Abstract
Angiogenin (ANG) is a 14 kDa angiogenic ribonuclease that is upregulated in a variety of human cancers. Accumulating evidence indicates that the angiogenic activity of ANG is related to its ability in regulating ribosomal RNA (rRNA) transcription. ANG is translocated to the nucleus of growth-stimulated endothelial cells where it accumulates in the nucleolus, binds to the promoter region of ribosomal DNA (rDNA), and stimulates rRNA transcription. This normally well-controlled process of nuclear translocation of ANG is hijacked by cancer cells that constitutively uptake ANG and translocate it into the nucleus so that rRNA is constantly transcribed to meet a higher metabolic requirement of this cells. Nuclear translocation of ANG therefore controls the rate of rRNA transcription and has been shown to be a molecular target for cancer drug development. Recently, ANG has also been shown to play a role in motor neuron physiology. Loss-of-function mutations in the coding region of ANG gene have been found in patients with amyotrophic lateral sclerosis (ALS). A deficiency in ANG function may result in insufficient rRNA transcription in motor neurons that require robust ribosome biogenesis due to the demand of long axonal transport. Haploinsufficiency of ANG has thus been implicated in ALS pathogenesis, and ANG has been shown to promote motor neuron survival both in vitro and in vivo. Promotion of ANG expression or activity has been recognized as a therapeutic opportunity for ALS treatment.
Keywords: Angiogenin, ANG, rRNA transcription, cancer, neurodegeneration, rDNA, amyotrophic lateral sclerosis, ALS
ANG
ANG was originally isolated from the conditioned medium of HT-29 human colon adenocarcinoma cells based solely on its angiogenic activity in the chicken embryo chorioallantoic membrane (CAM) angiogenesis assay [1]. Subsequently, it has been found to have a wide tissue distribution, with the liver to be the major source for circulating ANG in plasma [2]. ANG is the 5th member of the pancreatic ribonuclease (RNase) superfamily. It has a 33% amino acid identity and an overall homology of 56% to that of RNase-A, the 1st and prototypic member of this superfamily [3]. However, the ribonucleolytic activity of ANG is several orders of magnitude lower than that of RNase-A [4]. Strikingly, this low but unique ribonucleolytic activity is important for the biological activity of ANG [4]. The amino acid residues important for catalysis are conserved in all vertebrate ANG from fish to human [5]. Extensive work on site-directed mutagenesis has shown that ANG variants with reduced enzymatic activity all have reduced angiogenic activity [4, 6–13]. Some but not all of the variants having enhanced enzymatic activity have enhanced angiogenic activity [9, 14]. Structural work indicated that one of the reasons for ANG to have a reduced ribonucleolytic activity is that the side chain of Gln 117 occupies part of the enzymatic active site so that substrate binding is compromised [15].
ANG is angiogenic, whereas the prototype family member RNase-A is not. Two important structural differences between ANG and RNase-A are responsible for this feature. The first is that ANG has a receptor binding site comprising of amino acid residues 59 to 68 [8, 16] but RNase-A does not have this site as the corresponding amino acid sequence of this region is very different [15, 17]. Therefore ANG binds to its target cells (including endothelial cells, cancer cells and motor neurons) but RNase-A does not. Another structural difference between ANG and RNase-A is that ANG has a nuclear localization sequence (NLS) consisting of 30MRRRG34, whereas RNase-A does not [18]. Therefore ANG undergoes nuclear translocation in its target cells but RNase-A does not. Table 1 lists the differences and similarities between ANG and RNase-A.
Table 1.
Properties of ANG and RNase A
| Protein | MW (kDa) | PI | Enzyme Activity | Receptor Binding | Nuclear translocation | rRNA transcription | Angiogenic activity | Neuro protection |
|---|---|---|---|---|---|---|---|---|
| ANG | 14.4 | 10.1 | 1 | Yes | Yes | Yes | Yes | Yes |
| RNase A | 13.7 | 9.6 | ~10,000 | No | No | No | No | No |
Role of ANG in rRNA transcription
ANG has been shown to undergo nuclear translocation in endothelial cells [18–20], in cancer cells [21, 22], and in motor neurons [23–25]. Nuclear translocation of exogenous ANG is very fast and occurs through receptor-mediated endocytosis [18], is independent of microtubules and lysosomes [20]. ANG seems to enter the nuclear pore by the classic nuclear pore input route [18]. Upon arriving at the nucleus, ANG accumulates in the nucleolus [18] where ribosome biogenesis takes place. Nuclear ANG has been shown to bind to the promoter region of rDNA [26] and stimulates rRNA transcription [27, 28]. Thus, ANG acts as a transcription factor to promote rRNA transcription directly. An ANG binding element (ABE) has been identified from the non transcribed region of rDNA and shown to have ANG-dependent promoter activity in a luciferase reporter assay [26].
