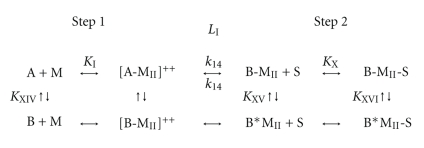Abstract
S100B is a calcium signaling protein that is a member of the S100 protein family. An important feature of S100B and most other S100 proteins (S100s) is that they often bind Ca2+ ions relatively weakly in the absence of a protein target; upon binding their target proteins, Ca2+-binding then increases by as much as from 200- to 400-fold. This manuscript reviews the structural basis and physiological significance of increased Ca2+-binding affinity in the presence of protein targets. New information regarding redundancy among family members and the structural domains that mediate the interaction of S100B, and other S100s, with their targets is also presented. It is the diversity among individual S100s, the protein targets that they interact with, and the Ca2+ dependency of these protein-protein interactions that allow S100s to transduce changes in [Ca2+]intracellular levels into spatially and temporally unique biological responses.
1. Introduction
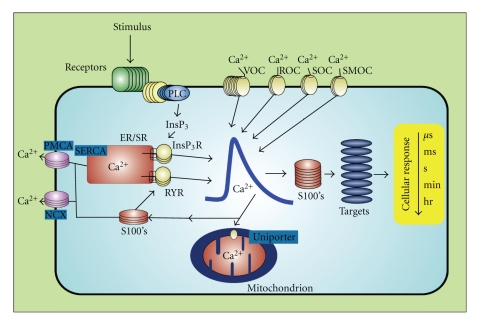
Ca2+ ions are important second messengers in all living cells [1]. Ca2+-binding proteins, including members of the calmodulin/troponin/S100 superfamily, maintain the integrity of the Ca2+ signal and transmit it in a temporally and spatially coordinated manner [2]. S100s were discovered in 1965 [3], and as with other EF-hand containing proteins, S100s also transduce changes in [Ca2+]intracellular levels (i.e., [Ca2+]i) into cellular responses by binding Ca2+ (Table 1), changing conformation, and then interacting with and modulating the activity of other proteins (target proteins) (Figure 1). The amino acid homology between the current family members ranges from approximately 20% to 55% [4]. Because of the extensive amino acid homology between S100B and S100A1, the first two family members identified, early models predicted that individual members were functionally redundant and essentially interchangeable. As the number of family members discovered has increased and differences in their cellular/subcellular localization, physical properties, and target proteins have expanded, additional models have arisen with specific S100 family members having unique biological functions [5–12]. This multigenic family now contains up to twenty-one members (humans) whose phylogenetic distribution is restricted to higher chordates. Oligomerization properties, affinities for divalent metal ions (Ca2+, Zn2+, Cu2+), and posttranslational modifications also contribute to diversity among S100 family members [7, 12]. Furthermore, each cell/tissue expresses a unique subset of family members [11, 12]. Collectively, these findings support the view that S100s often confer cell type specificity to Ca2+ signal transduction pathways in cells and tissues [11–16].
Table 1.
Dissociation of Ca2+ and Mn2+ from the EF-hand calcium-binding domains in wild-type and mutant S100 proteins.
| S100 protein | EF1 | EF2 |
|---|---|---|
| Ca2+ binding | ||
| S100B (wt) | >350 μMa,c | 56 ± 9 μMb,c |
| S100B (E31A) | >500 μMa | >500 μMa |
| S100B (E72A) | 480 ± 130 μMa | >500 μMa |
| S100B (E31A + E72A) | >2 mMa | >2 mMa |
| S100B (+p53) | — | 20 ± 3 μMa |
| S100B (E31A, +p53) | — | 21 ± 7 μMa |
| S100B (E72A, +p53) | — | 18 ± 4 μMa |
| S100B (E31A + E72A, +p53) | — | >300 μMa |
| S100B (wt, +TRTK12) | — | 12 ± 7 μMc |
| S100A1 (wt) | — | 27 ± 2 μMd |
| S100A1 (wt, +TRTK12) | — | 8 ± 3 μMe |
| S100A2 (wt) | — | 470 ± 50 μMf |
| S100A3 (wt) | — | ∼4 mMg |
| S100A4 (wt) | — | ∼2.6 μMh |
| S100A4 (wt, +p37) | — | ∼0.2 μMh |
| S100A5 (wt) | 160 μMi | ∼0.2 μMi |
| S100A6 (wt) | — | ∼3.0 μMj |
| S100A7 (wt) | — | ∼1.0 μMk,l |
| S100A11 (wt) | — | ∼0.5 mMm |
| S100A12 (wt) | — | ∼50 μMn |
| S100A13 (wt) | ∼400 μM | ∼8 μMo,p |
| S100A16 (wt) | no binding | 0.43 mMq |
| S100P (wt) | ∼800 μM | ∼2.0 μMr |
| S100Z (wt) | > 1 mM | ∼0.2 μMs |
|
| ||
| Mn2+ binding | ||
| S100B (wt) | — | 71 ± 12 μMa,c |
| S100B (wt, +p53) | — | 27 ± 4 μMa,c |
| S100B (wt, +TRTK12) | — | 6.0 ± 2.0 μMa,c |
aThe value listed is from previously published papers [109, 113], so direct comparisons of binding constants using similar methods/conditions could be made (+/− target, Figure 3). Several others report binding constants using different methods and varying conditions for EF1 (200 μM ≤KD≤ 500 μM) and for EF2 (10 μM ≤KD≤ 60 μM) [58, 78, 86, 87, 114–120].
bThe dissociation rate constant for wild-type S100B was determined via stopped-flow methods and is koff = 60 ± 22 s−1. The off-rate together with the KD enables the calculation of a macroscopic on-rate value of kon = 1.1 × 106 M−1 s−1 that includes calcium-association plus a large conformational change. The KD value for the mutants was also determined using competition studies of Ca2+ with the respective Tb3+-bound S100B mutant in the absence and presence of p53 peptide. The dissociation constants together with the calcium off-rate values measured for the E31A and E72A mutants of 7.1 ± 3.7 s−1 and 6.8 ± 2.0 s−1, respectively, were sufficient to calculate on-rate values of 3.4 ± 2.0 × 106 M−1 s−1 and 3.7 ± 1.3 × 106 M−1 s−1 for the mutants [109, 113].
cFrom Charpentier et al. (2010) [34].
dFrom Wright et al. (2005) [61].
eFrom Wright et al. (2009) [59]. S100A1 has also been shown to bind the full-length ryanodine receptor at 100 nM free Ca2+ [60, 67].
fFrom Franz et al. (1998) [89].
gFrom Fritz et al. (1998). A tenfold weaker affinity was reported when purified under aerobic conditions [90, 121].
hFrom Dukhanina et al. (1998). A weaker affinity was reported under different conditions in Pedrocchi et al. (1994) when S100A4 was originally discovered [122, 123].
iFrom Schäfer et al. (2000). For a direct comparison of Ca2+ and Zn2+ binding to S100A5 to those of other S100 proteins (i.e., S100B, S100A2, S100A3, S100A4, S1006, and S10011), under identical conditions and Methods, also see Schäfer et al., (2000) [124].
jFrom Kuznicki and Filipek (1987) and Mani and Kay (1990). Kordowska et al. also measured Ca2+-binding for S100A6 under different conditions (CaKD ∼18 μM) and found that binding to the target caldesmon (CaD) increased the affinity of S100A6 for Ca2+ by approximately 6-fold [96, 125, 126]. Other measurements under higher salt and other varying conditions are also reported with weaker affinities for S100A6 [68, 124].
kFrom Schäfer et al. (2000) [124]. Weaker binding to Ca2+ has also been reported for this protein in other conditions [127].
lNo data is available for S100A8/A9, and S100A10 does not bind Ca2+.
mFrom Allen et al. (1996) and Schäfer et al., (2000) [124, 128]. Note the affinity for Ca2+ increases by 10-fold upon the addition of a target molecule as found with other S100 proteins [128].
nFrom Dell'Angelica et al. (1994). Note that Zn2+-binding to S100A12 significantly increases Ca2+-binding affinity for this protein in the presence of Zn2+ (EF2: CaKD = 40 nM, EF1: CaKD = 15 μM) [129].
oFrom Ridinger et al. (2000). This protein is unique among S100 family members in that it does not bind to the hydrophobic binding dye, TNS, upon the addition of Ca2+ [130].
qFrom Sturchler et al. (2006). The value in the table is for human S100A16, mouse S100A16 bound one calcium too, only weaker (CaKD = 0.75 mM) [132].
Figure 1.
S100s function as Ca2+-signaling proteins. S100s bind and regulate protein targets as well as other Ca2+-signaling proteins in a Ca2+-dependent manner. S100s are distributed in a cell-specific manner to generate cell-type specific activities [1, 2, 10].
The 3-dimensional structure of numerous S100 proteins has been solved by NMR and X-ray crystallography techniques in the apo-, Ca2+-bound, Zn2+-bound, drug-bound, and target protein bound states, and with the exception of calbindinD9K, S100s such as S100B are typically symmetric dimers with each subunit containing two EF-hand calcium-binding domains, although, some higher-order oligomeric states have also been detected and discussed [17–64]. The first EF-hand motif (EF1) in each subunit has fourteen rather than twelve residues and is termed the “pseudo-EF-hand” or “S100 EF-hand” [31, 65]. Another unique feature of the S100-hand (EF1) is that calcium-coordination is achieved via backbone carbonyl oxygen atoms rather than sidechain carboxylate oxygen atoms, and it typically binds Ca2+ rather weakly (KD> 0.5 mM). The second EF-hand (EF2) is termed the “typical EF-hand” since it has the same number of residues as most EF-hand Ca2+-binding proteins (i.e., twelve), and it exactly matches the consensus EF-hand in both sequence and three-dimensional structure [66]. As with the helix-loop-helix Ca2+ binding domains of other members of the EF-hand superfamily (i.e., calmodulin, troponin C, etc.), the second EF-hand of S100B coordinates Ca2+ via a backbone carbonyl oxygen at position 7, sidechain oxygen atoms from Asp/Glu at positions 1, 3, and 5, and via bidentate coordination from oxygen atoms from a Glu sidechain at position 12 [66]. Position 9 of the coordination scheme is occupied by a water molecule that is, in turn, hydrogen-bonded to a Glu sidechain oxygen atom from the pseudo-EF-hand. Another feature of S100B and several other S100s is that several of them bind Ca2+ ions relatively weakly, relative to [Ca2+]i, in the absence of a protein target (KD≥ 1 μM). However, upon binding their full-length target protein, Ca2+-binding can then increase by as much as from 200- to 400-fold [60, 67]. This feature could allow for cells to contain high concentrations of some S100s, such as S100B (>1 μM), without depleting [Ca2+]i levels and “short-circuiting” Ca2+ oscillations necessary for signaling.
