Abstract
Utilizing the structure-activity relationship we have developed during the synthesis of the first two generations and mechanism of action studies that point to the interaction of these molecules with the key oncogenic protein Hsp90, we report here the design of 32 new Sansalvamide A derivatives and their synthesis. Our new structures, designed from previously reported potent compounds, were tested for cytotoxicity on the HCT116 colon cancer cell line, and their binding to the biological target was analyzed using computational studies involving blind docking of derivatives using Autodock. Further, we show new evidence that our molecules bind directly to Hsp90 and modulate Hsp90’s binding with client proteins. Finally, we demonstrate that we have integrated good ADME properties into a new derivative.
Keywords: Sansalvamide A, pancreatic cancer, macrocyclic peptides, macrocycles, cytotoxicity, growth inhibition, docking, Hsp90
1. Introduction
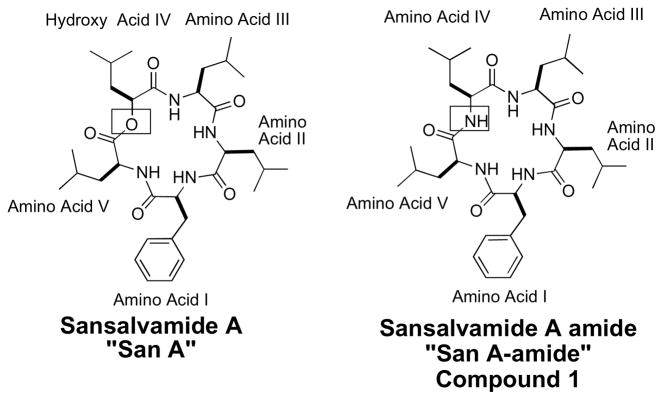
Compounds isolated from natural resources can provide novel structures that can be used in the development of new small molecules that have novel mechanisms of action. One such compound is Sansalvamide A (San A) (Figure 1). San A, which was isolated from a marine fungus (Fusarium ssp.), exhibits anti-tumor activity against multiple cancer cell lines.1–3 To date, the synthesis of 89 analogs have been reported by our lab4–6 and 11 by Silverman et. al.7 The natural product is a depsipeptide (Figure 1), which is prone to ring opening at the ester bond by esterases. Given the depsipeptide’s lability, Silverman and co-workers synthesized the natural product peptide, and found that the peptide was 10-fold more active in a cell-based cytotoxicity assay than the natural product depsipeptide, presumably because the peptide macrocycle was more stable within cells than the depsipeptide. 7, 8 Thus, to avoid degradation via ring-opening, all 89 derivatives reported by our laboratory were synthesized as derivatives of the San A peptide (San A-amide), where an amino acid replaced the alcohol acid at position IV (Figure 1, amino acid IV). Cytotoxicity of San A-amide derivatives against pancreatic,7–9 colon,3, 4,10, 11 breast, prostate, and melanoma cancers7 clearly indicate San A-amide’s potential as a new therapeutic lead structure in the treatment of various cancers and support further exploration of this class of compounds. Ten of the San A-amide derivatives prepared by Silverman and co-workers contained an N-methyl within the peptide structure and 3 were found to be more potent than San A-amide.7, 8, 12, 13 From our reported 89 derivatives, we have concluded that the most important structural motifs are the inclusion of 2 consecutive D-amino acids and an N-methyl moiety. This work has demonstrated that 3 compounds containing these motifs were significantly more potent than the natural product peptide, San A-amide.6 Our work has been validated by several current examples in the recent literature where cyclic peptides, and specifically pentapeptides, with both an N-methyl and D-amino acid lock the macrocycle into a single conformation.14–16 Data from these studies suggest that the compounds, once locked into a major conformation, will be appropriately positioned as a beta or gamma turn, which is likely to lead to a well-defined, high affinity interaction with the protein target.17, 18
Figure 1.
Structure of Sansalvamide A (San A) and Sansalvamide A-amide (San A-amide)
We report here the synthesis of 32 new Sansalvamide A derivatives. These compounds were designed using the structure-activity relationship (SAR) that we observed in earlier generations, and utilized specific features known to play a key role in compound potency, ie. the incorporation of several aromatic moieties, D-amino acids, and N-methyl amino acids. Further, San A-amide derivatives were shown to bind to Heat shock protein 90 (Hsp90).19 Given that Hsp90 is an oncogenic protein of interest,20–23 and that this new series of compounds expound on the SAR of previously reported potent derivatives by exploring new avenues for incorporating aromatic moieties, these data describe an important advance in the development of the San A-amide compound class as a potential drug lead.
Precedence has already been set for peptides to be used as drugs. To date, there are 617 peptide drugs or drug candidates, 24% of these are in clinical trials, 65% are in advanced preclinical phases, and 11% are on the market.24–26 These peptide drugs are used to treat a variety of diseases such as prostate and breast cancer, HIV infections, osteoporosis, acute coronary syndrome, and serve as immunosuppressants.27 Several key peptide-based drugs include: Cyclosporin A (MW=1185), Caspofungin (MW=1093), Vancomycin (MW=1431), and Fuzeon (MW= 4492). Cyclosporin A is an 11 amino acid macrocyclic peptide that is used to suppress the immune system after organ transplants.28 Caspofungin, Vancomycin, and Fuzeon are peptide-based antifungal, antibacterial, and anti-HIV drugs, respectively. Aplidine (MW=1067) is an 8 amino acid peptide-based cancer agent that is currently in clinical trials.29–31 Thus, peptides are successfully used to treat diseases, setting excellent precedence for San A-amide drug development (MW= ~600).32
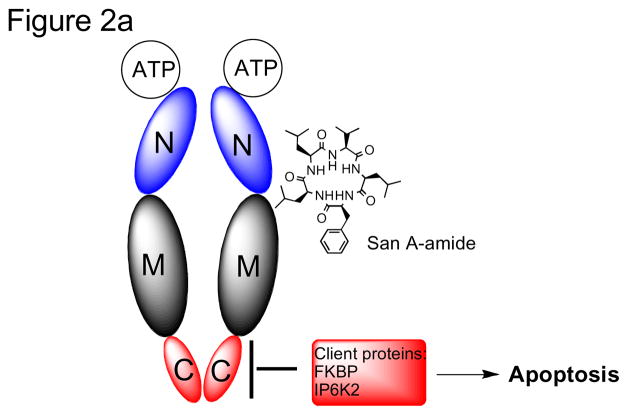
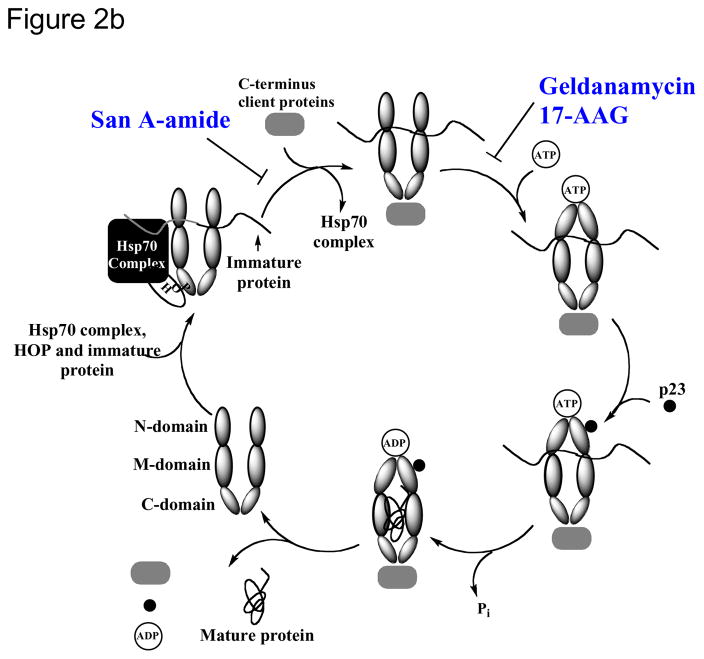
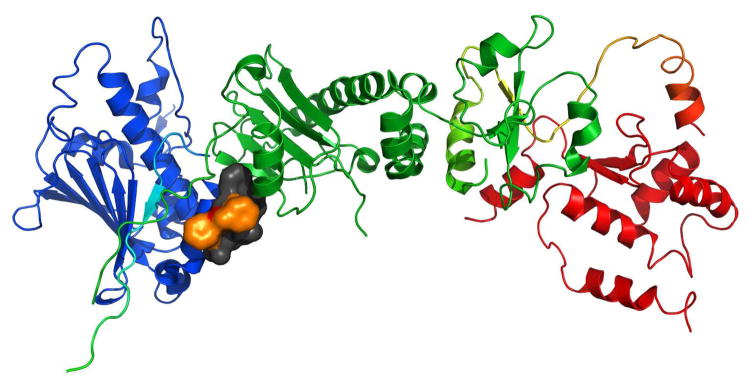
Recently we showed evidence that the target for San A-amide is heat shock protein 90 (Hsp90).19 Hsp90 functions as a molecular chaperone for intracellular signaling molecules,33–36 and it folds, assembles, and stabilizes proteins that regulate the growth of cells. It is also up-regulated in most cancers.33, 37–50 There are 3 distinct regions of Hsp90: the N-terminal, C-terminal, and middle domain, and it exists as a homodimer, connected via the C-terminal region.51–53 Its ATP binding site (located at the N-terminal domain) is the binding site for the 2 inhibitors currently in clinical trials, 17-DMAG and 17-AAG.23, 33, 39–44, 54–60 In our previous work,19 we show that San A-amide analogs bind to Hsp90 and inhibit its activity via an allosteric mechanism, where it binds to the N-middle domain, and inhibits, presumably via a conformational change, the binding of two C-terminal client proteins (figure 2). By inhibiting their binding to Hsp90, these two client proteins are now forced to remain in the cytosol, inducing apoptosis via their cytosolic pathways. San A-amide’s mechanism is unique from inhibitors that are currently in clinical development because San A-amide interferes with clients that interact with the C-terminus of Hsp90, as opposed to those currently under investigation that inhibit binding of client proteins to the N-terminal domain. This distinctive mechanism supports the further investigation of San A-amide compounds as potential new therapeutic drugs.
Figure 2.
a) Interaction of San A-amide with Hsp90 b) mechanism of San A-amide on Hsp90, inhibition of 2 C-terminal client proteins: IP6K2 and FKBP52 while binding to the N-Middle domain19
San A-amide derivatives have been tested extensively on numerous cancer cell lines, including several colon cancer cell lines.1, 3, 4, 10, 61, 62 Carcinogenesis in the colon rectum is thought to occur through two different pathways. The two pathways are usually referred to as having microsatellite stability (MSS) or microsatellite instability (MSI). Currently, only the MSS colon cancers are known to respond to chemotherapeutic drugs. Additionally, the drug of choice for treatment, 5-fluorouracil (5-FU) [IC50 = 5μM], has significant side effects, making it desirable to develop a drug with improved efficacy. Because MSI colon cancers do not respond to 5-FU, or to other current chemotherapeutic drugs,63, 64 finding new structures that target both cancer pathways is imperative. The 32 compounds and the derivatives from which they were designed were tested on the HCT116 colon cancer cell line. This cancer cell line was chosen not only because it is a commonly used cell line, found in the NCI 60 cell line panel, but it is also known to be microsatellite instable (MSI). Although major efforts have been made, few truly novel classes of compounds have been identified that have activity against drug-resistant (MSI) colon cancer tumors. This work reports our understanding of the complex structure-activity relationship of the 32 new compounds in a drug-resistant colon cancer cell line, establishes a phenotype for cytotoxicity in cell-based assays, and models these compounds bound to their biological target Hsp90.
2. Design and biological activity of new Sansalvamide A-amide derivatives
In order to explore the potency of this structural class we designed compounds based on several of the most interesting first and second generation structures,4–6, 10 which were tested against HCT-116 cancer cell lines.61, 62 Each new derivative was designed to examine the change in potency by altering the amino acid at one position relative to it’s “parent” compound(s). These alterations included a change in the stereochemistry of one of the aa’s to investigate the effect on potency as it relates to conformation or the replacement of one aa with another to investigate the effect as it relates to polarity at that position. Structural differences in the San A-amide derivatives can be easily identified by the reader in all subsequent figures as all L-amino acids are shown with wedged bonds, and D-amino acids are shown with dashed bonds.
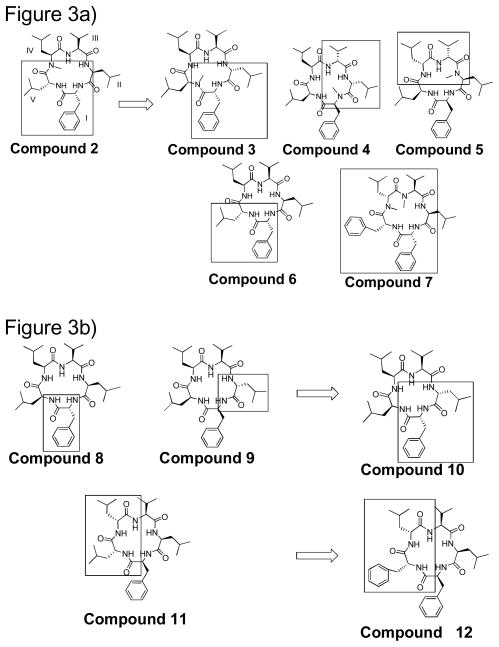
Several first generation structures were chosen as leads: 2, 8, 9, and 11 (Figure 3).9 These structures were chosen as they had resaonable potency in at least 1 of several cells lines in which they were tested.4–6 It was noted in our previous work that 2 consecutive D-amino acids and a single N-methyl were incorporated into the most potent structures. However, it was not known if the positioning of these structural features was important to the conformation, or if simply molecules containing these features were similiarly potent. Thus, a series of derivatives were made that incorporated these features and they were moved throughout the ring.
Figure 3.
The design of new derivatives based on potent first generation molecules: a) structures based on compound 2 b) structures based on compounds 8, 9, and 11.
Compounds 3–7 were all based on compound 2, where compounds 3–5 the N-methyl and 2 D-amino acids were rotated around the positions of the ring. Compounds 6 was identical to 2, except that it did not contain an N-methyl moiety at position IV. Derivative 7 involved the incorporation of an additional D-amino acid at IV, as well as a second N-methyl moiety on the N at III. Compound 10 has 2 D-amino acids, one at position I and one at position II based on the structures of 8 and 9. Compound 12 replaced the D-Leu of 11 with a D-Phe.
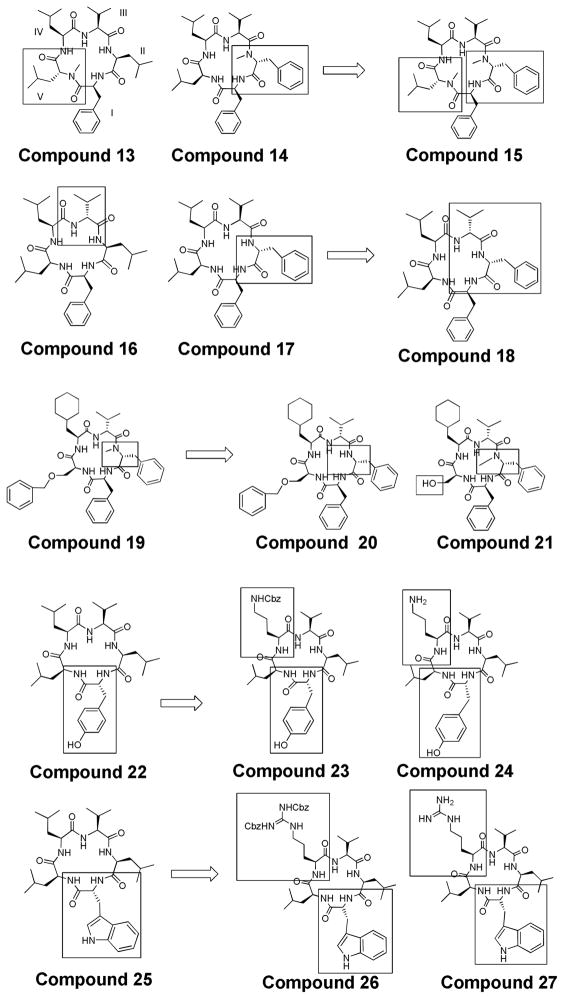
Noteworthy second-generation compounds chosen as leads included: 13, 14, 16, 17, 19, 22, and 25 (Figure 4).5, 6 The structures resulting from these leads include: 15, 18, 20, 21, 23, 24, 26, and 27. Compound 15 was derived from 13 and 14, where and N-methyl D-Phe was inserted at position II and an N-methyl D-Leu was inserted at position V. Derivative 18 was designed from 16 and 17, where a D-Phe and a D-Val were incorporated into positions II and III, respectively. Compound 20 and 21 was designed from 19, where an N-methyl moiety located at position II in 19 is not included in 20. The serine at V was a free alcohol in 21 as opposed to benzyl protected in 19. Derivatives 23 and 24 were based on 22, where a lysine (protected and unprotected, respectively) was included at position IV. Similarly 26 and 27 were based on 25, and included an arginine at position IV (protected and unprotected, respectively).
Figure 4.
The design of new derivatives based on the second-generation structures
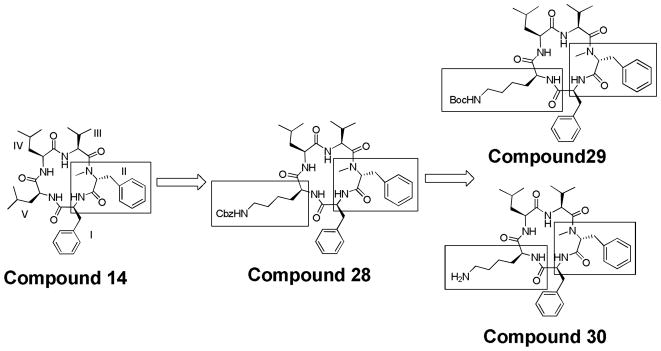
Compound 14 was seen as an excellent lead structure, demonstrating potency in both pancreatic (IC50 = 1.4 μM for PL-45)6, 65 and colon cancer cell lines (IC50 = 1.9 μM for HCT-116).5, 61 Thus, it was used as a template to design an additional new series of molecules to investigate the incorporation of a lysine residue to the structure. The first of these was compound 28, where the solution phase synthesis required the use of an orthogonal protecting group (Cbz) on the lysine side chain. It was found that 28 also had potent cytotoxic effects on two colon cancer cell lines (Table 1). Thus, we used 28 as a lead to make additional compounds that would explore the structure activity series of these molecules (Figure 5). The structures resulting from compounds 14 and 28 are compounds 29 and 30. In contrast to 28’s Cbz-protected lysine at position V, these compounds contain a Boc-protected lysine and a free lysine at position V respectively.
Table 1.
Bolded compounds are new structures, and compounds from which they were designed are non-bolded. Note: specific residues that were altered in the new structures are shown in bold. Cytotoxicity and cLogP data for all newly synthesized derivatives and parent compounds. IC50 values were determined for 8 of the most potent derivatives. cLogP values were calculated using software from ChemAxon
| Compound | Structure | cLogP | % growth inhibition | IC50 (μM) |
|---|---|---|---|---|
| 2 | D-Phe-Leu-Val-N-Me-Leu-D-Leu | 3.847 | 18 | - |
| 3 | D-Phe-D-Leu-Val-Leu-N-Me-Leu | 3.847 | 15 | - |
| 4 | N-Me-Phe-D-Leu-D-Val-Leu-Leu | 3.847 | 30 | - |
| 5 | Phe-N-Me-Leu-D-Val-D-Leu-Leu | 3.847 | 30 | - |
| 6 | D-Phe-Leu-Val-Leu-D-Leu | 3.624 | 7 | - |
| 7 | D-Phe-Leu-N-Me-Val-N-Me-D-Leu-D-Phe | 4.473 | 15 | - |
| 8 | D-Phe-Leu-Val-Leu-Leu | 3.624 | 26 | - |
| 9 | Phe-D-Leu-Val-Leu-Leu | 3.624 | 3 | - |
| 10 | D-Phe-D-Leu-Val-Leu-Leu | 3.624 | 14 | - |
| 11 | Phe-Leu-Val-D-Leu-D-Leu | 3.624 | 20 | - |
| 12 | Phe-Leu-Val-D-Leu-D-Phe | 4.026 | 28 | - |
| 13 | Phe-Leu-Val-Leu-N-Me-D-Leu | 3.847 | 32 | - |
| 14 | Phe-N-Me-D-Phe-Val-Leu-Leu | 4.249 | 99 | 1.9 |
| 15 | Phe-N-Me-D-Phe-Val-Leu-N-Me-D-Leu | 4.473 | 47 | - |
| 16 | Phe-Leu-D-Val-Leu-Leu | 3.624 | 8 | - |
| 17 | Phe-D-Phe-Val-Leu-Leu | 4.026 | 21 | - |
| 18 | Phe-D-Phe-D-Val-Leu-Leu | 4.026 | 4 | - |
| 19 | Phe-N-Me-D-Phe-D-Val-Cha-Ser(Bn) | 5.186 | 93 | 1.9 |
| 20 | Phe-D-Phe-D-Val-Cha-Ser(Bn) | 4.962 | 10 | - |
| 21 | Phe-N-Me-D-Phe-D-Val-Cha-Ser | 2.818 | 18 | - |
| 22 | D-Tyr-Leu-Val-Leu-Leu | 3.320 | 50 | - |
| 23 | D-Tyr-Leu-Val-Lys(Cbz)-Leu | 3.652 | 0 | - |
| 24 | D-Tyr-Leu-Val-Lys-Leu | 1.489 | 0 | - |
| 25 | D-Trp-Leu-Val-Leu-Leu | 3.452 | 30 | - |
| 26 | D-Trp-Leu-Val-Arg(Cbz)-Leu | 5.443 | 2 | - |
| 27 | D-Trp-Leu-Val-Arg-Leu | 1.288 | 0 | - |
| 14 | Phe-N-Me-D-Phe-Val-Leu-Leu | 4.249 | 99 | 1.9 |
| 28 | Phe-N-Me-D-Phe-Val-Leu-Lys(Cbz) | 5.026 | 99 | 3.9 |
| 29 | Phe-N-Me-D-Phe-Val-Leu-Lys(Boc) | 4.356 | 36 | - |
| 30 | Phe-N-Me-D-Phe-Val-Leu-Lys | 2.863 | 0 | - |
| 1 | Phe-Leu-Val-Leu-Leu | 3.624 | 35 | - |
| 31 | Phe-Leu-Val-Leu-Lys(Boc) | 3.730 | 0 | - |
| 32 | Phe-Leu-Val-Leu-Lys | 2.238 | 0 | - |
| 33 | Phe-Leu-Val-Leu-N-Me-Leu | 3.847 | 35 | - |
| 34 | Phe-Leu-Val-Leu-N-Me-Lys(Boc) | 3.953 | 58 | - |
| 35 | Phe-Leu-Val-Leu-N-Me-Lys | 2.461 | 0 | - |
| 16 | Phe-Leu-D-Val-Leu-Leu | 3.624 | 8 | - |
| 36 | Phe-Leu-D-Val-Leu-Lys(Boc) | 3.730 | 9 | - |
| 37 | Phe-Leu-D-Val-Leu-Lys | 2.238 | 0 | - |
| 38 | Phe-Leu-N-Me-Val-D-Lys(Cl-Cbz)-D-Phe | 5.630 | 69 | 7.6 |
| 39 | Phe-Leu-N-Me-Val-Lys(Cl-Cbz)-D-Phe | 5.630 | 58 | - |
| 40 | Phe-Leu-N-Me-Val-D-Lys(Cbz)-D-Phe | 5.026 | 36 | - |
| 41 | Phe-Leu-Val-N-Me-D-Lys(Cl-Cbz)-D-Phe | 5.630 | 36 | - |
| 42 | Phe-Leu-Val-D-Lys(Cl-Cbz)-N-Me-D-Phe | 5.630 | 98 | 2.9 |
| 43 | (R,R)β-OH(Bn)-Phe-Leu-N-Me-Val-D-Leu-D-Phe | 5.698 | 94 | 3.2 |
| 44 | (S,S)β-OH(Bn)-Phe-Leu-N-Me-Val-D-Leu-D-Phe | 5.698 | 64 | 5.8 |
| 45 | (S,R)β-OH(Bn)-Phe-Leu-N-Me-Val-D-Leu-D-Phe | 5.698 | 25 | - |
| 46 | (R,S)β-OH(Bn)-Phe-Leu-N-Me-Val-D-Leu-D-Phe | 5.698 | 63 | 8.9 |
Figure 5.
The design of new derivatives based on compound 14 and 28.
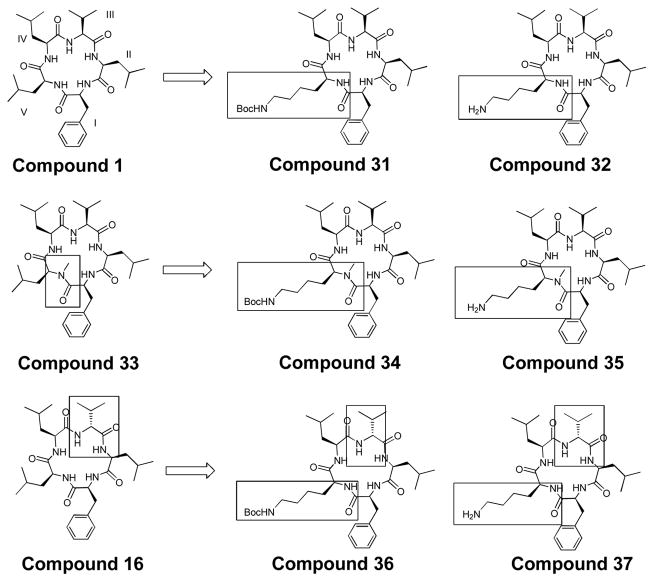
Given the success of compound 28, we chose 3 additional compounds from which we designed new structures with Boc-protected lysines and free lysines: 1, 16, and 33 (figure 6). Compound 1, San A-amide, was chosen as a “control” molecule, where 31 and 32 are related to the natural product peptide via a single change: a protected or free lysine at V. Compounds 16 and 33 have shown significant cytotoxicity in several types of cell lines, and as such were considered interesting leads.10, 61, 62 The structures that were designed from these leads were 34–37. In contrast to 28, 29, and 30, none of the 6 molecules in Figure 6 have the N-methyl D-Phe at II, but they still contain a Boc-protected lysine or a free lysine at V respectively. Molecules 34 and 35, similar to 33, contain an N-methyl at V in addition to the Boc-protected lysine and free lysine at V. Derivatives 36 and 37, similar to 16, incorporate a D-Val at III, as well as either a Boc-protected lysine or free lysine respectively at V.
Figure 6.
The design of new derivatives based on compound 1, 33, and 16.
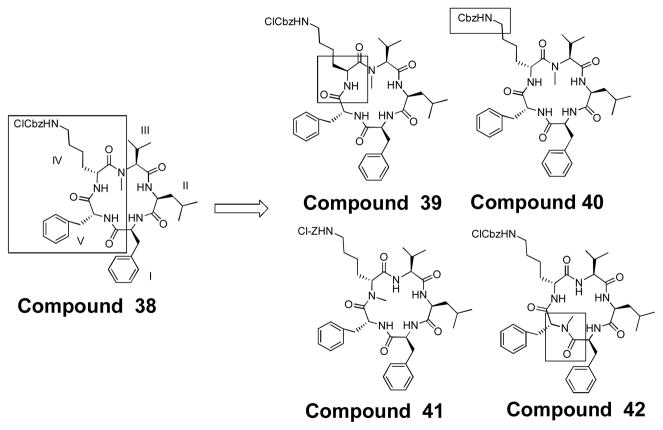
A recent publication describing the potency of compound 38 when tested against pancreatic cancer cell lines6 prompted us to design compounds that were based on the structure of this molecule. We created four compounds that would mimic 38 and explore the importance not only of stereochemistry but also of the protecting group on the lysine side-chain. These compounds are 39, 40, 41, and, 42 (figure 7). 39 contains an L-2-Chloro-Cbz-protected Lysine at IV rather than the D-2-Chloro-Cbz-protected lysine seen in 38. Compound 40 maintains the D stereochemistry at IV but utilizes a Cbz-protected lysine rather than a 2-chloro-Cbz lysine in order to investigate the effect of the 2-Chloro-Cbz on biological activity. Compound 41 maintains the stereochemistry, but moves the N-methyl in between the two D-amino acids. Finally, 42 has an N-methyl at position V rather than at III.
