Abstract
Objective
This study’s aims were to map loci linked to self-rating of the effects of alcohol and to determine if there was overlap with loci previously mapped for other substance dependence phenotypes in an American Indian community at high risk for substance dependence.
Methods
Each participant gave a blood sample and completed a structured diagnostic interview using the Semi Structured Assessment for the Genetics of Alcoholism (SSAGA). Retrospective report of responses to alcohol during the FIRST FIVE TIMES they had ever drank alcohol was estimated from the Self-Rating of the Effects of Alcohol (SRE) questionnaire for each participant. Genotypes were determined for a panel of 791 micro-satellite polymorphisms in 381 members of multiplex families using SOLAR.
Results
Analyses of multipoint variance component LOD scores, for the FIRST FIVE TIMES phenotype, revealed two loci that had a LOD score greater than 3.0 on chromosomes 6 and 9. Additionally, 3 locations were identified with LOD scores above 2.0 on chromosomes 10, 12, 17.
Conclusion
These results corroborate the importance of regions on chromosome 6 and 9 highlighted in prior segregation studies in this and other populations for substance dependence-related phenotypes, as well as an area on chromosome 10 previously identified for the FIRST FIVE TIMES phenotype in the collaborative study on the genetics of alcoholism. These studies additionally lend further support the construct that the SRE may represent an important endophenotype associated with alcohol and other substance dependence.
Keywords: American Indians, alcohol dependence, linkage studies, SRE
Introduction
Individual sensitivity to alcohol has been demonstrated to be an inherited factor that affects the likelihood of drinking and mediates the disposition for developing alcoholism (Schuckit, 1994). Heterogeneity in the intensity of the reported effects of alcohol between individuals has been widely endorsed (Morean and Corbin, 2009). The degree to which an individual responds to a given dose of alcohol is designated their “level of response” (see Schuckit, 2009) The heritability of a person’s level of response, which reflects individual differences in sensitivity to the pharmacological effects of alcohol, has been estimated to be between 0.4 and 0.6 (Heath et al., 1999; Schuckit et al., 2001a). In general, people at higher genetic risk for alcoholism are less sensitive to the effects of alcohol and people at lower genetic risk for alcoholism are more sensitive. Support for this theory is provided by many, but not all, studies directly examining the reaction to alcohol among children of alcoholics, who are at greatly elevated risk for developing alcoholism (Schuckit, 1984). Results have indicated that at moderate doses of alcohol, subjects who are family history positive for alcoholism and subjects who are family history negative for alcoholism attain equivalent blood alcohol concentrations, but most studies have found that subjects with a positive family history rate themselves as significantly less intoxicated than control subjects with a negative family history (Schuckit, 1984; O’Malley and Maisto, 1985; Savoie et al., 1988; Moss et al., 1989). Although not all studies agree (Newlin and Thomson, 1990), a meta-analyses focusing on subjective level of intoxication confirmed a diminished response to alcohol as a characteristic more frequently seen in subjects with a positive family history than in those with a negative family history (Pollock, 1992, Morean and Corbin, 2009). In addition, an 8-year follow-up of previously studied men with positive and negative family histories found that both a family history of alcoholism and a low response to alcohol were related to the development of alcohol-related problems (Schuckit and Smith, 1996).
Studies using similar alcohol challenge methodologies among groups at lower risk for alcoholism have provided additional support for the idea that individual sensitivity to alcohol might also mediate protection from developing alcoholism. Individuals of Asian heritage, who have mutations in the aldehyde dehydrogenase gene (ALDH2) (Wall et al., 1992), and individuals of Jewish decent (Monteiro et al., 1991), two groups with low rates of alcoholism, were found to have more intense, although not necessarily more negative, self reported responses to alcohol than matched control subjects of average alcoholism risk.