The large number of protein targets that S100B and other S100 family members bind and regulate provides yet another level of diversity/complexity to S100 signaling. Examination of the extensive lists of S100 target proteins provides important insights regarding S100 signaling [15, 68, 69]. First, S100s regulate a wide-range of cellular processes that includes energy metabolism, cytoskeleton organization, gene expression, and signal transduction pathways. Second, the list of in vitro target proteins for individual family members can be extensive, 23–25 different protein targets have been reported for S100B and S100A1 alone [5, 7, 13, 15, 68, 70]. Third, some target proteins interact with and have their activity modulated by multiple S100 family members suggesting functional redundancy among some family members. For example, both S100A1 and S100B interact with and activate aldolase A and aldolase C [71]. In contrast, S100A1 and S100B exhibit a Ca2+-dependent interaction with phosphoglucomtutase but this interaction results in differential effects on target protein activity: S100A1 stimulates phosphoglucomutase activity and S100B inhibits phosphoglucomutase activity [72]. Like phosphoglucomutase, both S100A1 and S100B bind glycogen phosphorylase a [73]. However, S100A1 inhibits and S100B has no effect on glycogen phosphorylase activity. Interestingly, S100A4, S100A1, and S100A6 interact with methionine aminopeptidase 2, but with different affinities [74]. The S100A6 (calcyclin) binding protein CACYBP also interacts with S100A1, S100A12, S100P, and S100B but not S100A4 [75]. And several S100 proteins (i.e., S100A1, S100A2, S100A4, S100A6, and S100B) bind to p53, p63, p73 [29, 76–80], and affect their biological functions [81–84]. Fifth, S100 activation of some target proteins is Ca2+-independent indicating that S100s sense changes in [Ca2+]i levels and have important intracellular functions at both resting and stimulated [Ca2+]i levels. Sixth, a subset of S100s have a conserved Zn2+-binding site that contributes to altered Ca2+-, target-, and in some cases, small molecule binding affinity in vitro [8, 20, 36, 40, 68, 85–99], which could potentially affect cellular functions. Seventh, the interaction of S100 family members with other family members as “target proteins” has been observed using yeast-two hybrid approaches [100–104]. Finally, S100 proteins may exert some of their effects via interaction with molecules other than proteins. For example, S100A8/S100A9 binds (poly) unsaturated fatty acids in a Ca2+-dependent manner [105, 106] and S100A1 binds IP3 (Baron, Coburn, and Zimmer, personal communication). New information regarding S100 family member specificity for individual target proteins and the structural motifs that mediate these interactions are detailed below. As discussed, the diversity among individual S100s, the protein targets that they interact with, and the Ca2+ dependency of these interactions are uniquely suited to conferring cell-type specificity to Ca2+-signaling pathways.
1.1. The S100 Calcium-Switch
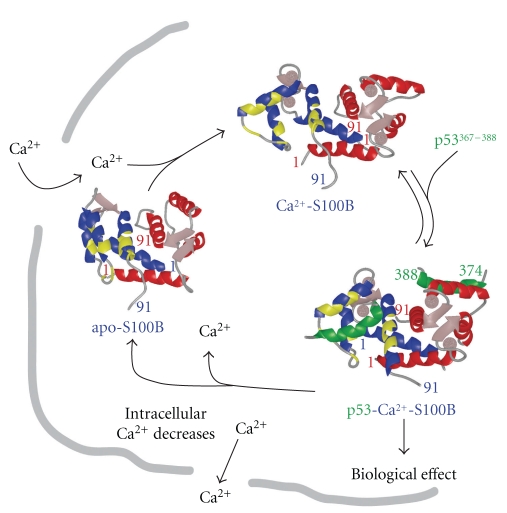
Biophysical and structural biology techniques are used to study molecular determinants involved in Ca2+-dependent S100-target interactions. A comparison of apo- and Ca2+-bound S100B (B-Ca2+) indicates that the pseudo -EF-hand (EF1, CaKD> 350 μM, Table 1) has minor structural changes when Ca2+ binds, whereas, the typical EF-hand (EF2, CaKD = 56 ± 9 μM, Table 1) has a large repositioning of several sidechain oxygen ligands during Ca2+ coordination (EF-2: D61, D63, D65, E67, D69, and E72) [23, 24, 26, 34, 78, 87, 107–109]. Much like S100A1, S100A4, and S100A5, Asp-61 and Asp-63, in positions 1 and 3 of the typical EF-hand, must rotate substantially to achieve a suitable Ca2+ binding orientation in S100B (Figures 2 and 4). That helix 4 is involved in the dimer interface explains why the entering helix 3 and not the exiting helix 4 moves significantly upon Ca2+ binding [23]. The conformational change, termed the “S100 Ca2+-switch”, is a feature unique to S100s and unlike other EF-hand proteins (i.e., calmodulin and troponin C) in which position 12 reorients the exiting helix upon Ca2+ binding [23, 26, 43, 44, 110, 111]. The conformational change of helix 3 involves breaking and forming hydrophobic contacts in several S100s with unique hydrophobic and/or hydrophilic residues becoming exposed for binding its protein target (Figures 2–4). However, S100A10 does not conform to other family members since it lacks a functional EF-hand Ca2+-binding domain, so its target protein interactions are all independent of Ca2+ [11, 112]. In several S100-target structures, much has been learned about specific target- and inhibitor-S100 interactions involving the “hinge (loop 2)”, the C-terminal loop, and the hydrophobic pocket involving helices 3/4 for several S100s (site 1), a second surface, nearby helix 4 and the Zn2+ site in S100B (i.e., site 2) was also discovered more recently in drug design studies aimed at inhibiting S100B [20, 21].
Figure 2.
The Ca2+-dependent S100-target protein interactions. In red/blue are subunits of S100B (dimerKD< 500 pM [1, 17]) with regions shaded (yellow) for residues that bind targets such as p53367-388 (green), p53 (p53321-346, KD = 24 ± 10 nM), or TRTK12 [10, 18].
Figure 4.
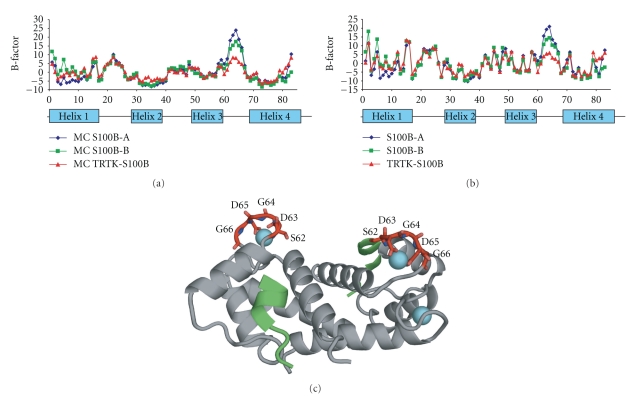
B-factors for X-ray structures of TRTK12-Ca2+-S100B (2.0 Å) and Ca2+-S100B (1.5 Å) from the PI's laboratory. (a) B-factors for backbone atoms for each subunit of Ca2+-S100B (blue, green) and for TRTK-Ca2+-S100B (red). (b) B-factors for sidechains with symbols as in (a). (c) Also shown is a ribbon diagram of the TRTK12-Ca2+-S100B structure with residues colored red in EF2 (residues 61–72), which display lower B-factors in the TRTK12-bound state (in panels (a) and (b)). These data are all published in Charpentier et al., 2010 [139].
However, several important aspects pertaining to the Ca2+- and target-binding properties of S100s are not yet answered. With S100A5 and S100Z as exceptions (CaKD~0.2 μM, Table 1), dimeric S100s typically have a low affinity for binding Ca2+ in vitro (CaKD≥ 1 μM, EF2) [10, 124, 134](Table 1), leading some to classifying this family of proteins as “calcium-buffers” while others predicted that S100s only bound Ca2+ in locations where Ca2+ ion concentrations were relatively high (endoplasmic reticulum and the extracellular space). While low in vitro binding affinities were first thought to eliminate a role for S100 Ca2+-binding in the cytosol, where [Ca2+]i typically oscillates between 0.1 and 1 μM [1, 2], we now know that for many S100s, their Ca2+-binding affinity goes up significantly when its target protein is bound [34, 60, 109] (Figure 3). In fact, for S100A1 and the full-length ryanodine receptor (Figure 3(c)), the complex was detected at resting cytosolic [Ca2+]i (i.e., at 100 nM) even though its Ca2+-binding affinity in vitro in the absence of target is ~300-fold higher (CaKD = 27 ± 2 μM) [60] (Table 1). One explanation for the tightening Ca2+-binding affinity is that target binding induces a structural change to provide a more optimal Ca2+-coordination geometry. Such an explanation was ruled out for S100B when the X-ray structures of Ca2+-S100B (±TRTK12) and found that the Ca2+-coordination in both EF-hands (EF1, EF2) was indistinguishable despite the fact that Cakoff is ~20-fold slower when TRTK12 is bound [34]. Thus, the mechanism for “tightening” of Ca2+-binding with target bound cannot be explained by structural data alone. Interestingly, B-factors from X-ray structures (Figure 4) and 15N NMR relaxation rate studies of S100s have given us some indication that dynamic properties of S100 proteins are involved in the “tightening effect” (Figure 5). Thus, “induced-fit” versus “selected-fit” binding models for both the binding of Ca2+ and target to S100s have been considered. The underlying premise of the “selected-fit” model is that in the absence of target, the Ca2+-S100 complex observed in the X-ray structure is in equilibrium with a dynamic state(s) that has a lower Ca2+-binding affinity; when target protein binds to an Ca2+-S100 complex, the equilibrium shifts so “low affinity states” are eliminated when target binds to give less conformational exchange and an increase in measured Ca2+-binding (i.e., less free Ca2+, slower Cakoff, Figure 3). Important for drug design, we found that S100B inhibitors can mimic target binding to also cause higher affinity Ca2+ binding (i.e., KD< 1 μM). It is also now understood that weak Ca2+-binding for S100s in the absence of target may also turn out to be biologically relevant (Figure 1). Since most target-free S100s have low affinity for Ca2+, this allows numerous stable S100s to be at high concentrations in the cell (>1 μM) without depleting [Ca2+]i levels inside the cell and “short-circuiting” Ca2+ oscillations. Thus, several highly stable S100s can be “poised and ready” in any given cell for when their specific target(s) are expressed, as necessary for them to regulate numerous functions in mammalian cells (Figure 1) [109].
Figure 3.
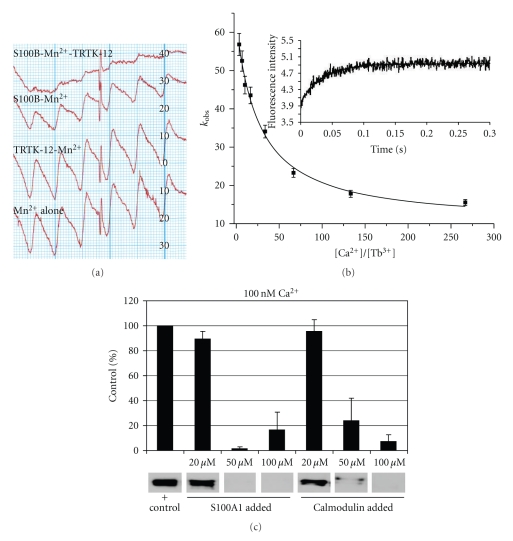
Metal ion and target binding properties of S100 proteins. (a) Binding studies with Mn2+ were completed since it is a good probe of the high affinity Ca2+ binding site on S100B (EF2) [113, 135]. Free Mn2+ was measured by electron paramagnetic resonance (EPR) in the absence and presence of S100B (+/− target peptide TRTK12) [34]. In all four traces, total [Mn2+] is identical (80 μM) with the bottom trace (4th trace) showing the signal for total [Mn2+]. TRTK12 alone (1 mM) has no effect on the EPR signal (3rd trace), whereas, the addition of S100B (65 μM) binds Mn2+ and reduces free [Mn2+] (2nd trace). The addition of the same amount of S100B (65 μM) plus TRTK12 (1 mM, top trace) has the least free [Mn2+] and indicates that TRTK12 binding to S100B-Mn2+ enhances Mn2+ binding (compare traces 1 and 2). A similar effect was observed for SBi1 (unpublished) and for p53367-388 [109]. As for p53367-388 and SBi1, TRTK12 increased the affinity of S100B for Ca2+ in competition studies with Mn2+ and via stopped-flow kinetic measurements of Cakoff as monitored in competition with Tb3+. (b) Plot of the decrease in kobs as a function of [Ca2+]/[Tb3+] as used to determine the off rate of Ca2+ from the 2nd EF-hand (EF2, Cakoff). The kobs values at each [Ca2+]/[Tb3+] ratio were calculated from kinetic traces of stopped-flow experiments where Tb3+ (syringe C) is mixed with S100B at varying Ca2+ concentrations (syringe A) and [Tb3+] signal is monitored as a function of time (λ ex = 230 nm, λ em = 545 nm). A Cakoff of 60 ± 8/sec was calculated from these experiments with S100B alone. When either TRTK12 or SBi1 is present, then the calculated Cakoff value for S100B is reduced to 5 ± 3/sec similar to that found for p53367-388 [109]. These studies demonstrated that TRTK12, p53367-388, or SBi1 increased the affinity of S100B for Ca2+ at least in part by decreasing Cakoff. In (c), S100A1 was found to bind the full length ryanodine receptor (RyR) at 100 nM free calcium. Specifically, S100A1 competed full-length RyR1 away from agarose-linked CaM beads as judged by a decreased RyR1 band in an anti-RyR Western blot. Free CaM, a positive control, also competed the RyR away from CaM-linked beads [60, 67].