Figure 7.
The design of new derivatives based on compound 38.
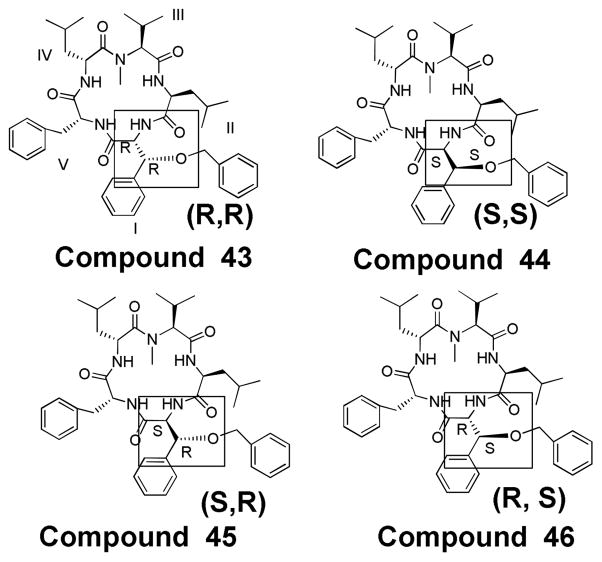
In previous work, we had noticed that in addition to the trend that two consecutive D-amino acids and an N-methyl were key to potency,6 there was a trend that the potent molecules typically contained 2–3 phenyl groups within the structure (similar to compounds 38 through 42). Thus, we designed 4 new compounds that would incorporate three phenyl rings, while maintaining the N-methyl and D-amino acids in the core structure. These compounds are: 43, 44, 45, and 46 (figure 8), where these 4 molecules are diastereomers of each other. Positions II–V are identical, and at position I, the alpha and beta carbons of the benzylated beta-hydroxy-phenylalanine have alternating stereochemistry.
Figure 8.
The design of new derivatives based on de novo design using SAR
3. Synthesis of Sansalvamide A-Amide derivatives
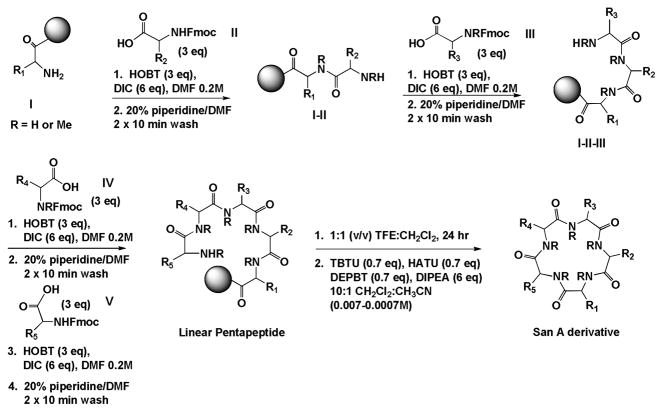
All thirty-two derivatives described here were constructed as the peptide analogs (Figures 3–8). Two synthetic protocols have been developed for the creation of these thirty-two derivatives: a convergent solution-phase strategy, which we have previously published,4, 66 as well as a solid-phase approach, which is described here the for the first time (Figure 9). Both routes have provided access to large milligram quantities of San A-amide derivatives. Ten of these new compounds were synthesized using solution phase (6, 7, 10, 12, 15, 18, 23, 24, 26, and 27), twenty-one of these compounds were synthesized using solid-phase 3, 4, 5, 20, 21, 29–32, 34–37, and 39–46, and one compound, 28, was made using both methods.
Figure 9.
Solid-Phase synthesis of San A-amide derivatives.
For the ten solution-phase compounds we used our previously published synthesis.5 The solid-phase synthesis compounds were synthesized using a preloaded chlorotrityl resin (where the first amino acid was already bound to the resin) (Figure 9). Sequential coupling then deprotection of four Fmoc protected amino acids using coupling conditions of Hydroxybenzotriazole (HOBT, 3 equivs), and N, N′-Diisopropylcarbodiimide (DIC, 3 equivs) in 0.2M DMF, and standard deprotection conditions of piperidine: DMF (20:80 ratio) yielded a resin-bound linear pentapeptide. Cleavage from the resin was accomplished using Trifluoroethanol (TFE) and DCM in a 1:1 ratio for 24 hours. After complete removal of residual TFE (to avoid trifluoroethyl esterification during the cyclization step) and confirmation of each linear pentapeptide via NMR and LCMS, cyclization was accomplished using our standard cyclization conditions employing a cocktail of 3 coupling agents (2-(1H-7-Azabenzotriazol-1-yl)--1,1,3,3-tetramethyl uronium hexafluorophosphate Methanaminium (HATU), 3-(Diethoxy-phosphoryloxy)-3H-benzo[d][1,2,3] triazin-4-one (DEPBT), and O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) 0.7 equivs each), N,N-Diisopropylethylamine (DIPEA), and run at 0.007M-0.0007M in ACN and DCM.66 These synthesis conditions generated a total of seventeen compounds, all in moderate yields (5–76%, average ~40%). This solid-phase route, although slightly more expensive, proves to be more efficient, quickly generating high purity linear precursors which result in cyclized compounds with overall significantly higher yields. The final purity of all compounds was verified by NMR and LCMS.66
4. Biological Data for compounds
All compounds were tested for their cytotoxicity on colon cancer cell line HCT-116 using 3H-thymidine incorporation assays. These new third generation compounds were compared to the potency of compounds from which they were designed, and cell proliferation was monitored by measuring how much 3H-thymidine was incorporated into a cell’s DNA. Lower thymidine incorporation is correlated to a decrease in cell proliferation in the presence of the compound, and hence the more toxic the compound. Data below is shown as a % growth inhibition, where the greater the % inhibition, the more cytotoxic the compound. Cytotoxicity data are shown by sequentially starting with first generation compounds and those designed from these structures, then the second generation molecules and the cytotoxicity of derivatives designed from these, and finally the toxicity of the de novo compounds.
Table 1 outlines the biological activity of new compounds and compares their vaules to the earlier generation structures from which they were designed. It is important to note that first generation compounds 1, 2, 8, 9, and 11 all have significantly higher % growth inhibition values in other cell lines: PL-45, BxPc3 (both pancreatic cancer cell lines) or SW480 and HT-29 (MSS colon cancer cell lines that respond to treatment with 5-FU), which is why they were initially chosen as lead structures.6, 9 However, given the enormous problems seen in treating drug-resistant colon cancer, we have chosen to focus on finding molecules that have a high percent growth inhibition against the drug-resistant (MSI) colon cancer cell line HCT-116. As the data shows, there was no significant improvement in the new compounds (bold) compared to the first generation leads (non-bold).
Next, we examine the biological activity of compounds that were designed from the second generation structures. As the data below shows, there was no significant improvement in the biological activity of the new compounds (bold) compared to the second-generation leads (non-bold). However, a very interesting structure-activity relationship is observed between compound 19 and 20, where 19 is significantly more potent than 20. Perhaps not surprisingly, 19 was also significantly more potent than derivative 21 This supports the conclusion we have published in a prior manuscript: an N-methyl is imperative for inducing an appropriate 3-dimensional structure. That is, we have shown that the most important structural motif is the inclusion of 2 consecutive D-amino acids with an N-methyl moiety, and we had demonstrated that 3 compounds were significantly more potent than the natural product peptide, San A-amide, when they followed this motif.6 This conclusion was validated by others who found that cyclic pentapeptides containing both an N-methyl and D-amino acids were fixed into a major or even single conformation.14, 15 Further, these data indicate that the compounds, if appropriately situated once locked, will have a well-defined, high affinity interaction with the protein target.17, 18 The cytotoxicity data describing compound 19 and 20 supports this conclusion. Second generation compounds 22 and 25 were reasonably potent in numerous colon cancer cell lines (HCT116, HCT15,61, 62 and HT-29,5 as well as pancreatic cancer cell lines6, 9) yet it is interesting to see that polar compounds 24 and 27, which were based on 22 and 25 but incorporated a polar residue that would improve solubility in aqueous media, and 23 and 26, with protecting groups on the respective polar residues, were significantly less potent than their lead structures in colon cancer cell lines..
The following series shows the biological activity of compounds that were designed based on a second-generation lead structure 14, as well as compounds designed from a new third generation structure that proved to be relatively cytotoxic, 28 (% growth inhibition at 10μM = 99%). As noted in earlier SAR discussions describing compound 14 and 19’s potency the inclusion of an N-methyl and D-Phenylalanine is favorable, which explains compound 28’s relative potency. It is interesting to note that structurally similar compound 29 is not nearly as potent, where there is an exchange of the Cbz to a Boc moiety. Given the poor cytotoxicity of 24 and 27, which contain a free lysine and free arginine respectively, it is not surprising that the free lysine-containing 30 is not active. Compounds 31, 34, and 36 were designed based on 29, 33, and 16 respectively. Compounds 32, 35, and 37 were then free lysine derivatives of 31, 34, and 36 respectively and, not surprisingly, were inactive.
We then show the biological activity of compounds that were designed based on a lead structure described in our most recent publication,6 38. Of the 3 compounds that were based on 38, we synthesized one compound, 42, with improved cytotoxicity over the second generation lead structure. It should be noted that the structure with a Chloro-carboxybenzyl protected L-Lysine that was substituted at position IV generated compound 39, which was less potent than the parent compound 38 that had the Chloro-carboxybenzyl protected D-Lysine was at this position. This data supports our hypothesis that the inclusion of 2 consecutive D-amino acids with an N-methyl moiety is important for potency. However, interestingly, the molecule where a carboxybenzyl protected D-lysine was placed at position IV (compound 40), which contained 2 consecutive D-amino acids with an N-methyl moiety, was significantly less potent than either 38 or 39, suggesting that the addition of the chloro substituent on the carboxybenzyl was crucial for improving binding to this molecule’s biological target. We noted the movement of the N-methyl to position V, compound 42, produced a highly potent compound that is more toxic than its lead structure 38, or its structurally related analog 41. It is very remarkable to note that although both 41 and 42 contain the chloro-carboxybenyl moiety, an N-methyl moiety, and 2-consecutive D-amino acids the placement of the N-methyl moiety is critical for potency as 41 is not very potent at all, but 42 shows remarkable cytotoxicity. These data support our hypothesis that 2-consecutive D-amino acids combined with an appropriately placed N-methyl moiety are important for inducing a favorable conformation. However, they also indicate that there may be a favorable electronic effect on binding induced by the inclusion of a chlorine in the structure.
Finally we look at the biological activity of compounds that were designed via a de-novo process. These 4 compounds, 43–46, included an N-methyl, at least 2 D-amino acids, and 3 phenyl moieties within the core structure. The most potent compound, 43, also has two consecutive D-amino acids and an N-methyl moiety, and then discretely places the benzyl protected phenyl threonine below the ring plane. Given that 43 has significantly great potency than the other three derivatives, it appears that this placement plays a key role in potency and demonstrates that the 3-D shape of the molecule is important for obtaining a tight binding to its target.
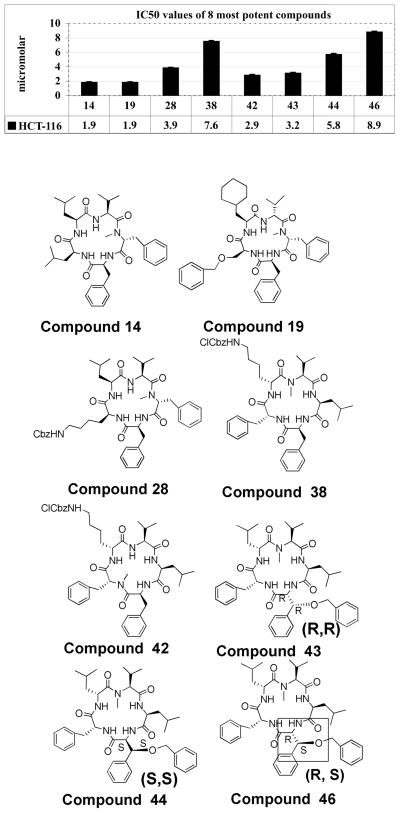
The most potent compounds, defined as ≥60% cytotoxicity against HCT-116 at 10μM, were then run in cytotoxicity assays and the IC50 values were calculated by plotting five concentrations (10, 3, 1, 0.3, and 0.1μM) and extracting data from the curves (Figure 10). There were 8 compounds that exhibited ≥60% growth inhibition, these included: 14, 19, 28, 38, 42, 43, 44, and 46. All relationships were exponential in nature, although it does appear that these compounds have limited solubility, which inhibits the compounds from dissolving at concentrations higher than 500nM (0.5μM). This indicates that for future studies we should include a polar moiety in order to improve solubility on an area of the molecule that will not interfere with binding to their biological target.
Figure 10.
IC50s of potent compounds. Each data point is an average of four wells run in three assays at 10, 3, 1, 0.3, 0.1 μM concentrations. Data represents the concentration required of each compound to inhibit 50% of viable cell growth in the assay using HCT-116 colon cancer cell lines. Inhibition is relative to 1% DMSO control.
5. Summary of SAR results
In summary, the most important features to emerge from this SAR study include the observation that the potent molecules contain 2 consecutive D-amino acids and an N-methyl moiety. In addition to understanding the importance of that structural feature, we also learn that a) a chloro-Cbz moiety improves cytotoxicity over a Cbz (38 versus 40), b) a Cbz generates a molecule with better cytotoxicity than one protected with a Boc group (28 versus 29), and c) an N-methyl positioned on the D-phenylalanine produces a molecule that is more potent than without the N-methyl (14 versus 17, 19 versus 20, and 42 versus 38). Finally, we learn that the benzylated beta-hydroxy-phenylalanine, when in the R, R configuration (43), affords a structure that is relatively potent compared to the other diastereomers (44, 45, and 46). Thus, it would appear that the ideal structure would have the following features: the benzylated beta-hydroxy-phenylalanine in with R, R stereochemistry (I), the L leucine (II), an amino acid with polar moiety in order to enhance solubility (III), a D-lysine protected with a chloro-Cbz (IV), and an N-methyl-D-Phenylalanine (V). It is noted that although structures 14, 19, and 28 incorporate an N-methyl-D-Phenylalanine at position II, this moiety is already incorporated at position V of the ideal structure, and therefore it seems unlikely that it should also be included at position II. Further, it will only increase the hydrophobicity of this molecule, which already lies outside the cLogP values that are quoted in Lipinski’s rules for enhancing drug like properties.67 Rather, leaving positions II and III open to modification with polar amino acids or peptidomimetic structures such as oxazoles or thiazoles seems like a better approach. These two options would decrease the hydrophobicity and move the molecule into more reasonable clogP values. One pro-drug approach is to place a methyl-protected acid side chain at II or III (i.e. glutamic acid or aspartic acid with a methyl ester on the side chain), which would allow the molecule to cross the hydrophobic cell wall and then be cleaved upon entering the cell. The other appropach is to incorporate thiazoles and oxazoles so as to increase the molecules hydrophilicity slightly, but still maintain a peptide-like backbone. Both of these approaches are now being pursued based on the above SAR.
6. Hsp90 competitive binding assay
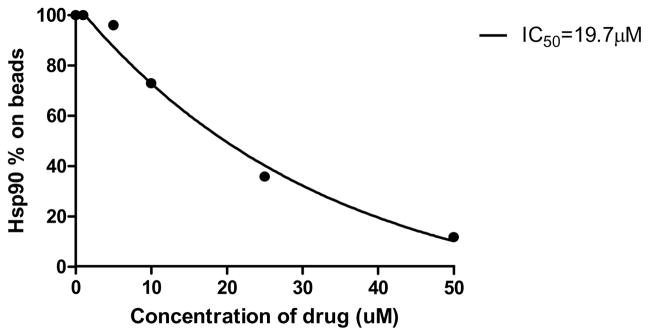
As described in our recent work,19 we identified Hsp90 as the target of the Sansalvamide A peptide (compound 1) using compound 1 tagged with biotin at position IV. Aware of the fact that this tagged derivative was no longer identical to compound 1 we wanted to confirm our findings. In order to do this, we have run a number of assays described in published work that shows our molecule inhibits the binding between several client proteins and Hsp90. However, we have not show data with a direct binding interaction between Hsp90 and compound 1-tag. Shown in Figure 11 is new, direct evidence that our molecule binds to Hsp90. This competitive binding assay was completed using compound 1, biotinylated compound 1 at position IV, and Hsp90. Increasing concentrations of compound 1 were incubated with Hsp90, followed by incubation with biotin-SanA1. It was found that compound 1 inhibited binding of biotin-SanA1 with an IC50 of 19.7μM (Figure 11), thus, confirming our findings that Hsp90 is a target of Sansalvamide A-amide (compound 1).
Figure 11.
Competitive binding affinity of SanA1 with tag at IV to Hsp90
7. Hsp90 client protein assays
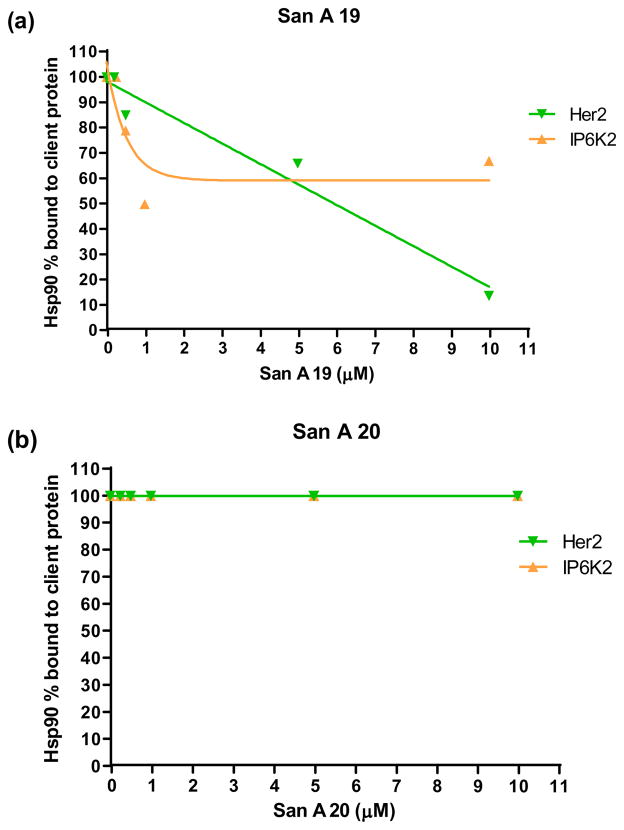
Our data indicated that the cytotoxicity of compound 1 was due, at least in part, to its ability to bind to Hsp9019 and inhibit client proteins and co-chaperones from binding. Thus, we anticipated that the cytotoxic effect of compound 19 was due, at least in part, to its ability to bind to also inhibit client proteins from binding to Hsp90. In order to test this hypothesis, we performed an in vitro binding assay, testing 19’s ability to inhibit binding between Hsp90 and two client proteins: Her2 and IP6K2. Her2 is a client protein that is associated with the N and M domains of Hsp90,68, 69 while IP6K2 is associated with the C-terminal domain and is a pro-apoptotic protein that is active when not bound to Hsp90.70 Compound 20 was used as the negative control as it exhibits little cytotoxic activity and only differs from 19 by a single N-methyl at position II. Excitingly, we found that compound 19 inhibited the binding of both IP6K2 and Her2 to Hsp90 (Figure 12). In contrast, compound 20 did not have any affect on the binding of IP6K2 or Her2 to Hsp90. These data suggest that compound 19 does bind to and modulate the function of Hsp90 and that the presence of an N-methyl is crucial for compound 19’s activity.
Figure 12.
In vitro binding assay: (a) San A 19 inhibits the binding of both IP6K2 and Her2 to Hsp90. (b) San A 20 does not affect Her2 and IP6K2 binding to Hsp90. Percent Hsp90 bound to client protein was quantified by densitometric scanning of Hsp90 protein on Western blot with normalization to client protein loading using Image J.
8. Docking to Hsp90 using Autodock
We have shown that the San A-amide, compound 1, binds to Hsp90 between the N-middle domain.19 Although we cannot assume that all of the potent compounds shown in figure 10 will bind to Hsp90, based on our published work, we investigated their binding affinity for this target at the N-middle domain. We used Autodock to visualize how our molecules may bind to Hsp90. This program is well established and is frequently used to dock small molecules to large protein targets via an automated prediction of ligand-binding sites.71–74 It generates an efficient docking of peptides and small molecules to proteins,72, 73, 75 and thus it is a powerful tool for visualizing protein inhibitors. As our recent work has shown, San A derivatives bind to Hsp90 at the N-Middle domain,19 thus we focused on binding our molecules to Hsp90 (pdb file 2CG9.pdb) in this region (Figures 13 through Figure 15). Using the Autoligand program, we identified two potential binding sites on the protein between these domains. Next, using Autodock 4.2, we docked four derivatives to Hsp90 and found one of the sites gave a much lower binding energy to all 4 molecules (-3.5 kcal/mol vs. −7.5 kcal/mol). Because San A-amide is a known micromolar inhibitor and thus reasonably potent, we chose to dock all molecules to the site that gave the lowest binding energy as it seemed to be an accurate reflection of the binding energy data obtained from Autodock. To examine the binding of our San A-amide derivatives, we docked each compound 250 times with the binding site identified on Hsp90. Docking modes that returned similar binding energies and conformations were clustered together to generate potential binding orientations for each derivative. The mode or orientation for each derivative that gave the lowest mean binding energy was selected and visualized using PyMol. This allowed us to visualize the conformation and relative orientation of each of the 8 potent derivatives bound to Hsp90.
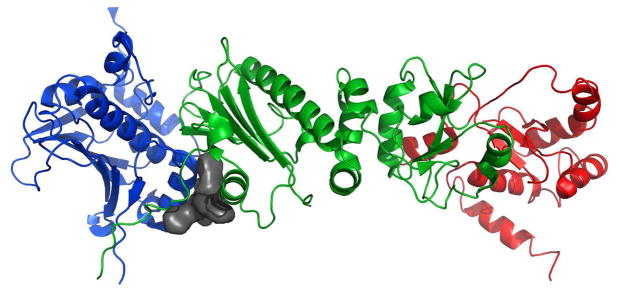
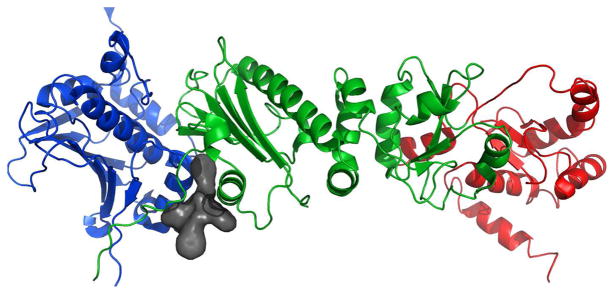
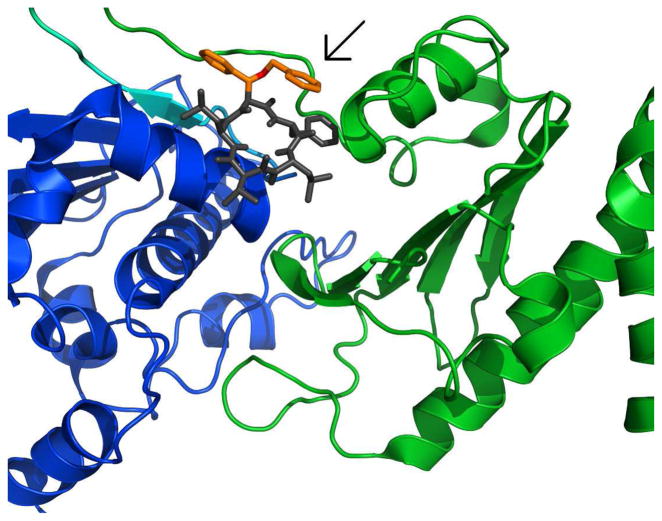
Figure 13.
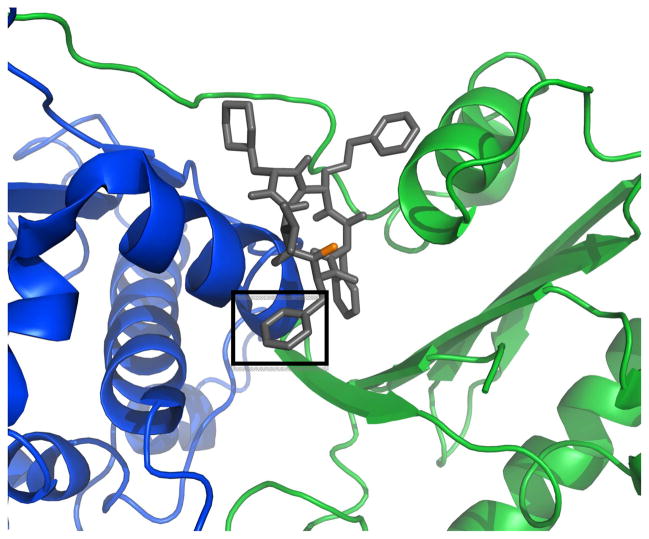
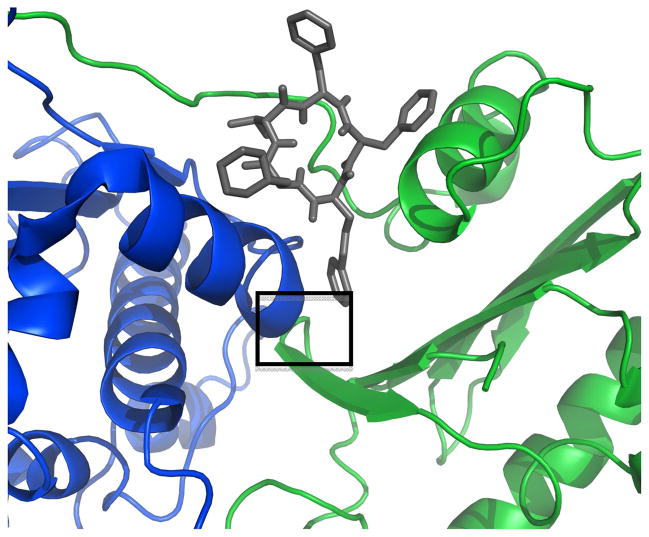
a) Full-length Hsp90 monomer with Compound 19 in the predicted binding site between the N-Middle domain. Blue, green and red are for the N, middle and C terminal domains respectively. 19 (space-filling, grey) is bound to the region between the N and Middle domains of full-length yeast Hsp90. This potent derivative adopts a conformation that fits well inside the binding pocket. b) 20 (space-filling, grey) is bound to the region between the N and Middle domains. Note how the differences in conformation prevent this non-potent derivative from inserting deep into the pocket, exposing a majority of the structure to solvent. The proposed binding site on Hsp90 was identified using AutoLigand correlated with pull-down assay results and the San A derivative binding mode was determined using AutoDock4.2. Molecular graphics were prepared using PyMol.