Cell growth requires the production of new ribosomes, a process involving rRNA transcription, processing of the pre-rRNA precursor and assembly of the mature rRNA with ribosomal proteins [29–31]. The rate-limiting step in ribosome biogenesis is rRNA transcription, which would be an important aspect of growth control. Ribosomes are also important for maintaining a normal cell function as proteins are required for essentially all cellular activities. ANG-stimulated rRNA transcription has been shown to be permissive for other angiogenic factors to induce angiogenesis [21]. For cancer cells and motor neurons, ANG-mediated rRNA synthesis is critical for sustained proliferation of cancer cells [21, 27, 32, 33] and for maintenance of normal physiological function of motor neurons [25, 34].
Up-regulation of ANG in cancer
ANG plays an important role in tumor angiogenesis [35, 36]. Its expression is up-regulated in many types of cancers [37], particularly in prostate cancer [22, 36, 38–40]. Olson et al used IHC to detect ANG protein level in the prostate and found that ANG expression is dramatically enhanced in the cancer tissues from all 10 patients as compared to the 7 normal subjects [40]. Majumder et al used ELISA to analyze the ANG level in the blood and reported that normal subjects (n=37), androgen-dependent (n=39) and androgen-independent (n=40) prostate cancer patients have an ANG level of 328 ± 20, 392 ± 17, and 436 ± 24 ng/ml, respectively [39]. There is a statistically significant difference in ANG levels between the controls and androgen-dependent prostate cancer patients (p<0.01) and between controls and androgen-independent prostate cancer patients (p<0.001). Following these two studies, Katona et al analyzed ANG expression level in a large cohort (n=107) of radical prostatectomy samples by IHC (with a polyclonal antibody) and reported that ANG expression is progressively upregulated as prostatic epithelial cells evolve from a benign to an invasive phenotype in the same patients [38]. We have used an anti-human ANG monoclonal antibody (mAb, 26-2F) to stain ANG in human prostate tissues from 23 prostate cancer, 20 benign prostate hyperplasia (BPH), and 10 normal patients, and found that strong to very strong nuclear staining of ANG is obvious in all 23 prostate cancer [22]. No ANG was detected in the cytoplasm and nucleus of the glandular epithelial cells in all 10 normal prostate tissue specimens. The staining on BPH samples was somewhere in between. It should be pointed out that the mAb 26-2F we used in this study is specific to human ANG and does not recognize any other human proteins [41].
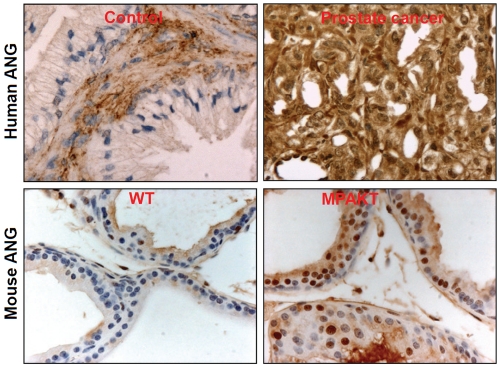
Mouse Ang1 is significantly up-regulated in AKT-induced prostate intraepithelial neoplasia (PIN) in the murine prostate-restricted AKT kinase transgenic (MPAKT) mice [39]. Figure 1 shows the representative staining of ANG in the prostates of normal human subjects and prostate cancer patients, and in the WT and MPAKT mice. In MAPKT mice, expression of AKT in the ventral prostate results in activation of the p70S6K pathway and induction of PIN [42]. Activated AKT promotes both cell growth and cell survival and its activity is frequently elevated in prostate cancers [43]. A critical downstream target of AKT is mTOR that mediates PI3K- and AKT-dependent oncogenesis, especially in the pathogenesis of prostate cancer [39, 44]. mTOR activates S6K [45] that phosphorylate small ribosomal protein S6 and results in enhanced translation of a specific class of mRNA termed with a TOP (a terminal oligopyrimidine track in the 5’ untranslated region) structure [46]. This class of mRNAs includes ribosomal proteins, elongation factors, and several other proteins involved in ribosome biogenesis or in translation control [47]. Thus, AKT activation will enhance ribosomal protein production. However, a missing link from AKT overexpression to enhanced ribosome biogenesis is how transcription of rRNA, which needs to be incorporated in an equal molar ratio, is proportionally elevated. We have recently shown that ANG-mediated rRNA transcription is this missing link that fulfills the growth requirement imposed by AKT activation [32,33].
Figure 1.
IHC detection of human and mouse ANG in normal and cancer tissues. Human ANG was detected by 26-2F, a human ANG-specific monoclonal antibody. Mouse ANG was detected by an affinity-purified polyclonal antibody.
ANG mediates both tumor angiogenesis and cancer cell proliferation
As an angiogenic protein, ANG has been shown to mediate rRNA transcription in endothelial cells [19, 26]. ANG-stimulated rRNA transcription is necessary for angiogenesis induced by other angiogenic factors including VEGF, bFGF, aFGF, and EGF [21]. Thus, ANG has been proposed as a permissive factor for angiogenesis and that ANG-induced rRNA transcription is a general requirement for angiogenesis [21]. ANG inhibitors have been shown to inhibit angiogenesis induced not only by ANG but also by other angiogenic factors [21]. Because these angiogenic factors have been shown to play a role in tumor angiogenesis [48–50], an essential role of ANG in tumor angiogenesis can be envisioned and has been demonstrated.