Figure 5.
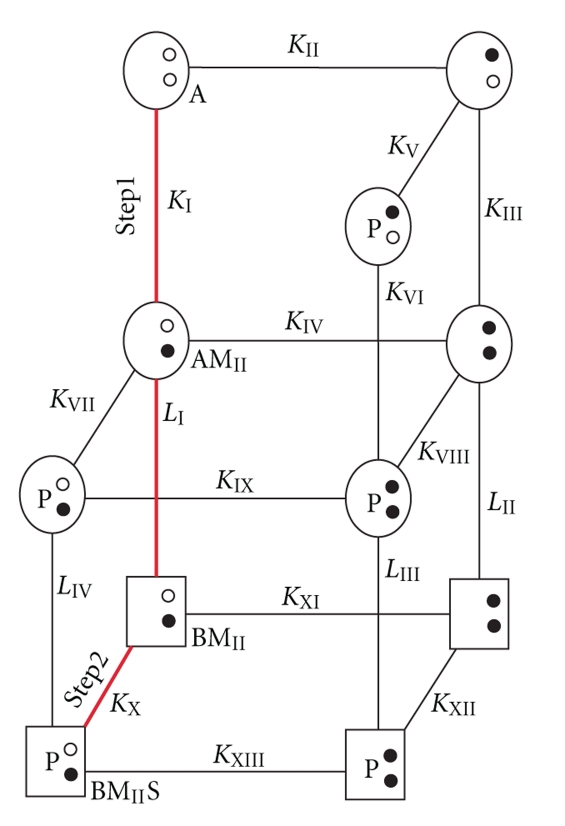
Models for Ca2+-binding and then target-binding to an S100 protein. (Top) A model for the Ca2+-dependent interaction of S100B with target proteins involves 13 equilibrium constants (KI to KXIII), 11 states, and 4 conformational changes (LI–LIV) [109]. The most highly populated states and the predominant pathway are colored red; this is due to weak Ca2+ binding in the pseudo-EF-hand (site I), which greatly simplifies this model (see Scheme 1). Specifically, the binding of Ca2+ to the pseudo- and typical EF-hand in each S100B subunit is described by six states (A, AMI, AMII, AMIMII, BMI, and BMII), five equilibrium constants (KI = [A][M]/[AMI], KII = [A][M]/[AMII], KIII = [AMI][M]/[AMI,II], KIV = [AMII][M]/[AMI,II], and KXI = [BMII][M]/[BMI,II], two conformational changes (LI: AMII↔ BMII, LII: AMI,II↔ BMI,II) with corresponding rate constants, respectively, where A = S100B prior to the 90° reorientation of helix three of S100B, B = S100B after 90° reorientation of helix three, MI = a Ca2+ ion bound to EF-hand I (pseudo-EF-hand), MII = a Ca2+ ion bound to EF-hand II (typical EF-hand), MI,II = Ca2+ ions bound to EF-hand I and EF-hand II. Upon the addition of p53 or another target (S), the model expands to 11 possible states, 13 dissociation constants, and four possible conformational changes. Whether additional equilibriums occur (KXIV, KXV, and KXVI) is considered in Scheme 1. (Bottom) In a second model (Scheme 1), state A is defined as the “closed” conformation observed in the apo-state (Figure 2), and state B is after a 90° reorientation of helix 3 termed the “open” conformation. In black, are states hypothesized to be populated. [A-MII]‡ and [B-MII]‡ represent short-lived intermediates, and L1 is the Ca2+-dependent conformational change involving helix 3 of S100B upon binding Ca2+ (Figure 2). Based on NMR relaxation rate data from the PI's lab [136], KXIV highly favors state A. States are also considered via KXV and KXVI which result in B* states that represent an ensemble of dynamic structures, of which, only a subset fully coordinate Ca2+ as observed in X-ray structures [9]. It is hypothesized that KXV favors the B*MII state(s), whereas, KXVI favors B-MII-S, explaining the apparent increase in Ca2+-binding affinity using equilibrium binding measurements that monitor free [metal ion] (Figure 3).
1.2. The S100 Model for Binding Calcium and Target
As a model system, Ca2+-dependent S100-target interactions are attractive, since S100s, in essence, have only one “functional” EF-hand (EF2, Figure 2) per subunit, which for S100B does not cooperate with the EF1 or EF1′/EF2′ of the other subunit in Ca2+-binding [10, 23, 31, 78, 87, 109]. This is very much unlike calmodulin (CaM) and troponin C (TnC), which have several functional and highly cooperative EF-hand Ca2+-binding domains [66]. Thus, a typical dimeric S100-target interaction is Ca2+-dependent and involves at least 11 possible states, 13 dissociation constants (K1–K13), four conformational changes (L1–L4), and the corresponding rate constants (k1–k17, k−1–k−17), per symmetric subunit [109] (Figure 5). As for Ca2+-binding, the conformational changes and the binding events for a single target are also symmetric and occur independently of those on the other subunit (i.e., without cooperativity), so only one S100 subunit needs to be considered in the thermodynamic scheme [10, 23, 31, 78, 87, 109]. At low [Ca2+] (Figure 5), the predominant kinetic pathway for target binding to S100s can be simplified to KI (k−1/k1, step 1), L1 (k−14/k14), and KX (k−10/k10, step 2) because an S100- or pseudo-EF-hand motif (EF1) binds Ca2+ about an order of magnitude more weakly than EF2 (KI ≪ KII, i.e., KII~ 0.5 mM) [109]. However, in addition to a two-step KI · L1 · KX pathway for target binding, we will also consider three additional equilibriums resulting from conformational exchange prior to Ca2+ binding (KXIV), prior to target binding (KXV), as well as for the S100-target bound state (KXIV, Figure 5, Scheme 1). In step 1 (Scheme 1), structural data provides support for the “induced-fit” aspect of Ca2+ binding to S100B (i.e., A + Ca2+↔ [A − Ca2+]‡↔ B − Ca2+) since the apo-S100B (state A) has a patch of negatively charged residues comprising residues in the typical EF-hand (D65, E67, D69, and E72) sufficient for a fast bimolecular interaction with Ca2+ [23, 24]. Furthermore, no conformational exchange (Rex) has yet been observed in 15N-relaxation rate NMR studies of apo-S100B for any residues in helix 3 or in either of the Ca2+-binding loops that subsequently undergo structural transitions when Ca2+ is added [136]. However, this needs to be examined more rigorously for apo-S100B and for other apo-S100s using ZZ-exchange and relaxation dispersion NMR methods (off-resonance R1ρ, rcCPMG). Nonetheless, the lack of detectable Rex in the apo-state currently provides an argument against the A ↔ B conversion (via KXIV) (Figure 5, Scheme 1, in green). In step 2, for target binding to an S100 (i.e., B −MII+ S ↔ B −MII− S, via KX), NMR data does exhibit Rex for both Ca2+-S100B and Ca2+-S100A1 [59, 60, 77, 136, 137]; these preliminary data are consistent with a model in which target binding selects a competent conformation(s) from an ensemble of dynamic states (via KXV, i.e., “selected-fit” model [138]). The selected-fit hypothesis is also supported by the loss of exchange broadening upon binding of p53 and other targets to Ca2+-S100B [77, 113]. Further, the rate of Ca2+ dissociation (Cakoff) from EF2 decreases from 60/s to 7/s when p53 binds as measured by stopped-flow methods [109]; similar results were found when other targets, including an S100B inhibitor (SBi1) bound to Ca2+-S100B (Cannon and Weber, unpublished results). We now have X-ray crystal structures of S100B-Ca2+ and S100B-Ca2+-TRTK12 that show Ca2+ coordination is indistinguishable in the two complexes [34], whereas, elevated B-factors were observed for S100B-Ca2+ for residues in EF2 in the absence of bound target (Figure 4). One interpretation of the elevated B-factors is that there is conformational dynamics affecting EF2 in the absence of target, however, other explanations cannot be ruled out such as the possibility that lattice contacts are different in the two structures giving rise to the changes in B-factors [34]. Thus, it is important to further examine the dynamics of Ca2+-S100s, including EF2, directly by NMR to determine whether the decreased B-factors in EF2 (Figure 4) were due to a loss of conformational exchange as represented in Figure 5.
Scheme 1.
Additional evidence supporting Scheme 1 (Figure 5) is from stopped-flow experiments with S100-Ca2+ and S100B-Ca2+-target complexes [109]. Here, a fast kinetic-step at the earliest time points in stopped-flow traces has been observed (i.e., biphasic, Cannon and Weber, unpublished results), indicative of KXV. Also satisfying is that KXV in Figure 5 provides a means for a single S100 protein to sample conformational space (i.e., at BMII), as may be necessary to bind more than one target protein, an observation made for several S100 proteins [10]. Structurally similar S100 proteins may also bind the same protein target (i.e., TRTK12), although koff (i.e., k−10) in these cases usually varies due to specific differences in the binding site that give S100-target protein complexes unique conformations and hence varying “lifetimes” inside the cell (i.e., different Cakoff values). These issues regarding specificity are important and require further examination. It is also necessary to remember that one assumption in this model (Figure 5) is that binding of Ca2+ to EF1 is not significant. While this assumption is valid based on existing KD values (and verified for S100B) [87]), weak Ca2+ binding to this EF-hand (EF1) in other S100s may slightly populate additional states, which together with cooperative binding effects observed in some cases [15] could complicate interpretations with this simple model (Figure 5).
1.3. S100 Family Members Exhibit Overlapping but Distinct Target Protein Binding Profiles
While structural, biochemical, and biophysical approaches yield important structural and mechanistic details, they cannot be used to screen simultaneously the interaction of multiple S100 family members with an extensive array of full-length target proteins. Therefore, we developed a quantitative assay that monitors the interaction of fluorophore labeled S100s with membrane-immobilized target proteins that could be used to efficiently identify/prioritize S100-target protein interactions and S100 domains for structural/mechanistic analyses. First, commercially available Alexa-Fluor 488-conjugated calmodulin (CaM-488) and two well-characterized calmodulin target proteins, calmodulin-dependent kinase II (CaM kinase II) and phosphorylase kinase, were used to determine if this methodology would yield data representative of published KDs and KAs (Figure 6). The interaction of CaM-488 with both target proteins was Ca2+ dependent. Immobilized CaM kinase II bound 5.4 pmoles CaM-488 in the presence of Ca2+ and 0.1 pmoles CaM-488 in the absence of Ca2+. Similarly, immobilized phosphorylase kinase bound 0.7 pmoles CaM-488 in the presence of Ca2+ and 0.3 pmoles CaM-488 in the absence of Ca2+. Furthermore, the 8-fold difference in bound CaM-488 (5.4 versus 0.7 pmoles) is in qualitative agreement with the higher affinity reported for CaM kinase II when compared to phosphorylase kinase [140, 141]. These data demonstrate the feasibility of using a membrane binding assay and Alexa-Fluor conjugated Ca2+-receptor proteins to monitor Ca2+ dependency of target protein interactions and to qualitatively compare the binding interaction.
