experimental evidence pointed to PPARα-involvement in several changed miR levels, in particular in the downregulation of miR let-7c by an as yet unidentified mechanism [104]. Let-7c controls c-Myc protein levels, a transcription factor regulating target genes involved in cell proliferation. Downregulation of let-7c stabilizes c-Myc mRNA leading to the expression of c-Myc target genes. This could be a reason for enhanced hepatocyte proliferation, that together with the induction of oxidative stress might lead to hepatocarcinogenesis in rodents. Induction of hepatocarcinogenesis seems to be restricted to rodents and is not documented in humans (extensively reviewed in [107]). Cancer formation after PPARα activation in tissues other than the liver has been described in rats and includes testicular (Ledig cell) and pancreatic acinar cell tumors [108]. However, if these findings are of significance for humans requires further in-depth risk assessments.
In summary, the hepatic response to PPARα activation is essential under fasting conditions. PPARα activation by FAs released from the adipose tissue leads to induction of several metabolic processes in mice: β-oxidation, ketogenesis, glycolysis/gluconeogenesis, with concomitant reduction of amino acid catabolism and an anti-inflammatory response. The changes result in an increased plasma concentration of glucose and ketone bodies and decreased urea and acute phase proteins. PPARα is important in both mice and humans for the regulation of lipid metabolism. In contrast to mice, humans show no effect on the glycolysis/gluconeogenesis pathway. One pathway specifically affected in humans and not in mice is apolipoprotein production. In humans treated with a PPARα activator, hepatic transcription activation leads to decreased VLDL production and plasma TAG levels, but increased HDL cholesterol, important parameters in the treatment for dyslipidemia, type 2 diabetes, or cardiometabolic disorders.
3. PPARα in Skeletal Muscle
In human skeletal muscles, three main muscle fiber types, type I (oxidative, slow twitch), IIA (intermediate) and IIX (glycolytic, fast twitch), can be delineated based on histochemical, functional and biochemical properties (reviewed in [109]). In human skeletal muscle cells in vitro, PPARα was shown to be induced early during myocyte differentiation [110, 111]. A correlation between the expression of PPARα, proportion of type I fibers and endurance exercise has been found in human skeletal muscle in vivo [112, 113]. The expression of PPARα (as well as of PPARδ and the PPARγ coactivator (PGC)- 1α and -1ß) in skeletal muscle was increased in athletes and reduced in spinal cord-injured subjects [113]. The observed increase of PPARα expression after endurance training [112, 114] was greater in type I fibers than in type IIA and IIX fibers [112]. Also in rat skeletal muscle, fiber-type specific PPARα activation was found. When treated with the PPARα agonist fenofibrate, 26 genes were identified that were significantly regulated in soleus (type I) but not in quadriceps femoris (type II) rat muscle [115]. The correlation of PPARα expression and exercise has not been found in animal studies. In rats, four weeks of exercise did not change the PPARα mRNA expression in skeletal muscle in control chow-fed animals, and in fat-fed rats exercise counteracted the diet-induced increase of PPARαexpression [116].
Both in human and rodent skeletal muscle, activation of PPARα affects lipid metabolism. Activation of PPARα by a potent agonist (GW7647) in differentiated human myotubes in vitro stimulated lipid oxidation [110, 117] and decreased accumulation of TAG [110]. Other, less potent PPARα agonists did not increase lipid oxidation in human myotubes [118]. In the same cell model, GW7647 upregulated the expression of pyruvate dehydrogenase kinase (PDK)4 [119]. PDK4 is an important isoenzyme regulating the activity of pyruvate dehydrogenase complex. The enzyme phosphorylates and inhibits the pyruvate dehydrogenase complex and thereby blocks the entry of carbohydrates into the mitochondria for oxidation (for reviews see [120, 121]. Pdk4 was also induced in rat gastrocnemius muscle after treatment of the animals with the PPARα agonist WY14643, by streptozotocin-induced diabetes, or by starvation, i.e. conditions where increased levels of long-chain fatty acids may activate PPARα [122]. Pathway analysis of the genes significantly regulated in soleus (type I), but not in quadriceps femoris (type II) muscle by fenofibrate in rats, revealed that the most significant function represented in the gene set was lipid metabolism [115]. Treatment with a potent PPARα agonist increased the expression of Cpt-1 in hamster soleus muscle [123].
Influence of PPARα on both lipid and glucose metabolism was highlighted in transgenic mice overexpressing PPARα in skeletal muscle [124]. In these animals many known PPARα target genes involved in cellular fatty acid import and binding, TAG synthesis, and mitochondrial and peroxisomal β-oxidation were activated, and genes involved in cellular glucose utilization were downregulated in skeletal muscle. Basal and insulin-stimulated glucose uptake was reduced in isolated skeletal muscle, and the transgenic animals developed glucose intolerance despite being protected from diet-induced obesity [124]. In contrast, in PPARα-null mice, glucose tolerance, insulin-stimulated glucose disposal and glucose uptake were increased in spite of high fat-induced weight gain and increased levels of TAGs in muscle [124]. In another study, fatty acid oxidation in skeletal muscle was found to be reduced by 28% in starved PPARα-null mice compared to wild type (WT) mice, however in fed animals fatty acid oxidation in PPARα-null and WT mice was similar [125]. TCA cycle intermediates, amino acids and short-chain acylcarnitine species were reduced in skeletal muscle of PPARα-null mice compared to WT mice, indicating impaired TCA cycle flux and increased protein catabolism combined with defects in fatty acid catabolism in PPARα-null mice [37].
In humans and mice, a negative side effect of PPARα activation in muscle is in rare cases (<1%) muscle weakness and pain (myopathy) or very seldom breakdown of muscle (rhabdomyolysis) [126–129]. In particular, type I fibers are affected by skeletal muscle toxicity in rats [115]. The exact mechanisms are unclear at present, but might include oxidative stress and tissue damage from elevated peroxisomal and mitochondrial β-oxidation [130].
PPARα also seems to exert a role in protecting against ischemic injury in skeletal muscle as well as in heart and liver [131]. Thus, in mouse skeletal muscle, loss of the oxygen sensor prolyl oxidase (PHD)1 was found to lower oxygen consumption by shifting to a more anaerobic glucose utilization through activation of PPARα-dependent genes [131].