ANG also plays a direct role in cancer cell proliferation by constitutive nuclear translocation in cancer cells where it also stimulates rRNA transcription [27]. Knocking-down ANG expression in PC-3 cells decreases rRNA transcription, ribosome biogenesis, cell proliferation and tumorigenecity both in vitro and in vivo [22]. Thus, ANG has a dual role in cancer progression. It stimulates cancer cell proliferation as well as mediates tumor angiogenesis. Consistently, ANG inhibitors including nuclear translocation blockers, enzymatic inhibitors, and lentivirusmediated siRNA all inhibit prostate cancer progression in both xenograft and transgenic animal models [32, 33].
Targeting nuclear translocation of ANG in cancer drug development
ANG needs to be physically in contact with rDNA in order to stimulate rRNA transcription. Since ANG is a secreted protein and is normally extracellular, nuclear translocation is the key for ANG to exert its biological activity. Aminoglycoside antibiotics neomycin and neamine have been shown to block nuclear translocation of ANG thereby abolishing the biological activity of ANG and inhibiting cancer cell proliferation as well as tumor angiogenesis [22, 32, 33, 51, 52]. To inhibit ANG activity, agents that block nuclear translocation of ANG are better choice than those that neutralize ANG protein directly for several reasons. ANG is normally circulating in the plasma at a concentration of ~250 ng/ml [53, 54]. Most of the circulating ANG in normal plasma is produced by the liver [2] rather than by the tumor tissues. The high basal level of ANG in the blood is an obstacle for directly targeting ANG as the amount of inhibitors needed may be unreachable. Targeting nuclear translocation will circumvent this caveat because it is not necessary to neutralize all the circulating ANG. More importantly, nuclear translocation occurs only in a cancer cells and in proliferating endothelial cells. Normal cells, including prostate epithelial cells, fibroblasts, and quiescent endothelial cells do not require ANG to survive or grow. Therefore, blocking nuclear translocation of ANG will selectively inhibit the growth and proliferation of cancer cells and proliferating endothelial cells but spare normal cells. The side-effects and toxicities of the agents that block nuclear translocation of ANG would be low. Neomycin was identified as an anti-prostate cancer agent from our effort to screen for compounds that block nuclear translocation of ANG [52]. It has been shown to inhibit prostate cancer growth both in a xenograft animal model [22] and in AKT transgenic animals [32]. Although neomycin is an FDA-approved antibiotic, it is nephro- and oto-toxic [55] that preclude its prolonged use as an anti-cancer agent to be delivered systemically. An extended effort has been put in searching for less toxic analogues and derivatives of neomycin. These efforts have resulted in the discovery of neamine [56], a much less toxic compound that is a degradation product of neomycin, also effectively blocks nuclear translocation of ANG [51]. Systemic treatment of neamine prevent and reverse AKT-induced PIN as well as inhibit xenograft growth of human prostate cancer cells in athymic mice [33]. The nephro- and oto-toxicity of neamine is ~5 and 6%, respectively, of that of neomycin [57, 58]. The acute LD50 (subcutaneous) in mice for neamine, neomycin, and streptomycin is 1,250, 220, and 600 mg/kg, respectively [55]. Neamine appears to be less toxic than streptomycin, an antibiotic that is currently in clinical use.
ANG mutations in ALS patients
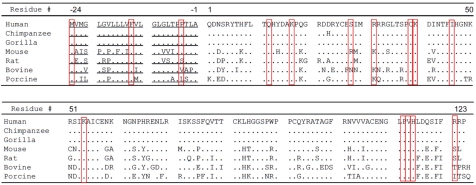
Since 2006, a total of 17 missense mutations (at 15 positions) in the coding region of ANG have been identified in 39 of the 4,774 ALS patients of the Irish, Scottish, Swedish [59], North American [25], Italian [60], French [61], Netherland [62], and German [63] populations. Thus, ANG seems to be the fourth member of the most frequently mutated gene found in ALS (after SOD1, FUS/TLS, TARDBP). Among ALS-associated ANG mutations, 3 occurred in the signal peptide regions and 12 in the mature protein. Figure 2 shows the amino acid sequences of primate and mammalian ANG with the ALS-related mutated amino acid bracketed in red. In the 10 sequencing efforts carried out so far, a total of 3,941 healthy controls were included and 2 mutations in the ANG gene were found in non-ALS controls [59, 60]. The first is a K17I mutation that was found in an apparent healthy 65-year-old male of European descent [59], and in 10 young (<50 years of age) asymptomatic members of a familiar ALS family [62]. The second is the I46V mutation that was found in 11 of the 1,568 healthy controls in Italian population [60, 64–66].
Figure 2.
Amino acid sequences of primate and mammalian ANG. By convention, the first amino acid in the mature protein is designated as 1. The signal peptide is underlined. Mutations identified so far are bracketed in red.