The estimated variation in response to a standard modest dose of alcohol has also been captured in larger more epidemiologically based studies using an instrument called the Self-Rating of the Effects of Alcohol (SRE) measure. This instrument asks study participants to retrospectively estimate the number of standard drinks containing ethanol that were required for that person to experience four potential effects of the drug: (1) feeling any effects, (2) feeling dizzy/trouble talking, (3) trouble thinking/walking, and (4) falling asleep when you didn’t want to. The instrument also queries an individual to rate these reactions during different time points in a person’s life including the first five times or so that they ever drank, during the most recent 3 months and during the period of heaviest alcohol intake (see Schuckit et al., 1997a,b). This measure is particularly suited to large populations where direct tests of responses to alcohol drinking may not be feasible due to logistical factors, underage status, or ineligibility due to a diagnosis of alcohol use or other medical disorder. The relationship between the SRE and results from direct alcohol challenge has also been investigated. In that study the average number of drinks endorsed on the SRE correlated significantly with a participants rating of their level of intoxication following drinking and correctly identified 79% of the participants whose level of response to alcohol fell into the lowest third of intensity during the alcohol challenge (Schuckit et al., 1996). The SRE has been utilized to estimate level of response to alcohol in a number of diverse populations including: middle age women (Schuckit et al., 2003), sons of alcoholics and matched controls (Schuckit et al., 1997b), participants in the collaborative study on the genetics of alcoholism (COGA) (Schuckit et al., 1997a), a military population (Schuckit et al., 2001b), and in African (Pedersen and McCarthy, 2009) and Asian Americans (Wall et al., 1999).
Genetic studies of complex phenotypes, such as level of response to alcohol, often have advantages when they are conducted in well-defined populations such as Native American tribes living on reservations (Lander and Schork, 1994). A once popular notion, called the firewater myth, proposed based on anecdotal observations, that Native American Indians are more sensitive to the effects of alcohol and thus get drunk more easily (see Leland, 1976). In one empirical study, Native American Mission Indians, like Caucasian sons of alcoholics, were actually found to have less intense objective and subjective effects of alcohol in an alcohol challenge paradigm. Additionally, participants with at least 50% Native American heritage reported less intense effects of alcohol than did those with less than 50% Native American heritage, despite equivalent blood alcohol concentrations (Garcia-Andrade et al., 1996, 1997; Ehlers et al., 1998, 1999). Therefore this population may be an important one for the study of the genetics of level of response to alcohol as a potential risk factor for the development of alcohol dependence.
The present report is part of a larger study exploring risk factors for substance dependence among Native American Indians (see Ehlers et al., 2001a,b,c,d; 2004a, 2008c; Gilder et al., 2004, 2006, 2007, 2009). The lifetime prevalence of substance dependence in this Indian population is high and evidence for heritability and linkage to specific chromosome locations and associations with candidate genes has been demonstrated (see Wall et al., 2003; Ehlers et al., 2004b, 2006b, 2007a,b,c, 2008a,b, 2009a,b, 2010a; Ehlers and Wilhelmsen, 2005, 2007; Wilhelmsen and Ehlers, 2005). The current study’s aims were to: (1) map loci linked to SRE phenotypes and (2) to determine if there was overlap of the loci identified for SRE phenotypes and loci previously mapped for alcohol and other substance dependence in this American Indian community.
Methods
Participants were recruited from eight geographically contiguous reservations, with a total population of about 3,000 individuals, using a combination of a venue-based method for sampling hard-to-reach populations (Kalton and Anderson, 1986; Muhib et al., 2001), as well as a respondent-driven procedure (Heckathorn, 1997) as previously described (Ehlers et al., 2004a; Gilder et al., 2004). The venues for recruitment included: tribal halls and culture centers, health clinics, tribal libraries, and stores on the reservations. A 10–25% rate of refusal was found depending on venue. Refusal rates were higher at tribal libraries and stores than health clinics and tribal halls/culture centers. Transportation from their home to The Scripps Research Institute was provided by the study.