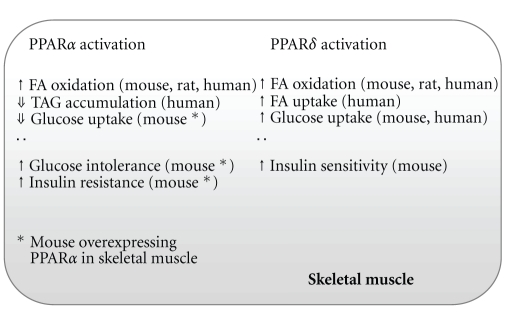
Another PPAR, PPARδ, is the most abundant PPAR isoform in skeletal muscle. Similar to PPARα, the expression of PPARδ has been described to be higher in type I fibers compared to type II fibers (reviewed in [132]). Also alike to PPARα, activation of PPARδ induces a number of genes involved in fatty acid import and oxidation, and increases lipid oxidation in skeletal muscle [125, 133–136], indicating redundancy in the functions of PPARα and δ as regulators of fatty acid metabolism [125]. However, in contrast to PPARα, activation of PPARδ has been shown to increase glucose uptake [136, 137] and prevent insulin resistance in skeletal muscle (Figure 2) [138].
Figure 2.
Examples of metabolic effects of PPARα or PPARδ activation in skeletal muscle. FA, fatty acid; TAG, triacylglycerol. For references, see the text.
In summary, PPARα has been shown to be involved in lipid and glucose metabolism in skeletal muscle. PPARα activation increases lipid oxidation and decreases TAG accumulation. Overexpression of PPARα in skeletal muscle causes reduced glucose uptake in muscle and glucose intolerance in the animals, while PPARα-null mice show increased glucose tolerance, increased insulin-stimulated glucose disposal and enhanced glucose uptake in skeletal muscle, in spite of high fat-induced weight gain and increased levels of TAGs in muscle. Thus, PPARα activation may potentially exert both beneficial and undesirable effects on skeletal muscle fuel metabolism. Activation of PPARα and PPARδ seems to have overlapping effects on fatty acid metabolism, but possibly different effects on glucose metabolism in skeletal muscle.
4. Concluding Remarks
The transcription factor PPARα influences metabolism through activation of many target genes in a variety of metabolically active tissues, in particular under fasting conditions. Cross-species prognostics are not always possible due to differences in metabolism, expression levels, or diet. While observations in rodents could have pointed to risks for human treatment with PPARα agonists (e.g., hepatocarcinogenesis, skeletal muscle insulin resistance, and myopathy) it has been shown that in humans, PPARα activation is a useful therapeutic target in treating metabolic disorders. Clinical studies on drug-induced PPARα activation include fibrates, statins, and more recently the combination of statins with fibrates. In humans, fibrates have the characteristic of reducing TAG levels and increasing HDL cholesterol, however not all trials show a vascular benefit. In some trials, clinical end-points like the rate of coronary heart disease in type 2 diabetes patients (VAHIT: Veterans affairs HDL intervention Trial, [139]) or the progression of atherosclerosis in young men after a first myocardial infarction (BECAIT: Bezafibrate Coronary Atherosclerosis Intervention Trial, [140]) could be reduced by treatment. Statin therapy shows more consistent benefits with decreased plasma LDL cholesterol levels and reduced vascular disorders and death [141]. The Action to Control Cardiovascular Risk in Diabetes (ACCORD) lipid study, addressed whether a fibrate (fenofibrate) and statin (simvastatin) combination would reduce the rate of cardiovascular events more than individual treatments in type 2 diabetes patients [142]. The combination treatment however did not influence the primary outcome significantly more than simvastatin alone, but instead showed a sex-dependent difference, with more benefits for men than women.
Rodent studies are mostly done in male animals, but the response of PPARα activation in male versus females was investigated in some studies and seems to be influenced by estrogen [143, 144]. This female hormone inhibits PPARα action and represses lipid regulatory pathways in the liver. Thus, in the treatment with PPARα agonists, gender-differences have to be taken into consideration and while therapy might be advantageous against lipid disorders in men and postmenopausal women with no interfering estrogen, premenopausal women might not benefit from the same treatment [145].
Acknowledgments
The authors thank Bodil Bjørndal, Jon Skorve, and Thomas Lundåsen for critical reading of the paper. This work was supported by a grant from NordForsk, Grant no. 070010, MitoHealth (to L. Burri and R. K. Berge).