Figure 6.
Characterization of CaM-488 target protein binding. Equimolar concentrations of CaM kinase II (CKII) phosphorylase kinase (PK) were immobilized on a PVDF membrane and incubated with 100 nM CaM-488 in the presence of Ca2+ or EDTA. Panel A contains a representative dot blot image. The histograms in Panel B represent the mean pmoles CaM-488 bound in the presence (black bars) and absence (gray bars) of Ca2+ assayed in triplicate in two independent experiments. Consistent with reported Kds and Kas, both targets exhibited Ca2+-dependent binding with the higher affinity target, CKII, binding more CaM-488 (7.0 pmoles) when compared to the lower affinity target PK (0.8 pmoles).
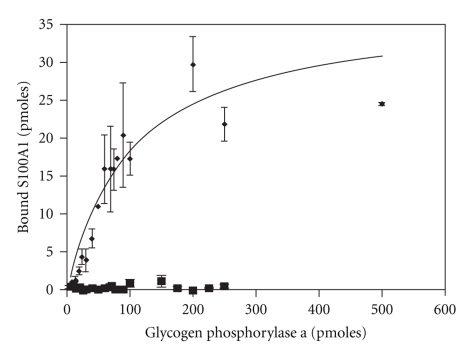
To verify that this methodology could also be used to monitor S100-target protein interactions, the binding of S100A1-488 to a previously characterized target protein, glycogen phosphorylase a, was evaluated [73]. Consistent with results of previous gel overlay and affinity chromatography experiments, S100A1-488 exhibited Ca2+-dependent binding to immobilized glycogen phosphorylase a at all points (Figure 7). Furthermore, S100A1-488 binding was saturable with a Bmax of 30.0 pmoles and EC50 for binding value of 37.5 pmoles. While this type of assay does not provide a KD, the Bmax and EC50 can be used to compare S100-target protein interactions. Nonetheless, full-binding curves do not permit the simultaneous characterization of multiple S100s interacting with numerous target proteins. Therefore, two targets which exhibit differential interactions with S100A1 and S100B were used to determine if a single point assay like the one used in Figure 6 to characterize CaM target protein interactions would accurately reflect S100 target protein interactions. The quantity of target protein (50 pmoles) and probe concentration (100 nM S100A1-488 or S100B-488) were based on the EC50 (~40 pmoles) for glycogen phosphorylase a in 100 nM S100A1-488 (Figure 8). S100A1-488 and S100B-488 exhibited Ca2+-dependent binding to phosphoglucomutase as well as glycogen phosphorylase, and the range of binding was similar to that observed for CaM target proteins. Glycogen phosphorylase a and b bound similar amounts of S100A1-488 (12–15 pmoles) and S100B-488 (15–20 pmoles). In contrast, phosphoglucomutase bound 5-fold more S100B-488 (14 pmoles) than S100A1-488 (2 pmoles). This differential binding was undetectable in previous gel overlay and affinity chromatography experiments despite the differential effects of S100A1 and S100B on phospohglucomutase activity [142]. The insensitivity/inability of the overlay and affinity chromatography experiments to detect differential binding is most likely attributable to the high levels of receptor and/or ligand present. Altogether, these results demonstrate that a single point fluorometric assay can be used to quickly assess the relative affinity and Ca2+-dependency of S100-target protein interactions.
Figure 7.
S100A1-488 binding curves for glycogen phosphorylase a. Membranes containing varying concentrations of glycogen phosphorylase were incubated in S100A1-488 in the presence (∙) or absence (■) of Ca2+. A standard curve of fluorescence intensity per mg of S100-488 was used to determine the experimental amount of labeled S100 per dot of target protein (n = 17).
Figure 8.
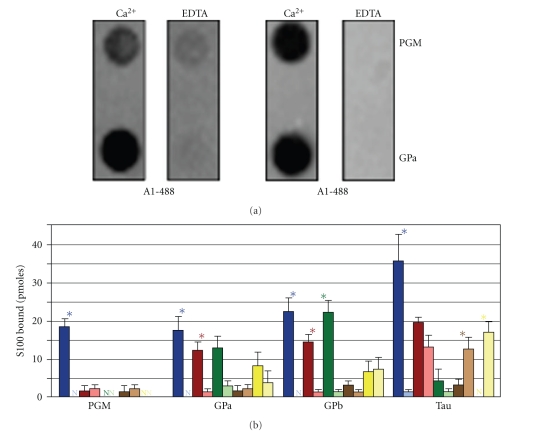
Target protein binding profiles for S100 family members. Membranes containing glycogen phosphorylase (a) (Gpa), glycogen phosphorylase (b) (Gpb), phosphoglucomutase (PGM), and tau (50 pmoles) were incubated in 100 nM Alexa Flour 488 labeled S100B (blue bars), S100A1 (red bars), S100P (green bars), S100A4 (brown bars), and S100A5 (yellow bars) in the presence (darker bars) or absence (lighter bars) of Ca2+. The histograms depict that the mean pmoles S100 bound ± the SEM and N's denote no detectable binding. Asterisks denote P ≤ .05 between the ±Ca2+ conditions.
Next, we compared the ability of different S100 family members (S100A1, S100A4, S100A5, and S100P) to interact with four previously reported S100B targets, glycogen phosphorylase a, glycogen phosphorylase b, tau, and phosphoglucomutase (Figure 8). As expected, all four target proteins bound S100B-488 (10–35 pmoles) in a Ca2+-dependent manner. While none of the other family members exhibited the same target protein binding profile as S100B, there did appear to be two distinct groups: one group (S100A1 and S100P) with profiles that were similar to and another group (S100A4 and S1005) that clearly distinct from S100B. Glycogen phosphorylase a and b bound similar levels of S100A1-488/S100P-488. However, phosphoglucomutase did not bind S100P-488 and 6-fold less S100A1-488 when compared to S100B-488. In addition, tau bound >10 fold less S100P-488 than S100B-488 and binding to S100A1-488 was Ca2+ independent. In the case of S100A4 and S1005, binding to all target proteins was below 10 pmoles. Finally, labeled S100A1, S100B, S100A4, and CaM (100 nM) did not bind to immobilized S100 family members (S100A1, S100B, S100A2, S100A4, S100A5, S100A11, and S100A13), calmodulin, or the negative control α-lactalbumin (75 pmoles) in the presence or absence of Ca2+ (data not shown). Collectively these data demonstrate that while there is extensive overlap among S100 family members with regard to the target proteins that they interact with, each S100 interacts with a unique compliment of target proteins. Nonetheless, there do appear to be family member-specific target proteins. For example, phospohglucomutase preferentially interacts with S100B. Furthermore, the relative affinity and Ca2+ dependency of the interaction can vary among family members. For example, tau is a target protein for multiple S100s including S100A1, S100A4, S100A5, and S100P, but only the interaction of tau with S100B is Ca2+-dependent.
1.4. The Linker Region Mediates Family Member-Specific Binding
The linker region, the amino acid sequence that links the two EF-hands, exhibits the greatest sequence diversity among family members and has been postulated to regulate family member-specific binding to target proteins such as phosphoglucomutase. Consistent with this hypothesis is the accessibility of this region to solvent in both the apo- and Ca2+-bound states of all family members for which 3D structures are available [10, 23, 57, 143]. To test this hypothesis, we checked the ability of a recombinant S100B-A1-B chimeric protein to bind to target proteins, in which the S100B linker region was replaced with comparable amino acid sequence from S100A1 (Figure 8). This chimeric protein was readily purified using similar procedures as used previously for S100B/S100A1, and the mutations to the hinge region did not exhibit any altered biochemical or biophysical properties tested. As anticipated, the interaction of the fluorophore-labeled chimeric protein with the S100A1/S100B targets glycogen phosphorylase a and b was indistinguishable from that of S100A1-488 and S100B-488, that is, Ca2+-dependent binding in the 10–25 pmole range. In contrast, phosphoglucomutase, a target protein that preferentially interacts with S100B, bound 2–4-fold less S100B-A1-B-488 when compared to S100B-488. Interestingly, the presence of the S100A1 linker region did not lower and/or reverse the Ca2+-dependent interaction with tau. Additional experiments will be needed to ascertain the linker region's contribution to other family-member specific target protein interactions as well as Ca2+-independent target protein binding. Nonetheless, this is the first demonstration that the linker region does confer family-specific binding for some Ca2+-dependent target proteins.
1.5. The Ca2+-Dependent Target Protein Binding Domain
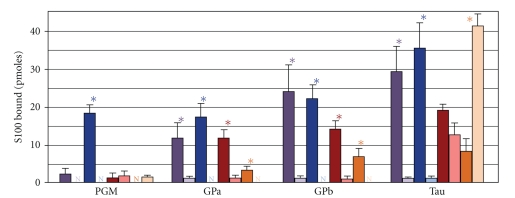
Carboxyl terminal aromatic residues (Phe88, Phe89, Trp90) of S100A1 have been previously shown to regulate Ca2+-dependent interaction of S100A1 with the TRTK peptide, GFAP, and tubulin [72, 144, 145]. Analogous residues are found in several other members including S100B, S100A4, and S100A10 [78, 146–149]. To determine if these residues are obligatory for Ca2+-dependent target protein interactions, we examined the interaction of the S100A1 (F88/89A-W90A)-488 with four additional S100A1 target proteins (Figure 9). As anticipated, mutant S100A1 binding to all four target proteins was decreased by ∼4-fold in the presence of Ca2+. Interestingly, there was a 4-fold increase in mutant S100A1 binding to tau in the absence of Ca2+. These results confirm that carboxy-terminal aromatic residues contribute to the Ca2+-dependent interaction of S100s with protein targets. However, it is not the only mechanism because not all family members that exhibit Ca2+-dependent target protein interactions have hydrophobic residues in their C-terminal extension [150].
Figure 9.
Interaction of wild-type and mutant S100s with target proteins. Membranes containing 50 pmoles glycogen phosphorylase a (Gpa), glycogen phosphorylase b (Gpb), phosphoglucomutase (PGM), and tau were incubated in 100 nM S100B-488 (blue bars), S100A1-488 (red bars), chimeric S100B-A1-B-488 (purple bars), or S100A1(F88/89A-W90A)-488 (orange bars) in the presence (darker bars) or absence (lighter bars) of Ca2+. The histograms depict that the mean pmoles S100 bound ± the SEM and N's denote no detectable binding. The asterisks denote P ≤ .05, and the N's denote no detectable binding between the ±Ca2+ conditions.
2. Summary
In summary, S100 family members have a distinct role in intracellular Ca2+ signaling. The complement of S100s and S100 target proteins expressed in an individual cell allows that cell to transduce a universal change in [Ca2+]i into a unique biological response. Furthermore, their unique metal binding properties and diverse lists of target proteins provide mechanisms for conferring Ca2+/metal sensitivity to cellular processes as well as integration and cross-talk among these processes. Thus, delineating S100-regulated processes in different cell types, ascertaining the relationship between intracellular and extracellular S100s, determining how S100-regulated processes are altered by Ca2+ dysregulation in disease states, and identifying that the molecular events involved are critical for understanding the function of this versatile protein family. The combination of structural, biochemical, molecular, cell biological, and in vivo techniques, which has been successful to date, will ultimately identify “inhibitors” of S100 function that can be used to normalize Ca2+ signaling in diseased cells.