To be included in the study, participants had to be an Indian indigenous to the catchment area, at least 1/16th Native American Heritage (NAH), between the age of 18 and 70 years, and be mobile enough to be transported from his or her home to The Scripps Research Institute (TSRI). The protocol for the study was approved by the Institutional Review Board (IRB) of TSRI, and the Indian Health Council, a tribal review group overseeing health issues for the reservations where recruitment was undertaken.
Potential participants first met individually with research staff to have the study explained and give written informed consent. During a screening period, participants had blood pressure and pulse taken, and completed a questionnaire that was used to gather information on demographics, personal medical history, ethnicity, and drinking history (Schuckit, 1985). Participants were asked to refrain from alcohol and drug usage for 24 hours prior to the testing. No individuals with detectable breath alcohol levels were included in the study dataset (n=3). During the screening period, the study coordinator also noted whether the participant was agitated, tremulous, or diaphoretic and their data were eliminated from subsequent analyses. Each participant also completed an interview with the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) and the family history assessment module (FHAM) (Bucholz et al., 1994), which was used to make substance use disorder and psychiatric disorder diagnoses according to Diagnostic and Statistical Manual (DSM-III-R) (American Psychiatric Association, 1987) criteria in the probands and their family members (American Psychiatric Association, 1987). The SSAGA is a semi-structured, poly-diagnostic psychiatric interview that has undergone both reliability and validity testing (Bucholz et al., 1994; Hesselbrock et al., 1999). It has been used in another Native American sample (Hesselbrock et al., 2000, 2003). Personnel from the Collaborative Study on the Genetics of Alcoholism (COGA) trained all interviewers. The SSAGA interview includes retrospective lifetime assessments of alcohol use, abuse, and dependence. A research psychiatrist/addiction specialist made all best final diagnoses.
Level of response to alcohol was estimated using a retrospective report instrument the Self-Rating of the Effects of Alcohol (SRE) measure. This instrument asks study participants to retrospectively estimate the number of standard drinks containing ethanol (e.g. the equivalence of 12 oz. of beer, 4 oz. of wine, and 1.5 oz. of 80 proof beverage) that were required for that person to experience four potential effects of the drug: (1) feeling any effects, (2) feeling dizzy/trouble talking, (3) trouble thinking/walking, and (4) falling asleep when you didn’t want to. The instrument also queries an individual to rate these reactions during different time points in a persons life including: (1) the FIRST FIVE TIMES or so that they ever drank, (2) during the most recent 3 months and (3) during the period of heaviest alcohol intake (see Schuckit et al., 1997a,b). The score is generated by counting up the number of drinks reported in each time period (FIRST FIVE, last 3 months, heaviest period) and dividing this score by the number of effects (any, dizzy, trouble thinking, falling asleep) endorsed. If a subject did not experience an effect during that time frame then that item is not counted. The analyses here focused on the number of drinks reported for the FIRST FIVE TIMES measure for the 4 effects. Selecting this measure increases the probability that participants refer more uniformly to a similar period in their lives and is most likely to result in a better estimate of “innate” tolerance rather than “acquired” tolerance. Additionally, it has been used previously in a linkage analysis utilizing the COGA dataset (see Schuckit et al., 2001a). Prior analyses have demonstrated that the relationship between SRE scores to alcohol challenge results remains statistically significant even after controlling for the number of effects of alcohol endorsed by the subjects (see Schuckit et al., 1997a,b).
One hundred and eighty-one pedigrees containing 1600 individuals were used in the genetic analyses. Sixty-six families have only a single individual with phenotype data. All these individuals were included within some analyses to the extent that they contribute information about trait means and variance and the impact of covariates. The family sizes for the remaining families ranged between 4 and 41 subjects (average 12.19 ± 8.19). Eighty-one families were genetically informative. The data includes 142 parent-child, 260 sibling, 53 half sibling, 11 grandparent-grandchild, 235 avuncular, and 240 cousin relative pairs. Only sibling, half-sibling, avuncular and cousin pairs were included as being potentially genetically informative. Several pedigrees contained large numbers of individuals and/or complex loops that could not be analyzed due to the high computational demands required. These pedigrees were thus broken using procedures originally described by Lange and Elston (1975), and treated as independent to allow for their inclusion in the linkage analysis.