References
- 1.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiological Reviews. 2006;86(2):465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 2.Kliewer SA, Forman BM, Blumberg B, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(15):7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt A, Endo N, Rutledge SJ, Vogel R, Shinar D, Rodan GA. Identification of a new member of the steroid hormone receptor superfamily that is activated by a peroxisome proliferator and fatty acids. Molecular Endocrinology. 1992;6(10):1634–1641. doi: 10.1210/mend.6.10.1333051. [DOI] [PubMed] [Google Scholar]
- 4.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68(5):879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 5.Houseknecht KL, Cole BM, Steele PJ. Peroxisome proliferator-activated receptor gamma (PPARγ) and its ligands: a review. Domestic Animal Endocrinology. 2002;22(1):1–23. doi: 10.1016/s0739-7240(01)00117-5. [DOI] [PubMed] [Google Scholar]
- 6.Bishop-Bailey D. Peroxisome proliferator-activated receptors in the cardiovascular system. British Journal of Pharmacology. 2000;129(5):823–834. doi: 10.1038/sj.bjp.0703149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howroyd P, Swanson C, Dunn C, Cattley RC, Corton JC. Decreased longevity and enhancement of age-dependent lesions in mice lacking the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) Toxicologic Pathology. 2004;32(5):591–599. doi: 10.1080/01926230490515283. [DOI] [PubMed] [Google Scholar]
- 8.Chinetti G, Fruchart J-C, Staels B. Peroxisome proliferator-activated receptors and inflammation: from basic science to clinical applications. International Journal of Obesity. 2003;27(supplement 3):S41–S45. doi: 10.1038/sj.ijo.0802499. [DOI] [PubMed] [Google Scholar]
- 9.Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochemistry and Biophysics. 2000;32:187–204. doi: 10.1385/cbb:32:1-3:187. [DOI] [PubMed] [Google Scholar]
- 10.Schoonjans K, Martin G, Staels B, Auwerx J. Peroxisome proliferator-activated receptors, orphans with ligands and functions. Current Opinion in Lipidology. 1997;8(3):159–166. doi: 10.1097/00041433-199706000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Tan NS, Michalik L, Desvergne B, Wahli W. Multiple expression control mechanisms of peroxisome proliferator-activated receptors and their target genes. Journal of Steroid Biochemistry and Molecular Biology. 2005;93(2–5):99–105. doi: 10.1016/j.jsbmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Mandard S, Müller M, Kersten S. Peroxisome proliferator-activated receptor α target genes. Cellular and Molecular Life Sciences. 2004;61(4):393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delerive P, Gervois P, Fruchart J-C, Staels B. Induction of IκBα expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-α activators. Journal of Biological Chemistry. 2000;275(47):36703–36707. doi: 10.1074/jbc.M004045200. [DOI] [PubMed] [Google Scholar]
- 14.Delerive P, De Bosscher K, Berghe WV, Fruchart J-C, Haegeman G, Staels B. DNA binding-independent induction of IκBα gene transcription by PPARα . Molecular Endocrinology. 2002;16(5):1029–1039. doi: 10.1210/mend.16.5.0826. [DOI] [PubMed] [Google Scholar]
- 15.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 16.Bünger M, van den Bosch HM, van der Meijde J, Kersten S, Hooiveld GJEJ, Müller M. Genome-wide analysis of PPARα activation in murine small intestine. Physiological Genomics. 2007;30(2):192–204. doi: 10.1152/physiolgenomics.00198.2006. [DOI] [PubMed] [Google Scholar]
- 17.Marx N, Mackman N, Schönbeck U, et al. PPARα activators inhibit tissue factor expression and activity in human monocytes. Circulation. 2001;103(2):213–219. doi: 10.1161/01.cir.103.2.213. [DOI] [PubMed] [Google Scholar]
- 18.Neve BP, Corseaux D, Chinetti G, et al. PPARα agonists inhibit tissue factor expression in human monocytes and macrophages. Circulation. 2001;103(2):207–212. doi: 10.1161/01.cir.103.2.207. [DOI] [PubMed] [Google Scholar]
- 19.Jones DC, Ding X, Daynes RA. Nuclear receptor peroxisome proliferator-activated receptor α (PPARα) is expressed in resting murine lymphocytes. Journal of Biological Chemistry. 2002;277(9):6838–6845. doi: 10.1074/jbc.M106908200. [DOI] [PubMed] [Google Scholar]
- 20.Berge RK, Tronstad KJ, Berge K, et al. The metabolic syndrome and the hepatic fatty acid drainage hypothesis. Biochimie. 2005;87(1):15–20. doi: 10.1016/j.biochi.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Kliewer SA, Sundseth SS, Jones SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ . Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ . Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor α (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(13):7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. Journal of Clinical Investigation. 1999;103(11):1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakhshandehroo M, Hooiveld G, Müller M, Kersten S. Comparative analysis of gene regulation by the transcription factor PPARα between mouse and human. PLoS ONE. 2009;4(8) doi: 10.1371/journal.pone.0006796. Article ID e6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gervois P, Torra IP, Chinetti G, et al. A truncated human peroxisome proliferator-activated receptor α splice variant with dominant negative activity. Molecular Endocrinology. 1999;13(9):1535–1549. doi: 10.1210/mend.13.9.0341. [DOI] [PubMed] [Google Scholar]
- 27.Patel DD, Knight BL, Wiggins D, Humphreys SM, Gibbons GF. Disturbances in the normal regulation of SREBP-sensitive genes in PPARα-deficient mice. Journal of Lipid Research. 2001;42(3):328–337. [PubMed] [Google Scholar]
- 28.Luci S, Giemsa B, Kluge H, Eder K. Clofibrate causes an upregulation of PPAR-α target genes but does not alter expression of SREBP target genes in liver and adipose tissue of pigs. American Journal of Physiology. 2007;293(1):R70–R77. doi: 10.1152/ajpregu.00603.2006. [DOI] [PubMed] [Google Scholar]