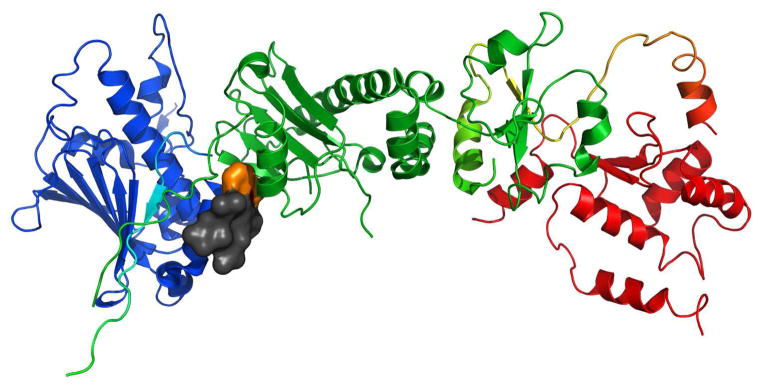
Figure 15.
a) Full-length Hsp90 with Compound 43 (space-filling, grey) docked to the N-middle domain. Blue, green and red are for the N, middle and C terminal domains respectively. The benzylated beta-hydroxy phenylalanine side chain is highlighted in orange. This derivative is predicted to adopt a conformation that orients the two aromatic groups of this side chain relatively coplanar with rest of the macrocycle and allows them to insert into the N-M pocket of the protein. b) 44 (space-filling, grey) is bound at the N-middle domain with the benzylated beta-hydroxy phenylalanine side chain highlighted in orange. This derivative is predicted to adopt a conformation that orients the aromatic groups of this side chain relatively perpendicular to the rest of the macrocycle. This prevents the key moiety from inserting into the pocket and forces the compound to bind in a much different orientation with the benzylated beta-hydroxy phenylalanine side chain pointed out into solution. The proposed binding site on Hsp90 was identified using AutoLigand correlated with pull-down assay results and the San A derivative binding mode was determined using AutoDock4.2. Molecular graphics were prepared using PyMol.
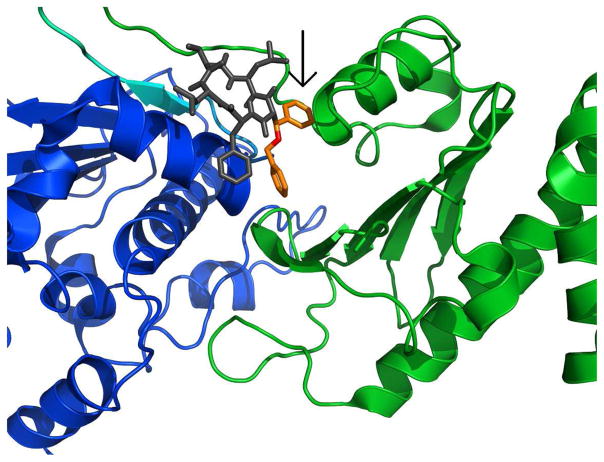
In figures 13 and 14, we show two structurally similar derivatives, 19 and 20, bound to the yeast variant of Hsp90. Compounds 19 and 20 differ only in the presence of an N-methyl group at position II. However, although structurally similar, compound 19 and 20’s cytotoxicity and ability to inhibit client proteins from binding to Hsp90 differ tremendously, and thus it is anticipated that their binding modes to Hsp90 may be different. Thus, it is not surprising that Autodock predicted separate binding modes for these two compounds. Compound 19’s potency is reflected in its greater binding affinity as predicted by Autodock. Figures 13a and 14a show that compound 19 adopts a conformation that allows it to insert into the binding pocket between the N-M domain (the anticipated binding site based on our published data),19 which results in an interaction between the aromatic side groups of the derivative and the sixth alpha-helix (blue) of Hsp90 (see boxed aromatic side chain) that is not present in compound 20 docked to Hsp90 (Figures 13b and 14b). It could be this contact with the helix that explains the ability of compound 19 to disrupt the binding of Hsp90 to its client protein Her2 and partially disrupt the binding of client protein IP6K2. The less potent compound 20, (Figures 13b and 14b) is shown to adopt a different conformation, preventing it from inserting into the binding pocket and engaging in the interactions with the helix that observed with 19.
Figure 14.
Close-up of docking results: (a) compound 19 and (b) compound 20 bound to Hsp90. Compound 19 contains a methyl group (orange) in the cyclic peptide backbone, which is not present in the peptide backbone of compound 20. The specific interaction that may be responsible for enhanced binding affinity of 19 is shown in the box, Figure 14a, and its absense is also highlighted, Figure 14b. Thus, the N-methyl has an obvious effect on the confirmation of the molecule and it’s binding to Hsp90.
Similarly, we show two homologous derivatives, 43 and 44, Figures 15 and 16 respectively. 43’s key feature is the benzylated beta-hydroxy-phenylalanine with R, R stereochemistry and 44 has this moiety with S, S stereochemistry. The predicted binding modes, in this case, showed the derivatives binding in much different orientations. It appears that the stereochemistry of the benzylated beta-hydroxy-phenylalanine of 43 allows the compound to adopt a conformation that results in the molecule binding with this moiety inserted into the binding pocket between the N-M domains. This same moiety on 44, with S, S stereochemistry, results in a conformation that prevents the compound from binding in the same orientation. The very different binding orientation predicted for these two compounds reflects the difference in cytotoxicity observed for them. These two models show how we can use the blind docking approach with Autodock to examine our derivatives bound to Hsp90 and use these images to help develop more potent derivatives.
Figure 16.
Close-up images of 43 (R, R) (top) and 44 (S, S) (bottom) as a line-bond structure with the benzylated beta-hydroxy phenylalanine side chain colored in orange (oxygen is red). Both molecules show an interaction between a phenyl group and the same α-helix as observed with 38 and 40. However, 43 is predicted to insert into the binding pocket in a orientation that allows all three of its aromatic groups to interact with the protein, whereas 44 adopts a conformation that forces it to bind with two of these aromatics relatively uninvolved with protein interactions. The interaction of multiple aromatic moieties with the protein target may explain the greater cytotoxicity demonstrated by compound 43 over 44.
9. ADME studies
Although the cytotoxic effects of lead compounds are thought to be primarily due to its ability to bind to Hsp90, other factors such as solubility, stability and/or efflux properties within the cell may also contribute. Therefore, we commissioned Biofocus, an outside company, to run ADME (Adsorption, Distribution, Metabolism, and Excretion) experiments. The two potent derivatives that were discussed in the modeling, compounds 19 and 43, were selected for ADME experiments. It was found that the de novo designed compound 43 had better overall ADME properties (Table 2). Compound 19 is hindered by very low aqueous solubility (<5uM). In comparison, compound 43 has good solubility, 7uM. Compound 19 showed a half life of 38 minutes, while compound 43 showed a half life of >172 minutes. Finally, the Caco-2 permeation study showed that compound 19 had a higher efflux ratio than 43 (25 to 3, respectively), where it is desirable to have an efflux ratio as close to 1 as possible. These data show that by using the SAR, we have improved the properties of a San A molecule, improving their drug-like character in this new series of derivatives.
Table 2.
Solubility, half-life and efflux data for compounds 19 and 43
| Compound | Kinetic solubility (μM) |
Half-life (minutes) |
Efflux ratio | ||||
|---|---|---|---|---|---|---|---|
| n1 | n2 | average | n1 | n2 | average | ||
| 19 | <5 | <5 | <5 | 45 | 30 | 38 | 25 |
| 43 | 7 | 7 | 7 | 172 | >200 | >172 | 3 |
10. Conclusion
For the first time, we report here the synthesis of 32 new Sansalvamide A structures and their activity against the drug resistant colon cancer cell line HCT-116. We have identified characteristics that are common to the potent molecules, and provided evidence that these characteristics play a role in their 3-D conformation. We have shown that the active molecules have unique docking interactions with the known biological target, Hsp90, compared to structurally related compounds that are inactive. We have also provided evidence that our molecules not only bind to Hsp90 directly, but that the potent molecule 19 inhibits 2 client proteins from binding to Hsp90, thus indicating its mode of action may inpart be due to modulating the function of Hsp90 via thes two client proteins. Finally, we have shown that our most promising lead structure in this series, 43, has improved ADME properties over compound 19, which indicates that we have built in some pharmacokinetic stability into the compounds. These data indicate that our molecules are cytotoxic, and act in part by modulating the activity of Hsp90. The synthesis of these new compounds and their evaluation in the context of their lead structures as well as their interactions with their potential protein target, Hsp90, provide insight into how this unique set of molecules induce cytotoxicity. Studies involving these compounds and their modulation of Hsp90’s function are ongoing and will be reported in due course.
EXPERIMENTAL PROCEDURES
Thymidine Uptake Assays
Proliferation of the HCT-116 colon cancer cells was tested in the presence and absence of the compounds using 3H-thymidine uptake assays. Cells treated with the compounds were compared to dimethyl sulfoxide (DMSO) controls for their ability to proliferate as indicated by the incorporation of 3H-thymidine into their DNA. Cells were cultured in 96 well plates at a concentration of 4000–5000 cells/well in DMEM (Gibco) supplemented with L-glutamine, 10% fetal bovine serum and 1% penicillin-streptomycin antibiotic. After overnight incubation, the compounds were added. The compounds were dissolved in DMSO at a final concentration of 1.0% and tested at the concentrations indicated in the manuscript. The DMSO control was also at 1.0%. After the cells had been incubated with the compounds for 54 h, 1mCi 3H-thymidine per well was added and the cells were cultured for an additional 18 h (for the cells to have a total of 72 h treatment), at which time the cells were harvested using a PHD cell harvester (Cambridge Technology Inc.). The samples were then counted (CPM) in a scintillation counter for 1.0 m. Decreases in 3H-thymidine incorporation, as compared to DMSO controls, are an indication that the cells are no longer progressing through the cell cycle or synthesizing DNA, as is shown in the studies presented. Mean growth inhibition (n=8–12) is the 1 minus CPM of compound-treated cells over DMSO-treated cells. IC50 were determined using 0, 0.1, 0.3, 1, 3, and 10 μM of compound (in 1% DMSO final concentration). All calculations including mean, SEM, and IC50 were performed on Excel.
Hsp90 binding constant assays
Purified, native Hsp90 (Stressgen) was incubated in PBS (without Ca/Mg) with or without SanA compounds for 1 hour at room temp, and then incubated with biotin-SanA for 1 hour at room temp. Strptavidin beads were added and incubated for 30 minutes at room temp followed by removal of the unbound supernatant. The beads were washed 3 times with PBS and heated for 15 minutes at 100°C in SDS-PAGE sample buffer. Samples were analyzed on SDS page protein gels (Invitrogen), and western blots done using Hsp90 antibodies. Bands in the western blots were quantified using ImageJ, and the percentage of Hsp90 still bound to the beads was calculated.
General Solution Phase Peptide Synthesis
All peptide coupling reactions were carried out under argon with dry solvent, using methylene chloride and acetonitrile (9:1) for dipeptide, tripeptide, and pentapeptide couplings. The amine (1.1 equivalents) and acid (1 equivalent) were weighed into a dry flask along with 4–8 equivalents of DIPEA and 1.1 equivalents of TBTU.* Anhydrous methylene chloride and acetonitrile was added to generate a 0.1M solution. The solution was stirred at room temperature and reactions were monitored by TLC. Reactions were run for 1 hour before checking via TLC. If reaction was not complete additional 0.25 equivalents of TBTU was added. If reaction was complete then work-up was done by washing with 10% aqueous hydrochloric acid and saturated sodium bicarbonate. After back extraction of aqueous layers with methylene chloride, organic layers were combined, dried over sodium sulfate, filtered and concentrated. Flash column chromatography using a gradient of ethyl actetate-hexane gave our desired peptide.
* Some coupling reactions would not go to completion using only TBTU and therefore 0.2–0.5 equivalents of HATU, and/or DEPBT were used. In a few cases up to 0.7 equivalents of all three coupling reagents were used.
General Solution Phase Amine Deprotection
Amines were deprotected using 20% TFA in methylene chloride (0.1M) with two equivalents of anisole. The reactions were monitored by TLC. Reactions were allowed to run for 1–2 hours and then concentrated in vacuo.
General Solution Phase Acid Deprotection
Acids were deprotected using 2 equivalents of lithium hydroxide with 3.4 equivalents of hydrogen peroxide in methanol (0.1 M). The peptide was dissolved in methanol and cooled to 0 °C. Hydrogen peroxide was added followed by lithium hydroxide. The reaction was monitored by TLC and usually done in 1–2 hours. Sodium thiosulfate (3.8 equivalents) was added to neutralize the peroxide and 5 % hydrochloric acid was added till the solution pH was 1. The aqueous solution was extracted five times with methylene chloride, and the combined organic layer was dried, filtered and concentrated in vacuo.
Macrocyclization procedure (in situ)
All pentapeptides were acid and amine deprotected using the general deprotection methods described above. Three coupling agents (DEPBT, HATU, and TBTU) were used at ~0.5 to 0.75 equivalents each. The dry double deprotected peptide (free acid and free amine) and coupling agents were dissolved in acetonitrile and methylene chloride (1:9 ratio) at a concentration of 0.1M to 0.007M. DIPEA (6–10 equivs in order to neutralize the pH) were then added to the reaction. TLC (macrocycle Rf similar to protected linear pentapeptide) and LCMS were used to monitor the reaction which was usually finished in 1–2 hours. If reaction was not complete in 2 hours, additional coupling agents were added. If reaction was complete then work-up was done by extracting with 10% aqueous hydrochloric acid and saturated sodium bicarbonate. After back extraction of aqueous layers with large quantities of methylene chloride, organic layers were combined, dried over sodium sulfate, filtered and concentrated. All macrocycles were first purified by flash column chromatography using an ethyl acetate/hexane gradient on silica gel. Finally, when necessary, reversed-phase HPLC was used for additional purification using a gradient of acetonitrile and deionized water with 0.1% TFA.
General Solid Phase Synthesis Remarks
Stepwise solid phase peptide synthesis was performed in a polypropylene solid-phase extraction cartridge fitted with a 20 μM polyethylene frit purchased from Applied Separations (Allentown, PA). 2-chlorotrityl resins were purchased in pre-loaded form with L-Phe, D-Phe, or L-Leu. Resins were swelled in DMF for 30 minutes prior to assembly of the linear five-residue peptide sequence. Solid-phase syntheses were performed on a 0.5 mmol scale based on resin-loading. All operations were performed at room temperature under open atmosphere unless stated otherwise.
General Solid Phase Peptide Synthesis
Fmoc-protected amino acids were coupled using 3 equivalents of amino acid, 3 equivalents of 1-hydroxybenzotriazole, and 6 equivalents of diisopropylcarbodiimide. Couplings were performed in DMF at 0.2 M with respect to the incoming Fmoc-protected amino acid. Couplings were allowed to proceed for a minimum of two hours, and were assayed via ninhydrin test to verify competition. Once complete, the coupling reaction solution was drained, and the resin subjected to Fmoc deprotection. (Note: Fmoc and N-methyl amino acids are coupled according to the cycle above, however for subsequent coupling onto the secondary amino terminus, 1-hydroxybenzotriazole was substituted with 1-hydroxy-7-azabenzotriazole and the coupling was allowed to proceed overnight).
General Solid Phase Amine Deprotectection
Following coupling completion, the peptide-resin was treated as follows for removal of the Fmoc protecting group: DMF wash (3 × 1 min), 20% Piperdine/DMF (1 × 5 min), 20% Piperdine/DMF (1 × 10 min), DMF wash (2 × 1 min), IPA wash (1 × 1 min), DMF was (1 × 1 min), IPA (1 × 1 min), DMF (3 × 1 min). A ninhydrin test was performed to verify completion.
General N-terminal Solid Phase Amine Deprotection
Once the final N-terminal amino acid residue had been coupled, the peptide-resin was treated as follows for removal of the Fmoc protecting group: DMF wash (3 × 1 min), 20% Piperdine/DMF (1 × 5 min), 20% Piperdine/DMF (1 × 10 min), DMF wash (3 × 1 min), IPA wash (3 × 1 min), MeOH (3 × 1 min). The fully-assembled peptide-resin was then drained and dried in vacuo overnight.
Cleavage of Linear Peptide
The full-length, linear peptide was cleaved from the resin by swelling and shaking the peptide-resin for 24 hours in a 1:1 (v:v) 2,2,2-Trifluoroethanol:CH2Cl2 (10 volumes/gram of dried resin). The cleavage solution was filtered through a Buchner filter, and the drained resin was washed with additional CH2Cl2 (5 volumes/gram of initial dried peptide-resin) to fully extract the cleaved peptide from the resin. Solvents in the combined filtrates were evaporated by rotary evaporation and the solids dried in vacuo overnight. The solids were then reconstituted in CH2Cl2, evaporated by rotary evaporation and dried in vacuo overnight again to remove residual entrapped TFE.
Macrocyclization procedure (syringe pump)
Three coupling agents (DEPBT, HATU, and TBTU) were used at ~0.5 to 0.75 equivalents each. These coupling agents were dissolved in ¾ of a calculated volume of dry methylene chloride that would give a 0.001 M to 0.0007 M overall concentration when included in the volume used for the deprotected peptide. The crude, dry, double deprotected peptide (free acid and free amine) was dissolved in the other ¼ solvent volume of methylene chloride. DIPEA (8 equivs) was then added to the solution containing coupling reagents dissolved in methylene chloride. The double deprotected peptide was then added to the bulk solution dropwise using a syringe pump at a rate of 30mL/hr. The reaction was monitored via LCMS and generally complete in 1–2 hours. Upon completion, the reaction was worked up by washing with aqueous HCl (pH 1) and saturated sodium bicarbonate. After back extraction of aqueous layers with large quantities of CH2Cl2, the organic layers were combined, dried, filtered and concentrated. All macrocycles were first purified by flash column chromatography using an ethyl acetate/hexane gradient on silica gel. Finally, when necessary, reversed-phase HPLC was used for additional purification using a gradient of acetonitrile and deionized water with 0.1% TFA.
Benzylation Procedure (for compounds 39–42)
The cyclized peptide was dissolved in 50% THF and 50% DMF to make a 0.1 M solution. The 60% NaH was used at 1.1 equivalents and dissolved in the 0.1M solution. Benzyl Bromide (2 equivalents) was then added to the reaction. After 2 hours, LC/MS indicated the reaction was developing. The reaction was completed in about 5 hours and then worked up by washing with deionized water. After that, the organic layer was collected, dried and preliminarily purified by flash column chromatography. Finally, reverse-phase HPLC was used for further purification by using a gradient of acetonitrile and deionized water with 0.1 % TFA.
METHODS OF CHROMATOGRAPHIC PURITY
Method A
|
Instrument: Agilent 1200 Series HPLC Agilent 62440A LC/MSD Trap | ||
|
Column: Zorbax SB-C18 2.1×30mm 3.5-Micron | ||
| Mobile Phase A: 0.1% (v/v) formic acid, 100% (v/v) water | ||
| Mobile Phase B: 0.1% (v/v) formic acid, 100% (v/v) acetonitrile | ||
| Gradient: | Time (min) | |
| Profile %A | Profile %B | |
| 0 | 80 | |
| 20 | ||
| 4.5 | 10 | |
| 90 | ||
| 4.6 | 10 | |
| 90 | ||
| 7.0 | 15 | |
| 85 | ||
| Flow Rate: 1.0 ml/min | ||
| Injection: 4μL | ||
| Solvent: 100% Methanol | ||
Method B
|
Instrument: Waters Flex Inject Waters 2487 Dual λ Absorbance Detector | ||
|
Column: Symmetry C18 3.5μm 4.6×75mm Column | ||
| Mobile Phase A: 0.1% (v/v) Trifluoroacetic acid, 100% (v/v) water | ||
| Mobile Phase B: 0.1% (v/v) Trifluoroacetic acid, 100% (v/v) acetonitrile | ||
| λ1: 215nm | ||
| λ2: 222nm | ||
| Gradient: | Time (min) | |
| Profile %A | Profile %B | |
| 0 | 70 | |
| 30 | ||
| 4.00 | 0 | |
| 100 | ||
| 13.00 | 0 | |
| 100 | ||
| 15.00 | 70 | |
| 30 | ||
| 16.00 | 70 | |
| 30 | ||
| Flow rate: 0.50 ml/min | ||
| Injection: 20μL | ||
| Solvent: 100% Methanol | ||
Synthesis of Compound 3
Dipeptide Fmoc-Val-D-Leu-O-Resin
Dipeptide Fmoc-D-Leu-Val-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1025.0 mg (.830 mmol, 1 equivalents) of NH2-D-Leu-O-Resin, the Val residue was incorporated using 844 mg of Fmoc-Val-OH (2.49 mmol, 3 equivalents), 331 mg (2.49 mmol, 3 equivalents) of HOBt, and 0.770 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-Val-D-Leu-O-Resin
Dipeptide NH2-D-Val-D-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Leu-Val-D-Leu-O-Resin
Tripeptide Fmoc-Leu-Val-D-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Val-D-Phe-O-Resin prepared above, the Leu residue was incorporated using 879.0 mg (2.49 mmol, 3 equivalents) of Fmoc-Leu-OH, 331 mg (2.49 mmol, 3 equivalents) of HOBt, and 0.770 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-Leu-Val-D-Leu-O-Resin
Tripeptide NH2-Leu-Val-D-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-NMe-Leu-Leu-Val-D-Leu-O-Resin
Tetrapeptide Fmoc-NMe-Leu-Leu-Val-D-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-Val-D-Leu-O-Resin prepared above, the Ser(Bzl) residue was incorporated using 915.0 mg (2.49 mmol, 3 equivalents) of Fmoc-NMe-Leu-OH, 381mg (2.49 mmol, 3 equivalents) of HOBt, and 0.770 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH-Me-Leu-Leu-Val-D-Leu-O-Resin
Tetrapeptide NH-Me-Leu-Leu-Val-D-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-D-Phe-NMe-Leu-Leu-Val-D-Leu-O-Resin
Pentapeptide Fmoc-D-Phe-NMe-Leu-Leu-Val-D-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH-Me-Leu-Leu-Val-D-Leu-O-Resin prepared above, the D-Phe residue was incorporated using 964.7 mg (2.49 mmol, 3 equivalents) of Fmoc-D-Phe-OH, 339 mg (2.2 mmol, 3 equivalents) of HOAt, 0.770 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-D-Phe-Nme-Leu-Leu-Val-D-Leu-O-Resin
Pentapeptide NH2-D-Phe-NMe-Leu-Leu-Val-D-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-D-Phe-NMe-Leu-Leu-Val-D-Leu-OH
Double Deprotected Pentapeptide NH2-D-Phe-NMe-Leu-Leu-Val-D-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1264.4 mg of dried NH2-D-Phe-NMe-Leu-Leu-Val-D-Leu-O-Resin, 6.5 mL of 2,2,2-trifluoroethanol and 6.5 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (436.3 mg, 85% yield)
Macrocycle D-Phe-D-Leu-Val-Leu-Leu-NMe
Macrocycle D-Phe-D-Leu-Val-Leu-Leu-NMe was synthesized following the “Macrocyclization procedure”. Utilizing 436 mg (0.706 mmols, 1.0 equivalents) of linear pentapeptide, 0.74 mL (6 equivalents) of DIPEA, 113.0 mg (0.353 mmols, 0.6 equivalents) of TBTU, 161 mg (0.424 mmols, 0.6 equivalents) HATU, and 129 mg (0.424 mmols, 0.6 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (16.9 mg, 3.4% yield).
1H NMR (400 MHz, CD3OD): δ 0.7–0.9 (m, 8H), 1.2–1.9 (m, 9H), 2.6 (s, 2H), 2.8–3.2 (m, 6H), 3.7 (m, αH), 3.8 (m, αH), 3.9 (m, αH), 4.1 (m, αH), 4.2 (m, αH), 5.4 (m, 2H), 7.0–7.3 (m, 5H), 7.6–8.2(d, 4H).
LCMS: m/z called for C33H53N5O5 (M+1) = 599.8, found 600.4
Synthesis of Compound 4
Dipeptide Fmoc-Leu-Leu-O-Resin
Dipeptide Fmoc-Leu-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1072.0 mg (.870 mmol, 1 equivalents) of NH2-Leu-O-Resin, the Leu residue was incorporated using 921 mg of Fmoc-Leu-OH (2.61 mmol, 3 equivalents), 399 mg (2.61 mmol, 3 equivalents) of HOBt, and 0.810 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-Leu-Leu-O-Resin
Dipeptide NH2-Leu-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-NMe-Phe-Leu-Leu-O-Resin
Tripeptide Fmoc-NMe-Phe-Leu-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-Leu-O-Resin prepared above, the NMe-Phe residue was incorporated using 1010.0 mg (2.61 mmol, 3 equivalents) of Fmoc-NMe-Phe-OH, 399 mg (2.61 mmol, 3 equivalents) of HOBt, and 0.810 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH-NMe-Phe-Leu-Leu-O-Resin
Tripeptide NH-NMe-Phe-Leu-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-D-Leu-NMe-Phe-Leu-Leu-O-Resin
Tetrapeptide Fmoc-D-Leu-NMe-Phe-Leu-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH-NMe-Phe-Leu-Leu-O-Resin prepared above, the D-Leu residue was incorporated using 921.3 mg (2.61 mmol, 3 equivalents) of Fmoc-D-Leu-OH, 355mg (2.61 mmol, 3 equivalents) of HOBt, and 0.810 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-D-Leu-NMe-Phe-Leu-Leu-O-Resin
Tetrapeptide NH2-D-Leu-NMe-Phe-Leu-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-D-Val-D-Leu-NMe-Phe-Leu-Leu-O-Resin
Pentapeptide Fmoc-D-Val-D-Leu-NMe-Phe-Leu-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Leu-NMe-Phe-Leu-Leu-O-Resin prepared above, the D-Val residue was incorporated using 885 mg (2.61 mmol, 3 equivalents) of Fmoc-D-Val-OH, 399mg (2.61 mmol, 3 equivalents) of HOAt, 0.810 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-D-Val-D-Leu-Nme-Phe-Leu-Leu-O-Resin
Pentapeptide NH2-D-Val-D-Leu-NMe-Phe-Leu-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-D-Val-D-Leu-NMe-Phe-Leu-Leu-OH
Double Deprotected Pentapeptide NH2-D-Val-D-Leu-NMe-Phe-Leu-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1286.4 mg of dried NH2-D-Val-D-Leu-NMe-Phe-Leu-Leu-O-Resin, 6.5 mL of 2,2,2-trifluoroethanol and 6.5 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (445.0 mg, 83% yield)
Macrocycle Phe-Nme-D-Leu-D-Val-Leu-Leu
Macrocycle Phe-NMe-D-Leu-D-Val-Leu-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 200 mg (0.324 mmols, 1.0 equivalents) of linear pentapeptide, 0.48 mL (6 equivalents) of DIPEA, 52.0 mg (0.162 mmols, 0.6 equivalents) of TBTU, 74 mg (0.194 mmols, 0.6 equivalents) HATU, and 58 mg (0.194 mmols, 0.6 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (17.2 mg, 8.8% yield).
1H NMR (400 MHz, CD3OD): δ 0.6–1.0 (m, 11H), 1.4–1.8 (m, 6H), 2.8 (s, 1H), 3.1 (s, 2H), 3.6 (m, αH), 3.9 (m, αH), 4.0 (m, αH), 4.2 (m, αH), 4.4 (m, αH), 5.3 (m, 1H), 5.7 (m, 1H) 7.1–7.3 (m, 5H) 7.4–8.2 (m, 4H).