DNA was isolated from whole blood using an automated DNA extraction procedure, genotyping was done as previously described (Wilhelmsen et al., 2003). Genotypes were determined for a panel of 791 autosomal microsatellite polymorphisms (Weber and May, 1989) using fluorescently labeled PCR primers under conditions recommended by the manufacturer (HD5 version 2.0; Applied Biosystems, Foster City, CA). The HD5 panel set has an average marker-to-marker distance of 4.6 cM, and an average heterozygosity of greater than 77% in a Caucasian population. Allele frequencies observed in the unrelated founders were used for linkage analysis. Gender and age accounted for greater than 5% of the phenotypic variance for the phenotype. Therefore, age and gender were included as covariates in the analyses.
Genotypes were determined for 410 subjects. The PREST software program, which assesses degree of allele sharing among relative-pairs, was used to identify potential errors in pedigree structure (McPeek and Sun, 2000). Six individuals were identified as problematic and removed from further analyses. Pedcheck was then used to detect non-Mendelian inheritance patterns (O’Connell and Weeks, 1998). When a Mendelian inconsistency was observed, genotypes for the nuclear family at that polymorphism were removed. This resulted in the removal of 772 genotypes. To further reduce errors, the maximum-likelihood error-checking algorithm implemented in Merlin (Abecasis et al., 2002) was used to identify genotypes that had a probability of less than 0.025 of being correct. A total of 508 genotypes were removed in this step. Ultimately 273,598 genotypes were accepted.
For linkage analysis, a variance components approach was used to calculate multipoint LOD scores at 1 cM intervals across the genome using SOLAR v2.0.4 (Almasy and Blangero, 1998; S.F.B.R, 2009). Variance components linkage analysis assumes that phenotypes are normally distributed, and violations of this assumption can result in inflated LOD scores. To protect against this possibility, simulations were conducted in which a single genetic locus was simulated under the null hypothesis of no linkage across 50,000 trials to derive empirical p-values. These p-values were used to determine the significance of the reported LOD scores (Blangero et al., 2000).
Results