- 29.Palmer CNA, Hsu M-H, Griffin KJ, Raucy JL, Johnson EF. Peroxisome proliferator activated receptor-α expression in human liver. Molecular Pharmacology. 1998;53(1):14–22. [PubMed] [Google Scholar]
- 30.Tugwood JD, Holden PR, James NH, Prince RA, Roberts RA. A peroxisome proliferator-activated receptor-alpha (PPARα) cDNA cloned from guinea-pig liver encodes a protein with similar properties to the mouse PPARα: Implications for species differences in responses to peroxisome proliferators. Archives of Toxicology. 1998;72(3):169–177. doi: 10.1007/s002040050483. [DOI] [PubMed] [Google Scholar]
- 31.Motojima K, Passilly P, Peters JM, Gonzalez FJ, Latruffe N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor α and γ activators in a tissue- and inducer-specific manner. Journal of Biological Chemistry. 1998;273(27):16710–16714. doi: 10.1074/jbc.273.27.16710. [DOI] [PubMed] [Google Scholar]
- 32.Poirier H, Niot I, Monnot M-C, et al. Differential involvement of peroxisome-proliferator-activated receptors α and δ in fibrate and fatty-acid-mediated inductions of the gene encoding liver fatty-acid-binding protein in the liver and the small intestine. Biochemical Journal. 2001;355(2):481–488. doi: 10.1042/0264-6021:3550481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aoyama T, Peters JM, Iritani N, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα) Journal of Biological Chemistry. 1998;273(10):5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 34.Gloerich J, Van Vlies N, Jansen GA, et al. A phytol-enriched diet induces changes in fatty acid metabolism in mice both via PPARα-dependent and -independent pathways. Journal of Lipid Research. 2005;46(4):716–726. doi: 10.1194/jlr.M400337-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Paul HS, Gleditsch CE, Adibi SA. Mechanism of increased hepatic concentration of carnitine by clofibrate. American Journal of Physiology. 1986;251(3, part 1):E311–E315. doi: 10.1152/ajpendo.1986.251.3.E311. [DOI] [PubMed] [Google Scholar]
- 36.McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. European Journal of Biochemistry. 1997;244(1):1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 37.Makowski L, Noland RC, Koves TR, et al. Metabolic profiling of PPARα-/- mice reveals defects in carnitine and amino acid homeostasis that are partially reversed by oral carnitine supplementation. FASEB Journal. 2009;23(2):586–604. doi: 10.1096/fj.08-119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Vlies N, Ferdinandusse S, Turkenburg M, Wanders RJA, Vaz FM. PPARα-activation results in enhanced carnitine biosynthesis and OCTN2-mediated hepatic carnitine accumulation. Biochimica et Biophysica Acta. 2007;1767(9):1134–1142. doi: 10.1016/j.bbabio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Ringseis R, Wege N, Wen G, et al. Carnitine synthesis and uptake into cells are stimulated by fasting in pigs as a model of nonproliferating species. Journal of Nutritional Biochemistry. 2009;20(11):840–847. doi: 10.1016/j.jnutbio.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 40.Tugwood JD, Issemann I, Anderson RG, Bundell KR, McPheat WL, Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5’ flanking sequence of the rat acyl CoA oxidase gene. EMBO Journal. 1992;11(2):433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas-Frances V, Dasari VK, Abruzzi E, Osumi T, Latruffe N. The peroxisome proliferator response element (PPRE) present at positions -681/-669 in the rat liver 3-ketoacyl-CoA thiolase B gene functionally interacts differently with PPARα and HNF-4. Biochemical and Biophysical Research Communications. 2000;269(2):347–351. doi: 10.1006/bbrc.2000.2249. [DOI] [PubMed] [Google Scholar]
- 42.Savas Ü, Machemer DEW, Hsu M-H, et al. Opposing roles of peroxisome proliferator-activated receptor α and growth hormone in the regulation of CYP4A11 expression in a transgenic mouse model. Journal of Biological Chemistry. 2009;284(24):16541–16552. doi: 10.1074/jbc.M902074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sérée E, Villard P-H, Pascussi J-M, et al. Evidence for a new human CYP1A1 regulation pathway involving PPAR-α and 2 PPRE sites. Gastroenterology. 2004;127(5):1436–1445. doi: 10.1053/j.gastro.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 44.Yu S, Rao S, Reddy JK. Peroxisome proliferator-activated receptors, fatty acid oxidation, steatohepatitis and hepatocarcinogenesis. Current Molecular Medicine. 2003;3(6):561–572. doi: 10.2174/1566524033479537. [DOI] [PubMed] [Google Scholar]
- 45.Castelein H, Gulick T, Declercq PE, Mannaerts GP, Moore DD, Baes MI. The peroxisome proliferator activated receptor regulates malic enzyme gene expression. Journal of Biological Chemistry. 1994;269(43):26754–26758. [PubMed] [Google Scholar]
- 46.Guillou H, Martin P, Jan S, et al. Comparative effect of fenofibrate on hepatic desaturases in wild-type and peroxisome proliferator-activated receptor α-deficient mice. Lipids. 2002;37(10):981–989. doi: 10.1007/s11745-006-0990-3. [DOI] [PubMed] [Google Scholar]
- 47.Miller CW, Ntambi JM. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9443–9448. doi: 10.1073/pnas.93.18.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang C, Cho HP, Nakamura MT, Clarke SD. Regulation of human Δ-6 desaturase gene transcription: identification of a functional direct repeat-1 element. Journal of Lipid Research. 2003;44(4):686–695. doi: 10.1194/jlr.M200195-JLR200. [DOI] [PubMed] [Google Scholar]
- 49.Watts GF, Barrett PHR, Ji J, et al. Differential regulation of lipoprotein kinetics by atorvastatin and fenofibrate in subjects with the metabolic syndrome. Diabetes. 2003;52(3):803–811. doi: 10.2337/diabetes.52.3.803. [DOI] [PubMed] [Google Scholar]
- 50.Vu-Dac N, Schoonjans K, Kosykh V, et al. Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. Journal of Clinical Investigation. 1995;96(2):741–750. doi: 10.1172/JCI118118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vu-Dac N, Chopin-Delannoy S, Gervois P, et al. The nuclear receptors peroxisome proliferator-activated receptor α and rev-erbα mediate the species-specific regulation of apolipoprotein A-I expression by fibrates. Journal of Biological Chemistry. 1998;273(40):25713–25720. doi: 10.1074/jbc.273.40.25713. [DOI] [PubMed] [Google Scholar]