3. Materials and Methods
3.1. Bacterial Expression Vectors for Wild-Type S100 Family Members
The pVex expression vector for human S100P was a generous gift of Dr. George Makhatadze [151]. Bacterial expression vectors for S100A1 and S100B have been previously described [152]. Plasmids encoding rat S100A4 (GenBank accession number AA997272) and S100A5 (GenBank accession number 1772854) were obtained through the IMAGE consortium library (Research Genetics, Huntsville, AL). The coding sequences were amplified using gene specific primers containing synthetic Nde I and Hind III sites at the 5′ and 3′ ends of the coding sequence, respectively. The nucleotide sequences for the primers were 5′-TTCCATATGGCGAGACCCTTGGAGGAG-3′ and 5′-CCCAAGCTTCACTTCTTCCGGGGCTCC-3′ (Lone Star Labs, Houston, TX) for S100A4, 5′-TTCCATATGGAGACTCCTCTTGAGAAG-3′ (Invitrogen, Carlsbad, CA) and 5′-CCCAAGCTTCACTTGTTGTCCTCTAAG-3′ (Lone Star Labs) for S100A5. Gene amplification was performed using an initial heat step (94°C, 5 minutes) followed by 30 cycles consisting of 1 minute at 94°C, 1 minute at 60°C, and 2 minutes at 68°C, and a final extension step of 7 minutes at 68°C. The resulting PCR products were subcloned into the TA cloning vector PCR2.1 (Invitrogen, Carlsbad, CA). The coding sequences were isolated from Nde I-Hind III digests of plasmid DNA and subcloned into pET21a+ (Novagen, San Diego, CA). The entire protein coding sequence was verified by DNA sequence analysis.
3.2. Bacterial Expression Vectors for Mutant S100 Proteins
The bacterial expression vector for the S100A1 triple point mutant (F88/89A-W90A) has been described previously [152]. A two-step PCR protocol followed by directional subcloning was used to generate the expression vector for the S100B-A1-B chimeric protein in which the amino acid sequence for the linker region in S100B (HFLEEIKEQ) was replaced with the S100A1 linker region (SFLDVQKDA). A sense oligonucleotide containing the amino terminal S100B sequence with an engineered Nde I restriction site (5′-CGCCATATGTCTGAACTCGAGAAAGCTG, Invitrogen) and a 3′ antisense oligonucleotide encoding the amino terminal half of the S100A1 linker region and an Xba I site (5′-CCTCTAGAAAGCTGCTAAGTTC, Invitrogen) were used to generate the 5′ half of the chimeric protein. The 3′ half of the chimeric protein was generated using the same template, a sense oligonucleotide encoding the carboxyl terminal half of the S100A1 linker region and an Xba I restriction enzyme site (5′-GCCATTTTCTAGACGTCCAGAAGGACGCGGAAGTTGTAGAC-3′, Integrated DNA Technologies, Coralville, IA) and an antisense oligonucleotide encoding the amino terminus of S100B with an engineered Hind III site (5′-CCCAAGCTTATTCATGTTCG, Intergrated DNA Technologies). The PCR program consisted of 30 cycles (1 minute denaturing at 94°C, 1 minute of annealing at 55°C, and 3 minutes of extension at 72°C). The resulting PCR products were ligated into the TA cloning vector PCR2.1. Purified insert DNA encoding the two halves of the chimeric protein was ligated together and subcloned into pET21a+ using the 5′ Nde I and 3′ Hind III restriction sites. Restriction enzyme digest and sequence analysis confirmed that the insert sequence encoded a protein with the appropriate amino acid sequence changes.
3.3. Expression/Purification of Recombinant S100 Proteins
Recombinant S100B, S100A1, and S100A1 triple point mutant were purified as previously described [152]. Minor modifications in this procedure were made to optimize purification for S100A4, S100A5, S100P, and the S100B-A1-B chimera. Unboiled bacterial lysates for S100A4, S100A5, and S100P were fractionated by ammonium sulfate precipitation. Prior to phenyl-Sepharose chromatography, solid ammonium sulfate was added up to 30% (w/v) to S100A4 [153] and up to 60% for S100P and S100A5. The unboiled S100B-A1-B lysate and the S100A4, S100A5, and S100P ammonium sulfate fractions were chromatographed on phenyl-Sepharose resin equilibrated with 50 mM Tris-Cl pH 7.4, 5 mM CaCl2, and 1 mM β-mercaptoethanol. The resin was washed with buffer containing high salt (50 mM Tris-Cl pH 7.4, 5 mM CaCl2, 1 mM β-mercaptoethanol, and 500 mM NaCl). Protein was eluted in 50 mM Tris-Cl, pH 7.4, 5 mM EDTA, and 1 mM DTT. Fractions containing S100 protein were identified by SDS-PAGE and pooled. The purity of the protein preparations was assessed by SDS-PAGE. Protein samples were considered to be >95% pure when only a single band of ~10,000 kDa was visible on the Coomassie-blue stained gel.
3.4. Alexa Fluor Conjugation
All wild-type and mutant S100 proteins (5.0 mg/ml in PBS) were labeled with the photostable fluorophore, Alexa Fluor 488 (Molecular Probes, Eugene, OR), according to the manufacturer's recommendations. Prior to labeling, the pH was adjusted to ~8.3 with 1 M sodium bicarbonate and EDTA was added to a final concentration of 5.0 mM. Alexa Fluor 488 dye (1 mg) dissolved in DMSO was added to the protein sample. The reaction was incubated at room temperature for one hour in the dark with continuous stirring. Unconjugated dye was hydrolyzed by incubation of the solution overnight in the dark at 4°C. After dilution with 1-2 volumes of water, the pH of the solution was adjusted to 7.4 by the addition of 2 M Tris-Cl pH 7.4. Free dye was separated from protein conjugates by Ca2+-dependent phenyl-Sepharose chromatography as described for protein purification. Next, we determined if this methodology could be used to quantify the binding of S100 proteins to their various target proteins. Alexa Fluor 488 labeled S100A1, S100B, S100A4, S100A5, and S100P exhibited physical characteristics that were indistinguishable from unlabeled proteins including Ca2+-dependent binding to phenyl-Sepharose (data not shown).
3.5. S100-Target Protein Binding Assay
Glycogen phosphorylase a, glycogen phosphorylase b, phosphoglucomutase (PGM), and phosphorylase kinase were purchased from Sigma Chemical Company (St. Louis, MO). Bovine brain tau and calmodulin-dependent kinase II (CaM kinase II) were generous gifts of Gloria Lee (University of Iowa) and Tom Soderling (Vollum Institute, Portland, OR), respectively. Varying concentrations of the target proteins were immobilized on Immobilon-P PVDF membrane (Millipore) using a 96-well dot-blot apparatus (BioRad, Hercules, CA) per supplier's instructions. After a 30-minute incubation, wells were washed twice with three volumes of 20 mM Tris-Cl pH 7.4. The membrane was removed from the dot-blot apparatus, cut into strips, and incubated overnight at room temperature in 1 μM Alexa Flour 488-labeled protein in 50 mM Tris-Cl, pH 7.4 with 200 mM NaCl (buffer A) containing either 5 mM EDTA or 1 mM CaCl2. The strips were rinsed three times in buffer A containing 5 mM EDTA or 1 mM CaCl2 and fluorescence quantified on a Fuji FLA-5000 Image Analysis System (Stamford, CT). A standard curve of fluorescence intensity versus pmoles of PVDF-immobilized S100-488 (or Cam-488) was used to determine the pmoles of labeled protein bound to that concentration of target protein (n ≥ 2). Data points for each target protein concentration were averaged together to yield the mean pmoles of bound S100-488 ± SEM which was calculated for each target protein. The student's t-test was used to determine the statistical significance (P < .05) of any measured differences between the means.
Acknowledgments
The authors would like to acknowledge Drs. Patti Sadosky, Tamiko Porter, Paul Wilder, Kristen Varney, Nathan Wright, Thomas Charpentier, and John May for technical assistance. These studies were supported by start-up funds from Texas A & M University (D. B. Zimmer) and grants from the NIH (GM58888, CA107331 to D. J. Weber).
References
- 1.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nature Reviews Molecular Cell Biology. 2003;4(7):517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ. Inositol trisphosphate and calcium oscillations. Biochemical Society symposium. 2007;(74):1–7. doi: 10.1042/BSS0740001. [DOI] [PubMed] [Google Scholar]
- 3.Moore BW. A soluble protein characteristic of the nervous system. Biochemical and Biophysical Research Communications. 1965;19(6):739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 4.Ravasi T, Hsu K, Goyette J, et al. Probing the S100 protein family through genomic and functional analysis. Genomics. 2004;84(1):10–22. doi: 10.1016/j.ygeno.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Donato R. Intracellular and extracellular roles of S100 proteins. Microscopy Research and Technique. 2003;60(6):540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 6.Donato R. RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Current Molecular Medicine. 2007;7(8):711–724. doi: 10.2174/156652407783220688. [DOI] [PubMed] [Google Scholar]
- 7.Heizmann CW. The multifunctional S100 protein family. Methods in Molecular Biology. 2002;172:69–80. doi: 10.1385/1-59259-183-3:069. [DOI] [PubMed] [Google Scholar]
- 8.Heizmann CW, Ackermann GE, Galichet A. Pathologies involving the S100 proteins and RAGE. Sub-Cellular Biochemistry. 2007;45:93–138. doi: 10.1007/978-1-4020-6191-2_5. [DOI] [PubMed] [Google Scholar]
- 9.Heizmann CW, Fritz G, Schäfer BW. S100 proteins: structure, functions and pathology. Frontiers in Bioscience. 2002;7:1356–1368. doi: 10.2741/A846. [DOI] [PubMed] [Google Scholar]
- 10.Weber DJ, Rustandi RR, Carrier F, Zimmer DB. Interaction of Dimeric S100B(bb) with the Tumor Suppressor Protein: A Model for Ca-Dependent S100-Target Protein Interactions. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. [Google Scholar]
- 11.Zimmer DB, Chaplin J, Baldwin A, Rast M. S100-mediated signal transduction in the nervous system and neurological diseases. Cellular and Molecular Biology. 2005;51(2):201–214. [PubMed] [Google Scholar]
- 12.Zimmer DB, Wright Sadosky P, Weber DJ. Molecular mechanisms of S100-target protein interactions. Microscopy Research and Technique. 2003;60(6):552–559. doi: 10.1002/jemt.10297. [DOI] [PubMed] [Google Scholar]
- 13.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. International Journal of Biochemistry and Cell Biology. 2001;33(7):637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 14.Heizmann CW. Ca2+-binding S100 proteins in the central nervous system. Neurochemical Research. 1999;24(9):1097–1100. doi: 10.1023/a:1020700117665. [DOI] [PubMed] [Google Scholar]
- 15.Heizmann CW, Cox JA. New perspectives on s100 proteins: a multi-functional Ca2+-, Zn2+- and Cu2+-binding protein family. BioMetals. 1998;11(4):383–397. doi: 10.1023/a:1009212521172. [DOI] [PubMed] [Google Scholar]