Three hundred eighty-one participants out of a larger population of 620 had completed SRE questionnaires and had genotyping data that were available for these analyses. Three hundred seventy-nine (61%) participants met criteria for Alcohol dependence. Demographics of this sample are presented in Table 1. There were no significant differences in the demographics between the participants with SRE data and genotyping available (e.g. the linkage sample, n=381) and the entire sample of participants in the study with valid SSAGA data (n=620) but no genotyping and/or SRE data.
Table 1.
Demographics
| Linkage Sample (n = 381) | Entire Sample (n = 620) | |
|---|---|---|
| Gender | Male = 149 Female = 232 |
Male = 251 Female = 369 |
| Married (n) | 81 | 110 |
| Employed (n) | 177 | 253 |
| Income ≥ $20,000 yr. (n) | 182 | 317 |
| Native American Heritage, n ≥ 50% | 157 | 257 |
| Age (yrs) | 30.1 ± 0.6 | 30.9 ± 0.5 |
| Education (yrs) | 11.6 ± 0.1 | 11.6 ± 0.1 |
| First Five SRE score (mean ± SE) | 5.8 ± 0.3 | 5.7 ± 0.2 |
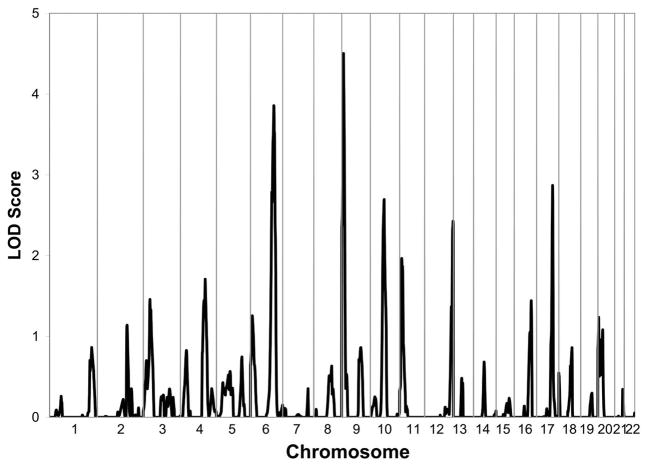
Analyses of multipoint variance component LOD scores, for the “FIRST FIVE” SRE phenotype, revealed two loci that had a LOD score greater than 3.0 on chromosomes 6 (LOD= 3.86) and 9 (LOD= 4.5). Figure 1 presents the linkage peaks generated by these analyses across the genome. Three additional locations were identified with LOD scores above 2.0 on chromosomes 10 (LOD=2.7), 12 (LOD=2.43), and 17 (LOD=2.87) for the “FIRST FIVE” SRE phenotype that are seen in Figure 2. Table 2 presents the peak LOD scores, the closest marker location for the loci identified, empirical p values, and additionally gives information of other findings in the literature for substance-related phenotypes observed at or near those locations.
Figure 1.
Multipoint Linkage Analysis for the SRE FIRST FIVE phenotype for the entire genome. Results for each chromosome are aligned end to end with the p terminus on the left. Log of the Odds (LOD) score is plotted on the Y-axis. Vertical lines indicate the boundaries between the chromosomes. The numbers above on the X-axis indicate the chromosome number.
Figure 2.
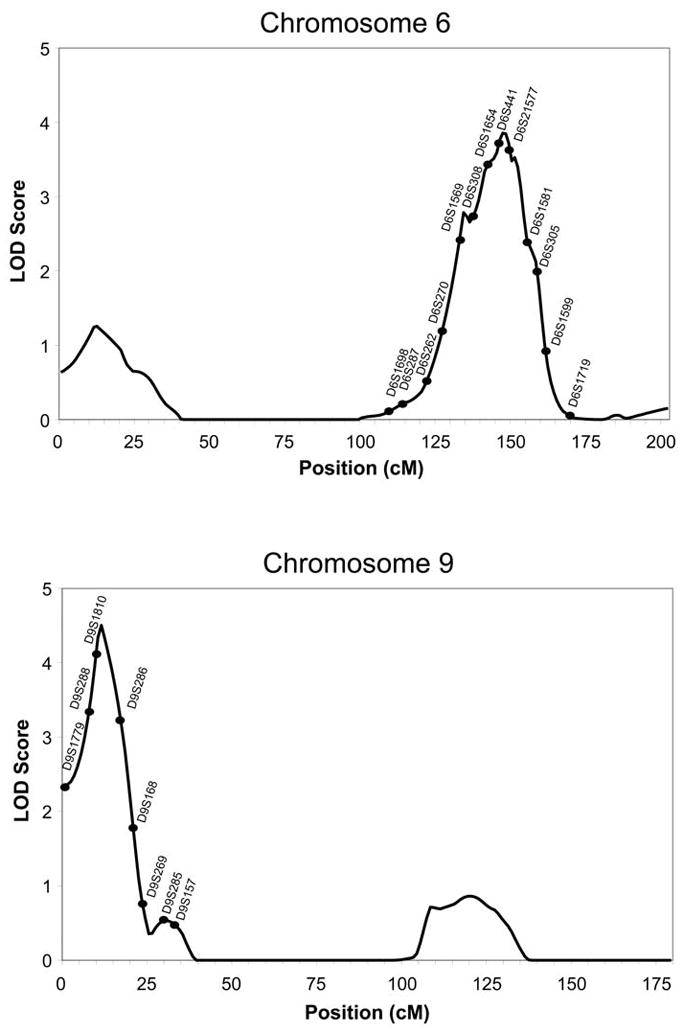
Multipoint Linkage Analysis for the SRE FIRST FIVE phenotype for chromosomes 6 and 9. Log of the Odds (LOD) score (Y-axis) is plotted for the chromosome location map (in centimorgans (cM), X-axis). Nearest markers to the peak are presented within the support interval.