- 52.Eriksson M, Carlson LA, Miettinen TA, Angelin B. Stimulation of fecal steroid excretion after infusion of recombinant proapolipoprotein A-I: potential reverse cholesterol transport in humans. Circulation. 1999;100(6):594–598. doi: 10.1161/01.cir.100.6.594. [DOI] [PubMed] [Google Scholar]
- 53.Schoonjans K, Peinado-Onsurbe J, Lefebvre A-M, et al. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO Journal. 1996;15(19):5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 54.Prieur X, Lesnik P, Moreau M, et al. Differential regulation of the human versus the mouse apolipoprotein AV gene by PPARalpha. Implications for the study of pharmaceutical modifiers of hypertriglyceridemia in mice. Biochimica et Biophysica Acta. 2009;1791(8):764–771. doi: 10.1016/j.bbalip.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 55.Vu-Dac N, Gervois P, Jakel H, et al. Apolipoprotein A5, a crucial determinant of plasma triglyceride levels, is highly responsive to peroxisome proliferator-activated receptor α activators. Journal of Biological Chemistry. 2003;278(20):17982–17985. doi: 10.1074/jbc.M212191200. [DOI] [PubMed] [Google Scholar]
- 56.Schultze AE, Alborn WE, Newton RK, Konrad RJ. Administration of a PPARα agonist increases serum apolipoprotein A-V levels and the apolipoprotein A-V/apolipoprotein C-III ratio. Journal of Lipid Research. 2005;46(8):1591–1595. doi: 10.1194/jlr.C500010-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Staels B, Vu-Dac N, Kosykh VA, et al. Fibrates downregulate apolipoprotein C-III expression independent of induction of peroxisomal acyl coenzyme A oxidase. A potential mechanism for the hypolipidemic action of fibrates. Journal of Clinical Investigation. 1995;95(2):705–712. doi: 10.1172/JCI117717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birjmohun RS, Hutten BA, Kastelein JJP, Stroes ESG. Efficacy and safety of high-density lipoprotein cholesterol-increasing compounds: a meta-analysis of randomized controlled trials. Journal of the American College of Cardiology. 2005;45(2):185–197. doi: 10.1016/j.jacc.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 59.Russell DW, Setchell KDR. Bile acid biosynthesis. Biochemistry. 1992;31(20):4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 60.Vlahcevic ZR, Heuman DM, Hylemon PB. Regulation of bile acid synthesis. Hepatology. 1991;13(3):590–600. [PubMed] [Google Scholar]
- 61.Cheema SK, Agellon LB. The murine and human cholesterol 7α-hydroxylase gene promoters are differentially responsive to regulation by fatty acids mediated via peroxisome proliferator-activated receptor α . Journal of Biological Chemistry. 2000;275(17):12530–12536. doi: 10.1074/jbc.275.17.12530. [DOI] [PubMed] [Google Scholar]
- 62.Hunt MC, Yang Y-Z, Eggertsen G, et al. The peroxisome proliferator-activated receptor α (PPARα) regulates bile acid biosynthesis. Journal of Biological Chemistry. 2000;275(37):28947–28953. doi: 10.1074/jbc.M002782200. [DOI] [PubMed] [Google Scholar]
- 63.Chiang JYL. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocrine Reviews. 2002;23(4):443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- 64.Marrapodi M, Chiang JYL. Peroxisome proliferator-activated receptor α (PPARα) and agonist inhibit cholesterol 7α-hydroxylase gene (CYP7A1) transcription. Journal of Lipid Research. 2000;41(4):514–520. [PubMed] [Google Scholar]
- 65.Patel DD, Knight BL, Soutar AK, Gibbons GF, Wade DP. The effect of peroxisome-proliferator-activated receptor-α on the activity of the cholesterol 7α-hydroxylase gene. Biochemical Journal. 2000;351(3):747–753. [PMC free article] [PubMed] [Google Scholar]
- 66.Post SM, Duez H, Gervois PP, Staels B, Kuipers F, Princen HMG. Fibrates suppress bile acid synthesis via peroxisome proliferator-activated receptor-α-mediated downregulation of cholesterol 7α-hydroxylase and sterol 27-hydroxylase expression. Arteriosclerosis, Thrombosis, and Vascular Biology. 2001;21(11):1840–1845. doi: 10.1161/hq1101.098228. [DOI] [PubMed] [Google Scholar]
- 67.Ståhlberg D, Reihnér E, Rudling M, Berglund L, Einarsson K, Angelin BO. Influence of bezafibrate on hepatic cholesterol metabolism in gallstone patients: reduced activity of cholesterol 7α-hydroxylase. Hepatology. 1995;21(4):1025–1030. doi: 10.1002/hep.1840210421. [DOI] [PubMed] [Google Scholar]
- 68.Ishida H, Kuruta Y, Gotoh O, Yamashita C, Yoshida Y, Noshiro M. Structure, evolution, and liver-specific expression of sterol 12α-hydroxylase P450 (CYP8B) Journal of Biochemistry. 1999;126(1):19–25. doi: 10.1093/oxfordjournals.jbchem.a022422. [DOI] [PubMed] [Google Scholar]
- 69.Hsu M-H, Savas Ü, Griffin KJ, Johnson EF. Identification of peroxisome proliferator-responsive human genes by elevated expression of the peroxisome proliferator-activated receptor α in HepG2 cells. Journal of Biological Chemistry. 2001;276(30):27950–27958. doi: 10.1074/jbc.M100258200. [DOI] [PubMed] [Google Scholar]
- 70.Rodríguez JC, Gil-Gómez G, Hegardt FG, Haro D. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. Journal of Biological Chemistry. 1994;269(29):18767–18772. [PubMed] [Google Scholar]
- 71.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metabolism. 2007;5(6):426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 72.Coskun T, Bina HA, Schneider MA, et al. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 2008;149(12):6018–6027. doi: 10.1210/en.2008-0816. [DOI] [PubMed] [Google Scholar]