LCMS: m/z called for C33H53N5O5 (M+1) = 599.8, found 600.3
Synthesis of Compound 5
Dipeptide Fmoc-Leu-D-Leu-O-Resin
Dipeptide Fmoc-Leu-D-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1014.0 mg (.821 mmol, 1 equivalents) of NH2-D-Leu-O-Resin, the Leu residue was incorporated using 868 mg of Fmoc-Leu-OH (2.45 mmol, 3 equivalents), 376 mg (2.45 mmol, 3 equivalents) of HOBt, and 0.760 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-Leu-D-Leu-O-Resin
Dipeptide NH2-Leu-D-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Phe-Leu-D-Leu-O-Resin
Tripeptide Fmoc-Phe-Leu-D-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-D-Leu-O-Resin prepared above, the Phe residue was incorporated using 952.0 mg (2.46 mmol, 3 equivalents) of Fmoc-NMe-Phe-OH, 376 mg (2.46 mmol, 3 equivalents) of HOBt, and 0.760 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH-Phe-Leu-D-Leu-O-Resin
Tripeptide NH-Phe-Leu-D-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-NMe-Leu-Phe-Leu-D-Leu-O-Resin
Tetrapeptide Fmoc-NMe-Leu-Phe-Leu-D-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH-Phe-Leu-D-Leu-O-Resin prepared above, the NMe-Leu residue was incorporated using 954 mg (2.46 mmol, 3 equivalents) of Fmoc-NMe-Leu-OH, 376mg (2.46 mmol, 3 equivalents) of HOBt, and 0.760 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-NMe-Leu-Phe-Leu-D-Leu-O-Resin
Tetrapeptide NH2-NMe-Leu-Phe-Leu-D-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-D-Val-NMe-Leu-Phe-Leu-D-Leu-O-Resin
Pentapeptide Fmoc-D-Val-NMe-Leu-Phe-Leu-D-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-NMe-Leu-Phe-Leu-D-Leu-O-Resin prepared above, the D-Val residue was incorporated using 834 mg (2.46 mmol, 3 equivalents) of Fmoc-D-Val-OH, 335mg (2.61 mmol, 3 equivalents) of HOAt, 0.760 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-D-Val-NMe-Leu-Phe-Leu-D-Leu-O-Resin
Pentapeptide NH2-D-Val-NMe-Leu-Phe-Leu-D-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-D-Val-NMe-Leu-Phe-Leu-D-Leu-OH
Double Deprotected Pentapeptide NH2-D-Val-NMe-Leu-Phe-Leu-D-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1956.0 mg of dried NH2-D-Val-NMe-Leu-Phe-Leu-D-Leu-O-Resin, 6.0 mL of 2,2,2-trifluoroethanol and 6.0 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (450.0 mg, 89% yield)
Macrocycle Phe-Leu-NMe-D-Val-D-Leu-Leu
Macrocycle Phe-Leu-NMe-D-Val-D-Leu-Leu-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 250 mg (0.405 mmols, 1.0 equivalents) of linear pentapeptide, 0.42 mL (6 equivalents) of DIPEA, 64.0 mg (0.20 mmols, 0.6 equivalents) of TBTU, 91 mg (0.24 mmols, 0.6 equivalents) HATU, and 72 mg (0.24 mmols, 0.6 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (9.2 mg, 4.7% yield).
1H NMR (400 MHz, CD3OD): δ 0.6–1.0 (m, 8H), 1.2–1.6 (m, 6H), 2.6 (s, 1H), 3.0 (s, 1H), 3.6 (m, αH), 3.8 (m, αH), 4.1 (m, αH), 4.2 (m, αH), 4.4 (m, αH),, 7.1–7.3 (m, 5H).
LCMS: m/z called for C33H53N5O5 (M+1) = 599.7, found 600.3
Synthesis of Compound 6
Dipeptide MeO-D-Phe-Leu-NHBoc
Dipeptide MeO-D-Phe-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 380.7 mg (1.8 mmols, 1.1 equiv.) of amine OMe-D-Phe-NH2, 400 mg (1.6 mmols, 1.0 equiv.) of acid HO-Leu-NHBoc, 3.0 mL (11 equivalents) of DIPEA, 566.7 mg (1.8 mmols, 1.1 equiv.) of TBTU, in 16 mL of Methylene Chloride. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (622.1 mg, 99% yield).
Rf: 0.9 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 6H), 1.4 (s, 9H), 1.4–1.7 (m, 3H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 4.0–4.2 (br, αH), 4.8 (d, 1H), 4.8–5.0 (q, αH), 6.6 (d, 1H), 7.1–7.4 (m, 5H)
Dipeptide MeO-D-Phe-Leu-NH2
Dipeptide MeO-D-Phe-Leu-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (464mg, 100% yield).
Tripeptide MeO-D-Phe-Leu-Val-NHBoc
Tripeptide MeO-D-Phe-Leu-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 464 mg (1.6 mmols, 1.0 equiv.) of amine MeO-D-Phe-Leu-NH2, 313 mg (1.4 mmols, 1.0 equiv.) of acid HO-Val-NHBoc, 2.6 mL (10 equiv.) of DIPEA, 509 mg (1.6 mmols, 1.1 equiv.) of TBTU, in 12 mL of methylene chloride and 3 mL of acetonitrile. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (614 mg, 87% yield).
Rf: 0.5 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 12H), 1.4 (s, 9H), 1.4–1.6 (m, 2H), 1.7 (s, 1H), 2.1 (m, 1H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 3.8 (dd, αH), 4.4 (dd, αH), 4.8 (dd, αH), 5.0 (d, 1H), 6.3 (d, 1H), 6.6 (d, 1H), 7.1–7.3 (m, 5H)
Tripeptide MeO-D-Phe-Leu-Val-NH2
Dipeptide MeO-D-Phe-Leu-Val-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (489mg, 100% yield).
Dipeptide MeO-Leu-D-Leu-NHBoc
Dipeptide MeO-Leu-D-Leu-NBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 321 mg (1.8 mmols, 1.1 equiv.) of amine MeO-Leu-NH2, 400 mg (1.6 mmols, 1.0 equiv.) of acid HO-D-Leu-NBoc, 2.2 mL (8 equiv.) of DIPEA, 567 mg (1.8 mmols, 1.1 equiv.) of TBTU, in 16 mL methylene chloride and 4 mL acetonitrile. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (530.3mg, 92% yield).
Rf: 0.8 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.9–1.0 (d, 12H), 1.4(s, 9H), 1.4–1.8 (m, 6H), 3.7 (s, 3H), 4.1 (br, αH), 4.6 (br, α H), 4.8 (br, 1H), 6.6 (d, 1H).
Dipeptide HO-Leu-D-Leu-NHBoc
Dipeptide HO-Leu-D-Leu-NHBoc was synthesized following the “General acid deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (491mg, 98% yield).
Pentapeptide MeO-D-Phe-Leu-Val-Leu-D-Leu-NHBoc
Pentapeptide MeO-D-Phe-Leu-Val-Leu-D-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 489 mg (1.3 mmols, 1.0 equivalents) of amine MeO-D-Phe-Leu-Val-NH2, 491 mg (1.4 mmols, 1.1 equiv.) of acid HO-Leu-D-Leu-NHBoc, 1.0 mL (5 equiv.) of DIPEA, 201 mg (0.63 mmols, 0.5 equiv.) of TBTU, 238 mg (0.63 mmols, 0.5 equiv.) of HATU, and 75 mg (0.25 mmols, 0.2 equiv.) of DEPBT, in 13 mL of methylene chloride and 2 mL acetonitrile. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (201 mg, 22% yield).
Rf: 0.4 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.8–1.0 (m, 24H),1.3 (m, 2H), 1.4 (s, 9H), 1.5 (m, 3H), 1.6 (m, 2H), 1.7 (m, 2H), 2.2 (m, 1H), 2.9–3.2 (m, 2H), 3.7 (s, 3H), 4.1 (m, αH), 4.2 (m, αH), 4.4 (m, 2αH), 4.6 (m, αH), 7.s-7.3 (m, 5H)
Macrocycle D-Phe-Leu-Val-Leu-D-Leu
Macrocycle D-Phe-Leu-Val-Leu-D-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 169.3 mg (0.28 mmols, 1.0 equivalents) of linear pentapeptide, 0.7 mL (15 equivalents) of DIPEA, 45 mg (0.14 mmols, 0.5 equivalents) of TBTU, 74.6 mg (0.2 mmols, 0.7 equivalents) HATU, and 41.9 mg (0.14 mmols, 0.5 equivalents) of DEPBT in 15 mL methylene chloride, 4 mL acetonitrile and 2 mL dimethyl formamide. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (12.3 mg, 7.5% yield).
Rf: 0.25 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.7–1.0 (m, 24H), 1.2–1.8 (m, 9H), 2.0 (m, 1H), 2.9–3.1 (m, 2H), 3.6 (m, αH), 3.8 (m, αH), 4.2 (m, αH), 4.5 (m, αH), 4.6 (m, αH), 7.1–7.3 (m, 5H), 6.6 (d, 1H), 7.0 (d, 1H), 7.5 (d, 1H), 7.6 (d, 1H), 8.0 (d, 1H)
LCMS: m/z calcd for C32H51N5O5 (M+1) = 586.4, found 587.5
Synthesis of Compound 7
Dipeptide MeO-D-Phe-Leu-Boc
Dipeptide MeO-D-Phe-Leu-Boc was synthesized following the “General peptide Synthesis” procedure. Utilizing 951 mg (4.4 mmols, 1.1 equivalents) of MeO-D-Phe-NH2, 1000 mg (4.0 mmols, 1.0 equivalents) of Boc-Leu-OH, 2.8 mL (4 equivalents) of DIPEA, 1545 mg (4.8 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (1381mg, 88% yield).
Rf: 0.35 (EtOAc: Hex 3:7)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 6H), 1.4 (s, 9H), 1.6 (m, 2H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 4.2 (m, α H), 4.7–5.0 (m, 2H), 6.5 (d, 1H), 7.0–7.3 (m, 5H)
Dipeptide MeO-D-Phe-Leu-NH2
Dipeptide MeO-D-Phe-Leu-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (1029mg, 100% yield).
Tripeptide MeO-D-Phe-Leu-N-Me-Val-Boc
Tripeptide MeO-D-Phe-Leu-N-Me-Val-Boc was synthesized following the “General peptide Synthesis” procedure. Utilizing 686 mg (2.34 mmols, 1.1 equivalents) of MeO-D-Phe-Leu-NH2, 493 mg (2.13 mmols, 1.0 equivalents) of Boc-N-Me-Val-OH, 1.49 mL (4 equivalents) of DIPEA, 324 mg (0.85 mmols, 0.4 equivalents) of HATU. 547 mg (1.7 mmols, 0.8 equivalents) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (1073 mg, 99.6% yield).
Rf: 0.75 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.9–1.1 (m, 9H), 1.5 (s, 9H), 1.6–1.8 (m, 3H), 2.3 (m, 2H), 2.8 (s, 3H), 3.1 (m, 3H), 3.7 (s, 3H), 4.0 (d, αH), 4.6 (m, αH), 4.8 (m, αH), 6.4 (d, 1H), 6.7 (d, 1H), 7.1–7.3 (m, 5H)
Tripeptide MeO-D-Phe-Leu-N-Me-Val-NH2
Tripeptide MeO-D-Phe-Leu-N-Me-Val-NH2 was synthesized following the “General amine deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (861 mg, 100% yield).
Tetrapeptide MeO-D-Phe-Leu-N-Me-Val-N-Me-D-Leu-Boc
Tetrapeptide MeO-D-Phe-Leu-N-Me-Val-N-Me-D-Leu-Boc was synthesized following the “General peptide Synthesis” procedure. Utilizing 562 mg (1.39 mmols, 1.1 equivalents) of MeO-D-Phe-Leu-N-Me-Val-NH2, 328 mg (1.26 mmols, 1.0 equivalents) of Boc-N-Me-D-Leu-OH, 1.32 mL (6 equivalents) of DIPEA, 162 mg (0.5 mmols, 0.4 equivalents) of TBTU, and 479 mg (1.26 mmols, 1.0 equivalents) of HATU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tetrapeptide (294 mg, 33.5% yield).
Rf: 0.55 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CDCl3): δ 0.7–1.0 (m, 18H), 1.2–1.4 (m, 6H), 1.5–1.6 (m, 9H), 2.2 (m, 2H), 3.0 (m, 3H), 3.1–3.2 (m, 2H), 3.7 (s, 3H), 4.4 (m, 2H), 4.7 (m, 2H), 5.0 (m, 1H), 5.2 (m, 1H), 7.1–7.3 (m, 5H)
Tetrapeptide MeO-D-Phe-Leu-N-Me-Val-N-Me-D-Leu-NH2
Tetrapeptide MeO-D-Phe-Leu-N-Me-Val-N-Me-D-Leu-NH2 was synthesized following the “General amine deprotection”. This tetrapeptide was taken on to the next reaction without further purification or characterization. (247 mg, 100% yield).
Pentapeptide MeO-D-Phe-Leu-N-Me-Val-N-Me-D-Leu-D-Phe-Boc
Pentapeptide MeO-D-Phe-Leu-N-Me-Val-N-Me-D-Leu-D-Phe-Boc was synthesized following the “General peptide Synthesis” procedure. Utilizing 294 mg (0.47 mmols, 1.1 equivalents) of MeO-D-Phe-Leu-N-Me-Val-N-Me-D-Leu-NH2, 112 mg (0.42 mmols, 1.0 equivalents) of Boc-D-Phe-OH, 0.6 mL (8 equivalents) of DIPEA, 81 mg (0.25 mmols, 0.6 equivalents) of TBTU, and 129 mg (0.34 mmols, 0.8 equivalents) HATU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (224mg, 68% yield).
Rf: 0.4 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.7–1.0 (m, 9H), 1.2–1.4 (m, 12H), 1.6–1.7 (m, 12H), 2.3 (m, 2H), 2.8 (s, 3H), 3.0 (s, 3H), 3.1–3.2 (m, 2H), 3.7 (s, 3H), 4.3–4.4 (m, 2αH), 4.8–4.9 (m, 2αH), 5.2 (m, 1H), 5.5 (t, 1H), 6.4–6.6 (dd, 2H), 7.0–7.4 (m, 10H)
Macrocycle D-Phe-Leu-N-Me-Val-N-Me-D-Leu-D-Phe
Macrocycle D-Phe-Leu-N-Me-Val-N-Me-D-Leu-D-Phe was synthesized following the “Macrocyclization procedure”. Utilizing 184 mg (0.24 mmols, 1.0 equivalents) of double deprotected linear pentapeptide, 0.42 mL (10 equivalents) of DIPEA, 62 mg (0.19 mmols, 0.8 equivalents) of TBTU, 73 mg (0.19 mmols, 0.8 equivalents) of HATU, and 29 mg (0.09 mmols, 0.4 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) yield the macrocycle (1 mg, 0.6% yield).
1H NMR (400 MHz, CD3OD): δ 0.7–1.0 (m, 9H), 1.2–1.4 (m, 12H), 1.6–1.7 (m, 6H), 2.2 (m, 2H), 2.7 (s, 3H), 2.9 (s, 3H), 3.1–3.2 (m, 2H), 4.2–4.4 (m, 2αH), 4.6–4.8 (m, 2αH), 5.0 (m, 1H), 5.4 (t, 1H), 7.0–7.4 (m, 10H)
LCMS: m/z calcd for C37H53N5O5= 647.85, found 647.6
Synthesis of Compound 10
Dipeptide MeO-D-Phe-D-Leu-NHBoc
Dipeptide MeO-D-Phe-D-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 951 mg (4.4 mmols, 1.1 equiv.) of amine OMe-D-Phe-NH2, 1.0 g (4.0 mmols, 1.0 equiv.) of acid HO-D-Leu-NHBoc, 2.8 mL (4 equivalents) of DIPEA, 1.54 g (4.8 mmols, 1.2 equiv.) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (1.57g, 98% yield).
Rf: 0.5 (EtOAc: Hex 1:4)
1H NMR (200 MHz, CDCl3): δ 0.9–1.0 (m, 6H), 1.4 (s, 9H), 1.6–1.8 (m, 3H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 4.0–4.1 (m, αH), 4.8–5.0 (m, αH), 4.8–4.9, 6.6 (d, 1H), 7.1–7.3 (m, 5H).
Dipeptide MeO-D-Phe-D-Leu-NH2
Dipeptide MeO-D-Phe-D-Leu-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (1.15 g, 100% yield).
Tripeptide MeO-D-Phe-D-Leu-Val-NHBoc
Tripeptide MeO-D-Phe-D-Leu-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1.15 g (3.95 mmols, 1.1 equiv.) of amine MeO-D-Phe-D-Leu-NH2, 780 mg (3.59 mmols, 1.0 equiv.) of acid HO-Val-NHBoc, 2.5 mL (4 equiv.) of DIPEA, 1.38 g (4.30 mmols, 1.2 equiv.) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (1.72 g, 97% yield).
Rf: 0.5 (EtOAc: Hex 1:1) 1H NMR (200 MHz, CDCl3): δ 0.8–1.1 (m, 12H), 1.4 (s, 9H), 1.6–1.8 (m, 3H), 2.1–2.2 (m, 1H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 3.8–4.0 (m, αH), 4.4–4.5 (m, αH), 4.7–4.9 (m, αH), 4.9 (br, 1H), 6.1–6.3 (d, 1H), 6.5–6.6 (br, 1H), 7.1–7.4 (m, 5H).
Dipeptide MeO-Leu-Leu-NHBoc
Dipeptide MeO-Leu-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 801 mg (4.4 mmols, 1.1 equiv.) of amine MeO-Leu-NH2, 1.0 g (4.0 mmols, 1.0 equiv.) of acid HO-Leu-NHBoc, 2.8 mL (4 equiv.) of DIPEA, 1.54 g (4.8 mmols, 1.2 equiv.) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (1.43 g, 98% yield).
Rf: 0.5 (EtOAc: Hex 1:3)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 12H), 1.2–1.3 (m, 4H), 1.4(m, 9H), 1.6–1.7 (m, 2H), 3.7 (s, 3H), 4.1–4.2 (m, αH), 4.6–4.7 (m, αH), 4.8–4.9 (br, 1H), 6.4–6.5 (br, 1H).
Tripeptide MeO-D-Phe-D-Leu-Val-NH2
Dipeptide MeO-D-Phe-D-Leu-Val-NH2 was synthesized following the “General amine deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (1.36 g, 100% yield).
Dipeptide HO-Leu-Leu-NHBoc
Dipeptide HO-Leu-Leu-NHBoc was synthesized following the “General acid deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (1.23 g, 90% yield).
Pentapeptide MeO-D-Phe-D-Leu-Val-Leu-Leu-NHBoc
Pentapeptide MeO-D-Phe-D-Leu-Val-Leu-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1.37 g (3.49 mmols, 1.1 equivalents) of amine MeO-D-Phe-D-Leu-Val-NH2, 1.09 g (3.17 mmols, 1.0 equiv.) of acid HO-Leu-Leu-NHBoc, 4.43 mL (8 equiv.) of DIPEA, 611 mg (1.90 mmols, 0.6 equiv.) of TBTU, 361 mg (0.95 mmols, 0.3 equiv.) HATU, and 284 mg (0.95 mmols, 0.3 equiv.) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (1.323 g, 58% yield).
Rf: 0.5 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.8–1.0 (m, 24H), 1.4 (s, 9H), 3.1–3.2 (m, 2H), 3.7 (s, 3H), 4.2 (m, 2αH), 4.4–4.5 (m, 2αH), 4.6–4.7 (m, αH), 7.2–7.3 (m, 5H).
Macrocycle D-Phe-D-Leu-Val-Leu-Leu
Macrocycle D-Phe-D-Leu-Val-Leu-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 163 mg (0.27 mmols, 1.0 equivalents) of linear pentapeptide, 0.38 mL (8 equivalents) of DIPEA, 43.1 mg (0.14 mmols, 0.5 equivalents) of TBTU, 51.1 mg (0.12 mmols, 0.5 equivalents) HATU, and 56.3 mg (0.19 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (8 mg, 5% yield).
Rf: 0.5 (EtOAc: Hex 4:1)
1H NMR (600 MHz, CD3OD): δ 0.8–1.0 (m, 24H), 1.2–1.4 (m, 3H), 1.5–1.7 (m, 6H), 2.9–3.1 (m, 1H), 3.2–3.4 (m, 1H), 3.6–3.8 (m, 1H), 4.0–4.1 (m, 1αH), 4.1–4.2 (m, 2αH), 4.2–4.4 (m, 1αH), 4.6–4.8 (m, 1αH), 7.2–7.4 (m, 5H).
LCMS: m/z calcd for C32H51N5O5 (M+23) = 608.39, found 608.6.
Synthesis of Compound 12
Dipeptide MeO-Phe-Leu-NHBoc
Dipeptide MeO-Phe-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 380.7 mg (1.8 mmols, 1.1 equiv.) of amine MeO-Phe-NH2, 400 mg (1.6 mmols, 1.0 equiv.) of acid HO-Leu-NHBoc, 3.0 mL (11 equivalents) of DIPEA, 566.7 mg (1.8 mmols, 1.1 equiv.) of TBTU, in 16 mL of Methylene Chloride. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (622.1 mg, 99% yield).
Rf: 0.9 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 6H), 1.4 (s, 9H), 1.4–1.7 (m, 3H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 4.0–4.2 (br, αH), 4.8 (d, 1H), 4.8–5.0 (q, αH), 6.6 (d, 1H), 7.1–7.4 (m, 5H)
Dipeptide MeO-Phe-Leu-NH2
Dipeptide MeO-Phe-Leu-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (464 mg, 100% yield).
Tripeptide MeO-Phe-Leu-Val-NHBoc
Tripeptide MeO-D-Phe-Leu-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 464 mg (1.6 mmols, 1.0 equiv.) of amine MeO-Phe-Leu-NH2, 313 mg (1.4 mmols, 1.0 equiv.) of acid HO-Val-NHBoc, 2.6 mL (10 equiv.) of DIPEA, 509 mg (1.6 mmols, 1.1 equiv.) of TBTU, in 12 mL of methylene chloride and 3 mL of acetonitrile. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (614 mg, 87% yield).
Rf: 0.5 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 12H), 1.4 (s, 9H), 1.4–1.6 (m, 2H), 1.7 (s, 1H), 2.1 (m, 1H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 3.8 (dd, αH), 4.4 (dd, αH), 4.8 (dd, αH), 5.0 (d, 1H), 6.3 (d, 1H), 6.6 (d, 1H), 7.1–7.3 (m, 5H)
Tripeptide MeO-Phe-Leu-Val-NH2
Tripeptide MeO-Phe-Leu-Val-NH2 was synthesized following the “General amine deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (464 mg, 100% yield).
Dipeptide MeO-D-Leu-D-Phe-NBoc
Dipeptide MeO-D-Leu-D-Phe-NBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 377 mg (2.0 mmols, 1.1 equiv.) of amine MeO-D-Leu-NH2, 500 mg (1.8 mmols, 1.0 equiv.) of acid HO-D-Phe-NBoc, 1.3 mL (4 equiv.) of DIPEA, and 724 mg (2.26 mmols, 1.2 equiv.) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (740 mg, 98% yield).
Rf: 0.8 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.9–1.0 (m, 6H), 1.4 (s, 9H), 2.5–2.6 (m, 3H), 3.0–3.1 (d, 2H), 3.7 (s, 3H), 4.3–4.4 (m, 1αH), 4.5–4.6 (m, 1αH), 6.2–6.3 (d, 1H), 7.2–7.4 (m, 5H).
Dipeptide HO-D-Leu-D-Phe-NHBoc
Dipeptide HO-D-Leu-D-Phe-NHBoc was synthesized following the “General acid deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (379 mg, 98% yield).
Pentapeptide MeO-Phe-Leu-Val-D-Leu-D-Phe-NHBoc
Pentapeptide MeO-Phe-Leu-Val-D-Leu-D-Phe-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 464 mg (1.19 mmols, 1.0 equivalents) of amine MeO-Phe-Leu-Val-NH2, 494 mg (1.30 mmols, 1.1 equiv.) of acid HO-D-Leu-D-Phe-NHBoc, 1.65 mL (8 equiv.) of DIPEA, 190 mg (0.59 mmols, 0.5 equiv.) of TBTU, 270 mg (0.71 mmols, 0.6 equiv.) HATU, and 141 mg (0.47 mmols, 0.4 equiv.) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (355 mg, 41% yield).
Rf: 0.5 (EtOAc: Hex 1:1)
1H NMR (600 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.4–1.5 (m, 2H), 1.5–1.6 (m, 2H), 2.0–2.1 (m, 2H), 2.9 (m, 1H), 3.0 (m, 2H), 3.1–3.2 (m, 2H), 4.0–4.1 (m, αH), 4.2 (m, αH), 4.3 (m, αH), 4.4 (m, αH), 4.6 (m, αH), 7.1–7.3 (m, 10H)
Macrocycle Phe-Leu-Val-D-Leu-D-Phe
Macrocycle Phe-Leu-Val-D-Leu-D-Phe was synthesized following the “Macrocyclization procedure”. Utilizing 219 mg (0.34 mmols, 1.0 equivalents) of linear pentapeptide, 0.47 mL (8 equivalents) of DIPEA, 77 mg (0.20 mmols, 0.6 equivalents) of TBTU, 65 mg (0.20 mmols, 0.6 equivalents) HATU, and 61 mg (0.20 mmols, 0.6 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (10 mg, 5% yield).
Rf: 0.5 (EtOAc: Hex 4:1)
1H NMR (400 MHz, (C4D8)O): δ 0.8–1.0 (m, 18H), 1.0–1.1 (m, 1H), 1.4–1.6 (m, 2H), 1.9–2.0 (m, 2H), 2.0–2.1(m, 1H), 2.6–2.7 (m, 1H), 2.8–3.0 (m, 4H), 3.9–4.0 (m, αH), 4.1–4.2 (m, αH), 5.3-4.4 (m, αH), 4.5–4.6 (m, αH), 4.6–4.7 (m, αH), 7.0–7.4 (m, 10H), 7.6–7.7 (br, 1H), 8.0–8.1 (br, 1H), 8.1–8.2 (br, 1H)
LCMS: m/z calcd for C35H49N5O5 (M+1) = 620.79, found 620.9
Synthesis of Compound 15
Dipeptide MeO-Phe-N-Me-D-Phe-Boc
Dipeptide MeO-Phe-N-Me-D-Phe-Boc was synthesized following the “General peptide Synthesis” procedure. Utilizing 295.5 mg (1.4 mmols, 1.1 equiv.) of amine OMe-Phe-NH2, 700 mg (1.6 mmols, 1.0 equiv.) of acid HO-N-Me-D-Phe-Boc, 0.87 mL (4 equivalents) of DIPEA, 321 mg (1.0 mmols, 0.8 equiv.) of TBTU, in 12.5 mL of Methylene Chloride. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (606.7 mg, 97% yield).
Rf: 0.8 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 6H), 1.4 (s, 9H), 1.4–1.7 (m, 3H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 4.0–4.2 (br, αH), 4.8 (d, 1H), 4.8–5.0 (q, αH), 6.6 (d, 1H), 7.1–7.4 (m, 5H)
Dipeptide MeO-Phe-N-Me-D-Phe-NH2
Dipeptide MeO-Phe-N-Me-D-Phe-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (464 mg, 100% yield).
Tripeptide MeO-Phe-N-Me-D-Phe-Val-NHBoc
Tripeptide MeO-Phe-N-Me-D-Phe-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 467 mg (1.3 mmols, 1.1 equiv.) of amine MeO-Phe-N-Me-D-Phe-NH2, 260 mg (1.2 mmols, 1.0 equiv.) of acid HO-Val-NHBoc, 1.2 mL (6 equiv.) of DIPEA, 231 mg (0.7 mmols, 0.6 equiv.) of TBTU, 319 mg (1.3 mmol, 0.7 equiv) in 12 mL of methylene chloride and 3. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (509 mg, 79% yield).