Table 2.
Genetic Loci for SRE traits in an American Indian Community
| CHR | Trait | LOC (cM) | LOD | Nearest Marker | Empirical p-value | Supporting References (phenotype) |
|---|---|---|---|---|---|---|
| 6 | First Five | 147 | 3.86 | D6S441 | .00074 | Ehlers and Wilhelmsen, 2007 (any drug dependence) |
| 9 | First Five | 11 | 4.50 | D9S1810 | .00030 | Long et al., 1998; Ehlers et al., 2009b (cannabis craving); Wang et al., 2005; Li et al., 2008 (smoking phenotypes) |
| 10 | First Five | 87 | 2.70 | D10S581/D10S210 | .00372 | Agrawal et al., 2008; Gelernter et al., 2009 (alcohol dependence), Schuckit et al., 2001a (FIRST FIVE SRE), Wilhelmsen et al., 2003; Schuckit et al., 2005 (response to alcohol challenge), Li et al., 2006 (smoking related phenotype) |
| 11 | First Five | 13 | 1.97 | D11S1760 | .01012 | Long et al., 1998 (alcohol dependence), Ehlers et al., 2010b (EEG alpha), Gelernter et al., 2006 (opioid dependence); Loukola et al., 2008; Li et al., 2008; Wang et al., 2005 (smoking phenotypes) |
| 12 | First Five | 179 | 2.43 | D12S1638 | .00528 | Ehlers et al., 2004b (alcohol drinking/dependence symptoms), Ehlers et al., 2008a (ASPD/CD), Kuo et al., 2006 (maximum drinking) |
| 17 | First Five | 101 | 2.87 | D17S1807 | .00304 | Ehlers et al., 2008a (ASPD/CD) |
Discussion
The CNS effects of alcohol range from mild euphoria (high), to impaired coordination, to ataxia, decreased mentation, labile mood, to poor judgment, slurred speech, nausea and vomiting, and finally to respiratory failure, coma and death, depending on the dose imbibed (see Schuckit, 1995). The final level of impairment appears to depend on a number of factors including a persons’ gender, age, weight, prior experience with alcohol and level of tolerance (Schuckit, 1994). Another source of variation in response to alcohol is individual variation in alcohol metabolism (see Wall et al., 1992, 1997, 2005; Wall and Ehlers, 1995; Ehlers et al., 2003; Duranceaux et al., 2006). Other sources of the genetic variation in sensitivity and tolerance to alcohol not attributed to differences in alcohol metabolism are less well understood. Several studies have found moderate heritability for level of response to alcohol. In one study, heritability was found to be 60% for a composite sensitivity measure that was used during an alcohol challenge in twins (Heath et al., 1999). Correlations of level of response to alcohol using body sway and the Subjective High Assessment Scale (SHAS) in an alcohol challenge paradigm using sibling pairs as participants was reported to be 0.36 (Wilhelmsen et al., 2003). Lower correlations were found in first-degree relatives using a retrospective self-report measure to assess level of response to alcohol (0.12–0.22).
Further evidence for a genetic component to level of response to alcohol was provided by a genome-wide segregation analysis that evaluated subjective response to alcohol challenge in sibpairs. In that study, nine chromosome regions were identified with LOD scores between 2.2 and 3.2 suggesting potential regions of interest in the genome that may contribute to the variance in alcohol responsivity (Wilhelmsen et al., 2003). An expanded dataset, collected in the same laboratory, also identified 5 areas of the genome with LOD scores between 2.2 and 2.6 for level of response to alcohol in sibpairs (Schuckit et al., 2005).