- 16.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochemical and Biophysical Research Communications. 2004;322(4):1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 17.Drohat AC, Nenortas E, Beckett D, Weber DJ. Oligomerization state of S100b at nanomolar concentration determined by large-zone analytical gel filtration chromatography. Protein Science. 1997;6(7):1577–1582. doi: 10.1002/pro.5560060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. Journal of Molecular Biology. 2002;324(5):1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42(49):14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 20.Charpentier TH, Wilder PT, Liriano MA, et al. Divalent metal ion complexes of S100B in the absence and presence of pentamidine. Journal of Molecular Biology. 2008;382(1):56–73. doi: 10.1016/j.jmb.2008.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charpentier TH, Wilder PT, Liriano MA, et al. Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography. Biochemistry. 2009;48(26):6202–6212. doi: 10.1021/bi9005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drohat AC, Amburgey JC, Abildgaard F, Starich MR, Baldisseri D, Weber DJ. Solution structure of rat Apo-S100B(β β) as determined by NMR spectroscopy. Biochemistry. 1996;35(36):11577–11588. doi: 10.1021/bi9612226. [DOI] [PubMed] [Google Scholar]
- 23.Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ. Solution structure of calcium-bound rat S100B(β β) as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37(9):2729–2740. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 24.Drohat AC, Tjandra N, Baldisseri DM, Weber DJ. The use of dipolar couplings for determining the solution structure of rat apo-S100B (β β) Protein Science. 1999;8(4):800–809. doi: 10.1110/ps.8.4.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilby PM, Van Eldik LJ, Roberts GCK. The solution structure of the bovine S100B protein dimer in the calcium-free state. Structure. 1996;4(9):1041–1052. doi: 10.1016/s0969-2126(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 26.Matsumura H, Shiba T, Inoue T, Harada S, Kai Y. A novel mode of target recognition suggested by the 2.0 Å structure of holo S100B from bovine brain. Structure. 1998;6(2):233–241. doi: 10.1016/s0969-2126(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 27.McClintock KA, Shaw GS. A novel S100 target conformation is revealed by the solution structure of the Ca2+-S100B-TRTK-12 complex. Journal of Biological Chemistry. 2003;278(8):6251–6257. doi: 10.1074/jbc.M210622200. [DOI] [PubMed] [Google Scholar]
- 28.Ostendorp T, Leclerc E, Galichet A, et al. Structural and functional insights into RAGE activation by multimeric S100B. EMBO Journal. 2007;26(16):3868–3878. doi: 10.1038/sj.emboj.7601805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(β β) Nature Structural Biology. 2000;7(7):570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 30.Wilder PT, Varney KM, Weiss MB, Gitti RK, Weber DJ. Solution structure of Zinc- and calcium-bound rat S100B as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44(15):5690–5702. doi: 10.1021/bi0475830. [DOI] [PubMed] [Google Scholar]
- 31.Amburgey JC, Abildgaard F, Starich MR, Shah S, Hilt DC, Weber DJ. 1H, 13C and 15N NMR assignments and solution secondary structure of rat Apo-S100β . Journal of Biomolecular NMR. 1995;6(2):171–179. doi: 10.1007/BF00211781. [DOI] [PubMed] [Google Scholar]
- 32.Brodersen DE, Etzerodt M, Madsen P, et al. EF-hands at atomic resolution: the structure of human psoriasin (S100A7) solved by MAD phasing. Structure. 1998;6(4):477–489. doi: 10.1016/s0969-2126(98)00049-5. [DOI] [PubMed] [Google Scholar]
- 33.Brodersen DE, Nyborg J, Kjeldgaard M. Zinc-binding site of an S100 protein revealed. Two crystal structures of Ca2+-bound human psoriasin (S100A7) in the Zn2+-loaded and Zn2+-free states. Biochemistry. 1999;38(6):1695–1704. doi: 10.1021/bi982483d. [DOI] [PubMed] [Google Scholar]
- 34.Charpentier TH, Thompson LE, Liriano MA, et al. The effects of CapZ peptide (TRTK-12) binding to S100B-Ca2+ as examined by NMR and X-ray crystallography. Journal of Molecular Biology. 2010;396:1227–1243. doi: 10.1016/j.jmb.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charpentier TH, Wilder PT, Varney KM, Toth EA, Weber DJ. Crystal structure of pentamidine bound to Ca2+-S100B. AACR Meeting Abstracts. 2006;2006 456-b- [Google Scholar]
- 36.Fritz G, Mittl PRE, Vasak M, Grütter MG, Heizmann CW. The crystal structure of metal-free human EF-hand protein S100A3 at 1.7-Å resolution. Journal of Biological Chemistry. 2002;277(36):33092–33098. doi: 10.1074/jbc.M200574200. [DOI] [PubMed] [Google Scholar]
- 37.Groves P, Finn BE, Kuźnicki J, Forsén S. A model for target protein binding to calcium-activated S100 dimers. FEBS Letters. 1998;421(3):175–179. doi: 10.1016/s0014-5793(97)01535-4. [DOI] [PubMed] [Google Scholar]
- 38.Itou H, Yao M, Fujita I, et al. The crystal structure of human MRP14 (S100A9), a Ca2+-dependent regulator protein in inflammatory process. Journal of Molecular Biology. 2002;316(2):265–276. doi: 10.1006/jmbi.2001.5340. [DOI] [PubMed] [Google Scholar]
- 39.Kilby PM, Van Eldik LJ, Robert GCK. Nuclear magnetic resonance assignments and secondary structure of bovine S100β protein. FEBS Letters. 1995;363(1-2):90–96. doi: 10.1016/0014-5793(95)00296-l. [DOI] [PubMed] [Google Scholar]
- 40.Korndörfer IP, Brueckner F, Skerra A. The crystal structure of the human (S100A8/S100A9)2 heterotetramer, calprotectin, illustrates how conformational changes of interacting α-helices can determine specific association of two EF-hand proteins. Journal of Molecular Biology. 2007;370(5):887–898. doi: 10.1016/j.jmb.2007.04.065. [DOI] [PubMed] [Google Scholar]
- 41.Kouno T, Mizuguchi M, Sakaguchi M, et al. The structure of S100A11 fragment explains a local structural change induced by phosphorylation. Journal of Peptide Science. 2008;14(10):1129–1138. doi: 10.1002/psc.1050. [DOI] [PubMed] [Google Scholar]
- 42.Malashkevich VN, Varney KM, Garrett SC, et al. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47(18):5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mäler L, Potts BCM, Chazin WJ. High resolution solution structure of apo calcyclin and structural variations in the S100 family of calcium-binding proteins. Journal of Biomolecular NMR. 1999;13(3):233–247. doi: 10.1023/a:1008315517955. [DOI] [PubMed] [Google Scholar]
- 44.Mäler L, Sastry M, Chazin WJ. A structural basis for S100 protein specificity derived from comparative analysis of apo and Ca2+-calcyclin. Journal of Molecular Biology. 2002;317(2):279–290. doi: 10.1006/jmbi.2002.5421. [DOI] [PubMed] [Google Scholar]
- 45.Malik S, Revington M, Smith SP, Shaw GS. Analysis of the structure of human apo-S100B at low temperature indicates a unimodal conformational distribution is adopted by calcium-free S100 proteins. Proteins. 2008;73(1):28–42. doi: 10.1002/prot.22037. [DOI] [PubMed] [Google Scholar]
- 46.Mittl PRE, Fritz G, Sargent DF, Richmond TJ, Heizmann CW, Grütter MG. Metal-free MIRAS phasing: structure of apo-S100A3. Acta Crystallographica Section D. 2002;58(8):1255–1261. doi: 10.1107/s0907444902008430. [DOI] [PubMed] [Google Scholar]
- 47.Moroz OV, Antson AA, Dodson EJ, et al. The structure of S100A12 in a hexameric form and its proposed role in receptor signalling. Acta Crystallographica Section D. 2002;58(3):407–413. doi: 10.1107/s0907444901021278. [DOI] [PubMed] [Google Scholar]
- 48.Moroz OV, Antson AA, Grist SJ, et al. Structure of the human S100A12-copper complex: implications for host-parasite defence. Acta Crystallographica D. 2003;59(5):859–867. doi: 10.1107/s0907444903004700. [DOI] [PubMed] [Google Scholar]
- 49.Moroz OV, Antson AA, Murshudov GN, et al. The three-dimensional structure of human S100A12. Acta Crystallographica Section D. 2001;57(1):20–29. doi: 10.1107/s090744490001458x. [DOI] [PubMed] [Google Scholar]
- 50.Moroz OV, Dodson GG, Wilson KS, Lukanidin E, Bronstein IB. Multiple structural states of S100A12: a key to its functional diversity. Microscopy Research and Technique. 2003;60(6):581–592. doi: 10.1002/jemt.10300. [DOI] [PubMed] [Google Scholar]
- 51.Otterbein LR, Kordowska J, Witte-Hoffmann C, Wang C-LA, Dominguez R. Crystal structures of S100A6 in the Ca2+-free and Ca2+-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution. Structure. 2002;10(4):557–567. doi: 10.1016/s0969-2126(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 52.Potts BCM, Carlström G, Okazaki K, Hidaka H, Chazin WJ. 1H NMR assignments of apo calcyclin and comparative structural analysis with calbindin d(9k) and s 100β . Protein Science. 1996;5(11):2162–2174. doi: 10.1002/pro.5560051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potts BCM, Smith J, Akke M, et al. The structure of calcyclin reveals a novel homodimeric fold for S100 Ca2+-binding proteins. Nature Structural Biology. 1995;2(9):790–796. doi: 10.1038/nsb0995-790. [DOI] [PubMed] [Google Scholar]
- 54.Réty S, Osterloh D, Arié J-P, et al. Structural basis of the Ca2+-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure. 2000;8(2):175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 55.Réty S, Sopkova J, Renouard M, et al. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nature Structural Biology. 1999;6(1):89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 56.Rustandi RR, Baldisseri DM, Inman KG, et al. Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR. Biochemistry. 2002;41(3):788–796. doi: 10.1021/bi0118308. [DOI] [PubMed] [Google Scholar]
- 57.Smith SP, Shaw GS. A change-in-hand mechanism for S100 signalling. Biochemistry and Cell Biology. 1998;76(2-3):324–333. doi: 10.1139/bcb-76-2-3-324. [DOI] [PubMed] [Google Scholar]
- 58.Wilder PT, Baldisseri DM, Udan R, Vallely KM, Weber DJ. Location of the Zn2+-binding site on S100B as determined by NMR spectroscopy and site-directed mutagenesis. Biochemistry. 2003;42(46):13410–13421. doi: 10.1021/bi035334q. [DOI] [PubMed] [Google Scholar]
- 59.Wright NT, Cannon BR, Wilder PT, et al. Solution structure of S100A1 bound to the CapZ peptide (TRTK12) Journal of Molecular Biology. 2009;386(5):1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright NT, Prosser BL, Varney KM, Zimmer DB, Schneider MF, Weber DJ. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. Journal of Biological Chemistry. 2008;283(39):26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright NT, Varney KM, Ellis KC, et al. The three-dimensional solution structure of Ca2+-bound S100A1 as determined by NMR spectroscopy. Journal of Molecular Biology. 2005;353(2):410–426. doi: 10.1016/j.jmb.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H, Wang G, Ding Y, et al. The crystal structure at 2 Å resolution of the Ca2+-binding protein S100P. Journal of Molecular Biology. 2003;325(4):785–794. doi: 10.1016/s0022-2836(02)01278-0. [DOI] [PubMed] [Google Scholar]
- 63.Zhukov I, Ejchart A, Bierzyński A. Structural and motional changes induced in apo-S100A1 protein by the disulfide formation between its Cys 85 residue and β-mercaptoethanol. Biochemistry. 2008;47(2):640–650. doi: 10.1021/bi701762v. [DOI] [PubMed] [Google Scholar]
- 64.Wilder PT, Charpentier TH, Liriano M. In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction. International Journal of High Throughput Screening. 2010;1:109–126. doi: 10.2147/IJHTS.S8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kligman D, Hilt DC. The S100 protein family. Trends in Biochemical Sciences. 1988;13(11):437–443. doi: 10.1016/0968-0004(88)90218-6. [DOI] [PubMed] [Google Scholar]