Rf: 0.5 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 12H), 1.4 (s, 9H), 1.4–1.6 (m, 2H), 1.7 (s, 1H), 2.1 (m, 1H), 3.0–3.2 (m, 2H), 3.7 (s, 3H), 3.8 (dd, αH), 4.4 (dd, αH), 4.8 (dd, αH), 5.0 (d, 1H), 6.3 (d, 1H), 6.6 (d, 1H), 7.1–7.3 (m, 5H)
Tripeptide MeO-Phe-N-Me-D-Phe-Val-NH2
Tripeptide MeO-Phe-N-Me-D-Phe-Val-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (509 mg, 100% yield).
Dipeptide MeO-Leu-N-Me-D-Leu-Boc
Dipeptide MeO-Leu-N-Me-D-Leu-Boc was synthesized following the “General peptide Synthesis” procedure. Utilizing 326 mg (2.2 mmols, 1.1 equiv.) of amine MeO-Leu-NH2, 500 mg (2.0 mmols, 1.0 equiv.) of acid HO-N-Me-D-Leu-Boc, 2.0 mL (6 equiv.) of DIPEA, 722 mg (2.2 mmols, 1.1 equiv.) of TBTU, in 20 mL methylene chloride. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (709.3 mg, 93% yield).
Rf: 0.8 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.9–1.0 (d, 12H), 1.4(s, 9H), 1.4–1.8 (m, 6H), 3.7 (s, 3H), 4.1 (br, αH), 4.6 (br, αH), 4.8 (br, 1H), 6.6 (d, 1H).
Dipeptide HO-Leu-N-Me-D-Leu-Boc
Dipeptide HO-Leu-N-Me-D-Leu-Boc was synthesized following the “General acid deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (614 mg, 91% yield).
Pentapeptide MeO-Phe-N-Me-D-Phe-Val-Leu-N Me-D-Leu-Boc
Pentapeptide MeO-Phe-N-Me-D-Phe-Val-Leu-N-Me D-Leu-Boc was synthesized following the “General peptide Synthesis” procedure. Utilizing 439 mg (0.94 mmols, 1.1 equivalents) of amine MeO-Phe-N Me-D-Phe-Val-NH2, 301 mg (0.84 mmols, 1.0 equiv. of acid HO-Leu-N-Me-D-Leu-Boc, 1.2 mL (8 equiv.) of DIPEA, 161 mg (0.5 mmols, 0.6 equiv.) of TBTU, 223 mg (0.58 mmols, 0.7 equiv.) of HATU, and 100 mg (0.34 mmols, 0.4 equiv.) of DEPBT, in 8.4 mL of methylene chloride. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (420 mg, 40% yield).
Rf: 0.4 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.8–1.0 (m, 24H), 1.3 (m, 2H), 1.4 (s, 9H), 1.5 (m, 3H), 1.6 (m, 2H), 1.7 (m, 2H), 2.2 (m, 1H), 2.9–3.2 (m, 2H), 3.7 (s, 3H), 4.1 (m, αH), 4.2 (m, αH), 4.4 (m, 2αH), 4.6 (m, αH), 7.s-7.3 (m, 5H)
Macrocycle Phe-N-Me-D-Phe-Val-Leu-N-Me-D-Leu
Macrocycle Phe-N-Me-D-Phe-Val-Leu-N-Me-D-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 179 mg (0.28 mmols, 1.0 equivalents) of linear pentapeptide, 0.4 mL (8 equivalents) of DIPEA, 54 mg (0.17 mmols, 0.6 equivalents) of TBTU, 85.1 mg (0.2 mmols, 0.8 equivalents) HATU, and 67.0 mg (0.17 mmols, 0.8 equivalents) of DEPBT in 2.3 mL methylene chloride, 2.3 mL acetonitrile. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (12.3 mg, 7.5% yield).
Rf: 0.25 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.7–1.0 (m, 24H), 1.2–1.8 (m, 9H), 2.0 (m, 1H), 2.9–3.1 (m, 2H), 3.6 (m, αH), 3.8 (m, αH), 4.2 (m, αH), 4.5 (m, αH), 4.6 (m, αH), 7.1–7.3 (m, 5H), 6.6 (d, 1H), 7.0 (d, 1H), 7.5 (d, 1H), 7.6 (d, 1H), 8.0 (d, 1H)
LCMS: m/z calcd for C32H51N5O5 (M+1) = 586.4, found 587.5
Synthesis of Compound 18
Dipeptide MeO-Phe-D-Phe-NHBoc
Dipeptide MeO-Phe-D-Phe-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 450 mg (2.0 mmols, 1.1 equiv.) of amine OMe-Phe-NH2, 500 mg (1.9 mmols, 1.0 equiv.) of acid HO-D-Phe-NHBoc, 1.3 mL (4 equivalents) of DIPEA, 724 mg (2.3 mmols, 1.2 equiv.) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (798 mg, 99% yield).
Rf: 0.5 (EtOAc: Hex 1:4)
1H NMR (200 MHz, CDCl3): δ 1.4 (s, 9H), 2.9–3.1 (m, 4H), 3.7 (s, 3H), 4.3–4.4 (m, αH), 4.8–5.0 (m, αH), 6.3–6.4 (m, 1H), 6.9–7.0 (m, 1H), 7.1–7.3 (m, 10H).
Dipeptide MeO-Phe-D-Phe-NH2
Dipeptide MeO-Phe-D-Phe-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (610 mg, 100% yield).
Tripeptide MeO-Phe-D-Phe-D-Val-NHBoc
Tripeptide MeO-Phe-D-Phe-D-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 610 mg (1.87 mmols, 1.1 equiv.) of amine MeO-Phe-D-Phe-NH2, 369 mg (1.7 mmols, 1.0 equiv.) of acid HO-D-Val-NHBoc, 1.5 mL (4 equiv.) of DIPEA, 655 mg (4.30 mmols, 1.2 equiv.) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (890 mg, 98% yield).
Rf: 0.5 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.7–0.9 (m, 6H), 1.4 (s, 9H), 2.0–2.1 (m, 1H), 2.9–3.1 (m, 4H), 3.7 (s, 3H), 3.8–4.0 (m, αH), 4.6–4.9 (m, 2αH), 4.8–4.9 (m, 1H), 6.4–6.5 (m, 2H), 7.1–7.3 (m, 10H).
Dipeptide MeO-Leu-Leu-NHBoc
Dipeptide MeO-Leu-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 402 mg (2.2 mmols, 1.1 equiv.) of amine MeO-Leu-NH2, 500 mg (2.0 mmols, 1.0 equiv.) of acid HO-Leu-NHBoc, 1.4 mL (4 equiv.) of DIPEA, 774 mg (2.4 mmols, 1.2 equiv.) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (664 mg, 92% yield).
Rf: 0.5 (EtOAc: Hex 1:3)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 12H), 1.2–1.3 (m, 4H), 1.4(m, 9H), 1.6–1.7 (m, 2H), 3.7 (s, 3H), 4.1–4.2 (m, αH), 4.6–4.7 (m, αH), 4.8–4.9 (br, 1H), 6.4–6.5 (br, 1H).
Tripeptide MeO-Phe-D-Phe-D-Val-NH2
Dipeptide MeO-Phe-D-Phe-D-Val-NH2 was synthesized following the “General amine deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (724 mg, 100% yield).
Dipeptide HO-Leu-Leu-NHBoc
Dipeptide HO-Leu-Leu-NHBoc was synthesized following the “General acid deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (573 mg, 90% yield).
Pentapeptide MeO-Phe-D-Phe-D-Val-Leu-Leu-NHBoc
Pentapeptide MeO-Phe-D-Phe-D-Val-Leu-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 724 mg (1.7 mmols, 1.1 equivalents) of amine MeO-Phe-D-Phe-D-Val-NH2, 532 mg (1.5 mmols, 1.0 equiv.) of acid HO-Leu-Leu-NHBoc, 2.2 mL (8 equiv.) of DIPEA, 346 mg (1.1 mmols, 0.7 equiv.) of TBTU, and 505 mg (1.3 mmols, 0.8 equiv.) HATU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (931 mg, 80% yield).
Rf: 0.5 (EtOAc: Hex 3:1)
1H NMR (400 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.4 (s, 9H), 1.5–1.8 (m, 4H), 1.9–2.0 (m, 1H), 2.0–2.1 (m, 1H), 2.7–2.8 (m, 1H), 2.9–3.0 (m, 2H), 3.0–3.1 (m, 2H), 3.7 (s, 3H), 4.0–4.2 (m, 2αH), 4.4–4.5 (m, αH), 4.6–4.7 (m, 2αH), 7.1–7.3 (m, 10H).
Macrocycle Phe-D-Phe-D-Val-Leu-Leu
Macrocycle Phe-D-Phe-D-Val-Leu-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 163 mg (0.27 mmols, 1.0 equivalents) of linear pentapeptide, 0.38 mL (8 equivalents) of DIPEA, 43.1 mg (0.14 mmols, 0.5 equivalents) of TBTU, 51.1 mg (0.12 mmols, 0.5 equivalents) HATU, and 56.3 mg (0.19 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (8 mg, 5% yield).
Rf: 0.5 (EtOAc: Hex 4:1)
1H NMR (600 MHz, CD3OD): δ 0.8–1.0 (m, 24H), 1.2–1.4 (m, 3H), 1.5–1.7 (m, 6H), 2.9–3.1 (m, 1H), 3.2–3.4 (m, 1H), 3.6–3.8 (m, 1H), 4.0–4.1 (m, 1αH), 4.1–4.2 (m, 2αH), 4.2–4.4 (m, 1αH), 4.6–4.8 (m, 1αH), 7.2–7.4 (m, 5H).
LCMS: m/z calcd for C32H51N5O5 (M+23) = 608.39, found 608.6.
Synthesis of Compound 20
Dipeptide Fmoc-D-Val-D-Phe-O-Resin
Dipeptide Fmoc-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1021.6 mg (.735 mmol, 1 equivalents) of NH2-D-Phe-O-Resin, the D-Val residue was incorporated using 748 mg of Fmoc-D-Val-OH (2.2 mmol, 3 equivalents), 338 mg (2.2 mmol, 3 equivalents) of HOBt, and 0.680 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-D-Val-D-Phe-O-Resin
Dipeptide NH2-D-Val-D-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Cha-D-Val-D-Phe-O-Resin
Tripeptide Fmoc-Cha-D-Val-D-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Val-D-Phe-O-Resin prepared above, the Cha residue was incorporated using 866.9 mg (2.2 mmol, 3 equivalents) of Fmoc-Cha-OH, 338 mg (2.2 mmol, 3 equivalents) of HOBt, and 0.680 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-Cha-D-Val-D-Phe-O-Resin
Tripeptide NH2-Cha-D-Val-D-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin
Tetrapeptide Fmoc-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Cha-D-Val-D-Phe-O-Resin prepared above, the Ser(Bzl) residue was incorporated using 920.0 mg (2.2 mmol, 3 equivalents) of Fmoc-Ser(Bzl)-OH, 338 mg (2.3 mmol, 3 equivalents) of HOBt, and 0.680 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin
Tetrapeptide NH2-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-Phe-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin
Pentapeptide Fmoc-Phe-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin prepared above, the Phe residue was incorporated using 853.7 mg (2.2 mmol, 3 equivalents) of Fmoc-Phe-OH, 338 mg (2.2 mmol, 3 equivalents) of HOBt, 0.680 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-Phe-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin
Pentapeptide NH2-Phe-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-Phe-Ser(Bzl)-Cha-D-Val-D-Phe-OH
Double Deprotected Pentapeptide NH2-Phe-Ser(Bzl)-Cha-D-Val-D-Phe-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1353.5 mg of dried NH2-Phe-Ser(Bzl)-Cha-D-Val-D-Phe-O-Resin, 6.7 mL of 2,2,2-trifluoroethanol and 6.7 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (273 mg, 82% yield)
Macrocycle Phe-D-Phe-D-Val-Cha-Ser(Bzl)
Macrocycle Phe-D-Phe-D-Val-Cha-Ser(Bzl) was synthesized following the “Macrocyclization procedure”. Utilizing 160 mg (0.21 mmols, 1.0 equivalents) of linear pentapeptide, 0.3 mL (8 equivalents) of DIPEA, 41.5 mg (0.13 mmols, 0.6 equivalents) of TBTU, 49.2 mg (0.13 mmols, 0.6 equivalents) HATU, and 38.7 mg (0.13 mmols, 0.6 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (7.9 mg, 5.1% yield).
1H NMR (400 MHz, CD3OD): δ 0.7–0.9 (m, 6H), 1.0–1.4 (m, 11H), 1.9 (s, 2H), 2.6 (s, 1H), 3.2–3.6 (m, 6H), 4.4–4.5 (m, αH), 4.6 (m, 2H), 7.0–7.3 (m, 15H).
LCMS: m/z called for C42H53N5O6 (M+1) = 724.9, found 727.0
Synthesis of Compound 21
Dipeptide Fmoc-N-Me-D-Phe-Phe-O-Resin
Dipeptide Fmoc-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 2510 mg (1.62 mmol, 1 equivalents) of NH2-Phe-O-Resin, the N-Me-D-Phe residue was incorporated using 1930 mg of Fmoc-N-Me-Phe-OH (4.8 mmol, 3 equivalents), 750 mg (4.8 mmol, 3 equivalents) of HOBt, and 1.50 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH-N-Me-D-Phe-Phe-O-Resin
Dipeptide NH2-Leu-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-D-Val-N-Me-D-Phe-Phe-O-Resin
Tripeptide Fmoc-D-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH-N-Me-D-Phe-Phe-O-Resin prepared above, the D-Val residue was incorporated using 1620 mg (4.8 mmol, 3 equivalents) of Fmoc-D-Val-OH, 750 mg (4.8 mmol, 3 equivalents) of HOAt, and 1.50 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-D-Val-N-Me-D-Phe-Phe-O-Resin
Tripeptide NH2-D-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin
Tetrapeptide Fmoc-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Val-N-Me-D-Phe-Phe-O-Resin prepared above, the Cha residue was incorporated using 1860 mg (4.8 mmol, 3 equivalents) of Fmoc-Cha-OH, 750 mg (4.8 mmol, 3 equivalents) of HOBt, and 1.50 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin
Tetrapeptide NH2-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-Ser(Bzl)-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin
Pentapeptide Fmoc-Ser(Bzl)-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin prepared above, the Ser(Bzl) residue was incorporated using 2000 mg (4.8 mmol, 3 equivalents) of Fmoc-Ser(Bzl)-OH, 750 mg (4.8 mmol, 3 equivalents) of HOBt, 1.50 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-Ser(Bzl)-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin
Pentapeptide NH2-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-Ser(Bzl)-Cha-D-Val-N-Me-D-Phe-Phe-OH
Double Deprotected Pentapeptide NH2-Ser(Bzl)-Cha-D-Val-N-Me-D-Phe-Phe-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 3830.1 mg of dried NH2-Ser(Bzl)-Cha-D-Val-N-Me-D-Phe-Phe-O-Resin, 18.5 mL of 2,2,2-trifluoroethanol and 18.5 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (894 mg, 82% yield)
Macrocycle Phe-N-Me-D-Phe-D-Val-Cha-Ser(Bzl)
Macrocycle Phe-N-Me-D-Phe-D-Val-Cha-Ser(Bzl) was synthesized following the “Macrocyclization procedure”. Utilizing 300 mg (0.397 mmols, 1.0 equivalents) of linear pentapeptide, 0.543 mL (8 equivalents) of DIPEA, 67.0 mg (0.21 mmols, 0.7 equivalents) of TBTU, 80.0 mg (0.21 mmols, 0.7 equivalents) HATU, and 62.5 mg (0.21 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (42.3 mg, 16.2% yield).
1H NMR (400 MHz, CD3OD): δ 0.6–1.0 (m, 8H), 1.2 (m, 11H), 1.4–1.6 (m, 3H), 1.7–1.8 (m, 4H), 2.0 (m, 1H), 3.4 (m, 3H), 4.1 (s, αH), 4.2 (s, αH), 4.5 (s, αH), 5.2 (m, 2H), 7.0–7.2 (m, 22H),
Macrocycle Phe-N-Me-D-Phe-D-Val-Cha-Ser
Macrocycle Phe-N-Me-D-Phe-D-Val-Cha-Ser was synthesized utilizing 20 mg (0.08 mmols, 1.0 equivalents) of macrocycle Phe-N-Me-D-Phe-D-Val-Cha-Ser(Bzl). The compound was dissolved in 0.8 mL EtOH (0.1 M) and was hydrogenated using a catalytic amount of Pd/C and excess H2 for 24 hours. The reaction was filtered over celite and afforded 18 mg (93% yield) of pure macrocycle Phe-N-Me-D-Phe-D-Val-Cha-Ser.
1H NMR (400 MHz, CD3OD): δ 0.7–0.9 (m, 8H), 1.2 (m, 11H), 1.4–1.6 (m, 3H), 1.7–1.8 (m, 4H), 2.0 (m, 1H), 3.4 (m, 3H), 4.2–4.3 (s, αH), 4.2 (s, αH), 4.5 (s, αH), 5.2 (m, 2H), 7.2–7.4 (m, 14H),
LCMS: m/z called for C36H49N5O6 (M+1) = 648.4, found 648.3
Synthesis of Compounds 23 and 24
Dipeptide MeO-D-Tyr-Leu-NHBoc
Dipeptide MeO-D-Tyr-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1026.9 mg (4.43 mmols, 1.1 equivalents) of amine MeO-D-Tyr-NH2, 1000.9 mg (4.01 mmols, 1.0 equivalent) of acid HO-Leu-NHBoc, 2.8 mL (4 equivalents) of DIPEA, and 1440 mg (4.81 mmols, 1.2 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (1526 mg, 93% yield).
Rf: 0.65 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.9–1.0 (d, 6H), 1.4 (s, 9H), 1.6 (br, 1H), 2.0 (s, 2H), 3.0 (m, 2H), 3.7 (s, 3H), 4.1 (dd, αH), 4.8 (dd, αH), 5.0 (s, 1H), 6.6–6.8 (d, 2H), 7.0 (d, 2H), 7.2(s, 2H)
Dipeptide MeO-D-Tyr-Leu-NH2
Dipeptide MeO-D-Tyr-Leu-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (1153 mg, 100% yield).
Tripeptide MeO-D-Tyr-Leu-Val-NHBoc
Tripeptide MeO-D-Tyr-Leu-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1153 mg (3.73 mmols, 1.1 equivalents) of amine MeO-D-Tyr-Leu-NH2, 750 mg (3.4 mmols, 1.0 equivalents) of acid HO-Val-NHBoc, 2.4 mL (4 equivalents) of DIPEA, and 1240 mg (4.07 mmols, 1.2 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (1754 mg, 73 % yield).
Rf: 0.65 (EtOAc: Hex, 3:1)
1H NMR (200 MHz, CD3OD): δ 0.8–0.9 (m, 12H), 1.4 (s, 9H), 1.9 (br, 1H), 2.0 (s, 2H), 2.8 (m, 1H), 3.0 (m, 2H), 3.7 (s, 3H), 3.8 (dd, αH), 4.4 (dd, αH), 4.6 (dd, 2 αH), 5.0 (s, 1H), 6.6–6.8 (d, 2H), 7.0 (d, 2H)
Tripeptide MeO-D-Tyr-Leu-Val-NH2
Tripeptide MeO-D-Tyr-Leu-Val-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (1021 mg, 100% yield).
Dipeptide MeO-Lys(CBz)-Leu-NHBoc
Dipeptide MeO-Lys(CBz)-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1459 mg (4.41 mmols, 1.1 equivalents) of amine MeO-Lys(CBz)-NH2, 1000 mg (4.01 mmols, 1.0 equivalent) of acid HO-Leu-NHBoc, 2.8 mL (4 equivalents) of DIPEA, and 1545 mg (4.8 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (1182 mg, 97% yield).
Rf: 0.65 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 1.0–1.2 (d, 6H), 1.6 (s, 9H), 1.7–2.2 (m, 9H), 3.3 (m, 2H), 3.7 (s, 3H), 4.3 (dd, αH), 4.6 (dd, αH), 5.3 (s, 2H), 7.5 (d, 5H)
Dipeptide HO-Lys(CBz)-Leu-NHBoc
Dipeptide HO-Lys(CBz)-Leu-NHBoc was synthesized following the “General acid deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (1089 mg, 95% yield).
Pentapeptide MeO-D-Tyr-Leu-Val-Lys(CBz)-Leu-NHBoc
Pentapeptide MeO-D-Tyr-Leu-Val-Lys(CBz)-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1021 mg (2.5 mmols, 1.1 equivalents) of amine MeO-D-Tyr-Leu-Val-NH2, 1089 mg (2.3 mmols, 1.0 equivalent) of acid HO-Lys(CBz)-Leu-NHBoc, 1.6 mL (4 equivalents) of DIPEA, and 826 mg (2.76 mmols, 1.2 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (192 mg, 20% yield).
Rf: 0.6 (EtOAc: Hex 3:1)
1H NMR (500 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.3 (m, 2H), 1.5 (s, 9H), 1.6–1.8 (m, 6H), 2.0–2.2 (m, 6H), 2.9 (m, 2H), 3.1 (m, 4H), 3.7 (s, 3H), 4.2 (m, 2αH), 4.4 (m, 2αH), 4.6 (m, αH), 5.0 (s, 2H), 6.7–7.0 (d, 4H), 7.4 (d, 5H), 7.8 (m, 2H), 8.0–8.2 (m, 4H).
Macrocycle D-Tyr-Leu-Val-Lys(CBz)-Leu (Cyclized protected)
Macrocycle D-Tyr-Leu-Val-Lys(CBz)-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 169 mg (0.22 mmols, 1.0 equivalent) of linear pentapeptide, 0.3 mL (8 equivalents) of DIPEA, 35 mg (0.11 mmols, 0.5 equivalents) of TBTU, 42 mg (0.11 mmols, 0.5 equivalents) HATU, and 33 mg (0.11 mmols, 0.5 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (30 mg, 18% yield).
Rf: 0.6 (EtOAc: Hex 1:0)
1H NMR (500 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.2–1.4 (m, 4H), 1.5–1.8 (m, 8H), 2.6 (m, αH), 3.1 (m, 4H), 3.2 (m, 2H), 4.1 (m, 2αH), 4.3 (m, 2αH), 4.5 (m, αH), 5.1 (s, 2H), 6.7 (dd, 2H), 7.0 (dd, 2H), 7.2–7.4 (d, 5H), 7.5 (d, 1H), 7.7 (d, 1H), 8.1 (d, 1H), 8.3 (d, 1H), 8.5 (d, 1H), 8.7 (d, 1H)
LCMS: m/z calcd for C34H52N6O5 (M+1) = 750.4, found 752
Removal of Cbz group
Macrocycle D-Tyr-Leu-Val-Lys-Leu
Macrocyclic pentapeptide D-Tyr-Leu-Val-Lys(CBz)-Leu was further deprotected to remove Cbz protecting group of the lysine residue. The compound was synthesized by utilizing 16 mg (0.021 mmols, 1.0 equivalent) of cyclic pentapeptide, 5.0 mL of Ethanol, 8.0 mg 10% wt palladium on carbon. The crude reaction was purified by reverse phase-HPLC to yield the deprotected macrocycle D-Tyr-Leu-Val-Lys-Leu (5.0 mg, 38% yield).
Rf: 0.5 (DCM: MeOH 98:2)
1H NMR (500 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.2–1.4 (m, 4H), 1.5–1.8 (m, 8H), 2.0 (m, 2H), 2.6 (m, αH), 3.0 (m, 4H), 3.2 (m, 2H), 4.1 (m, 2αH), 4.3 (m, 2αH), 4.5 (m, αH), 6.7 (dd, 2H), 7.0 (dd, 2H), 7.5 (d, 1H), 7.7 (d, 1H), 8.1 (d, 1H), 8.3 (d, 1H), 8.5 (d, 1H), 8.7 (d, 1H)
LCMS: m/z calcd for C34H52N6O5 (M+1) = 617.
Synthesis of Compounds 26 and 27
Dipeptide MeO-D-Trp-Leu-NHBoc
Dipeptide MeO-D-Trp-Leu-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1126.3 mg (4.4 mmols, 1.1 equivalents) of amine MeO-D-Trp-NH2, 1008.9 mg (4.01 mmols, 1.0 equivalents) of acid HO-Leu-NHBoc, 2.8 mL (4 equivalents) of DIPEA, and 1446 mg (4.83 mmols, 1.2 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (1692 mg, 98% yield).
Rf: 0.65 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.9–1.0 (d, 6H), 1.4 (s, 9H), 1.5–1.7 (m, 2H), 2.0 (m, 1H), 3.3 (d, 2H), 3.6 (s, 3H), 4.0 (m, αH), 4.9 (m, αH), 6.6 (br, 1H), 7.0–7.5 (m, 4H), 8.1 (br, 1H)
Dipeptide MeO-D-Trp-Leu-NH2
Dipeptide MeO-D-Trp-Leu-NH2 was synthesized following the “General amine deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (1298 mg, 100% yield).
Tripeptide MeO-D-Trp-Leu-Val-NHBoc
Tripeptide MeO-D-Trp-Leu-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 1298 mg (3.91 mmols, 1.1 equivalents) of amine MeO-D-Trp-Leu-NH2, 771 mg (3.55 mmols, 1.0 equivalents) of acid HO-Val-NHBoc, 2.5 mL (4 equivalents) of DIPEA, and 1274 mg (4.26 mmols, 1.2 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tripeptide (1465 mg, 78% yield).
Rf: 0.55 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.7–1.0 (dd, 12H), 1.4 (s, 9H), 2.0 (m, 2H), 3.2–3.3 (m, 3H), 3.6 (s, 3H), 3.8 (m, αH), 4.4 (m, αH), 4.8 (m, αH), 6.5 (br, 1H), 7.0–7.6 (m, 5H), 8.2 (br, 1H)
Tripeptide MeO-D-Trp-Leu-Val-NH2
Tripeptide MeO-D-Trp-Leu-Val-NH2 was synthesized following the “General amine deprotection”. This tripeptide used 503 mg (0.95 mmols, 1.0 equivalents) was taken on to the next reaction without further purification or characterization. (410 mg, 100% yield).
Dipeptide MeO-Arg(CBz)-Val-NHBoc
Dipeptide MeO-Arg(CBz)-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 799 mg (4.4 mmols, 1.1 equivalents) of amine MeO-Arg(CBz)-NH2, 1 g (4.0 mmols, 1.0 equivalent) of acid HO-Val-NHBoc, 2.8 mL (4 equivalents) of DIPEA, and 1.5 g (1.4 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the dipeptide (660 mg, 98% yield).
Rf: 0.5 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ1.4 (s, 9H), 1.6–2.0 (m, 10H), 3.4 (s, 2H), 3.6 (s, 3H), 4.0 (dd, αH), 4.6–4.8 (dd, 2αH), 5.0 (s, 2H), 5.1 (s, 2H), 7.2–7.6 (m, 10H), 7.8 (s, 2H)
Dipeptide HO-Arg(CBz)-Val-NHBoc
Dipeptide HO-Arg(CBz)-Val-NHBoc was synthesized following the “General acid deprotection”. This dipeptide was taken on to the next reaction without further purification or characterization. (564 mg, 89% yield).
Pentapeptide MeO-D-Trp-Leu-Val-Arg(CBz)-Val-NHBoc
Pentapeptide MeO-D-Trp-Leu-Val-Arg(CBz)-Val-NHBoc was synthesized following the “General peptide Synthesis” procedure. Utilizing 362 mg (0.84 mmols, 1.1 equivalents) of amine MeO-D-Trp-Leu-Val-NH2, 501 mg (0.76 mmols, 1.0 equivalent) of acid HO-Arg(CBz)-Val-NHBoc, 0.53 mL (4 equivalents) of DIPEA, and 274 mg (0.92 mmols, 1.2 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (430 mg, 47% yield).