Several mutant ANG proteins had been prepared and their ribonucleolytic activity [25, 67], nuclear translocation capacity [25], and angiogenic activity [25, 67] have been characterized. All of them have impaired angiogenic activity in both Matrigel tubule formation assay [25] and in rat aortic ring assay [67]. Among the 9 ALS variants that have been tested for the ribonucleolytic activity, only R31K has a comparable activity (69%) to that of the WT ANG [67]. All of the other mutant proteins have severely impaired ribonucleolytic activity ranging from <1% (K40I) to 19% (K17E) of the WT ANG. Among the 3 mutant ANG proteins (K17I, S28N, P112L) that have been tested in the nuclear translocation assay, S28N and P112L do not undergo nuclear translocation and K17I has a reduced capacity [25]. These results indicate that ANG mutations identified in ALS patients are associated with a functional loss of the angiogenic activity of the ANG protein due to a deficient in ribonucleolysis, nuclear translocation, or both. ANG appears as the first “loss-of-function” gene so far identified in ALS patients.
ANG is also the second angiogenic factor associated with ALS pathogenesis. The first is VEGF. The link between VEGF and ALS was uncovered when transgenic mice with a deletion in the hypoxia responsive element of VEGF gene, which results in low VEGF levels, develop an adultonset, progressive neurodegenerative disorders similar to ALS [68]. Studies in ALS animal models have indicated that VEGF exerts neuroprotective activities on motor neurons not only as an angiogenic factor by increasing neurovascular perfusion, but also via direct effects on the neuron themselves [69]. Since ANG-mediated rRNA transcription is essential for VEGF to stimulate angiogenesis, it is possible that a deficiency in ANG function may also impair the physiological role of VEGF toward motor neurons.
Expression of ANG in the CNS
Mouse Ang1 has been reported to be strongly expressed in the developing mouse nervous system both in the brain and in the spinal cord [24]. IHC and IF staining show that Ang1 expression is the strongest in the forebrain, midbrain, hindbrain and spinal cord at 9.5 day pc [24]. At 11.5 day pc, Ang1 expression remains high in the telencephalon, mesencephalon and mylencephalon as well as in the spinal cord, spinal ganglia and choroids plexus [24]. Until midgestation, Ang1 expression is stronger in the nervous system than in any other tissues. Costaining with Peripherin and Islet1 demonstrated that Ang1 is expressed in mouse motor neurons.
IHC with an anti-ANG mAb indicate that human ANG is expressed in normal spinal cords obtained from fetal (ranging from 15 to 30 weeks gestation) and adult human subjects. Strong ANG staining was observed in the motor neurons of the ventral horn of both fetal and adult human spinal cord [25]. ANG was also detected in the extracellular matrix and interstitial tissues in all cases, consistent with it being a secreted and angiogenic protein. ANG expression in the spinal cord appears to be down-regulated as development proceeds but is still strongly expressed in the adulthood. Strong cytoplasmic and nuclear accumulation of ANG in motor neurons of both prenatal and adult spinal cords suggest a physiological role of ANG, both early in development and later in adulthood.
Double IF with an anti-ANG mAb 26-2F and antivon Willebrand factor (vWF) polyclonal antibody (pAb) showed that ANG is localized in both vasculatures and motor neurons of spinal cord tissues [25], suggesting that ANG abnormalities may have a direct (through motor neurons) and indirect (through angiogenesis) role in ALS pathogenesis. The dual role of ANG in ALS involves decreased activity in endothelial cells and motor neurons, whereas the dual role of ANG in cancer involves increased activity in endothelial cells and cancer cells.
ANG enhances motor neuron survival in vitro and in S0D1G93A mice
A recent publication demonstrated that ANG promotes motor neuron survival and is beneficial to S0D1G93A mice that develop ALS like symptoms [23]. In motor neuron cultures (NSC34 cells or primary mouse spinal cord motor neurons), exogenously added ANG protein or ANG overexpression protect motor neuron apoptosis induced by trophic factor deprivation (serum free), excitotoxicity (AMPA treatment) or ER stress (tunicamycin treatment). More importantly, ip delivery of ANG protein (1 μg per mouse per day) after disease onset enhances muscle strength of S0D1G93A mice and prolongs their survival by 12 days. The protective role of ANG, both in vitro and in vivo, is related to its activation of Akt and Erk survival signaling pathways.
Role of ANG in stress response
In mammalian cells, stress-induced phosphorylation of elF2α inhibits global protein synthesis to conserve anabolic energy for the repair of stress-induced damage [70]. However, stress-induced translational silencing is also observed in cells expressing a nonphosphorylatable elF2α mutant (S51A) [71], indicating the existence of a phosphor-elF2α independent pathway of translational control. Stress-induced cleavage of tRNA is a phenomenon first described as a starvation response in Tetrahymena thermophila [72], and later observed in bacteria [73], fungi [74], and mammalian cells [75]. ANG has been shown to be responsible for stress-induced cleavage of tRNA [76]. This class of small RNA has been designated as tiRNA (tRNA-derived, stress-induced small RNA) and shown to inhibit protein translation in an elF2α independent manner [76]. It is therefore possible that both the neuroprotective and the cancer-promoting activity of ANG are related to its ability in mediating the production of tiRNA. In case of ALS, accumulation of misfolded protein aggregates, a hallmark of ALS and several other neurodegenerative diseases, are known to increase the stress of endoplasmic reticulum (ER) and lead to apoptosis. In case of cancer, hypoxic stress is also known to lead to cell death. It would be an intriguing hypothesis that ANG-mediated production of tiRNAs in response to stress results in reprogramming of protein translation thereby promoting damage repairs and cell survival.