- 66.Strynadka NCJ, James MNG. Crystal structures of the helix-loop-helix calcium-binding proteins. Annual Review of Biochemistry. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 67.Prosser BL, Wright NT, Hernãndez-Ochoa EO, et al. S100A1 binds to the calmodulin-binding site of ryanodine receptor and modulates skeletal muscle excitation-contraction coupling. Journal of Biological Chemistry. 2008;283(8):5046–5057. doi: 10.1074/jbc.M709231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donato R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochimica et Biophysica Acta. 1999;1450(3):191–231. doi: 10.1016/s0167-4889(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 69.Wilder PT, Lin J, Bair CL, et al. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochimica et Biophysica Acta. 2006;1763(11):1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 70.Donato R, Sorci G, Riuzzi F, et al. S100B’s double life: intracellular regulator and extracellular signal. Biochimica et Biophysica Acta. 2009;1793(6):1008–1022. doi: 10.1016/j.bbamcr.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 71.Zimmer DB, Van Eldik LJ. Identification of a molecular target for the calcium-modulated protein S100. Fructose-1,6-bisphosphate aldolase. Journal of Biological Chemistry. 1986;261(24):11424–11428. [PubMed] [Google Scholar]
- 72.Landar A, Caddell G, Chessher J, Zimmer DB. Identification of an S100A1/S100B target protein: phosphoglucomutase. Cell Calcium. 1996;20(3):279–285. doi: 10.1016/s0143-4160(96)90033-0. [DOI] [PubMed] [Google Scholar]
- 73.Zimmer DB, Dubuisson JG. Identification of an S100 target protein: glycogen phosphorylase. Cell Calcium. 1993;14(4):323–332. doi: 10.1016/0143-4160(93)90053-9. [DOI] [PubMed] [Google Scholar]
- 74.Endo H, Takenaga K, Kanno T, Satoh H, Mori S. Methionine aminopeptidase 2 is a new target for the metastasis-associated protein, S100A4. Journal of Biological Chemistry. 2002;277(29):26396–26402. doi: 10.1074/jbc.M202244200. [DOI] [PubMed] [Google Scholar]
- 75.Filipek A, Jastrzebska B, Nowotny M, Kuznicki J. CacyBP/SIP, a calcyclin and Siah-1-interacting protein, binds EF-hand proteins of the S100 family. Journal of Biological Chemistry. 2002;277(32):28848–28852. doi: 10.1074/jbc.M203602200. [DOI] [PubMed] [Google Scholar]
- 76.Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence JJ. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(23):11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rustandi RR, Baldisseri DM, Drohat AC, Weber DJ. Structural changes in the C-terminus of Ca2+-bound rat S100B(β β) upon binding to a peptide derived from the C-terminal regulatory domain of p53. Protein Science. 1999;8(9):1743–1751. doi: 10.1110/ps.8.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca2+-dependent interaction of S100B(β β) with a peptide derived from p53. Biochemistry. 1998;37(7):1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 79.Van Dieck J, Brandt T, Teufel DP, Veprintsev DB, Joerger AC, Fersht AR. Molecular basis of S100 proteins interacting with the p53 homologs p63 and p73. Oncogene. 2010;29(14):2024–2035. doi: 10.1038/onc.2009.490. [DOI] [PubMed] [Google Scholar]
- 80.van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. Journal of Biological Chemistry. 2009;284(20):13804–13811. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin J, Blake M, Tang C, et al. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. Journal of Biological Chemistry. 2001;276(37):35037–35041. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- 82.Lin J, Yang Q, Yan Z, et al. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. Journal of Biological Chemistry. 2004;279(32):34071–34077. doi: 10.1074/jbc.M405419200. [DOI] [PubMed] [Google Scholar]
- 83.Scotto C, Deloulme JC, Rousseau D, Chambaz E, Baudier J. Calcium and S100B regulation of p53-dependent cell growth arrest and apoptosis. Molecular and Cellular Biology. 1998;18(7):4272–4281. doi: 10.1128/mcb.18.7.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scotto C, Delphin C, Deloulme JC, Baudier J. Concerted regulation of wild-type p53 nuclear accumulation and activation by S100B and calcium-dependent protein kinase C. Molecular and Cellular Biology. 1999;19(10):7168–7180. doi: 10.1128/mcb.19.10.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barber KR, McClintock KA, Jamieson GA, Jr., Dimlich RVW, Shaw GS. Specificity and Zn2+ enhancement of the S100B binding epitope TRTK-12. Journal of Biological Chemistry. 1999;274(3):1502–1508. doi: 10.1074/jbc.274.3.1502. [DOI] [PubMed] [Google Scholar]
- 86.Baudier J, Haglid K, Haiech J, Gerard D. Zinc ion binding to human brain calcium binding proteins, calmodulin and S100b protein. Biochemical and Biophysical Research Communications. 1983;114(3):1138–1146. doi: 10.1016/0006-291x(83)90681-2. [DOI] [PubMed] [Google Scholar]
- 87.Chaudhuri D, Horrocks WD, Jr., Amburgey JC, Weber DJ. Characterization of lanthanide ion binding to the EF-hand protein S100β by luminescence spectroscopy. Biochemistry. 1997;36(32):9674–9680. doi: 10.1021/bi9704358. [DOI] [PubMed] [Google Scholar]
- 88.Davey GE, Murmann P, Heizmann CW. Intracellular Ca2+ and Zn2+ levels regulate the alternative cell density-dependent secretion of S100B in human glioblastoma cells. Journal of Biological Chemistry. 2001;276(33):30819–30826. doi: 10.1074/jbc.M103541200. [DOI] [PubMed] [Google Scholar]
- 89.Franz C, Durussel I, Cox JA, Schäfer BW, Heizmann CW. Binding of Ca2+ and Zn2+ to human nuclear S100A2 and mutant proteins. Journal of Biological Chemistry. 1998;273(30):18826–18834. doi: 10.1074/jbc.273.30.18826. [DOI] [PubMed] [Google Scholar]
- 90.Fritz G, Heizmann CW, Kroneck PMH. Probing the structure of the human Ca2+- and Zn2+-binding protein S100A3: spectroscopic investigations of its transition metal ion complexes, and three-dimensional structural model. Biochimica et Biophysica Acta. 1998;1448(2):264–276. doi: 10.1016/s0167-4889(98)00138-4. [DOI] [PubMed] [Google Scholar]
- 91.Gläser R, Harder J, Lange H, Bartels J, Christophers E, Schröder J-M. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nature Immunology. 2005;6(1):57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 92.Gribenko AV, Makhatadze GI. Oligomerization and divalent ion binding properties of the S100P protein: a Ca2+/Mg2+-switch model. Journal of Molecular Biology. 1998;283(3):679–694. doi: 10.1006/jmbi.1998.2116. [DOI] [PubMed] [Google Scholar]
- 93.Heierhorst J, Mann RJ, Kemp BE. Interaction of the recombinant S100A1 protein with twitchin kinase, and comparison with other Ca2+-binding proteins. European Journal of Biochemistry. 1997;249(1):127–133. doi: 10.1111/j.1432-1033.1997.00127.x. [DOI] [PubMed] [Google Scholar]
- 94.Hoyaux D, Alao J, Fuchs J, et al. S100A6, a calcium- and zinc-binding protein, is overexpressed in SOD1 mutant mice, a model for amyotrophic lateral sclerosis. Biochimica et Biophysica Acta. 2000;1498(2-3):264–272. doi: 10.1016/s0167-4889(00)00101-4. [DOI] [PubMed] [Google Scholar]
- 95.Kerkhoff C, Vogl T, Nacken W, Sopalla C, Sorg C. Zinc binding reverses the calcium-induced arachidonic acid-binding capacity of the S100A8/A9 protein complex. FEBS Letters. 1999;460(1):134–138. doi: 10.1016/s0014-5793(99)01322-8. [DOI] [PubMed] [Google Scholar]
- 96.Kordowska J, Stafford WF, Wang C-LA. Ca2+ and Zn2+ bind to different sites and induce different conformational changes in human calcyclin. European Journal of Biochemistry. 1998;253(1):57–66. doi: 10.1046/j.1432-1327.1998.2530057.x. [DOI] [PubMed] [Google Scholar]
- 97.Mbele GO, Deloulme JC, Gentil BJ, et al. The zinc- and calcium-binding S100B interacts and co-localizes with IQGAP1 during dynamic rearrangement of cell membranes. Journal of Biological Chemistry. 2002;277(51):49998–50007. doi: 10.1074/jbc.M205363200. [DOI] [PubMed] [Google Scholar]
- 98.Ogoma Y, Kobayashi H, Fujii T, et al. Binding study of metal ions to S100 protein: 43Ca, 25Mg, 67Zn and 39K n.m.r. International Journal of Biological Macromolecules. 1992;14(5):279–286. doi: 10.1016/s0141-8130(05)80041-8. [DOI] [PubMed] [Google Scholar]
- 99.Ogoma Y, Shimizu T, Kobayashi H, et al. Effects of Ca2+ and Zn2+ on trifluoperazine-S100 proteins interactions: induced circular dichroism and fluorescence spectra. Biochimica et Biophysica Acta. 1989;997(3):188–192. doi: 10.1016/0167-4838(89)90185-4. [DOI] [PubMed] [Google Scholar]
- 100.Deloulme JC, Assard N, Mbele GO, Mangin C, Kuwano R, Baudier J. S100A6 and S100A11 are specific targets of the calcium- and zinc-binding S100B protein in vivo. Journal of Biological Chemistry. 2000;275(45):35302–35310. doi: 10.1074/jbc.M003943200. [DOI] [PubMed] [Google Scholar]
- 101.Deloulme JC, Gentil BJ, Baudier J. Monitoring of S100 homodimerization and heterodimeric interactions by the yeast two-hybrid system. Microscopy Research and Technique. 2003;60(6):560–568. doi: 10.1002/jemt.10298. [DOI] [PubMed] [Google Scholar]
- 102.Gribenko AV, Hopper JE, Makhatadze GI. Molecular characterization and tissue distribution of a novel member of the S100 family of EF-hand proteins. Biochemistry. 2001;40(51):15538–15548. doi: 10.1021/bi0114731. [DOI] [PubMed] [Google Scholar]
- 103.Wang G, Rudland PS, White MR, Barraclough R. Interaction in vivo and in vitro of the metastasis-inducing S100 protein, S100A4 (p9Ka) with S100A1. Journal of Biological Chemistry. 2000;275(15):11141–11146. doi: 10.1074/jbc.275.15.11141. [DOI] [PubMed] [Google Scholar]
- 104.Yang Q, O’Hanlon D, Heizmann CW, Marks A. Demonstration of heterodimer formation between S100B and S100A6 in the yeast two-hybrid system and human melanoma. Experimental Cell Research. 1999;246(2):501–509. doi: 10.1006/excr.1998.4314. [DOI] [PubMed] [Google Scholar]
- 105.Hagens G, Masouyé I, Augsburger E, Hotz R, Saurat J-H, Siegenthaler G. Calcium-binding protein S100A7 and epidermal-type fatty acid-binding protein are associated in the cytosol of human keratinocytes. Biochemical Journal. 1999;339(2):419–427. [PMC free article] [PubMed] [Google Scholar]
- 106.Kerkhoff C, Klempt M, Kaever V, Sorg C. The two calcium-binding proteins, S100A8 and S100A9, are involved in the metabolism of arachidonic acid in human neutrophils. Journal of Biological Chemistry. 1999;274(46):32672–32679. doi: 10.1074/jbc.274.46.32672. [DOI] [PubMed] [Google Scholar]
- 107.Baudier J, Gérard D. Ions binding to S100 proteins: structural changes induced by calcium and zinc on S100a and S100b proteins. Biochemistry. 1983;22(14):3360–3369. doi: 10.1021/bi00283a009. [DOI] [PubMed] [Google Scholar]
- 108.Baudier J, Gerard D. Ions binding to S100 proteins. II. Conformational studies and calcium-induced conformational changes in S100α α protein: the effect of acidic pH and calcium incubation on subunit exchange in S100a (α β) protein. Journal of Biological Chemistry. 1986;261(18):8204–8212. [PubMed] [Google Scholar]