Rf: 0.55 (EtOAc: Hex 3:1)
1H NMR (500 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.4 (m, 9H), 1.5–1.8 (m, 10H), 2.1 (m, 1H), 3.2 (m, 3H), 3.4 (m, 2H), 3.7 (s, 3H), 4.1 (d, αH), 4.6 (m, 4αH), 5.1 (s, 2H), 7.0–7.5 (m, 15H), 7.8 (d, 1H), 8.0 (d, 1H), 8.1 (d, 1H), 8.3 (d, 1H)
Macrocycle D-Trp-Leu-Val-Arg(CBz)-Val (Cyclized Protected)
Macrocycle D-Trp-Leu-Val-Arg(CBz)-Val was synthesized following the “Macrocyclization procedure”. Utilizing 430 mg (0.45 mmols, 1.0 equivalents) of linear pentapeptide, 0.3 mL (4 equivalents) of DIPEA, 72.5 mg (0.22 mmols, 0.5 equivalents) of TBTU, 85.5 mg (0.224 mmols, 0.5 equivalents) HATU, and 67.3 mg (0.225 mmols, 0.5 equivalents) of DEPBT. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (212 mg, 50% yield).
Rf: 0.65 (EtOAc: MeOH 98:2)
1H NMR (500 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.2–1.4 (m, 6H), 1.5–1.7 (m, 4H), 2.0 (m, 1H), 2.8 (m, αH) 3.1–3.2 (m, 2H), 4.1 (d, αH), 4.4 (m, 2αH), 4.8 (m, 2 αH), 5.3 (s, 2H), 7.0–7.5 (m, 15H), 7.5 (d, 1H), 8.2 (d, 1H), 8.8 (d, 1H)
LCMS: m/z calcd for C34H52N6O5 (M+1) = 802, found 802
Removal of Cbz group
Macrocycle D-Trp-Leu-Val-Arg-Val
Macrocyclic pentapeptide D-Trp-Leu-Val-Arg(CBz)-Val was further deprotected to remove Cbz protecting group of the arginine residue. The compound was synthesized by utilizing 106 mg (0.113 mmols, 1.0 equivalent) of cyclic pentapeptide, 2.5 mL of Ethanol (0.05 M), 45.0 mg 10% wt palladium on carbon. The crude reaction was purified by reverse phase-HPLC to yield the deprotected macrocycle D-Trp-Leu-Val-Arg-Val (6.0 mg, 48% yield).
Rf: 0.5 (DCM: MeOH 98:2)
1H NMR (500 MHz, CD3OD): δ 0.8–1.0 (m, 18H), 1.2–1.4 (m, 6H), 1.5–1.7 (m, 4H), 2.0 (m, 3H), 2.8 (m, αH) 3.1–3.2 (m, 2H), 4.2 (d, αH), 4.4 (m, 2αH), 4.8 (m, 2 αH), 6.8 (m, 1H), 7.0–7.5 (m, 14H), 7.8 (d, 1H), 8.2 (d, 1H), 8.3 (d, 1H), 8.5 (d, 1H), 8.8 (d, 1H)
LCMS: m/z calcd for C34H52N6O5 (M+1) = 668.
Synthesis of Compound 28
Dipeptide MeO-Phe-D-N-Me-Phe-NBoc
Dipeptide MeO-Ia-IIc-NBoc was synthesized using the “General Peptide Synthesis” procedure. Utilizing 309 mg (1.43 mmols, 1.1 equivalents) of amine Ia, 600 mg (1.30 mmols, 1.0 equivalents) of acid IIc, 0.91 mL (4 equivalents) of DIPEA, and 502 mg (1.56 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by an acid-base wash to yield the dipeptide (574 mg, 99% yield).
Rf: 0.5 (EtOAc: Hex 1:3)
1H NMR (200 MHz, CD3OD): δ 1.2–1.4 (s, 9H), 2.7 (s, 3H), 2.8 (m, 1H), 2.9 (m, 1H), 3.0–3.2 (m, 3H), 3.7 (s, 3H), 4.8–5.0 (m, 2αH), 7.1–7.3 (m, 10H), 7.8 (d, 1H)
Dipeptide MeO-Phe-D-N-Me-Phe-NH
Dipeptide MeO-Phe-D-N-Me-Phe-NH was synthesized using the “General Amine Deprotection” procedure. The dipeptide was taken on to the next reaction without further purification or characterization. (439 mg, 100% yield).
Tripeptide MeO-Phe-D-N-Me-Phe-Val-NHBoc
Tripeptide MeO-Phe-D-N-Me-Phe-Val-NHBoc was constructed following the procedure outlined “General Peptide Synthesis”. Utilizing 439 mg (1.29 mmols, 1.1 equivalents) of amine MeO-Ia-IIc-NH2, 254 mg (1.17 mmols, 1.0 equivalents) of acid IIIa, 1.92 mL (8 equivalents) of DIPEA, and 452 mg (1.41 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by an acid-base wash and column chromatography (silica gel, EtOAc/Hex) to yield the desired tripeptide (667 mg, 96% yield).
Rf = 0.5 (EtOAc: Hex 3:2)
1H NMR (200 MHz, CDCl3): δ 0.6 (d, 6H), 1.4 (s, 9H), 1.6–1.8 (m, 4H), 2.9 (m, 3H), 3.0–3.2 (dd, 1H), 3.3 (s, 1H), 3.7 (m, 3H), 4.2 (dd, 1αH), 4.7–4.9 (dd, 1αH), 5.0 (d, 1H), 5.5 (dd, 1H), 6.9 (d, 1H), 7.0–7.4 (m, 10H)
Tripeptide MeO-Phe-D-N-Me-Phe-Val-NH
Tripeptide MeO-Ia-IIc-IIIa-NH was synthesized using the “General Amine Deprotection” procedure. The tripeptide was taken on to the next reaction without further purification or characterization. (620 mg, 100% yield).
Dipeptide MeO-Leu-Lys(CBz)-NHBoc
Dipeptide MeO-Leu-Lys(CBz)-NHBoc was constructed following the procedure outlined “General Peptide Synthesis”. Utilizing 315 mg (1.74 mmols, 1.1 equivalents) of amine IVa, 600 mg (1.57 mmols, 1.0 equivalents) of acid Vd, 1.10 mL (4 equivalents) of DIPEA, and 605 mg (1.88 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by an acid-base wash to yield the dipeptide (715 mg, 90% yield).
Rf = 0.4 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 6H), 1.4 (s, 9H), 1.5–1.7 (m, 6H), 1.8 (m, 1H), 3.1 (q, 2H), 3.7 (s, 3H), 4.0–4.2 (m, 1αH), 4.5–4.7 (m, 1αH), 5.0 (m, 1H), 5.1 (s, 2H), 6.4 (d, 1H), 7.2–7.4 (m, 5H)
Dipeptide HO-Leu-Lys(CBz)-NHBoc
Dipeptide HO-Leu-Lys(CBz)-NHBoc was synthesized using the “General Acid Deprotection” procedure. The dipeptide was taken on to the next reaction without further purification or characterization. (650 mg, 86% yield).
Pentapeptide MeO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc
Pentapeptide MeO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc was constructed using the “General Peptide Synthesis” procedure. Utilizing 283 mg (0.65 mmols, 1.1 equivalents) of MeO-Ia-IIc-IIIa-NH2, 289 mg (0.59 mmols, 1.0 equivalents) of HO-IVa-Vd-NHBoc, 178 mg (0.49 mmols, 0.8 equivalents) HATU, 70.1 mg (0.23 mmols, 0.4 equivalents) DEPBT, and 0.82 mL (8 equivalents) of DIPEA. The crude material was purified using column chromatography (silica gel, EtOAc/Hex) to yield 415 mg (78% yield) of the pentapeptide.
Rf = 0.5 (EtOAc: Hex 7:3)
1H NMR (500 MHz, CDCl3): δ 0.5 (d, 2H), 0.6 (d, 2H), 0.9 (m, 4H), 1.2–1.4 (m, 1H), 1.4 (s, 9H), 1.5 (m, 1H), 1.6 (m, 1H), 1.7 (m, 1H), 2.9 (s, 3H), 3.2 (m, 2H), 3.3 (dd, 1H), 3.7 (s, 3H), 4.1 (s, 1αH), 4.4 (m, 2αH), 4.8 (dd, 1αH), 5.1 (s, 3H), 5.6 (m, 1H), 6.5 (d, 1H), 6.7 (d, 1H), 7.0 (d, 1H), 7.1–7.4 (m, 15H)
LCMS: m/z calculated for C50H70N5O10 (M+1) = 915.6, found 915.6
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc was synthesized using the “General Acid Deprotection” procedure. The pentapeptide was taken on to the next reaction without further purification or characterization. (346 mg, 87% yield).
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NH
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NH was synthesized using the “General Amine Deprotection” procedure. The pentapeptide was taken on to the next reaction without further purification. (298 mg, 100% yield).
LCMS: m/z calculated for C44H60N6O8 (M+1) = 802, found 801.5
Macrocycle Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)
Macrocycle Phe-D-N-Me-Phe-Val-Leu-Lys(CBz) was constructed using the “General Macrocyclization Procedure”. Utilizing 149 mg (0.19 mmols, 1.0 equivalents) of the double deprotected linear pentapeptide, 30.0 mg (0.09 mmols, 0.5 equivalents) TBTU, 35.2 mg (0.09 mmols, 0.5 equivalents) HATU, 28.4 mg (0.09 mmols, 0.5 equivalents) DEPBT, and 0.27 mL (8 equivalents) of DIPEA. The crude material was purified using column chromatography (silica gel, EtOAc/Hex) to yield 19.2 mg (17% yield) of the macrocycle.
Rf = 0.5 (EtOAc: Hex 4:1)
1H NMR (400 MHz, CD3OD): δ 0.5 (d, 1H), 0.6 (d, 4H), 0.7 (m, 4H), 0.8–1.0 (m, 6H), 1.2–1.4 (m, 6H), 1.5 (m, 2H), 1.7 (m, 2H), 1.8 (m, 2H), 2.9 (s, 3H), 3.0 (m, 2H), 3.1 (m, 2H), 3.9 (t, 1αH), 4.1 (t, 1αH), 4.4 (t, 1αH), 4.6 (t, 1αH), 5.1 (s, 2H), 5.3 (s, 1H), 7.1–7.4 (m, 15H)
LCMS: m/z calculated for C44H58N6O7 (M+1) = 784.1, found 784.2
Synthesis of Compound 29
Dipeptide Fmoc-N-Me-D-Phe-Phe-O-Resin
Dipeptide Fmoc-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 510.4 mg (0.326 mmol, 1 equivalents) of H-Phe-O-Resin, the N-Me-D-Phe residue was incorporated using 392.6 mg of Fmoc-N-Me-D-Phe-OH (0.978 mmol, 3 equivalents), 150 mg (0.978 mmol, 3 equivalents) of HOBt, and 0.303 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide H-N-Me-D-Phe-Phe-O-Resin
Dipeptide H-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Val-N-Me-D-Phe-Phe-O-Resin
Tripeptide Fmoc-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-N-Me-D-Phe-Phe-O-Resin prepared above, the Val residue was incorporated using 332 mg (0.978 mmol, 3 equivalents) of Fmoc-Val-OH, 133 mg (0.978 mmol, 3 equivalents) of HOAt, and 0.303 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-Val-N-Me-D-Phe-Phe-O-Resin
Tripeptide NH2-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-Leu-Val-N-Me-D-Phe-Phe-O-Resin
Tetrapeptide Fmoc-Leu-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Val-N-Me-D-Phe-Phe-O-Resin prepared above, the Leu residue was incorporated using 346 mg (0.978 mmol, 3 equivalents) of Fmoc-Leu-OH, 150 mg (0.978 mmol, 3 equivalents) of HOBt, and 0.303 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-Leu-Val-N-Me-D-Phe-Phe-O-Resin
Tetrapeptide NH2-Leu-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe-O-Resin
Pentapeptide Fmoc-Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-Leu-Val-N-Me-D-Phe-Phe-O-Resin prepared above, the Lys(Boc) residue was incorporated using 458 mg (0.978 mmol, 3 equivalents) of Fmoc-Lys(Boc)-OH, 150 mg (0.978 mmol, 3 equivalents) of HOBt, 0.303 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe-O-Resin
Pentapeptide NH2-Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe-OH
Double Deprotected Pentapeptide NH2-Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 420 mg of dried NH2-Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe-O-Resin, 2.5 mL of 2,2,2-trifluoroethanol and 2.5 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (203 mg, 92% yield)
Macrocycle Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe
Macrocycle Lys(Boc)-Leu-Val-N-Me-D-Phe-Phe was synthesized following the “Macrocyclization procedure”. Utilizing 203 mg (0.30 mmols, 1.0 equivalents) of linear pentapeptide, 0.36 mL (8 equivalents) of DIPEA, 66.3 mg (0.206 mmols, 0.8 equivalents) of TBTU, 86.1 mg (0.206 mmols, 0.8 equivalents) HATU, and 46.4 mg (0.155 mmols, 0.6 equivalents) of DEPBT. The crude reaction was purified by silica based column to yield the macrocycle (17.5 mg, 7.8% yield).
Rf: 0.65 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.6 (d, 12H), 0.7–1.0 (m, 4H), 1.2 (s, 9H), 1.6 (m, 3H), 1.7 (m 2H), 1.9 (m, 1H), 2.8 (s, 1H), 2.9–3.0 (m, 6H), 3.8 (t, αH), 3.9 (t, αH), 4.5 (t, αH), 5.1 (t, αH) 7.0–7.3 (m, 10H)
LCMS: m/z calcd for C41H60N6O7 (M+1) = 749.0, found 750.8
Synthesis of Compound 30
Dipeptide MeO-Phe-D-N-Me-Phe-NBoc
Dipeptide MeO-Phe-D-N-Me-Phe-NBoc was synthesized using the “General Peptide Synthesis” procedure. Utilizing 309 mg (1.43 mmols, 1.1 equivalents) of amine MeO-Phe-NH2, 600 mg (1.30 mmols, 1.0 equivalents) of acid HO-N-Me-Phe-NBoc, 0.91 mL (4 equivalents) of DIPEA, and 502 mg (1.56 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by an acid-base wash to yield the dipeptide (574 mg, 99% yield).
Rf: 0.5 (EtOAc:Hex 1:3)
1H NMR (200 MHz, CD3OD): δ 1.2–1.4 (s, 9H), 2.7 (s, 3H), 2.8 (m, 1H), 2.9 (m, 1H), 3.0–3.2 (m, 3H), 3.7 (s, 3H), 4.8–5.0 (m, 2αH), 7.1–7.3 (m, 10H), 7.8 (d, 1H)
Dipeptide MeO-Phe-D-N-Me-Phe-NH
Dipeptide MeO-Phe-D-N-Me-Phe-NH was synthesized using the “General Amine Deprotection” procedure. The dipeptide was taken on to the next reaction without further purification or characterization. (439 mg, 100% yield).
Tripeptide MeO-Phe-D-N-Me-Phe-Val-NHBoc
Tripeptide MeO-Phe-D-N-Me-Phe-Val-NHBoc was constructed following the procedure outlined “General Peptide Synthesis”. Utilizing 439 mg (1.29 mmols, 1.1 equivalents) of amine MeO-Phe-D-N-Me-Phe-NH2, 254 mg (1.17 mmols, 1.0 equivalents) of acid HO-Val-NHBoc, 1.92 mL (8 equivalents) of DIPEA, and 452 mg (1.41 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by an acid-base wash and column chromatography (silica gel, EtOAc/Hex) to yield the desired tripeptide (667 mg, 96% yield).
Rf = 0.5 (EtOAc: Hex 3:2)
1H NMR (200 MHz, CDCl3): δ 0.6 (d, 6H), 1.4 (s, 9H), 1.6–1.8 (m, 4H), 2.9 (m, 3H), 3.0–3.2 (dd, 1H), 3.3 (s, 1H), 3.7 (m, 3H), 4.2 (dd, 1αH), 4.7–4.9 (dd, 1αH), 5.0 (d, 1H), 5.5 (dd, 1H), 6.9 (d, 1H), 7.0–7.4 (m, 10H)
Tripeptide MeO-Phe-D-N-Me-Phe-Val-NH2
Tripeptide MeO-Phe-D-N-Me-Phe-Val-NH2 was synthesized using the “General Amine Deprotection” procedure. The tripeptide was taken on to the next reaction without further purification or characterization. (620 mg, 100% yield).
Dipeptide MeO-Leu-Lys(CBz)-NHBoc
Dipeptide MeO-Leu-Lys(CBz)-NHBoc was constructed following the procedure outlined “General Peptide Synthesis”. Utilizing 315 mg (1.74 mmols, 1.1 equivalents) of amine MeO-Leu-NH2, 600 mg (1.57 mmols, 1.0 equivalents) of acid HO-Lys(CBz)-NHBoc, 1.10 mL (4 equivalents) of DIPEA, and 605 mg (1.88 mmols, 1.2 equivalents) of TBTU. The crude reaction was purified by an acid-base wash to yield the dipeptide (715 mg, 90% yield).
Rf = 0.4 (EtOAc: Hex 1:1)
1H NMR (200 MHz, CDCl3): δ 0.8–1.0 (m, 6H), 1.4 (s, 9H), 1.5–1.7 (m, 6H), 1.8 (m, 1H), 3.1 (q, 2H), 3.7 (s, 3H), 4.0–4.2 (m, 1αH), 4.5–4.7 (m, 1αH), 5.0 (m, 1H), 5.1 (s, 2H), 6.4 (d, 1H), 7.2–7.4 (m, 5H)
Dipeptide HO-Leu-Lys(CBz)-NHBoc
Dipeptide HO-Leu-Lys(CBz)-NHBoc was synthesized using the “General Acid Deprotection” procedure. The dipeptide was taken on to the next reaction without further purification or characterization. (650 mg, 86% yield).
Pentapeptide MeO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc
Pentapeptide MeO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc was constructed using the “General Peptide Synthesis” procedure. Utilizing 283 mg (0.65 mmols, 1.1 equivalents) of MeO-Phe-D-N-Me-Phe-Val-NH2, 289 mg (0.59 mmols, 1.0 equivalents) of HO-Leu-Lys(CBz)-NHBoc, 178 mg (0.49 mmols, 0.8 equivalents) HATU, 70.1 mg (0.23 mmols, 0.4 equivalents) DEPBT, and 0.82 mL (8 equivalents) of DIPEA. The crude material was purified using column chromatography (silica gel, EtOAc/Hex) to yield 415 mg (78% yield) of the pentapeptide.
Rf = 0.5 (EtOAc: Hex 7:3)
1H NMR (500 MHz, CDCl3): δ 0.5 (d, 2H), 0.6 (d, 2H), 0.9 (m, 4H), 1.2–1.4 (m, 1H), 1.4 (s, 9H), 1.5 (m, 1H), 1.6 (m, 1H), 1.7 (m, 1H), 2.9 (s, 3H), 3.2 (m, 2H), 3.3 (dd, 1H), 3.7 (s, 3H), 4.1 (s, 1αH), 4.4 (m, 2αH), 4.8 (dd, 1αH), 5.1 (s, 3H), 5.6 (m, 1H), 6.5 (d, 1H), 6.7 (d, 1H), 7.0 (d, 1H), 7.1–7.4 (m, 15H)
LCMS: m/z calculated for C50H70N5O10 (M+1) = 915.6, found 915.6
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NHBoc was synthesized using the “General Acid Deprotection” procedure. The pentapeptide was taken on to the next reaction without further purification or characterization. (346 mg, 87% yield).
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NH2
Pentapeptide HO-Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)-NH was synthesized using the “General Amine Deprotection” procedure. The pentapeptide was taken on to the next reaction without further purification. (298 mg, 100% yield).
LCMS: m/z calculated for C44H60N6O8 (M+1) = 802, found 801.5
Macrocycle Phe-D-N-Me-Phe-Val-Leu-Lys(CBz)
Macrocycle Phe-D-N-Me-Phe-Val-Leu-Lys(CBz) was constructed using the “General Macrocyclization Procedure”. Utilizing 149 mg (0.19 mmols, 1.0 equivalents) of the double deprotected linear pentapeptide, 30.0 mg (0.09 mmols, 0.5 equivalents) TBTU, 35.2 mg (0.09 mmols, 0.5 equivalents) HATU, 28.4 mg (0.09 mmols, 0.5 equivalents) DEPBT, and 0.27 mL (8 equivalents) of DIPEA. The crude material was purified using column chromatography (silica gel, EtOAc/Hex) to yield 19.2 mg (17% yield) of the macrocycle.
Rf = 0.5 (EtOAc: Hex 4:1)
1H NMR (400 MHz, CD3OD): δ 0.5 (d, 1H), 0.6 (d, 4H), 0.7 (m, 4H), 0.8–1.0 (m, 6H), 1.2–1.4 (m, 6H), 1.5 (m, 2H), 1.7 (m, 2H), 1.8 (m, 2H), 2.9 (s, 3H), 3.0 (m, 2H), 3.1 (m, 2H), 3.9 (t, 1αH), 4.1 (t, 1αH), 4.4 (t, 1αH), 4.6 (t, 1αH), 5.1 (s, 2H), 5.3 (s, 1H), 7.1–7.4 (m, 15H)
LCMS: m/z calculated for C44H58N6O7 (M+1) = 784.1, found 784.2
Macrocycle Phe-D-N-Me-Phe-Val-Leu-Lys
Macrocycle Phe-D-N-Me-Phe-Val-Leu-Lys was synthesized using “General Carboxybenzyl Removal Procedure Using HBr”. The crude product was purified by reverse phase HPLC. (15.2 mg, 33% yield).
Rf = 0.3 (MeOH: Hex 4:1)
1H-NMR (500 MHz, CD3OD): δ 0.7 (d, 2H), 0.8–1.0 (m, 12H), 1.2 (m, 2H), 1.3 (m, 4H), 1.5 (m, 2H), 1.6 (m, 2H), 1.7 (m, 2H), 1.8 (m, 2H), 1.9 (m, 2H), 2.7 (m, 1H), 2.8–3.1 (m, 2H), 3.7 (m, 1H), 3.9 (m, 1αH), 4.1 (m, 1αH), 4.4 (m, 1αH), 4.6 (m, 1αH), 5.2 (m, 1H), 7.1–7.4 (m, 10H)
LCMS: m/z calculated for C36H52N6O5 (M+1) = 649.8, found 649.5
Synthesis of Compound 31 and 32
Dipeptide Fmoc-Leu(Boc)-Phe-O-Resin
Dipeptide Fmoc-Leu(Boc)-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1005.0 mg (0.40 mmol, 1 equivalents) of NH2-Phe-O-Resin, the Leu residue was incorporated using 679 mg of Fmoc-Leu-OH (1.92 mmol, 3 equivalents), 294.0 mg (1.92 mmol, 3 equivalents) of HOBt, and 0.60 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-Leu-Phe-O-Resin
Dipeptide NH2-Leu-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Val-Leu-Phe-O-Resin
Tripeptide Fmoc-Val-Leu-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-Phe-O-Resin prepared above, the Phe residue was incorporated using 640.0 mg (1.92 mmol, 3 equivalents) of Fmoc-Val-OH, 294 mg (1.92 mmol, 3 equivalents) of HOBt, and 0.60 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-Val-Leu-Phe-O-Resin
Tripeptide NH2-Val-Leu-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-Leu-Val-Leu-Phe-O-Resin
Tetrapeptide Fmoc-Leu-Val-Leu-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-Val-Leu-Phe-O-Resin prepared above, the Leu residue was incorporated using 340.0 mg (0.96 mmol, 3 equivalents) of Fmoc-Leu-OH, 147.0 mg (0.96 mmol, 3 equivalents) of HOBt, and 0.3 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-Leu-Val-Leu-Phe-O-Resin
Tetrapeptide NH2-Leu-Val-Leu-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-Lys(Boc)-Leu-Val-Leu-Phe-O-Resin
Pentapeptide Fmoc-Lys(Boc)-Leu-Val-Leu-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-Val-Leu-Phe-O-Resin prepared above, the Lys(Boc) residue was incorporated using 450.0 mg (0.96 mmol, 3 equivalents) of Fmoc-Lys(Boc)-OH, 147.0 mg (0.96 mmol, 3 equivalents) of HOBt, 0.3 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-Lys(Boc)-Leu-Val-Leu-Phe-O-Resin
Pentapeptide NH2-Lys(Boc)-Leu-Val-Leu-Phe-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-Lys(Boc)-Leu-Val-Leu-Phe-OH
Double Deprotected Pentapeptide NH2-Lys(Boc)-Leu-Val-Leu-Phe-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 691.3 mg of dried NH2-Lys(Boc)-Leu-Val-Leu-Phe-O-Resin, 3.5 mL of 2,2,2-trifluoroethanol and 3.5 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (186 mg, 66% yield)
Macrocycle Lys(Boc)-Leu-Val-Leu-Phe
Macrocycle Lys(Boc)-Leu-Val-Leu-Phe was synthesized following the “Macrocyclization procedure”. Utilizing 40 mg (0.056 mmols, 1.0 equivalents) of linear pentapeptide, 0.06 mL (6 equivalents) of DIPEA, 12.6 mg (0.04 mmols, 0.7 equivalents) of TBTU, 14.9 mg (0.04 mmols, 0.7 equivalents) HATU, and 11.7 mg (0.04 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (13 mg, 40% yield).
Rf: 0.8 (100% EtOAc)
1H NMR (400 MHz, CD3OD): δ 0.8–1.2 (m, 18H), 1.4 (s, 9H), 1.6 (m, 2H), 1.8 (m, 2H), 2.2 (m, 1H), 3.1–3.2 (m, 2H), 3.6 (m, 2H), 3.8 (m, αH), 4.1 (m, αH), 4.4 (m, α H), 4.5 (m, αH), 4, 7 (m, αH), 7.1–7.3 (m, 5H)
LCMS: m/z calcd for C37H60N6O7 (M+Na) = 724.3, found 724.3
Macrocycle Lys-Leu-Val-Leu-Phe
Macrocycle Lys-Leu-Val-Leu-Phe was synthesized following the “General amine deprotection”. Utilizing 11.0 mg (0.015 mmols, 1.0 equivalents) of Macrocycle D-Val-Leu-Phe-Lys(Boc)-Leu, 1.20 mL of methylene chloride, 0.3 mL of TFA (20% TFA, 0.1M) to remove the Boc protecting group on the Lysine. The crude reaction was taken on to the next reaction without further purification or characterization (68.7 mg, quantitative yield).
1H NMR (400 MHz, CD3OD): δ 0.8–1.2 (m, 18H), 1.3 (m, 2H), 1.6 (m, 2H), 1.8 (m, 2H), 2.2 (m, 1H), 2.9–3.3 (m, 4H), 3.9–4.1 (m, αH), 4.1 (m, αH), 4.4 (m, 2αH), 4.5 (m, αH), 4, 7 (m, αH), 7.1–7.3 (m, 5H)
LCMS: m/z calcd for C32H52N6O5 (M+1) = 601.79, found 601.7
Synthesis of Compound 34 and 35
Dipeptide Fmoc-N-Me-Lys(Boc)-Leu-O-Resin
Dipeptide Fmoc-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1233.0 mg (0.99 mmol, 1 eqiv) of H-Leu-O-Resin, the N-Me-Lys residue was incorporated using 935 mg of Fmoc-N-Me-Lys(Boc)-OH, 458 mg of HOBt, 0.756 mL of DIC, and 5 mL of DMF. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the Fmoc-protected resin-bound dipeptide.
Dipeptide NH-N-Me-Lys(Boc)-Leu-O-Resin
Dipeptide NH-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc deprotection.