Stress-induced phosphorylation of elF2α inhibits translation and promotes the assembly of stress granules (SGs), cytoplasmic foci at which untranslated mRNPs are transiently concentrated [77]. SGs contain stalled 48S preinitiation complexes that remain after elongating ribosomes have run off their transcripts [78]. The ability of tiRNAs to inhibit translation in a phosphor-elF2α independent manner [76] suggests that they may also inhibit 48S scanning and that ANG may promote SG assembly. Indeed, ANG has been found to promote the formation of SG when cells are subjected to stresses [79]. Neither WT ANG nor the inactive P112L variant (identified from an ALS patient in the North American cohort) [25] induces SG assembly in the absence of stress. In contrast, WT ANG, but not P112L, significantly promotes the assembly of arsenite- and pateamine-induced SGs. Arsenite and pateamine induce SG assembly in an elF-2α-dependent and independent manner, respectively [80]. These results therefore indicate that ANG potentiates both phosphor-elF2α-dependent and independent inducers of SGs. 5'tiRNA, but not 3'tiRNA also promotes SA-induced SG assembly [79]
WT ANG, but not P12L variant, significantly inhibits protein synthesis [76]. The combination of ANG and SA inhibits protein synthesis more than either treatment alone. The finding that ANG inhibits protein synthesis [76] but does not induce SG assembly [79] suggests that a second signal is required for untranslated mRNAs to aggregate at SGs. Nevertheless, these results strongly implicate ANG as an effector of stress-induced translational repression. Ribonuclease/ANG inhibitor 1 (RNH1) inhibits angiogenic activity of ANG and ANG-mediated production of tiRNA[76]. It also plays a role in regulating ANG-induced promotion of arsenite-induced SG assembly. RNH1 over-expression has been shown to prevent ANG-mediated enhancement of arsenite-induced SG assembly [79].
Summary
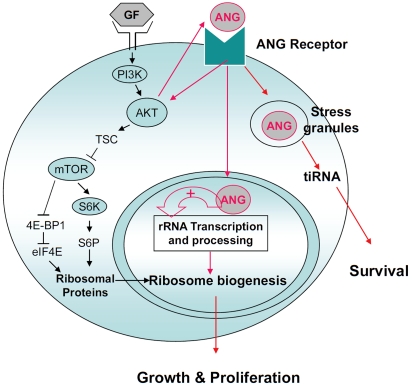
Based on these findings, we have proposed three distinct modes of action of ANG in mediating cell proliferation and survival (Figure 3). ANG has three types of target cells (endothelial cells, cancer cells, and motor neurons). These cells respond to ANG stimulation but exhibit some subtle difference probably due to the receptor expression profile. Endothelial cells are the first type of responsive cells that have been used extensively for studying ANG biology. The activity of ANG in endothelial cells is strictly dependent on the cell density [81]. ANG receptor is expressed only in sparsely cultured endothelial cells. More recently, ANG was shown to undergo nuclear translocation in cancer cells in a cell density-independent manner. Constitutive nuclear translocation of ANG in cancer cells is a driving force for cancer progression. Thus, ANG has been demonstrated to play a dual role in cancer progression: it mediates tumor angiogenesis as well as directly stimulates cancer cell proliferation. Recently, motor neurons have been shown to be the third type of ANG responsive cells. ANG is strongly expressed in the spinal cord both during development and in the adulthood [25]. Loss-of-function mutations in the coding region of ANG have been found in ALS patients and ANG has been shown to enhance motor neuron survival.
Figure 3.
Current understanding of the mechanism of ANG.
The signal transduction pathway triggered by ANG-Receptor interaction is currently unknown as the identity of a functional ANG receptor has not been fully determined. However, ANG has been shown to activate Akt [23, 82]. Akt activation is known to upregulate Ang so there is a positive feedback loop between ANG and AKT. This crosstalk between ANG and AKT may thus coordinate an orchestrated synthesis of ribosomes. Ribosome biogenesis requires both ribosome proteins and rRNAs. The production of ribosomal proteins is mediated by the AKT-mTOR pathway, and the production of rRNAs is likely mediated by ANG. VEGF and other angiogenic proteins stimulate nuclear translocation of ANG. ANG inhibitors inhibit angiogenesis induced by these angiogenic factors. Thus ANG is a permissive factor for other angiogenic factors. ANG has also been shown to mediate the production of tiRNA, promote SG assembly, and reprogram protein translation of the cells under stress conditions thereby promoting their survival.