Six areas of the genome provided LOD scores suggesting evidence for linkage to the FIRST FIVE SRE phenotype in this Indian population. One location was on chromosome 6 at 147 cM that had a LOD score of 3.86. A previous study in this Indian population also found suggestive evidence for linkage on chromosome 6q24-25 for “any drug dependence/regular tobacco use” as well as Body Mass Index suggesting genes in that location may be associated with risk for substance dependence and other consumption-related phenotypes (Ehlers and Wilhelmsen, 2007). Additionally, this locus on chromosome 6 is centered only 2 cM from the location of the mu opioid receptor gene OPRM1. In an association study in this Indian population several SNPs in OPRM1 were found to be significantly associated with a level of response to alcohol phenotype (Ehlers et al., 2008b). Endorsing the expectation of a more intense response to alcohol after imbibing 2–3 drinks was significantly associated with having at least one minor allele for at least one of 7 SNPs (p<0.01) in the OPRM1 receptor gene.
A second area of the genome that was identified in the present study for the SRE FIRST FIVE phenotype was on chromosome 9 @11 cM (LOD score 4.5). This site is near a location (35 cM) that has been reported previously to be linked to cannabis craving (Ehlers et al., 2009b) in the San Francisco Family Alcoholism study. Additionally, a location near this area on chromosome 9 was identified in a South West Indian population for an alcohol dependence phenotype (Long et al., 1998). This site has also been identified for smoking phenotypes (Wang et al., 2005; Li et al., 2008).
A locus was identified on chromosome 10 at 87 cM with a LOD score of 2.7 that has also been identified previously in several genome scans for a number of substance abuse related traits. For instance this area was found to be linked (LOD= 4.17) to smoking phenotypes in the Midsouth Tobacco Family Study in African Americans (Li et al., 2006), as well as to alcohol dependence in the COGA sample (Agrawal et al., 2008) and in a dense genomewide linkage scan for alcohol dependence in African Americans (Gelernter et al., 2009). Interestingly, this site appears to be within 30cM of an area identified in two studies investigating the low response to alcohol as assessed using an alcohol challenge study (Wilhelmsen et al., 2003; Schuckit et al., 2005) as well as a study of COGA participants using the SRE FIRST FIVE phenotype (Schuckit et al., 2001a). It has been suggested by Schuckit et al., (2005) that one potentially promising gene in the support region on chromosome 10 is KCNMAI, which has an important role in the effects of alcohol on potassium channels (Davies et al., 2003). The concordance between studies in identifying several loci in the genome that are associated with substance dependence phenotypes and the SRE further suggests that the search for candidate genes within these locations may be productive in identifying some general mechanisms that may underlie variation in a number of phenotypes.
Another area that was identified on chromosome 11 at 13 cM with a LOD score of 1.97 for the FIRST FIVE SRE phenotype was also within 17 cM of a site identified previously for an EEG alpha power phenotype in this Indian population at @30 cM with a LOD score of 2.98. This site also appears to be close to a site identified in a genome scan for opioid dependence in a sample of small nuclear families recruited from 4 populations of participants (Gelernter et al., 2006), and also overlaps with a site identified for alcohol dependence in a Southwest Indian tribe (Long et al., 1998) as well as several smoking phenotypes (Wang et al., 2005; Li et al., 2008; Loukola et al., 2008).
Additionally, the area on chromosome 12 that was identified in the present linkage analyses for the “FIRST FIVE” SRE phenotype was also found previously in a genome scan for a the severity of alcohol dependence drinking symptomatology phenotype (Ehlers et al., 2004b) and also an antisocial personality disorder/conduct disorder phenotype in this Indian population (Ehlers et al., 2008a). A region near this site was also identified in a genome scan in the Irish Affected Sib Pair Study for a maximum drinking phenotype (Kuo et al., 2006).