- 73.Xu J, Lloyd DJ, Hale C, et al. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes. 2009;58(1):250–259. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inagaki T, Dutchak P, Zhao G, et al. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metabolism. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 75.Lundåsen T, Hunt MC, Nilsson L-M, et al. PPARα is a key regulator of hepatic FGF21. Biochemical and Biophysical Research Communications. 2007;360(2):437–440. doi: 10.1016/j.bbrc.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 76.Kharitonenkov A, Shiyanova TL, Koester A, et al. FGF-21 as a novel metabolic regulator. Journal of Clinical Investigation. 2005;115(6):1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu J, Xiao G, Tirujillo C, et al. Peroxisome proliferator-activated receptor α (PPARα) influences: substrate utilization for hepatic glucose production. Journal of Biological Chemistry. 2002;277(52):50237–50244. doi: 10.1074/jbc.M201208200. [DOI] [PubMed] [Google Scholar]
- 78.Patsouris D, Mandard S, Voshol PJ, et al. PPARα governs glycerol metabolism. Journal of Clinical Investigation. 2004;114(1):94–103. doi: 10.1172/JCI20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bandsma RHJ, van Dijk TH, ter Harmsel A, et al. Hepatic de novo synthesis of glucose 6-phosphate is not affected in peroxisome proliferator-activated receptor α-deficient mice but is preferentially directed toward hepatic glycogen stores after a short term fast. Journal of Biological Chemistry. 2004;279(10):8930–8937. doi: 10.1074/jbc.M310067200. [DOI] [PubMed] [Google Scholar]
- 80.Genolet R, Kersten S, Braissant O, et al. Promoter rearrangements cause species-specific hepatic regulation of the glyoxylate reductase/hydroxypyruvate reductase gene by the peroxisome proliferator-activated receptor α . Journal of Biological Chemistry. 2005;280(25):24143–24152. doi: 10.1074/jbc.M502649200. [DOI] [PubMed] [Google Scholar]
- 81.Kersten S, Mandard S, Escher P, et al. The peroxisome proliferator-activated receptor α regulates amino acid metabolism. FASEB Journal. 2001;15(11):1971–1978. doi: 10.1096/fj.01-0147com. [DOI] [PubMed] [Google Scholar]
- 82.Sheikh K, Camejo G, Lanne B, Halvarsson T, Landergren MR, Oakes ND. Beyond lipids, pharmacological PPARα activation has important effects on amino acid metabolism as studied in the rat. American Journal of Physiology. 2007;292(4):E1157–E1165. doi: 10.1152/ajpendo.00254.2006. [DOI] [PubMed] [Google Scholar]
- 83.Dierkes J, Westphal S, Martens-Lobenhoffer J, Luley C, Bode-Böger SM. Fenofibrate increases the L-arginine: ADMA ratio by increase of L-arginine concentration but has no effect on ADMA concentration. Atherosclerosis. 2004;173(2):239–244. doi: 10.1016/j.atherosclerosis.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 84.Kobayashi R, Murakami T, Obayashi M, et al. Clofibric acid stimulates branched-chain amino acid catabolism by three mechanisms. Archives of Biochemistry and Biophysics. 2002;407(2):231–240. doi: 10.1016/s0003-9861(02)00472-1. [DOI] [PubMed] [Google Scholar]
- 85.Gervois P, Kleemann R, Pilon A, et al. Global suppression of IL-6-induced acute phase response gene expression after chronic in vivo treatment with the peroxisome proliferator-activated receptor-α activator fenofibrate. Journal of Biological Chemistry. 2004;279(16):16154–16160. doi: 10.1074/jbc.M400346200. [DOI] [PubMed] [Google Scholar]
- 86.Belfort R, Berria R, Cornell J, Cusi K. Fenofibrate reduces systemic inflammation markers independent of its effects on lipid and glucose metabolism in patients with the metabolic syndrome. Journal of Clinical Endocrinology and Metabolism. 2010;95(2):829–836. doi: 10.1210/jc.2009-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee M-W, Chanda D, Yang J, et al. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metabolism. 2010;11(4):331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 88.Zhang K, Shen X, Wu J, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124(3):587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 89.Yu C, Wang F, Jin C, Wu X, Chan W-K, McKeehan WL. Increased carbon tetrachloride-induced liver injury and fibrosis in FGFR4-deficient mice. American Journal of Pathology. 2002;161(6):2003–2010. doi: 10.1016/S0002-9440(10)64478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Echtay KS, Roussel D, St-Plerre J, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415(6867):96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 91.Wu Q, Gong D, Tian N, et al. Protection of regenerating liver after partial hepatectomy from carbon tetrachloride hepatotoxicity in rats: roles of mitochondrial uncoupling protein 2 and ATP stores. Digestive Diseases and Sciences. 2009;54(9):1918–1925. doi: 10.1007/s10620-008-0650-y. [DOI] [PubMed] [Google Scholar]
- 92.Anderson SP, Howroyd P, Liu J, et al. The transcriptional response to a peroxisome proliferator-activated receptor α agonist includes increased expression of proteome maintenance genes. Journal of Biological Chemistry. 2004;279(50):52390–52398. doi: 10.1074/jbc.M409347200. [DOI] [PubMed] [Google Scholar]
- 93.Ashby J, Brady A, Elcombe CR, et al. Mechanistically-based human hazard assessment of peroxisome proliferator-induced hepatocarcinogenesis. Human and Experimental Toxicology. 1994;13(supplement 2):S1–S117. doi: 10.1177/096032719401300201. [DOI] [PubMed] [Google Scholar]
- 94.Bentley P, Calder I, Elcombe C, Grasso P, Stringer D, Wiegand H-J. Hepatic peroxisome proliferation in rodents and its significance for humans. Food and Chemical Toxicology. 1993;31(11):857–907. doi: 10.1016/0278-6915(93)90225-n. [DOI] [PubMed] [Google Scholar]
- 95.Hays T, Rusyn I, Burns AM, et al. Role of peroxisome proliferator-activated receptor-α (PPARα) in bezafibrate-induced hepatocarcinogenesis and cholestasis. Carcinogenesis. 2005;26(1):219–227. doi: 10.1093/carcin/bgh285. [DOI] [PubMed] [Google Scholar]
- 96.Peters JM, Cheung C, Gonzalez FJ. Peroxisome proliferator-activated receptor-α and liver cancer: where do we stand? Journal of Molecular Medicine. 2005;83(10):774–785. doi: 10.1007/s00109-005-0678-9. [DOI] [PubMed] [Google Scholar]
- 97.Rao MS, Reddy JK. An overview of peroxisome proliferator-induced hepatocarcinogenesis. Environmental Health Perspectives. 1991;93:205–209. doi: 10.1289/ehp.9193205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reddy JK, Rao MS. Malignant tumors in rats fed nafenopin, a hepatic peroxisome proliferator. Journal of the National Cancer Institute. 1977;59(6):1645–1650. doi: 10.1093/jnci/59.6.1645. [DOI] [PubMed] [Google Scholar]