References
- [1].Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- [2].Weiner HL, Weiner LH, Swain JL. Science. 1987;237:280–282. doi: 10.1126/science.2440105. [DOI] [PubMed] [Google Scholar]
- [3].Strydom DJ, Fett JW, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Biochemistry. 1985;24:5486–5494. doi: 10.1021/bi00341a031. [DOI] [PubMed] [Google Scholar]
- [4].Shapiro R, Riordan JF, Vallee BL. Biochemistry. 1986;25:3527–3532. doi: 10.1021/bi00360a008. [DOI] [PubMed] [Google Scholar]
- [5].Riordan JF. Methods Enzymol. 2001;341:263–273. doi: 10.1016/s0076-6879(01)41157-8. [DOI] [PubMed] [Google Scholar]
- [6].Curran TP, Shapiro R, Riordan JF, Vallee BL. Biochim BiophysActa. 1993;1202:281–286. doi: 10.1016/0167-4838(93)90017-l. [DOI] [PubMed] [Google Scholar]
- [7].Hallahan TW, Shapiro R, Strydom DJ, Vallee BL. Biochemistry. 1992;31:8022–8029. doi: 10.1021/bi00149a036. [DOI] [PubMed] [Google Scholar]
- [8].Hallahan TW, Shapiro R, Vallee BL. Proc Natl Acad Sci U S A. 1991;88:2222–2226. doi: 10.1073/pnas.88.6.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harper JW, Fox EA, Shapiro R, Vallee BL. Biochemistry. 1990;29:7297–7302. doi: 10.1021/bi00483a020. [DOI] [PubMed] [Google Scholar]
- [10].Shapiro R, Fox EA, Riordan JF. Biochemistry. 1989;28:1726–1732. doi: 10.1021/bi00430a045. [DOI] [PubMed] [Google Scholar]
- [11].Shapiro R, Vallee BL. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- [12].Shapiro R, Vallee BL. Biochemistry. 1992;31:12477–12485. doi: 10.1021/bi00164a026. [DOI] [PubMed] [Google Scholar]
- [13].Shapiro R, Strydom DJ, Olson KA, Vallee BL. Biochemistry. 1987;26:5141–5146. doi: 10.1021/bi00390a037. [DOI] [PubMed] [Google Scholar]
- [14].Harper JW, Vallee BL. Proc Natl Acad Sci U S A. 1988;85:7139–7143. doi: 10.1073/pnas.85.19.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Russo N, Shapiro R, Acharya KR, Riordan JF, Vallee BL. Proc Natl Acad Sci U S A. 1994;91:2920–2924. doi: 10.1073/pnas.91.8.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hu GF, Chang SI, Riordan JF, Vallee BL. Proc Natl Acad Sci U S A. 1991;88:2227–2231. doi: 10.1073/pnas.88.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Acharya KR, Shapiro R, Riordan JF, Vallee BL. Proc Natl Acad Sci U S A. 1995;92:2949–2953. doi: 10.1073/pnas.92.7.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moroianu J, Riordan JF. Proc Natl Acad Sci U S A. 1994;91:1677–1681. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hu G, Xu C, Riordan JF. J Cell Biochem. 2000;76:452–462. doi: 10.1002/(sici)1097-4644(20000301)76:3<452::aid-jcb12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- [20].Li R, Riordan JF, Hu G. Biochem Biophys Res Commun. 1997;238:305–312. doi: 10.1006/bbrc.1997.7290. [DOI] [PubMed] [Google Scholar]
- [21].Kishimoto K, Liu S, Tsuji T, Olson KA, Hu GF. Oncogene. 2005;24:445–456. doi: 10.1038/sj.onc.1208223. [DOI] [PubMed] [Google Scholar]
- [22].Yoshioka N, Wang L, Kishimoto K, Tsuji T, Hu GF. Proc Natl Acad Sci USA. 2006;103:14519–14524. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kieran D, Sebastia J, Greenway MJ, King MA, Connaughton D, Concannon CG, Fenner B, Hardiman O, Prehn JH. J Neurosci. 2008;28:14056–14061. doi: 10.1523/JNEUROSCI.3399-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Subramanian V, Feng Y. Hum Mol Genet. 2007;16:1445–1453. doi: 10.1093/hmg/ddm095. [DOI] [PubMed] [Google Scholar]
- [25].Wu D, Yu W, Kishikawa H, Folkerth RD, lafrate AJ, Shen Y, Xin W, Sims K, Hu GF. Ann Neurol. 2007 doi: 10.1002/ana.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu ZP, Tsuji T, Riordan JF, Hu GF. Biochemistry. 2003;42:121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- [27].Tsuji T, Sun Y, Kishimoto K, Olson KA, Liu S, Hiru kawa S, Hu GF. Cancer Res. 2005;65:1352–1360. doi: 10.1158/0008-5472.CAN-04-2058. [DOI] [PubMed] [Google Scholar]
- [28].Xu ZP, Tsuji T, Riordan JF, Hu GF. Biochem Biophys Res Commun. 2002;294:287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- [29].Comai L Braz. J Med Biol Res. 1999;32:1473–1478. doi: 10.1590/s0100-879x1999001200004. [DOI] [PubMed] [Google Scholar]