- 109.Markowitz J, Rustandi RR, Varney KM, et al. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 2005;44(19):7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- 110.Yap KL, Ames JB, Swindells MB, Ikura M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins: Structure, Function and Genetics. 1999;37(3):499–507. doi: 10.1002/(sici)1097-0134(19991115)37:3<499::aid-prot17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 111.Smith SP, Shaw GS. A novel calcium-sensitive switch revealed by the structure of human S100B in the calcium-bound form. Structure. 1998;6(2):211–222. doi: 10.1016/s0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 112.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochemical Journal. 2006;396(2):201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca2+-dependent interaction of S100B(β β) with a peptide derived from p53. Biochemistry. 1998;37(7):1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 114.Baudier J, Cole RD. The Ca2+-binding sequence in bovine brain S100b protein β-subunit. A spectroscopic study. Biochemical Journal. 1989;264(1):79–85. doi: 10.1042/bj2640079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baudier J, Glasser N, Gerard D. Ions binding to S100 proteins. I. Calcium- and zinc-binding properties of bovine brain S100α α, S100a (α β), and S100b (β β) protein: Zn2+ regulates Ca2+ binding on S100b protein. Journal of Biological Chemistry. 1986;261(18):8192–8203. [PubMed] [Google Scholar]
- 116.Baudier J, Labourdette G, Gerard D. Rat brain S100b protein: purification, characterization, and ion binding properties. A comparison with bovine S100b protein. Journal of Neurochemistry. 1985;44(1):76–84. doi: 10.1111/j.1471-4159.1985.tb07115.x. [DOI] [PubMed] [Google Scholar]
- 117.Durussel I, Van Eldik LJ, Cox JA. Ion-binding properties of recombinant S100β and two derivatives with either an inactivated Ca2+ site II or a normalized Ca2+ site I. Biochimica et Biophysica Acta. 1997;1343(2):139–143. doi: 10.1016/s0167-4838(97)00106-4. [DOI] [PubMed] [Google Scholar]
- 118.Marshak DR, Watterson DM, Van Eldik LJ. Calcium-dependent interaction of S100b, troponin C, and calmodulin with an immobilized phenothiazine. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(11):6793–6797. doi: 10.1073/pnas.78.11.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mely Y, Gerard D. Structural and ion-binding properties of an S100b protein mixed disulfide: comparison with the reappraised native S100b protein properties. Archives of Biochemistry and Biophysics. 1990;279(1):174–182. doi: 10.1016/0003-9861(90)90478-h. [DOI] [PubMed] [Google Scholar]
- 120.Pozdnyakov N, Goraczniak R, Margulis A, et al. Structural and functional characterization of retinal calcium-dependent guanylate cyclase activator protein (CD-GCAP): identity with S100β protein. Biochemistry. 1997;36(46):14159–14166. doi: 10.1021/bi971792l. [DOI] [PubMed] [Google Scholar]
- 121.Fohr UG, Heizmann CW, Engelkamp D, Schafer BW, Cox JA. Purification and cation binding properties of the recombinant human S100 calcium-binding protein A3, an EF-hand motif protein with high affinity for zinc. Journal of Biological Chemistry. 1995;270(36):21056–21061. doi: 10.1074/jbc.270.36.21056. [DOI] [PubMed] [Google Scholar]
- 122.Dukhanina EA, Dukhanin AS, Lomonosov MY, Lukanidin EM, Georgiev GP. Spectral studies on the calcium-binding properties of Mts 1 protein and its interaction with target protein. FEBS Letters. 1997;410(2-3):403–406. doi: 10.1016/s0014-5793(97)00576-0. [DOI] [PubMed] [Google Scholar]
- 123.Pedrocchi M, Schafer BW, Durussel I, Cox JA, Heizmann CW. Purification and characterization of the recombinant human calcium-binding S100 proteins CAPL and CACY. Biochemistry. 1994;33(21):6732–6738. doi: 10.1021/bi00187a045. [DOI] [PubMed] [Google Scholar]
- 124.Schäfer BW, Fritschy J-M, Murmann P, et al. Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. Journal of Biological Chemistry. 2000;275(39):30623–30630. doi: 10.1074/jbc.M002260200. [DOI] [PubMed] [Google Scholar]
- 125.Kuźnicki J, Filipek A. Purification and properties of a novel Ca2+-binding protein (10.5 kDa) from Ehrlich-ascites-tumour cells. Biochemical Journal. 1987;247(3):663–667. doi: 10.1042/bj2470663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mani RS, Kay CM. Isolation and characterization of a novel molecular weight 11 000 Ca2+-binding protein from smooth muscle. Biochemistry. 1990;29(6):1398–1404. doi: 10.1021/bi00458a009. [DOI] [PubMed] [Google Scholar]
- 127.Vorum H, Madsen P, Rasmussen HH, et al. Expression and divalent cation binding properties of the novel chemotactic inflammatory protein psoriasin. Electrophoresis. 1996;17(11):1787–1796. doi: 10.1002/elps.1150171118. [DOI] [PubMed] [Google Scholar]
- 128.Allen BG. Characterization of the Ca2+-binding properties of calgizzarin (S100C) isolated from chicken gizzard smooth muscle. Biochemistry and Cell Biology. 1996;74(5):687–694. doi: 10.1139/o96-075. [DOI] [PubMed] [Google Scholar]
- 129.Dell’Angelica EC, Schleicher CH, Santome JA. Primary structure and binding properties of calgranulin C, a novel S100- like calcium-binding protein from pig granulocytes. Journal of Biological Chemistry. 1994;269(46):28929–28936. [PubMed] [Google Scholar]
- 130.Ridinger K, Schäfer BW, Durussel I, Cox JA, Heizmann CW. S100A13. Biochemical characterization and subcellular localization in different cell lines. Journal of Biological Chemistry. 2000;275(12):8686–8694. doi: 10.1074/jbc.275.12.8686. [DOI] [PubMed] [Google Scholar]
- 131.Marenholz I, Lovering RC, Heizmann CW. An update of the S100 nomenclature. Biochimica et Biophysica Acta. 2006;1763(11):1282–1283. doi: 10.1016/j.bbamcr.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 132.Sturchler E, Cox JA, Durussel I, Weibel M, Heizmann CW. S100A16, a novel calcium-binding protein of the EF-hand superfamily. Journal of Biological Chemistry. 2006;281(50):38905–38917. doi: 10.1074/jbc.M605798200. [DOI] [PubMed] [Google Scholar]
- 133.Becker T, Gerke V, Kube E, Weber K. S100P, a novel Ca2+-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. European Journal of Biochemistry. 1992;207(2):541–547. doi: 10.1111/j.1432-1033.1992.tb17080.x. [DOI] [PubMed] [Google Scholar]
- 134.Hilt DC, Kligman D. The S100 protein family: a biochemical and functional overview. In: Heizmann C, editor. Novel Calcium Binding Proteins. Berlin, Germany: Springer; 1991. pp. 65–103. [Google Scholar]
- 135.Mildvan AS, Loeb LA. The role of metal ions in the mechanisms of DNA and RNA polymerases. CRC Critical Reviews in Biochemistry. 1979;6(3):219–244. doi: 10.3109/10409237909102564. [DOI] [PubMed] [Google Scholar]
- 136.Inman KG, Baldisseri DM, Miller KE, Weber DJ. Backbone dynamics of the calcium-signaling protein apo-S100B as determined by 15N NMR relaxation. Biochemistry. 2001;40(12):3439–3448. doi: 10.1021/bi0027478. [DOI] [PubMed] [Google Scholar]
- 137.Wright NT, Inman KG, Levine JA, Cannon BR, Varney KM, Weber DJ. Refinement of the solution structure and dynamic properties of Ca2+-bound rat S100B. Journal of Biomolecular NMR. 2008;42(4):279–286. doi: 10.1007/s10858-008-9282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang C, Karpowich N, Hunt JF, Rance M, Palmer AG. Dynamics of ATP-binding cassette contribute to allosteric control, nucleotide binding and energy transduction in ABC transporters. Journal of Molecular Biology. 2004;342(2):525–537. doi: 10.1016/j.jmb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 139.Charpentier TH, Thompson LE, Liriano M, et al. The effects of the CapZ peptide (TRTK-12) binding to S100B-Ca2+ as examined by NMR and X-ray crystallography. Journal of Molecular Biology. 2010;396(5):1227–1243. doi: 10.1016/j.jmb.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cohen P, Picton C, Klee CB. Activation of phosphorylase kinase from rabbit skeletal muscle by calmodulin and troponin. FEBS Letters. 1979;104(1):25–30. doi: 10.1016/0014-5793(79)81078-9. [DOI] [PubMed] [Google Scholar]
- 141.Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science. 1992;256(5060):1199–1202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- 142.Landar A, Caddell G, Chessher J, Zimmer DB. Identification of an S100A1/S100B target protein: phosphoglucomutase. Cell Calcium. 1996;20(3):279–285. doi: 10.1016/s0143-4160(96)90033-0. [DOI] [PubMed] [Google Scholar]
- 143.McClintock KA, Van Eldik LJ, Shaw GS. The C-terminus and linker region of S100B exert dual control on protein-protein interactions with TRTK-12. Biochemistry. 2002;41(17):5421–5428. doi: 10.1021/bi011732m. [DOI] [PubMed] [Google Scholar]
- 144.Garbuglia M, Verzini M, Rustandi RR, et al. Role of the C-terminal extension in the interaction of S100A1 with GFAP, tubulin, the S100A1- and S100B-inhibitory peptide, TRTK-12, and a peptide derived from p53, and the S100A1 inhibitory effect on GFAP polymerization. Biochemical and Biophysical Research Communications. 1999;254(1):36–41. doi: 10.1006/bbrc.1998.9881. [DOI] [PubMed] [Google Scholar]
- 145.Osterloh D, Ivanenkov VV, Gerke V. Hydrophobic residues in the C-terminal region of S100A1 are essential for target protein binding but not for dimerization. Cell Calcium. 1998;24(2):137–151. doi: 10.1016/s0143-4160(98)90081-1. [DOI] [PubMed] [Google Scholar]
- 146.Kube E, Becker T, Weber K, Gerke V. Protein-protein interaction studied by site-directed mutagenesis. Journal of Biological Chemistry. 1992;267(20):14175–14182. [PubMed] [Google Scholar]
- 147.Wilder PT, Rustandi RR, Drohat AC, Weber DJ. S100b(β β) inhibits the protein kinase C-dependent phosphorylation of a peptide derived from p53 in a Ca2+-dependent manner. Protein Science. 1998;7(3):794–798. doi: 10.1002/pro.5560070330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang S, Wang G, Fernig DG, et al. Interaction of metastasis-inducing S100A4 protein in vivo by fluorescence lifetime imaging microscopy. European Biophysics Journal. 2005;34(1):19–27. doi: 10.1007/s00249-004-0428-x. [DOI] [PubMed] [Google Scholar]
- 149.Zhang S, Wang G, Liu D, et al. The C-terminal region of S100A4 is important for its metastasis-inducing properties. Oncogene. 2005;24(27):4401–4411. doi: 10.1038/sj.onc.1208663. [DOI] [PubMed] [Google Scholar]
- 150.Choi J, Chang J-S, Song M-S, et al. Association of hepatitis B virus polymerase with promyelocytic leukemia nuclear bodies mediated by the S100 family protein p11. Biochemical and Biophysical Research Communications. 2003;305(4):1049–1056. doi: 10.1016/s0006-291x(03)00881-7. [DOI] [PubMed] [Google Scholar]
- 151.Gribenko A, Lopez MM, Richardson JM, III, Makhatadze GI. Cloning, overexpression, purification, and spectroscopic characterization of human S100P. Protein Science. 1998;7(1):211–215. doi: 10.1002/pro.5560070123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Landar A, Rustandi RR, Weber DJ, Zimmer DB. S100A1 utilizes different mechanisms for interacting with calcium-dependent and calcium-independent target proteins. Biochemistry. 1998;37(50):17429–17438. doi: 10.1021/bi9817921. [DOI] [PubMed] [Google Scholar]
- 153.Vallely KM, Rustandi RR, Ellis KC, Varlamova O, Bresnick AR, Weber DJ. Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy. Biochemistry. 2002;41(42):12670–12680. doi: 10.1021/bi020365r. [DOI] [PubMed] [Google Scholar]