- 99.Bayly AC, Roberts RA, Dive C. Suppression of liver cell apoptosis in vitro by the non-genotoxic hepatocarcinogen and peroxisome proliferator nafenopin. Journal of Cell Biology. 1994;125(1):197–203. doi: 10.1083/jcb.125.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cunningham ML, Soliman MS, Badr MZ, Matthews HB. Rotenone, an anticarcinogen, inhibits cellular proliferation but not peroxisome proliferation in mouse liver. Cancer Letters. 1995;95(1-2):93–97. doi: 10.1016/0304-3835(95)03869-x. [DOI] [PubMed] [Google Scholar]
- 101.James NH, Soames AR, Roberts RA. Suppression of hepatocyte apoptosis and induction of DNA synthesis by the rat and mouse hepatocarcinogen diethylhexylphlathate (DEHP) and the mouse hepatocarcinogen 1,4-dichlorobenzene (DCB) Archives of Toxicology. 1998;72(12):784–790. doi: 10.1007/s002040050574. [DOI] [PubMed] [Google Scholar]
- 102.Oberhammer FA, Qin H-M. Effect of three tumour promoters on the stability of hepatocyte cultures and apoptosis after transforming growth factor-β1. Carcinogenesis. 1995;16(6):1363–1371. doi: 10.1093/carcin/16.6.1363. [DOI] [PubMed] [Google Scholar]
- 103.Reddy JK, Rao MS. Oxidative DNA damage caused by persistent peroxisome proliferation: its role in hepatocarcinogenesis. Mutation Research. 1989;214(1):63–68. doi: 10.1016/0027-5107(89)90198-x. [DOI] [PubMed] [Google Scholar]
- 104.Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor α regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Molecular and Cellular Biology. 2007;27(12):4238–4247. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biology. 2005;3(3, article e85) doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 107.Balfour JA, McTavish D, Heel RC. Fenofibrate. A review of its pharmacodynamic and pharmacokinetic properties and therapeutic use in dyslipidaemia. Drugs. 1990;40(2):260–290. doi: 10.2165/00003495-199040020-00007. [DOI] [PubMed] [Google Scholar]
- 108.Klaunig JE, Babich MA, Baetcke KP, et al. PPARα agonist-induced rodent tumors: modes of action and human relevance. Critical Reviews in Toxicology. 2003;33(6):655–780. doi: 10.1080/713608372. [DOI] [PubMed] [Google Scholar]
- 109.Bottinelli R, Reggiani C. Human skeletal muscle fibres: molecular and functional diversity. Progress in Biophysics and Molecular Biology. 2000;73(2–4):195–262. doi: 10.1016/s0079-6107(00)00006-7. [DOI] [PubMed] [Google Scholar]
- 110.Muoio DM, Way JM, Tanner CJ, et al. Peroxisome proliferator-activated receptor-α regulates fatty acid utilization in primary human skeletal muscle cells. Diabetes. 2002;51(4):901–909. doi: 10.2337/diabetes.51.4.901. [DOI] [PubMed] [Google Scholar]
- 111.Kase ET, Andersen B, Nebb HI, Rustan AC, Hege Thoresen G. 22-Hydroxycholesterols regulate lipid metabolism differently than T0901317 in human myotubes. Biochimica et Biophysica Acta. 2006;1761(12):1515–1522. doi: 10.1016/j.bbalip.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 112.Russell AP, Feilchenfeldt J, Schreiber S, et al. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 2003;52(12):2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 113.Krämer DK, Ahlsén M, Norrbom J, et al. Human skeletal muscle fibre type variations correlate with PPARα, PPARδ and PGC-1α mRNA. Acta Physiologica. 2006;188(3-4):207–216. doi: 10.1111/j.1748-1716.2006.01620.x. [DOI] [PubMed] [Google Scholar]
- 114.Horowitz JF, Leone TC, Feng W, Kelly DP, Klein S. Effect of endurance training on lipid metabolism in women: a potential role for PPARα in the metabolic response to training. American Journal of Physiology. 2000;279(2):E348–E355. doi: 10.1152/ajpendo.2000.279.2.E348. [DOI] [PubMed] [Google Scholar]
- 115.De Souza AT, Cornwell PD, Dai X, Caguyong MJ, Ulrich RG. Agonists of the peroxisome proliferator-activated receptor alpha induce a fiber-type-selective transcriptional response in rat skeletal muscle. Toxicological Sciences. 2006;92(2):578–586. doi: 10.1093/toxsci/kfl019. [DOI] [PubMed] [Google Scholar]
- 116.Kannisto K, Chibalin A, Glinghammar B, Zierath JR, Hamsten A, Ehrenborg E. Differential expression of peroxisomal proliferator activated receptors alpha and delta in skeletal muscle in response to changes in diet and exercise. International Journal of Molecular Medicine. 2006;17(1):45–52. [PubMed] [Google Scholar]
- 117.Djouadi F, Aubey F, Schlemmer D, Bastin J. Peroxisome proliferator activated receptor δ (PPARδ) agonist but not PPARα corrects carnitine palmitoyl transferase 2 deficiency in human muscle cells. Journal of Clinical Endocrinology and Metabolism. 2005;90(3):1791–1797. doi: 10.1210/jc.2004-1936. [DOI] [PubMed] [Google Scholar]
- 118.Løvås K, Røst TH, Skorve J, et al. Tetradecylthioacetic acid attenuates dyslipidaemia in male patients with type 2 diabetes mellitus, possibly by dual PPAR-α/δ activation and increased mitochondrial fatty acid oxidation. Diabetes, Obesity and Metabolism. 2009;11(4):304–314. doi: 10.1111/j.1463-1326.2008.00958.x. [DOI] [PubMed] [Google Scholar]
- 119.Abbot EL, McCormack JG, Reynet C, Hassall DG, Buchan KW, Yeaman SJ. Diverging regulation of pyruvate dehydrogenase kinase isoform gene expression in cultured human muscle cells. FEBS Journal. 2005;272(12):3004–3014. doi: 10.1111/j.1742-4658.2005.04713.x. [DOI] [PubMed] [Google Scholar]
- 120.Pilegaard H, Neufer PD. Transcriptional regulation of pyruvate dehydrogenase kinase 4 in skeletal muscle during and after exercise. Proceedings of the Nutrition Society. 2004;63(2):221–226. doi: 10.1079/pns2004345. [DOI] [PubMed] [Google Scholar]
- 121.Sugden MC, Holness MJ. Mechanisms underlying regulation of the expression and activities of the mammalian pyruvate dehydrogenase kinases. Archives of Physiology and Biochemistry. 2006;112(3):139–149. doi: 10.1080/13813450600935263. [DOI] [PubMed] [Google Scholar]