Tripeptide Fmoc-Phe-N-Me-Lys(Boc)-Leu-O-Resin
Tripeptide Fmoc-Phe-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-N-Me-Lys(Boc)-Leu-O-Resin prepared above, the Phe residue was incorporated using 1150 mg (2.9 mmol, 3 equiv) of Fmoc-Phe-OH, 407 mg (2.9 mmol, 3 equiv) of HOAt, 0.756 mL of DIC, and 5 mL of DMF. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the Fmoc-protected resin-bound tripeptide.
Tripeptide NH2-Phe-N-Me-Lys(Boc)-Leu-O-Resin
Tripeptide NH2-Phe-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc deprotection.
Tetrapeptide Fmoc-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin
Tetrapeptide Fmoc-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-Phe-N-Me-Lys(Boc)-Leu-O-Resin prepared above, the Leu residue was incorporated using 1056 mg (2.9 mmol, 3 equiv) of Fmoc-Leu-OH, 458 mg (2.9 mmol, 3 equiv) of HOBt, 0.756 mL of DIC, and 5 mL of DMF. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the Fmoc-protected resin-bound tetrapeptide.
Tetrapeptide NH2-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin
Tetrapeptide NH2-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc deprotection.
Pentapeptide Fmoc-Val-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin
Pentapeptide Fmoc-Val-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin prepared above, the Val residue was incorporated using 1010 mg (2.9 mmol, 3 equiv) of Fmoc-Val-OH, 458 mg (2.9 mmol, 3 equiv) of HOBt, 0.756 mL of DIC, and 5 mL of DMF. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the Fmoc-protected resin-bound pentapeptide.
Pentapeptide H-Val-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin
Pentapeptide H-Val-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc deprotection. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-Val-Leu-Phe-N-Me-Lys(Boc)-Leu-OH
Double Deprotected Pentapeptide NH2-Val-Leu-Phe-N-Me-Lys(Boc)-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1708.3 mg of dried NH2-Val-Leu-Phe-N-Me-Lys(Boc)-Leu-O-Resin, 10 mL of 2,2,2-trifluoroethanol and 10 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (565 mg, 77% yield)
Macrocycle Val-Leu-Phe-N-Me-Lys(Boc)-Leu
Macrocycle Leu-Phe-N-Me-Lys(Boc)-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 188 mg (0.25 mmols, 1.0 equivalents) of linear pentapeptide, 0.34 mL (8 equivalents) of DIPEA, 57 mg (0.14 mmols, 0.6 equivalents) of TBTU, 58 mg (0.18 mmols, 0.6 equivalents) HATU, and 46 mg (0.15 mmols, 0.6 equivalents) of DEPBT. The crude reaction of cyclization was purified by Column Chromatography to yield the macrocycle Leu-Phe-N-Me-Lys(Boc)-Leu (65 mg, 36% yield).
Rf: 0.2 (75% EtOAc/25% Hexane)
1H NMR (400 MHz, CD3OD: δ 0.8–1 (m, 18H), 1.1–1.2 (s, 9H), 1.3–1.4 (m, 2H), 1.4–1.5 (m, 2H), 1.6–1.7 (m, 2H), 1.8–2.0 (m, 2H), 2.0–2.1 (m, 2H), 2.1–2.2 (m, 2H), 2.8 (m, 1H), 2.9–3.0 (m, 1H), 3.0–3.1 (m, 1H), 3.1–3.2 (m, 1H), 3.2–3.4 (m, 1H), 3.2 (s, 3H) 3.9 (m, 1αH), 4.0–4.1 (m, 1αH), 4.2–4.3 (m, 1αH), 4.6 (m, 1αH), 5.0 (m, 1αH), 7.1–7.3 (m, 5H), 7.7 (m, 1H), 8.0 (m, 1H), 8.1 (m 1H), 8.3 (m, 1H)
LCMS: m/z calcd C38H62N6O7 = 714.93, found 716.1
Macrocycle Val-Leu-Phe-N-Me-Lys-Leu
Macrocycle Val-Leu-Phe-N-Me-Lys-Leu was synthesized following the “General amine deprotection.” The crude reaction of Boc removal was purified by reverse phase HPLC to yield the macrocycle Leu-Phe-N-Me-Lys-Leu (1.3 mg, 27% yield)
1H NMR (400 MHz, CD3OD: δ 0.8–1 (m, 18H), 1.2–1.4 (m, 4H), 1.5–1.7 (m, 2H), 1.8–2.0 (m, 2H), 2.1–2.2 (m, 2H), 2.1–2.2 (m, 2H), 2.6 (m, 1H), 2.7–2.8 (m, 1H), 2.8–2.9 (m, 1H), 3.0–3.1 (m, 1H), 3.4–3.5 (m, 1H), 3.6 (s, 3H) 3.9 (m, 1αH), 4.2–4.4 (m, 1αH), 4.4–4.6 (m, 1αH), 5.0 (m, 1αH), 5.3 (m, 1αH), 7.2–7.4 (m, 5H), 7.8 (m, 1H), 8.0 (m, 1H), 8.4 (m 1H), 8.6 (m, 1H)
LCMS: m/z calcd C33H54N6O5 = 614.82, found 615.7
Synthesis of Compound 36 and 37
Dipeptide Fmoc-Lys(Boc)-Leu-O-Resin
Dipeptide Fmoc-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1003.0 mg (0.81 mmol, 1 equivalents) of NH2-Leu-O-Resin, the Lys residue was incorporated using 1138.6 mg of Fmoc-Lys(Boc)-OH (2.43 mmol, 3 equivalents), 371.8 mg (2.43 mmol, 3 equivalents) of HOBt, and 0.756 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-Lys(Boc)-Leu-O-Resin
Dipeptide NH2-Lys(Boc)-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Phe-Lys(Boc)-Leu-O-Resin
Tripeptide Fmoc-Phe-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Lys(Boc)-Leu-O-Resin prepared above, the Phe residue was incorporated using 941.48 mg (2.43 mmol, 3 equivalents) of Fmoc-Phe-OH, 371.8 mg (2.43 mmol, 3 equivalents) of HOBt, and 0.756 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-Phe-Lys(Boc)-Leu-O-Resin
Tripeptide NH2-Phe-Lys(Boc)-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-Leu-Phe-Lys(Boc)-Leu-O-Resin
Tetrapeptide Fmoc-Leu-Phe-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Phe-Lys(Boc)-Leu-O-Resin prepared above, the Leu residue was incorporated using 858.8 mg (2.43 mmol, 3 equivalents) of Fmoc-Leu-OH, 371.8 mg (2.43 mmol, 3 equivalents) of HOBt, and 0.756 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-Leu-Phe-Lys(Boc)-Leu-O-Resin
Tetrapeptide NH2-Leu-Phe-Lys(Boc)-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-D-Val-Leu-Phe-Lys(Boc)-Leu-O-Resin
Pentapeptide Fmoc-D-Val-Leu-Phe-Lys(Boc)-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-Phe-Lys(Boc)-Leu-O-Resin prepared above, the Val residue was incorporated using 824.5 mg (2.43 mmol, 3 equivalents) of Fmoc-D-Val-OH, 371.8 mg (2.43 mmol, 3 equivalents) of HOBt, 0.756 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-D-Val-Leu-Phe-Lys(Boc)-Leu-O-Resin
Pentapeptide NH2-D-Val-Leu-Phe-Lys(Boc)-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-D-Val-Leu-Phe-Lys(Boc)-Leu-OH
Double Deprotected Pentapeptide NH2-D-Val-Leu-Phe-Lys(Boc)-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1391.3 mg of dried NH2-D-Val-Leu-Phe-Lys(Boc)-Leu-O-Resin, 7 mL of 2,2,2-trifluoroethanol and 7 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (386 mg, 66% yield)
Macrocycle D-Val-Leu-Phe-Lys(Boc)-Leu
Macrocycle D-Val-Leu-Phe-Lys(Boc)-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 232 mg (0.32 mmols, 1.0 equivalents) of linear pentapeptide, 0.33 mL (6 equivalents) of DIPEA, 72.5 mg (0.22 mmols, 0.7 equivalents) of TBTU, 85.8 mg (0.22 mmols, 0.7 equivalents) HATU, and 67.5 mg (0.22 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (100.8 mg, 44.5% yield).
Rf: 0.8 (100% EtOAc)
1H NMR (400 MHz, CD3OD): δ 0.8–1.2 (m, 20H), 1.3–1.7 (m, 18H), 1.8 (m, 1H), 2.0 (m, 2H), 2.9 (m, 3H), 3.1 (m, 1H), 3.6 (m, αH), 3.8 (m, αH), 4.2 (m, αH), 4.5 (m, αH), 4.6 (m, αH), 7.1–7.3 (m, 5H)
LCMS: m/z calcd for C37H60N6O7 (M+1) = 701.91, found 702.1
Macrocycle D-Val-Leu-Phe-Lys-Leu
Macrocycle D-Val-Leu-Phe-Lys-Leu was synthesized following the “General amine deprotection”. Utilizing 80.2 mg (0.11 mmols, 1.0 equivalents) of Macrocycle D-Val-Leu-Phe-Lys(Boc)-Leu, 1.83 mL of methylene chloride, 0.45 mL of TFA (20% TFA, 0.05 M) to remove the BOC protecting group on the Lysine. The crude reaction was taken on to the next reaction without further purification or characterization (68.7 mg, quantitative yield).
1H NMR (400 MHz, CD3OD): δ 0.8–1.1 (m, 20H), 1.2 (m, 2H), 1.4 (m, 2H), 1.5–1.7 (m, 5H), 1.8 (m, 1H), 2.0 (m, 2H), 2.8 (m, 3H), 3.1 (m, 1H), 3.2 (m, 1H), 3.7 (m, α H), 3.8 (m, αH), 4.2 (m, αH), 4.5 (m, αH), 4.7 (m, αH), 7.1–7.3 (m, 5H), 7.6 (d, 1H), 8.0 (d, 1H), 8.6 (d, 1H), 8.7 (d, 1H)
LCMS: m/z calcd for C32H52N6O5 (M+1) = 601.79, found 601.7
Synthesis of Compound 39
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1610.3 mg (1.3 mmol, 1 equivalents) of H-Leu-O-Resin, the N-Me-Val residue was incorporated using 1378 mg of Fmoc-N-Me-Val-OH (3.9 mmol, 3 equivalents), 597 mg (3.9 mmol, 3 equivalents) of HOBt, and 1.21 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide H-N-Me-Val-Leu-O-Resin
Dipeptide H-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin
Tripeptide Fmoc-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-N-Me-Val-Leu-O-Resin prepared above, the Lys(2-Cl-Z) residue was incorporated using 1050 mg (1.95 mmol, 3 equivalents) of Fmoc-Lys(2-Cl-Z)-OH, 265 mg (1.95 mmol, 3 equivalents) of HOAt, and 0.604 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin
Tripeptide NH2-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin
Tetrapeptide Fmoc-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin prepared above, the D-Phe residue was incorporated using 755 mg (1.95 mmol, 3 equivalents) of Fmoc-D-Phe-OH, 298 mg (1.95 mmol, 3 equivalents) of HOBt, and 0.604 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin
Tetrapeptide NH2-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin
Pentapeptide Fmoc-Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin prepared above, the Phe residue was incorporated using 755 mg (1.95 mmol, 3 equivalents) of Fmoc-Phe-OH, 298 mg (1.95 mmol, 3 equivalents) of HOBt, 0.604 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin
Pentapeptide NH2-Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-OH
Double Deprotected Pentapeptide NH2-Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 2078.6 mg of dried NH2-Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin, 10.39 mL of 2,2,2-trifluoroethanol and 10.39 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (323 mg, 59.5% yield)
Macrocycle Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu
Macrocycle Phe-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 160 mg (0.19 mmols, 1.0 equivalents) of linear pentapeptide, 0.335 mL (10 equivalents) of DIPEA, 58.3 mg (0.153 mmols, 0.8 equivalents) of TBTU, 49.3 mg (0.153 mmols, 0.8 equivalents) HATU, and 46 mg (0.153 mmols, 0.8 equivalents) of DEPBT. The crude reaction was purified by column purification to yield the macrocycle (15.4 mg, 9.79% yield).
Rf: 0.25 (EtOAc: Hex 3:1)
1H NMR (400 MHz, CD3OD): δ 0.7–1.0 (m, 12H), 1.3–1.4 (m, 2H), 1.5–1.6 (m, 4H), 1.7–1.8 (m, 2H), 2.7 (m, 1H), 2.9 (m, 2H), 3.1 (t, 4H), 3.2 (s, 1H), 4.2 (t, αH), 4.4 (t, αH), 4.7 (t, αH), 5.1 (s, 1H), 7.0–7.2 (m, 13H), 7.4 (d, 1H)
LCMS: m/z calcd for C44H57ClN6O7 (M+1) = 817, found 818.5
Synthesis of Compound 40
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1610.3 mg (1.3 mmol, 1 equivalents) of H-Leu-O-Resin, the N-Me-Val residue was incorporated using 1378 mg of Fmoc-N-Me-Val-OH (3.9 mmol, 3 equivalents), 597 mg (3.9 mmol, 3 equivalents) of HOBt, and 1.21 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide H-N-Me-Val-Leu-O-Resin
Dipeptide H-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-D-Lys(Z)-N-Me-Val-Leu-O-Resin
Tripeptide Fmoc-D-Lys(Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-N-Me-Val-Leu-O-Resin prepared above, the D-Lys(Z) residue was incorporated using 980 mg (1.95 mmol, 3 equivalents) of Fmoc-D-Lys(Z)-OH, 265 mg (1.95 mmol, 3 equivalents) of HOAt, and 0.604 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-D-Lys(Z)-N-Me-Val-Leu-O-Resin
Tripeptide NH2-D-Lys(Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin
Tetrapeptide Fmoc-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Lys(Z)-N-Me-Val-Leu-O-Resin prepared above, the D-Phe residue was incorporated using 755 mg (1.95 mmol, 3 equivalents) of Fmoc-D-Phe-OH, 298 mg (1.95 mmol, 3 equivalents) of HOBt, and 0.604 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin
Tetrapeptide NH2-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin
Pentapeptide Fmoc-Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Phe-Lys(2-Cl-Z)-N-Me-Val-Leu-O-Resin prepared above, the Phe residue was incorporated using 755 mg (1.95 mmol, 3 equivalents) of Fmoc-Phe-OH, 298 mg (1.95 mmol, 3 equivalents) of HOBt, 0.604 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin
Pentapeptide NH2-Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu-OH
Double Deprotected Pentapeptide NH2-Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 2094 mg of dried NH2-Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu-O-Resin, 10.47 mL of 2,2,2-trifluoroethanol and 10.47 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (301 mg, 57.9% yield)
Macrocycle Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu
Macrocycle Phe-D-Phe-D-Lys(Z)-N-Me-Val-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 150 mg (0.187 mmols, 1.0 equivalents) of linear pentapeptide, 0.294 mL (9 equivalents) of DIPEA, 42.1 mg (0.131 mmols, 0.7 equivalents) of HATU, 49.9 mg (0.131 mmols, 0.7 equivalents) TBTU, and 39.3 mg (0.131 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC purification to yield the macrocycle (8.1 mg, 5.52% yield).
Rf: 0.3 (EtOAc: Hex 1:1)
1H NMR (400 MHz, CD3OD): δ 0.7–1.0 (m, 12H), 1.3–1.4 (m, 2H), 1.5–1.6 (m, 4H), 1.7–1.8 (m, 2H), 2.7 (m, 1H), 2.9 (m, 2H), 3.1 (t, 4H), 3.2 (s, 1H), 4.2 (t, αH), 4.4 (t, αH), 4.7 (t, αH), 5.1 (s, 1H), 7.0–7.2 (m, 13H), 7.4 (d, 1H)
LCMS: m/z calcd for C44H57ClN6O7 (M+1) = 783, found 806.8
Synthesis of Compound 41
Dipeptide Fmoc-Leu-Phe-O-Resin
Dipeptide Fmoc-Leu-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 14002 mg (.896 mmol, 1 equivalents) of NH2-Phe-O-Resin, the Leu residue was incorporated using 917 mg of Fmoc-Leu-OH (2.69 mmol, 3 equivalents), 410 mg (2.69 mmol, 3 equivalents) of HOBt, and 0.80 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-Leu-Phe-O-Resin
Dipeptide NH2-Leu-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-Val-Leu-Phe-O-Resin
Tripeptide Fmoc-Val-Leu-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Leu-Phe-O-Resin prepared above, the Val residue was incorporated using 911.0 mg (2.69 mmol, 3 equivalents) of Fmoc-Val-OH, 410 mg (2.69 mmol, 3 equivalents) of HOBt, and 0.80 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-Val-Leu-Phe-O-Resin
Tripeptide NH2-Cha-D-Val-D-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin
Tetrapeptide Fmoc-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Val-Leu-Phe-O-Resin prepared above, the N-Me-D-Lys(2-Cl-Cbz) residue was incorporated using 860.0 mg (2.69 mmol, 3 equivalents) of Fmoc-N-Me-D-Lys(2-Cl-Cbz)-OH, 410 mg (2.69 mmol, 3 equivalents) of HOBt, and 0.680 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin
Tetrapeptide NH-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Pentapeptide Fmoc-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin
Pentapeptide Fmoc-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin prepared above, the Phe residue was incorporated using 10410 mg (2.69 mmol, 3 equivalents) of Fmoc-D-Phe-OH, 410 mg (2.69 mmol, 3 equivalents) of HOAt, 0.80 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Pentapeptide NH2-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin
Pentapeptide NH2-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Pentapeptide NH2-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-OH
Double Deprotected Pentapeptide NH2-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1672.1 mg of dried NH2-D-Phe-N-Me-D-Lys(2-Cl-Cbz)-Val-Leu-Phe-O-Resin, 8.4 mL of 2,2,2-trifluoroethanol and 8.4 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (394 mg, 79% yield)
Macrocycle Phe-Leu-Val-N-Me-D-Lys(2-Cl-Cbz)-D-Phe
Macrocycle Phe-Leu-Val-N-Me-D-Lys(2-Cl-Cbz)-D-Phe was synthesized following the “Macrocyclization procedure”. Utilizing 250 mg (0.30 mmols, 1.0 equivalents) of linear pentapeptide, 0.417 mL (8 equivalents) of DIPEA, 67.0 mg (0.21 mmols, 0.7 equivalents) of TBTU, 80.0 mg (0.21 mmols, 0.7 equivalents) HATU, and 62.5 mg (0.21 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (8.0 mg, 7.2% yield).
1H NMR (400 MHz, CD3OD): δ 0.7–1.0 (m, 12H), 1.2–1.4 (m, 2H), 1.4–1.6 (m, 3H), 1.7–1.8 (m, 4H), 2.0 (m, 1H), 3.0–3.1 (m, 2H), 3.1–3.2 (m, 2H), 4.1 (s, αH), 4.2 (s, αH), 4.4 (s, αH), 4.6 (s, αH), 5.2 (m, 2H), 7.0–7.4 (m, 14H), 8.2 (m, 1H)
LCMS: m/z called for C44H59ClN6O8 (M+1) = 818.4, found 817.4
Synthesis of Compound 42
Dipeptide Fmoc-Val-Leu-O-Resin
Dipeptide Fmoc-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 1002.2 mg (.812 mmol, 1 equivalents) of NH2-Leu-O-Resin, the Val residue was incorporated using 825.5 mg of Fmoc-Val-OH (2.4 mmol, 3 equivalents), 377 mg (2.4 mmol, 3 equivalents) of HOBt, and 0.753 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-Val-Leu-O-Resin
Dipeptide NH2-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Boc-D-Lys(2-Cl-Z)-Val-Leu-O-Resin
Tripeptide Boc-D-Lys(2-Cl-Z)-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-Val-Leu-O-Resin prepared above, the Lys residue was incorporated using 1012.6 mg (2.4 mmol, 3 equivalents) of Boc-D-Lys(2-Cl-Z)-OH, 373 mg (2.4 mmol, 3 equivalents) of HOBt, and 0.753 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide Boc-D-Lys(2-Cl-Z)-Val-Leu-OH
Tripeptide Boc-D-Lys(2-Cl-Z)-Val-Leu was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 1390.2 mg of dried Boc-D-Lys(2-Cl-Z)-Val-Leu-O-Resin, 7.0 mL of 2,2,2-trifluoroethanol and 7.0 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (613 mg, 92% yield)
Tripeptide Boc-D-Lys(2-Cl-Z)-Val-Leu-OMe
Tripeptide Boc-D-Lys(2-Cl-Z)-Val-Leu-OMe was synthesized following the “General Acid Protection” procedure. Utilizing the 613.6 mg Boc-D-Lys(2-Cl-Z)-Val-Leu-OH peptide prepared above, the ester was incorporated using .8 mL TMSD (7.1 equivalents) in 9.78 mL 3:1 (v/v) Benzene/Methanol mixture and stirred for two hours. The crude reaction was purified via column chromatography (EtOAc/Hex) to yield the double protected tripeptide. (494.3 mg, 79% yield) (Rf 0.3 (EtOAc: Hex 1:1)
Tripeptide NH2-D-Lys(2-Cl-Z)-Val-Leu-OMe
Tripeptide NH2-D-Lys(2-Cl-Z)-Val-Leu-OMe was synthesized following the “General amine deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (608.6 mg, 100% yield).
Tetrapeptide Boc-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OMe
Tetrapeptide Boc-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OMe was synthesized following the “General peptide Synthesis” procedure. Utilizing 608.6 mg (1.12 mmols, 1.1 equiv.) of amine NH2-D-Lys(2-Cl-Z)-Val-Leu-OMe, 285.7 mg (1.0 mmols, 1.0 equiv.) of acid Boc-N-Me-D-Phe-OH, .712 mL (4 equiv.) of DIPEA, 131.4 mg (.4 mmols, .4 equiv.) of TBTU, 311.1 mg (0.8 mmol, .8 eq) HATU. The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the tetrapeptide (188 mg, 21% yield).
Rf: 0.3 (EtOAc: Hex 1:1)
Tetrapeptide HN-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OMe
Tetrapeptide HN-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OMe was synthesized following the “General amine deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (251 mg, 100% yield).
Pentapeptide Boc-Phe-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OMe
Tetrapeptide Boc-Phe-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OMe was synthesized following the “General peptide Synthesis” procedure. Utilizing 251 mg (.358 mmols, 1.1 equiv.) of amine HN-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OMe, 104 mg (0.4 mmols, 1.0 equiv.) of acid Boc-Phe-OH, .272 mL (4 equiv.) of DIPEA, 63.4 mg (.2 mmols, .5 equiv.) of TBTU, 104.8 mg (0.28 mmol, .7 eq) HATU, 23.5 mg (.08 mmol, .2 eq). The crude reaction was purified by column chromatography (silica gel, EtOAc/Hex) to yield the pentapeptide (174.7 mg, 73% yield).
Rf: 0.3 (EtOAc: Hex 3:2)
1H NMR (400 MHz, CD3OD): δ 0.8–1.0 (m, 6H), 1.2 (m, 2H), 1.2–1.4 (m, 10H), 1.4–1.5 (m, 3H), 1.5–1.6 (m, 4H), 2.8 (s, 1H), 3.0–3.3 (m, 7H), 3.6 (s, 3H), 4.0 (s, αH), 4.2 (s, αH), 5.1 (s, 2H), 7.1–7.4 (m, 14H)
LCMS: m/z called for C50H69ClN6O10 (M+1) = 950.6, found 950.9
Pentapeptide Boc-Phe-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OH
Pentapeptide Boc-Phe-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OH was synthesized following the “General acid deprotection” procedure. The pentapeptide was taken on to the next reaction without further purification or characterization. (165 mg, 96% yield).
Deprotected Pentapeptide NH2-Phe-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OH
Pentapeptide NH2-Phe-N-Me-D-Phe-D-Lys(2-Cl-Z)-Val-Leu-OH was synthesized following the “General amine deprotection”. This tripeptide was taken on to the next reaction without further purification or characterization. (182.6 mg, 100% yield).
LCMS: m/z called for C44H59ClN6O8 (M+1) = 836.4, found 836.1
Macrocycle Phe-Leu-Val-D-Lys(2-Cl-Z)-N-Me-D-Phe
Macrocycle Leu-Val-D-Lys(2-Cl-Z)-N-Me-D-Phe-Phe was synthesized following the “Macrocyclization procedure”. Utilizing 165 mg (0.19 mmols, 1.0 equivalents) of linear pentapeptide, 0.27 mL (8 equivalents) of DIPEA, 44.3 mg (0.14 mmols, 0.7 equivalents) of TBTU, 52.5 mg (0.14 mmols, 0.7 equivalents) HATU, and 41.3 mg (0.14 mmols, 0.7 equivalents) of DEPBT. The crude reaction was purified by reverse phase-HPLC to yield the macrocycle (21.8 mg, 13.2% yield).
1H NMR (400 MHz, CD3OD): δ 0.8–1.0 (m, 12H), 1.2–1.4 (m, 2H), 1.4–1.6 (m, 3H), 1.7–1.8 (m, 4H), 2.1 (m, 1H), 3.0–3.1 (m, 2H), 3.1–3.2 (m, 2H), 3.2–3.4 (m, 7H), 4.0 (m, αH), 4.2 (m, αH), 4.7 (m, αH), 5.4 (m, 2H), 7.0–7.5 (m, 14H), 8.2 (m, 1H)
LCMS: m/z called for C44H57ClN6O7 (M+1) = 818.4, found 818.1
Synthesis of Compound 43 and 44
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 2032.0 mg (1.64 mmol, 1 equivalents) of NH2-Leu-O-Resin, the N-Me-Val residue was incorporated using 1740 mg of Fmoc-D-Val-OH (4.9 mmol, 3 equivalents), 734 mg (4.9 mmol, 3 equivalents) of HOBt, and 1.48 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide NH2-N-Me-Val-Leu-O-Resin
Dipeptide NH2-N-Me0Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-D-Leu-N-Me-Val-Leu-O-Resin
Tripeptide Fmoc-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-N-Me-Val-Leu-O-Resin prepared above, the D-Leu residue was incorporated using 1736 mg (4.9 mmol, 3 equivalents) of Fmoc-D-Leu-OH, 669 mg (4.9 mmol, 3 equivalents) of HOAt, and 1.48 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-D-Leu-N-Me-Val-Leu-O-Resin
Tripeptide NH2-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Tetrapeptide Fmoc-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Leu-N-Me-Val-Leu-O-Resin prepared above, the D-Phe residue was incorporated using 1900 mg (4.9 mmol, 3 equivalents) of Fmoc-D-Phe-OH, 743 mg (4.9 mmol, 3 equivalents) of HOBt, and 1.48 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Tetrapeptide NH2-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Racemic Pentapeptide Fmoc-β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Racemic Pentapeptide Fmoc-β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin prepared above, the racemic β-OH-Phe residue was incorporated using 992 mg (4.9 mmol, 3 equivalents) of (2R, 3R)/(2S, 3S)- racemic Fmoc- β-OH-Phe-OH, 743 mg (4.9 mmol, 3 equivalents) of HOBt, 1.48 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Racemic Pentapeptide NH2-β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Racemic pentapeptide NH2-β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Racemic Pentapeptide NH2-β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu-OH
Double Deprotected Racemic Pentapeptide NH2-β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 2511.2 mg of dried NH2-β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin, 13 mL of 2,2,2-trifluoroethanol and 13 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (719 mg, 67% yield)
Racemic Macrocycle β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu
Racemic Macrocycle β-OH-Phe-D-Phe-D-Leu-N-Me-Val-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 160 mg (0.23 mmols, 1.0 equivalents) of linear pentapeptide, 0.32 mL (8 equivalents) of DIPEA, 52 mg (0.13 mmols, 0.6 equivalents) of TBTU, 44 mg (0.13 mmols, 0.6 equivalents) HATU, and 41 mg (0.13 mmols, 0.6 equivalents) of DEPBT. The crude reaction of cyclization was purified by Column Chromatography to yield the racemic macrocycle-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu (115.4 mg, 76% yield).