- [30].Melese T, Xue Z. Curr Opin Cell Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- [31].Stoykova AS, Dabeva MD, Dimova RN, Had Jiolov AA. J Neurochem. 1985;45:1667–1676. doi: 10.1111/j.1471-4159.1985.tb10521.x. [DOI] [PubMed] [Google Scholar]
- [32].Ibaragi S, Yoshioka N, Kishikawa H, Hu JK, Sadow PM, Li M, Hu G. Mol Cancer Res. 2009;7:415–424. doi: 10.1158/1541-7786.MCR-08-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ibaragi S, Yoshioka N, Li S, Hu MG, Hirukawa S, Sadow PM, Hu G f. Clin Cancer Res. 2009;15:1981–1988. doi: 10.1158/1078-0432.CCR-08-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kishikawa H, Wu D, Hu GF. Expert Opin Ther Targets. 2008;12:1229–1242. doi: 10.1517/14728222.12.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Olson KA, Fett JW, French TC, Key ME, Vallee BL. Proc Natl Acad Sci U S A. 1995;92:442–446. doi: 10.1073/pnas.92.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Olson KA, Byers HR, Key ME, Fett JW. Clin Cancer Res. 2001;7:3598–3605. [PubMed] [Google Scholar]
- [37].Tello –Montoliu A, Patel JV, Lip GY. J Thromb Haemost. 2006;4:1864–1874. doi: 10.1111/j.1538-7836.2006.01995.x. [DOI] [PubMed] [Google Scholar]
- [38].Katona TM, Neubauer BL, Iversen PW, Zhang S, Baldridge LA, Cheng L. Clin Cancer Res. 2005;11:8358–8363. doi: 10.1158/1078-0432.CCR-05-0962. [DOI] [PubMed] [Google Scholar]
- [39].Majumder PK, Yeh JJ, George DJ, Febbo PG, Kum J, Xue Q, Bikoff R, Ma H, Kantoff PW, Golub TR, Loda M, Sellers WR. Proc Natl Acad Sci U S A. 2003;100:7841–7846. doi: 10.1073/pnas.1232229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Olson KA, Byers HR, Key ME, Fett JW. Int J Cancer. 2002;98:923–929. doi: 10.1002/ijc.10282. [DOI] [PubMed] [Google Scholar]
- [41].Chavali GB, Papageorgiou AC, Olson KA, Fett JW, Hu G, Shapiro R, Acharya KR. Structure. 2003;11:875–885. doi: 10.1016/s0969-2126(03)00131-x. [DOI] [PubMed] [Google Scholar]
- [42].Di Cristofano A, Pesce B, Cordon–Cardo C, Pandolfi PP. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- [43].Sun M, Wang G, Paciga JE, Feldman Rl, Yuan ZQ, Ma XL, Shelley SA, Jove R, Tsichlis PN, Nicosia SV, Cheng JQ. Am J Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J,, McDonnell TJ, Golub TR, Loda M, Lane HA and, Sellers WR. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- [45].Aoki M, Blažek E, Vogt PK. Proc Natl Acad Sci USA. 2001;98:136–141. doi: 10.1073/pnas.011528498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Terada N, Patel HR, Takase K, Kohno K, Nairn AC, Gelfand EW. Proc Natl Acad Sci U S A. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Campbell SC. J Urol. 1997;158:1663–1674. doi: 10.1016/s0022-5347(01)64090-4. [DOI] [PubMed] [Google Scholar]
- [49].Russell PJ, Bennett S, Strieker P. Clin Chem. 1998;44:705–723. [PubMed] [Google Scholar]
- [50].Ferrer FA, Miller U, Andrawis Rl, Kurtzman SH,, Albertsen PC, Laudone VP, Kreutzer DLJ. Urol. 1997;157:2329–2333. [PubMed] [Google Scholar]
- [51].Hirukawa S, Olson KA, Tsuji T, Hu GF. Clin Cancer Res. 2005;11:8745–8752. doi: 10.1158/1078-0432.CCR-05-1495. [DOI] [PubMed] [Google Scholar]
- [52].Hu GF. Proc Natl Acad Sci U S A. 1998;95:9791–9795. doi: 10.1073/pnas.95.17.9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shimoyama S, Gansauge F, Gansauge S, Negri G, Oohara T, Beger HG. Cancer Res. 1996;56:2703–2706. [PubMed] [Google Scholar]
- [54].Miyake H, Hara I, Yamanaka K, Gohji K, Arakawa S, Kamidono S. Cancer. 1999;86:316–324. [PubMed] [Google Scholar]
- [55].Glasby JS. Encyclopedia of antibiotics. 3rd edition. John Wiley &Sons Ltd; New York NY: 1993. [Google Scholar]
- [56].Ford JH, Bergy ME, Brooks AA, Garrett ER, Alberti J, Dyer JR, Carter HE. J Am Chem Soc. 1955;77:5311–5314. [Google Scholar]
- [57].Au S, Weiner N, Schacht J. Antimicrob Agents Chemother. 1986;30:395–397. doi: 10.1128/aac.30.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Williams PD, Bennett DB, Gleason CR, Hot–tendorf GH. Antimicrob Agents Chemother. 1987;31:570–574. doi: 10.1128/aac.31.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Jr. Hardiman 0. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- [60].Gellera C, Colombrita C, Ticozzi N, Castellotti B, Bragato C, Ratti A, Taroni F, Silani V. Neurogenetics. 2008;9:33–40. doi: 10.1007/s10048-007-0111-3. [DOI] [PubMed] [Google Scholar]