- 122.Wu P, Inskeep K, Bowker-Kinley MM, Popov KM, Harris RA. Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes. 1999;48(8):1593–1599. doi: 10.2337/diabetes.48.8.1593. [DOI] [PubMed] [Google Scholar]
- 123.Minnich A, Tian N, Byan L, Bilder G. A potent PPARα agonist stimulates mitochondrial fatty acid β-oxidation in liver and skeletal muscle. American Journal of Physiology. 2001;280(2):E270–E279. doi: 10.1152/ajpendo.2001.280.2.E270. [DOI] [PubMed] [Google Scholar]
- 124.Finck BN, Bernal-Mizrachi C, Han DH, et al. A potential link between muscle peroxisome proliferator-activated receptor-α signaling and obesity-related diabetes. Cell Metabolism. 2005;1(2):133–144. doi: 10.1016/j.cmet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 125.Muoio DM, MacLean PS, Lang DB, et al. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) α knock-out mice. Evidence for compensatory regulation by PPARδ . Journal of Biological Chemistry. 2002;277(29):26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- 126.Hodel C. Myopathy and rhabdomyolysis with lipid-lowering drugs. Toxicology Letters. 2002;128(1–3):159–168. doi: 10.1016/s0378-4274(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 127.Magarian GJ, Lucas LM, Colley C. Gemfibrozil-induced myopathy. Archives of Internal Medicine. 1991;151(9):1873–1874. [PubMed] [Google Scholar]
- 128.Langer T, Levy RI. Acute muscular syndrome associated with administration of clofibrate. The New England Journal of Medicine. 1968;279(16):856–858. doi: 10.1056/NEJM196810172791604. [DOI] [PubMed] [Google Scholar]
- 129.Rush P, Baron M, Kapusta M. Clofibrate myopathy: a case report and a review of the literature. Seminars in Arthritis and Rheumatism. 1986;15(3):226–229. doi: 10.1016/0049-0172(86)90019-3. [DOI] [PubMed] [Google Scholar]
- 130.Faiola B, Falls JG, Peterson RA, et al. PPAR alpha, more than PPAR delta, mediates the hepatic and skeletal muscle alterations induced by the PPAR agonist GW0742. Toxicological Sciences. 2008;105(2):384–394. doi: 10.1093/toxsci/kfn130. [DOI] [PubMed] [Google Scholar]
- 131.Aragonés J, Schneider M, Van Geyte K, et al. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nature Genetics. 2008;40(2):170–180. doi: 10.1038/ng.2007.62. [DOI] [PubMed] [Google Scholar]
- 132.Ehrenborg E, Krook A. Regulation of skeletal muscle physiology and metabolism by peroxisome proliferator-activated receptor δ . Pharmacological Reviews. 2009;61(3):373–393. doi: 10.1124/pr.109.001560. [DOI] [PubMed] [Google Scholar]
- 133.Dressel U, Allen TL, Pippal JB, Rohde PR, Lau P, Muscat GEO. The peroxisome proliferator-activated receptor beta/delta agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Molecular Endocrinology. 2003;17(12):2477–2493. doi: 10.1210/me.2003-0151. [DOI] [PubMed] [Google Scholar]
- 134.Holst D, Luquet S, Nogueira V, Kristiansen K, Leverve X, Grimaldi PA. Nutritional regulation and role of peroxisome proliferator-activated receptor δ in fatty acid catabolism in skeletal muscle. Biochimica et Biophysica Acta. 2003;1633(1):43–50. doi: 10.1016/s1388-1981(03)00071-4. [DOI] [PubMed] [Google Scholar]
- 135.Tanaka T, Yamamoto J, Iwasaki S, et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Krämer DK, Al-Khalili L, Guigas B, Leng Y, Garcia-Roves PM, Krook A. Role of AMP kinase and PPARδ in the regulation of lipid and glucose metabolism in human skeletal muscle. Journal of Biological Chemistry. 2007;282(27):19313–19320. doi: 10.1074/jbc.M702329200. [DOI] [PubMed] [Google Scholar]
- 137.Krämer DK, Al-Khalili L, Perrini S, et al. Direct activation of glucose transport in primary human myotubes after activation of peroxisome proliferator-activated receptor δ . Diabetes. 2005;54(4):1157–1163. doi: 10.2337/diabetes.54.4.1157. [DOI] [PubMed] [Google Scholar]
- 138.Coll T, Álvarez-Guardia D, Barroso E, et al. Activation of peroxisome proliferator-activated receptor-δ by GW501516 prevents fatty acid-induced nuclear factor-κB activation and insulin resistance in skeletal muscle cells. Endocrinology. 2010;151(4):1560–1569. doi: 10.1210/en.2009-1211. [DOI] [PubMed] [Google Scholar]
- 139.Rubins HB, Robins SJ, Collins D, et al. Diabetes, plasma insulin, and cardiovascular disease: subgroup analysis from the Department of Veterans Affairs High-density Lipoprotein Intervention Trial (VA-HIT) Archives of Internal Medicine. 2002;162(22):2597–2604. doi: 10.1001/archinte.162.22.2597. [DOI] [PubMed] [Google Scholar]
- 140.Ruotolo G, Ericsson C-G, Tettamanti C, et al. Treatment effects on serum lipoprotein lipids, apolipoproteins and low density lipoprotein particle size and relationships of lipoprotein variables to progression of coronary artery disease in the Bezafibrate Coronary Atherosclerosis Intervention Trial (BECAIT) Journal of the American College of Cardiology. 1998;32(6):1648–1656. doi: 10.1016/s0735-1097(98)00442-2. [DOI] [PubMed] [Google Scholar]
- 141.Baigent C, Keech A, Kearney PM, et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. The Lancet. 2005;366(9493):1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 142.Ginsberg HN, Elam MB, Lovato LC, et al. Effects of combination lipid therapy in type 2 diabetes mellitus. The New England Journal of Medicine. 2010;362(17):1563–1574. doi: 10.1056/NEJMoa1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leuenberger N, Pradervand S, Wahli W. Sumoylated PPARα mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. Journal of Clinical Investigation. 2009;119(10):3138–3148. doi: 10.1172/JCI39019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang X, Kilgore MW. Signal cross-talk between estrogen receptor alpha and beta and the peroxisome proliferator-activated receptor gamma1 in MDA-MB-231 and MCF-7 breast cancer cells. Molecular and Cellular Endocrinology. 2002;194(1-2):123–133. doi: 10.1016/s0303-7207(02)00154-5. [DOI] [PubMed] [Google Scholar]
- 145.Yoon M. The role of PPARα in lipid metabolism and obesity: focusing on the effects of estrogen on PPARα actions. Pharmacological Research. 2009;60(3):151–159. doi: 10.1016/j.phrs.2009.02.004. [DOI] [PubMed] [Google Scholar]