Rf: 0.1 (35% EtOAc/65% Hexane)
LCMS: m/z calcd for C36H51N5O6 (M+1) = 650.82, found 651.2 (earlier retention time, 3.3 min) and 651.6 (later retention time, 3.4 min)
Macrocycle β-benzoxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu
Macrocycle β-benzoxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu was synthesized following the “Benzylation procedure”. Utilizing 115.4 mg (0.17 mmols, 1.0 equivalent) of Racemic Macrocycle β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu, 7.1 mg of 60% NaH, 0.04 mL of benzyl bromide (0.35 mmol, 3.0 equivalents), 0.88 mL of THF and 0.88 mL of DMF. The crude reaction was purified by reverse phase-HPLC to yield the two diastereomic macrocycles (4 mg for compound 38-earlier retention time (RT=4.0 min), 3%/3 mg for compound 39-later retention time (RT=4.9 min), 2%).
For compound 43-earlier retention time
Rf: 0.2 (35% EtOAc/65% Hexane)
1H NMR (400 MHz, (CD3)2SO): δ 0.8–1 (m, 18H), 1.2 (m, 1H), 1.4 (m, 1H), 1.5 (m, 1H), 1.6 (m, 1H), 1.7 (m, 1H), 2.0 (m, 1H), 2.8 (m, 1H), 3.1 (m, 1H), 3.4 (s, 3H), 4.1 (m, 1αH), 4.2 (m, 1αH), 4.4–4,5 (m, 2H), 4.5 (m, 1αH) 4.6 (m, 1αH), 5.3 (m, 1αH), 6.6 (m, 1H), 7.0–7.4 (m, 15H), 7.6 (m, 1H), 8.1 (m, 1H), 8.2 (m, 1H)
LCMS: m/z calcd for C43H57N5O6 = 739.94, found 764.7 (+23)
For compound 44-later retention time
Rf: 0.2 (35% EtOAc/65% Hexane)
1H NMR (400 MHz, CD3OD): δ 0.8–1 (m, 18H), 1.2 (m, 1H), 1.4 (m, 1H), 1.5 (m, 1H), 1.6 (m, 1H), 1.9 (m, 1H), 2.2 (m, 1H), 3.0 (m, 1H), 3.1 (m, 1H), 3.5 (s, 3H), 4.1 (m, 1αH), 4.3 (m, 1αH), 4.4–4.5 (m, 2H), 4.5 (m, 1αH) 4.7 (m, 1αH), 5.3 (m, 1αH), 7.0 (m, 1H), 7.1–7.3 (m, 15H), 7.8 (m, 1H), 8.2 (m, 1H), 8.3 (m, 1H)
LCMS: m/z calcd for C43H57N5O6 = 739.94, found 763.9 (+23)
Synthesis of Compound 45 and 46
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin
Dipeptide Fmoc-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using 2.032 g (1.64 mmol, 1 equivalent) of NH2-Leu-O-Resin, the N-Me-Val residue was incorporated using 1.74 g of Fmoc-N-Me-Val-OH (4.92 mmol, 3 equivalents), 743 mg (4.92 mmol, 3 equivalents) of HOBt, and 1.48 mL (6 equivalents) of DIC. Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound dipeptide.
Dipeptide H-N-Me-Val-Leu-O-Resin
Dipeptide H-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tripeptide Fmoc-D-Leu-N-Me-Val-Leu-O-Resin
Tripeptide Fmoc-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the H-N-Me-Val-Leu-O-Resin prepared above, the D-Leu residue was incorporated using 1.736 g (4.92 mmol, 3 equivalents) of Fmoc-D-Leu-OH, 669 mg (4.92 mmol, 3 equivalents) of HOAt, and 1.48 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tripeptide.
Tripeptide NH2-D-Leu-N-Me-Val-Leu-O-Resin
Tripeptide NH2-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Tetrapeptide Fmoc-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Tetrapeptide Fmoc-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Leu-N-Me-Val-Leu-O-Resin prepared above, the D-Phe residue was incorporated using 1.906 g (4.92 mmol, 3 equivalents) of Fmoc-D-Phe-OH, 743 mg (4.92 mmol, 3 equivalents) of HOBt, and 1.48 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound tetrapeptide.
Tetrapeptide NH2-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Tetrapeptide NH2-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal.
Racemic Pentapeptide Fmoc-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Racemic Pentapeptide Fmoc-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General solid-phase Peptide Synthesis” procedure. Using the NH2-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin prepared above, the racemic Fmoc-β-hydroxy-Phe residue was incorporated using 1.98 g (4.92 mmol, 3 equivalents) of (2R, 3S)/(2S, 3R)-Racemic-Fmoc-beta-hydroxy-phenylalanine, 743 mg (4.92 mmol, 3 equivalents) of HOBt, 1.48 mL of DIC (6 equivalents). Completion of the coupling reaction was verified by a negative ninhydrin test. The reaction mixture was then drained to leave the amine-protected resin-bound pentapeptide.
Racemic Pentapeptide NH2-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin
Racemic Pentapeptide NH2-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin was synthesized following the “General N-terminal Solid Phase Amine Deprotection” procedure. A positive ninhydrin test served to verify Fmoc removal. The resin was then dried in vacuo for 24 hours in a vacuum desiccator.
Double Deprotected Racemic Pentapeptide NH2-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-OH
Double Deprotected Racemic Pentapeptide NH2-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-OH was synthesized following the “Cleavage of Linear Peptide” procedure. Utilizing the 2.6313 g of dried racemic NH2-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-O-Resin, 14 mL of 2,2,2-trifluoroethanol and 14 mL of CH2Cl2. The resin slurry was stirred for 24 hours, after which it was filtered, washed with additional CH2Cl2, and dried in vacuo for 24 hours. (860 mg, 78.5% yield)
Racemic Macrocycle β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu
Racemic Macrocycle β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu was synthesized following the “Macrocyclization procedure”. Utilizing 160 mg (0.23 mmols, 1.0 equivalent) of racemic linear pentapeptide NH2-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu-OH, 0.32 mL (6 equivalents) of DIPEA, 52 mg (0.14 mmols, 0.6 equivalents) of TBTU, 44 mg (0.14 mmols, 0.6 equivalents) HATU, and 41 mg (0.14 mmols, 0.6 equivalents) of DEPBT. The crude reaction of cyclization was purified by Column Chromatography to yield the racemic macrocycle-β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu (110.2 mg, 73.9% yield).
Rf: 0.1 (35% EtOAc/65% Hexane)
LCMS: m/z calcd for C36H51N5O6 (M+1) = 650.82, found 651.3 (earlier retention time) and 650.7 (later retention time)
Macrocycle β-benzoxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu
Macrocycle β-benzoxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu was synthesized following the “Benzylation procedure”. Utilizing 110.2 mg (0.17 mmols, 1.0 equivalent) of Racemic Macrocycle β-hydroxy-Phe-D-Phe-D-Leu-N-Me-Val-Leu, 26.8 mg of 60% NaH, 0.08 mL of benzyl bromide (0.68 mmol, 4.0 equivalents), 0.85 mL of THF and 0.85 mL of DMF. The crude reaction was purified by reverse phase-HPLC to yield the two diastereomic macrocycles (0.6 mg for compound 40-earlier retention time, 0.5%/2.3 mg for compound 41-later retention time, 1.8%).
For compound 45-earlier retention time
Rf: 0.2 (35% EtOAc/65% Hexane)
1H NMR (400 MHz, CD3OD): δ 0.8–1.1 (m, 9H), 1.2–1.4 (m, 9H), 1.5–1.7 (m, 4H), 2.0 (m, 2H), 2.2–2.4 (m, 2H), 3.0 (m, 1H), 3.5–3.6 (m, 5H), 4.1 (m, 1H), 4.2 (m, 1H), 4.3 (m, 1H), 4.4 (m, 1H), 4.5 (m, 1H), 6.7 (d, 1H), 7.0 (d, 1H)7.2–7.5 (m, 15H), 7.6 (m, 1H), 7.7 (m, 1H), 8.0 (m, 1H)
LCMS: m/z calcd for C43H57N5O6 = 739.94, found 740.35
For compound 46-later retention time
Rf: 0.2 (35% EtOAc/65% Hexane)
1H NMR (400 MHz, CD3OD): δ 0.8–1.1 (m, 18H), 1.2–1.4 (m, 5H), 1.5–1.7 (m, 4H), 2.0 (m, 1H), 2.2–2.3 (m, 2H), 2.7–2.9 (m, 5H), 3.6 (m, 1H), 4.1 (t, 1H), 4.2 (m, 1H), 4.4 (m, 1H), 4.5 (d, 1H), 4.6 (d, 1H), 4.7 (d, 1H), 6.8–7.3 (m, 15H)
LCMS: m/z calcd for C43H57N5O6 = 739.94, found 739.85
Docking to Hsp90 using Autodock
The Yeast HSP90 protein crystal structure (PDB code: 2CG9)76 was downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) and prepared for docking by removing all water molecules, the ATP analog, and all protein chains except the A chain. AutoDockTools (ADT)77 was used to add polar hydrogens and gasteiger charges. Ligands were prepared by drawing the 2D structure in CambridgeSoft’s ChemDraw, and converted to PDB format using Chem3D. Ligands were then energy minimized with the UFF forcefield from OpenBabel before using ADT to add polar hydrogens and gasteiger charges.
AutoLigand was used to identify two putative binding sites that have contact to both the N-terminal domain and middle domain of HSP90. When four Sansalvamide A (SanA) compounds (1, 2, 16, 33) were docked using AutoDock 4.2 to each site, one site returned energy values of −7 kcal/mol to −8 kcal/mol, while the other site gave a lowest value of −3.5 kcal/mol (data not shown). The former site was used for the following docking studies.
SanA derivatives were each docked 250 times, using AutoDock 4.2, to a 26 X 32 X 36 Å3 box centered on the chosen AutoLigand fill using 2.5 X 106 energy evaluations per run and a grid map spacing of 0.375Å. The number of active torsions was set to the maximum available for each ligand, which range from nine for San A-amide to fifteen for SanA 43.
Similar docking modes were grouped in the 250 results per compound using a clustering method based on root mean squared deviation (RMSD). An in-house program was written to perform this clustering, which calculated the RMSD value (using Bio3d78) for all pairwise combinations. Similar compounds were included in this calculation so that structures which have the same number and types of atoms, and which only differ by stereochemistry could be clustered together. Such groupings included compounds 43–46 and 31, 34, 36 and 38–42, and 19, 20. A matrix containing the RMSD values was imported into python for clustering work and visual analysis. The clustering was performed by finding all dockings within a 2.0Å–3.75Å cutoff at 0.25Å increments. Those groups that had an intersection of at least one docked compound were then joined to form clusters. The four largest clusters were further analyzed. Visualization of clusters was performed in PyMol (http://www.pymol.org/) by superimposing each compound within a cluster in conjunction with HSP90. The minimal software predicted energy of each cluster was determined by parsing the Autodock result for each member of the cluster. The lowest energy of this conformation for each compound was the binding mode used for further studies.
Supplementary Material
Acknowledgments
We thank the Frasch foundation (658-HF07) for support of RPS, LDA, VAJ, CCL, EKS, MRD and CMP. We thank NIH 1U54CA132379–01A1 for support of LDA, VAJ, and SRM and 1R01CA137873 for support of RPS, LDA, VAJ, CMP, and SRM. We thank NIH MIRT program for support of LDA, RPS, VAJ, and EKS. We thank the Howell Foundation for support of TS. Finally, we are extremely grateful to Dr. David Vander Velde at Caltech who helped us evaluate the stereochemistry of the four diastereomers 43–46.
Abbreviations
- San A
Sansalvamide A
- San A-amide
Sansalvamide A-amide
- Hsp90
Heat shock protein 90
Footnotes
Supporting Information Available A table that includes the description of all changes made to compound structures in text format is included in the supporting information. IC50 data curves are supplied in the supplementary material. In addition, all spectral data gathered while synthesizing new compounds is shown. This material is available free of charge on the Internet at http://pubs.acs.org.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cueto M, Jensen PR, Fenical W. Phytochemistry. 2000;55:223. doi: 10.1016/s0031-9422(00)00280-6. [DOI] [PubMed] [Google Scholar]
- 2.Hwang Y, Rowley D, Rhodes D, Gertsch J, Fenical W, Bushman F. Molecular Pharmacology. 1999;55:1049. doi: 10.1124/mol.55.6.1049. [DOI] [PubMed] [Google Scholar]
- 3.Belofsky GN, Jensen PR, Fenical W. Tetrahedron Lett. 1999;40:2913. [Google Scholar]
- 4.Styers TJ, Kekec A, Rodriguez RA, Brown JD, Cajica J, Pan PS, Parry E, Carroll CL, Medina I, Corral R, Lapera S, Otrubova K, Pan CM, Mcguire KL, Mcalpine SR. Bioorganic and Medicinal Chemistry. 2006;14:5625. doi: 10.1016/j.bmc.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez RA, Pan PS, Pan CM, Ravula S, Lapera SA, Singh EK, Styers TJ, Brown JD, Cajica J, Parry E, Otrubova K, Mcalpine SR. J Org Chem. 2007;72:1980. doi: 10.1021/jo061830j. [DOI] [PubMed] [Google Scholar]
- 6.Pan PS, Vasko RC, Lapera SA, Johnson VA, Sellers RP, Lin CC, Pan CM, Davis MR, Ardi VC, Mcalpine SR. Bio Org Med Chem. 2009;17:5806. doi: 10.1016/j.bmc.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Gu W, DL, Ding X-Z, Ujiki M, Adrian TE, Soff GA, Silverman RB. J Med Chem. 2005;48:3630. doi: 10.1021/jm048952t. [DOI] [PubMed] [Google Scholar]
- 8.Ujiki M, Milam B, Ding XZ, Roginsky AB, Salabat MR, Talamonti MS, Bell RH, Gu W, Silverman RB, Adrian TE. Biochemical and Biophysical Research Communications. 2006;340:1224. doi: 10.1016/j.bbrc.2005.12.131. [DOI] [PubMed] [Google Scholar]
- 9.Pan PS, Mcguire K, Mcalpine SR. Bioorg & Med Chem Lett. 2007;17:5072. doi: 10.1016/j.bmcl.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 10.Otrubova K, Styers TJ, Pan PS, Rodriguez R, Mcguire KL, Mcalpine SR. Chem Commun. 2006:1033. doi: 10.1039/b517434a. [DOI] [PubMed] [Google Scholar]
- 11.Carroll CL, Johnston JVC, Kekec A, Brown JD, Parry E, Cajica J, Medina I, Cook KM, Corral R, Pan PS, Mcalpine SR. Org Lett. 2005;7:3481. doi: 10.1021/ol051161g. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Silverman RB. Org Lett. 2000;2:3743. doi: 10.1021/ol0002830. [DOI] [PubMed] [Google Scholar]
- 13.Gu W, Liu S, Silverman RB. Org Lett. 2002;4:4171. doi: 10.1021/ol0269392. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee J, Mierke DF, Kessler H. J Am Chem Soc. 2006;128:15164. doi: 10.1021/ja063123d. [DOI] [PubMed] [Google Scholar]
- 15.Heller M, Sukopp M, Tsomaia N, John M, Mierke DF, Reif B, Kessler H. J Am Chem Soc. 2006;128:13806. doi: 10.1021/ja063174a. [DOI] [PubMed] [Google Scholar]
- 16.Yongye AB, Li YM, Giulianotti MA, Yu YP, Houghten RA, Martinez-Mayorga K. J Comput-Aided Mol Des. 2009;23:677. doi: 10.1007/s10822-009-9295-y. ISI Document Delivery No.: 486PE Times Cited: 1 Cited Reference Count: 41 Yongye, Austin B. Li, Yangmei Giulianotti, Marc A. Yu, Yongping Houghten, Richard A. Martinez-Mayorga, Karina SPRINGER. [DOI] [PubMed] [Google Scholar]
- 17.Tyndall JD, Pfieiffer B, Abbenante G, Fairlie DP. Chem Rev. 2005;105:793. doi: 10.1021/cr040689g. [DOI] [PubMed] [Google Scholar]
- 18.Viles JH, Mitchell JB, LGS, Doyle PM, Harris CJ, Sadler PJ, Thornton JM. Eur J Biochem. 1996;242:352. doi: 10.1111/j.1432-1033.1996.0352r.x. [DOI] [PubMed] [Google Scholar]
- 19.Vasko RC, Rodriguez RA, Cunningham CN, Ardi VC, Agard DA, Mcalpine SR. ACS Med Chem Lett. 2010 doi: 10.1021/ml900003t. published 1/4/2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnelly A, Blagg BSJ. Curr Med Chem. 2008;15:2702. doi: 10.2174/092986708786242895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messaoudi S, Peyrat JF, Brion JD, Alami M. Anti-cancer agents in Med Chem. 2008;8:761. doi: 10.2174/187152008785914824. [DOI] [PubMed] [Google Scholar]
- 22.Gava LM, Ramos CHI. Current Chem Bio. 2009;3:330. [Google Scholar]
- 23.Johnson VA, Singh EK, Nazarova LA, Alexander LD, Mcalpine SR. Current Topics in Medicinal Chemistry. 2010 doi: 10.2174/156802610792232088. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis LM. C and E News. 2006 [Google Scholar]
- 25.Marx V. C and E News. 2005;83:17 . http://pubs.acs.org/cen/business/83/i11/8311bus1.html.
- 26.Danho W, Swistok J, Khan W, Chu X, Cheung A, Fry D, Sun H, Kurylko G, Rumennik L, Cefalu J, Cefalu G, Nunn P. In: Peptides for youth: The proceedings of the 20th american peptide symposium. Susan Del Valle EEaWDL., editor. Vol. 611. Springer; New York: 2009. p. 467. [DOI] [PubMed] [Google Scholar]
- 27.Loffet A. European Peptide Society. 2002;8:1. doi: 10.1002/psc.366. [DOI] [PubMed] [Google Scholar]
- 28.Starzl TE, Klintmalm GB, Porter KA, Iwatsuki S, Schroter GP. New England Journal of Medicine. 1981;305:266. doi: 10.1056/NEJM198107303050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faivre S, Chieze S, Delbaldo C, Ady-Vago N, Guzman C, Lopez-Lazaro L, Lozahic S, Jimeno J, Pico F, Armand JP, Martin JA, Raymond E. J Clin Oncol. 2005;23:7871. doi: 10.1200/JCO.2005.09.357. [DOI] [PubMed] [Google Scholar]
- 30.Maroun JA, Belanger K, Seymour L, Matthews S, Roach J, Dionne J, Soulieres D, Stewart D, Goel R, Charpentier D, Goss G, Tomiak E, Yau J, Jeimeno J, Chiritescu G. Ann Oncol. 2006;17:1371. doi: 10.1093/annonc/mdl165. [DOI] [PubMed] [Google Scholar]
- 31.Le Tourneau C, Raymond E, Faivre S. Curr Pharm Des. 2007;13:3427. [PubMed] [Google Scholar]
- 32.Corey EJ, Czako B, Kurti L. Molecules and medicine. Hoboken, NJ: John Wiley and Sons; 2007. [Google Scholar]
- 33.Neckers L. Trends Mol Med. 2002;8:S55. doi: 10.1016/s1471-4914(02)02316-x. [DOI] [PubMed] [Google Scholar]
- 34.Neckers L, PIS Curr Opin Oncol. 2003;15:419. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Jolly C, Morimoto RI. J Natl Cancer Inst. 2000;92:1564. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- 36.Sarto C, Binz PA, Mocarelli P. Electrophoresis. 2000;21:1218. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1218::AID-ELPS1218>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 37.Chiosis G, Jhuezo H, Rosen N, Mimgaugh E, Whitesell L, Neckers L. Mol Cancer Ther. 2003;2:123. [PubMed] [Google Scholar]
- 38.Ehrenfried JA, Herron BE, Townsend CM, Evers BM. Surgical oncology. 1995;4:197. doi: 10.1016/s0960-7404(10)80036-2. [DOI] [PubMed] [Google Scholar]
- 39.Hollingshead MG, Alley M, Burger AM, Borgel S, Pacula-Cox C, Fiebig HH, Sausville EA. Cancer Chemother Pharmacol. 2005;56:115. doi: 10.1007/s00280-004-0939-2. [DOI] [PubMed] [Google Scholar]
- 40.Senju M, Sueoka N, Sato A, Iwanaga K, Sakao Y, Tomimitsu S, Tominaga M, Irie K, Hayashi S, Sueoka E. J Cancer Res Clin Oncol. 2006;132:150. doi: 10.1007/s00432-005-0047-7. [DOI] [PubMed] [Google Scholar]
- 41.Chang YS, Lee LC, Sun FC, Chao CC, Fu HW, Lai YK. J Cell Biochem. 2006;97:156. doi: 10.1002/jcb.20623. [DOI] [PubMed] [Google Scholar]
- 42.Matei D, Satpathy M, Cao L, Lai YK, Nakshatri H, Donner DB. J Biol Chem. 2007;282:445. doi: 10.1074/jbc.M607012200. [DOI] [PubMed] [Google Scholar]
- 43.Orosz A, Szabo A, Szeman G, Janaky T, Somlai C, Penke B, Bodor A, Perczel A. Inter J Biochem Cell Biol. 2006;38:1352. doi: 10.1016/j.biocel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Ju W, Renouard J, Aden J, Belinsky SA, Lin Y. Cancer Res. 2006;66:1089. doi: 10.1158/0008-5472.CAN-05-2698. [DOI] [PubMed] [Google Scholar]
- 45.Shu CW, Cheng NL, Chang WM, Tseng TL, Lai YK. J Cell Biochem. 2005;94:1199. doi: 10.1002/jcb.20348. [DOI] [PubMed] [Google Scholar]
- 46.Bull EEA, Khideaki D, Brady KJ, Burgan WE, Carter DJ, Cerra MA, Oswald KA, Hollingshead MG, Camphausen K, Tofilon PJ. Clinical Cancer Research. 2004;10:8077. doi: 10.1158/1078-0432.CCR-04-1212. [DOI] [PubMed] [Google Scholar]
- 47.Shen G, Blagg BSJ. Org Lett. 2005;7:2157. doi: 10.1021/ol050580a. [DOI] [PubMed] [Google Scholar]
- 48.Bai J, Sui J, Demirjian A, Vollmer CM, Marasco W, Gcaller MP. Cancer Res. 2005;65:2344. doi: 10.1158/0008-5472.CAN-04-3502. [DOI] [PubMed] [Google Scholar]
- 49.Masao O, Zenya N, Chigeo T, Yuhkichi M, Goro A. J Nippon Medical School. 2000;67:177. [Google Scholar]
- 50.Dote H, Cerna D, Burgan WE, Camphausen K, Tofilon PJ. Cancer Res. 2005;65:6967. doi: 10.1158/0008-5472.CAN-05-1304. [DOI] [PubMed] [Google Scholar]
- 51.Maruya M, Sameshima M, Nemoto T, Yahara I. J Mol Bio. 1999;285:903. doi: 10.1006/jmbi.1998.2349. [DOI] [PubMed] [Google Scholar]
- 52.Nemoto T, Ohara-Nemoto Y, Ota M, Takagi T, Yokoyama K. Eur J Biochem. 1995;233:1. doi: 10.1111/j.1432-1033.1995.001_1.x. [DOI] [PubMed] [Google Scholar]
- 53.Fang L, Ricketson D, Getubig L, Darimont B. Proc Natl Acad Sci. 2006;103:18487. doi: 10.1073/pnas.0609163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banerji U. Proc Am Assoc Cancer Res. 2003;44:677. Hsp90. [Google Scholar]
- 55.Sausville EA. Curr Cancer Drug Targets. 2003;3:377. doi: 10.2174/1568009033481831. Hsp90. [DOI] [PubMed] [Google Scholar]
- 56.Dehner A, Furrer J, Richter K, Schuster I, Buchner J, Kessler H. Chem Bio Chem. 2003;4:870. doi: 10.1002/cbic.200300658. [DOI] [PubMed] [Google Scholar]
- 57.Hagn FX, Richter K, Buchner J, Kessler H. Proc. experimental nuclear magnetic resonance conference; 2005. [Google Scholar]
- 58.Jez JM, Chen JC, Rastelli G, Stroud RM, Santi DV. Chemistry and Biology. 2003;10:361. doi: 10.1016/s1074-5521(03)00075-9. [DOI] [PubMed] [Google Scholar]
- 59.Whitesell L, Mimnaugh EG, De Costa B, Meyers CE, Neckers LM. Proc Natl Acad Sci U S A. 1994;91:8324. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sydor JR, Normant E, Pien CS, Porter JR, Ge J, Grenier L, Pak RH, Ali JA, Dembski MS, Hudak J, Patterson J, Penders C, Pink M, Read MA, Sang J, Woodward C, Zhang Y, Grayzel DS, Wright J, Barrett JA, Palombella VJ, Adams J, Tong JK. Proc Natl Acad Sci U S A. 2006;103:17408. doi: 10.1073/pnas.0608372103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otrubova K, Lushington GH, Vander Velde D, Mcguire KL, Mcalpine SR. J Med Chem. 2008;51:530. doi: 10.1021/jm070731a. [DOI] [PubMed] [Google Scholar]
- 62.Otrubova K, Mcguire KL, Mcalpine SR. J Med Chem. 2007;50:1999. doi: 10.1021/jm070088s. [DOI] [PubMed] [Google Scholar]
- 63.Boland CR, Sinicrope FA, Brenner DE, Carethers JM. Gastroenterology. 2000;118:S115. doi: 10.1016/s0016-5085(00)70010-2. [DOI] [PubMed] [Google Scholar]
- 64.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Gastroenterology. 1999;117:123. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez RA, Pan PS, Vasko RC, Pan CM, Disman W, Mcalpine SR. J Mex Chem Soc, V. 2008:201–211. [Google Scholar]; J Mex Chem Soc. 2008;52:201. [Google Scholar]
- 66.Styers TJ, Rodriguez RA, Pan PS, Mcalpine SR. Tetrahedron Lett. 2006;47:515. [Google Scholar]
- 67.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Dev Rev. 1997;23:3. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 68.Citri A, Gan J, Mosesson Y, Vereb G, Szollosi J, Yarden Y. Embo Rep. 2004:1165–70. doi: 10.1038/sj.embor.7400300. [DOI] [PMC free article] [PubMed] [Google Scholar]; EMBO J. 2004;5:1165. [Google Scholar]
- 69.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y, Neckers L. J Biol Chem. 2001;276:3702. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 70.Chakraborty A, Koldobskiy MA, Sixt KM, Juluri K, Mustafa AK, Snowman AM, Van Rossum DB, Patterson RL, Snyder SH. Proc Natl Acad Sci. 2008;105:1134. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J Comput Chem. 1998;19:1639. autodock. [Google Scholar]
- 72.Hetenyi C, Van Der Spoel D. Protein science. 2002;11:1729. doi: 10.1110/ps.0202302. autodock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hetenyi C, Van Der Spoel D. FEBS Letters. 2006;580:1447. doi: 10.1016/j.febslet.2006.01.074. autodock. [DOI] [PubMed] [Google Scholar]
- 74.Harris R, Goodsell DS, Olson AJ. Proteins. 2007;69:1506. doi: 10.1002/prot.21645. autodock. [DOI] [PubMed] [Google Scholar]
- 75.Li C, Xu L, Wolan DW, Wilson IA, Olson AJ. J Med Chem. 2004;47:6681. doi: 10.1021/jm049504o. [DOI] [PubMed] [Google Scholar]
- 76.Ali MMU, Roe SM, Vaughan CK, Meyer P, Panareto B, Piper PW, PCP, LH Nature. 2006;440:1013. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morris GM, Huey R, Olson AJ. Current Protocols in Bioinformatics. 2008;8:14. doi: 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
- 78.Grant BJ, Rodrigues AP, Elsawy KM, JAM, Caves LS. Bioinformatics. 2006;22:2695. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.