- [61].Paubel A, Violette J, Amy M, Praline J, Meininger V, Camu W, Corcia P, Andres CR, Vourc'h P. Arch Neurol. 2008;65:1333–1336. doi: 10.1001/archneur.65.10.1333. [DOI] [PubMed] [Google Scholar]
- [62].van Es MA, Diekstra FP, Veldink JH, Baas F, Bourque PR, Schelhaas HJ, Strengman E, Henne–kam EA, Lindhout D, Ophoff RA, van den Berg LH. Neurology. 2009;72:287–288. doi: 10.1212/01.wnl.0000339487.84908.00. [DOI] [PubMed] [Google Scholar]
- [63].Fernandez–Santiago R, Hoenig S, Lichtner P, Sperfeld AD, Sharma M, Berg D, Weichenrieder 0, lllig T, Eger K, Meyer T, Anneser J, Munch C, Zierz S, Gasser T, Ludolph A. J Neurol. 2009;256:1337–1342. doi: 10.1007/s00415-009-5124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Conforti FL, Sprovieri T, Mazzei R, Ungaro C, La Bella V, Tessitore A, Patitucci A, Magariello A, Gabriele AL, Tedeschi G, Simoně IL, Majorána G, Valentino P, Condino F, Bono F, Monsurro MR, Muglia M, Quattrone A. Neuromuscul Disord. 2008;18:68–70. doi: 10.1016/j.nmd.2007.07.003. [DOI] [PubMed] [Google Scholar]
- [65].Corrado L, Battistini S, Penco S, Bergamaschi L, Testa L, Ricci C, Giannini F, Greco G, Patrosso MC, Pileggi S, Causarano R, Mazzini L, Momigliano–Richiardi P, D'Alfonso S. J Neurol Sci. 2007;258:123–127. doi: 10.1016/j.jns.2007.03.009. [DOI] [PubMed] [Google Scholar]
- [66].Del Bo R, Scarlato M, Ghezzi S, Martinelli–Boneschi F, Corti S, Locatelli F, Santoro D, Prelle A, Briani C, Nardini M, Siciliano G, Mancuso M, Murri L, Bresolin N, Comi GP. Neurobiol Aging. 2006 doi: 10.1016/j.neurobiolaging.2006.10.008. [DOI] [PubMed] [Google Scholar]
- [67].Crabtree B, Thiyagarajan N, Prior SH, Wilson P, Iyer S, Ferns T, Shapiro R, Brew K, Subramanian V, Acharya KR. Biochemistry. 2007;46:11810–11818. doi: 10.1021/bi701333h. [DOI] [PubMed] [Google Scholar]
- [68].Oosthuyse B, Moons L, Storkebaum E, Beck H, Nuyens D, Brusselmans K, Van Dorpe J, Hellings P, Gorselink M, Heymans S, Theilmeier G, Dewerchin M, Laudenbach V, Vermylen P, Raat H, Acker T, Vleminckx V, Van Den, Bosch L, Cashman N, Fujisawa H, Drost MR, Sciot R, Bruyninckx F, Hicklin DJ, Ince C, Gressens P, Lupu F, Plate KH, Robberecht W, Herbert JM, Collen D, Carmeliet P. Nat Genet. 2001;28:131–138. doi: 10.1038/88842. [DOI] [PubMed] [Google Scholar]
- [69].Storkebaum E, Lambrechts D, Dewerchin M, Moreno–Murciano MP, Appelmans S, Oh H, Van Damme P, Rutten B, Man WY, De Mol M, Wyns S, Manka D, Vermeulen K, Van Den Bosch L, Mertens N, Schmitz C, Robberecht W, Conway EM, Collen D, Moons L, Carmeliet P. Nat Neurosci. 2005;8:85–92. doi: 10.1038/nn1360. [DOI] [PubMed] [Google Scholar]
- [70].Yamasaki S, Anderson P. Curr Opin Cell Biol. 2008;20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. J Biol Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- [72].Lee SR, Collins K. J Biol Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- [73].Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jochl C, Rederstorff M, Hertel J, Stadier PF, Ho–facker IL, Schrettl M, Haas H, Huttenhofer A. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Thompson DM, Lu C, Green PJ, Parker R. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yamasaki S, Ivanov P, Hu GF, Anderson P. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Anderson P, Kedersha N. Curr Biol. 2009;19:R397–398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- [78].Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Mol Biol Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. J Biol Chem 285 10959. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. J Biolr Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- [81].Hu GF, Riordan JF, Vallee BL. Proc Natl Acad Sci U S A. 1997;94:2204–2209. doi: 10.1073/pnas.94.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kim HM, Kang DK, Kim SS, Chang SI. Biochem Biophys Res Commun. 2007;352:509–513. doi: 10.1016/j.bbrc.2006.11.047. [DOI] [PubMed] [Google Scholar]