Finally, on chromosome 17 a location was identified at 101cM with a LOD score of 2.87 that was also found in this Indian population for an ASPD/CD phenotype (Ehlers et al., 2008a). Thus it appears that areas on chromosomes 6,9,10,11,12,17 may harbor genes for both the SRE FIRST FIVE phenotype as well as a number of substance related traits observed in this Indian population as well as in other population samples.
In conclusion, these data represent the first linkage analysis of a level of response to alcohol phenotype in American Indians. The results suggest that several areas of the genome may harbor genes that modulate an individual’s level of response to alcohol. Loci highlighted in prior studies in this population as well as other populations for substance dependence phenotypes were identified including a site on chromosome 10 previously identified for this FIRST FIVE SRE phenotype in the COGA study. The results of this study should, however, be interpreted in the context of several limitations. Level of response to alcohol was evaluated using the SRE, and a more direct measure of intoxication using and alcohol challenge protocol combined with objective and subjective measures of intoxication may produce more reliable results. Additionally, the findings may not generalize to other Native Americans or represent all Indians of the tribes studied, and comparisons of linkage findings to non-Indian populations may be limited by differences in a host of potential genetic and environmental variables. Despite these limitations, this report represents an important step in an ongoing investigation to understand the genetic determinants associated with the development of substance use disorders in this high risk and understudied ethnic group.
Acknowledgments
This research was supported by a grant from the National Institutes of Health (NIH) from the National Institute on Alcoholism and Alcohol Abuse grant (NIAAA) and the National Center on Minority Health and Health Disparities (NCMHD) (5R37 AA010201) (CLE), T32 AA007573 (IRG) and by funds provided by the University of North Carolina (KCW). The authors wish to acknowledge the technical support of Heidi Feiler, Evie Phillips, Linda Corey, David Gilder, James Lee, Samantha Segal, Michelle Dixon, Lilach Harris, Gina Stouffer, Shirley Sanchez and Philip Lau.
Sources of Support: This research was supported by a grant from the National Institutes of Health (NIH) from the National Institute on Alcoholism and Alcohol Abuse grant (NIAAA) and the National Center on Minority Health and Health Disparities (NCMHD) (5R37 AA010201) (CLE), T32 AA007573 (IRG) and by funds provided by the University of North Carolina (KCW).
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, et al. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2008;93:12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnosis and statistical manual of mental disorders (DSM-III-R) Washington, DC: American Psychiatric Association; 1987. [Google Scholar]
- Blangero J, Williams JT, Almasy L. Robust LOD scores for variance component-based linkage analysis. Genet Epidemiol. 2000;19 (Suppl 1):S8–14. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, et al. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- Duranceaux NC, Schuckit MA, Eng MY, Robinson SK, Carr LG, Wall TL. Associations of variations in alcohol dehydrogenase genes with the level of response to alcohol in non-Asians. Alcohol Clin Exp Res. 2006;30:1470–1478. doi: 10.1111/j.1530-0277.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Garcia-Andrade C, Wall TL, Sobel DF, Phillips E. Determinants of P3 amplitude and response to alcohol in Native American Mission Indians. Neuropsychopharmacology. 1998;18:282–292. doi: 10.1016/S0893-133X(97)00160-7. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Garcia-Andrade C, Wall TL, Cloutier D, Phillips E. Electroencephalographic responses to alcohol challenge in Native American Mission Indians. Biol Psychiatry. 1999;45:776–787. doi: 10.1016/s0006-3223(98)00113-9. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Garcia-Andrade C, Phillips E. EEG asymmetry: relationship to mood and risk for alcoholism in Mission Indian youth. Biol Psychiatry. 2001a;50:129–136. doi: 10.1016/s0006-3223(01)01132-5. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Garcia-Andrade C, Phillips E. Visual P3 findings in Mission Indian youth: relationship to family history of alcohol dependence and behavioral problems. Psychiatry Res. 2001b;105:67–78. doi: 10.1016/s0165-1781(01)00313-4. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Garcia-Andrade C, Phillips E. Effects of age and parental history of alcoholism on EEG findings in mission Indian children and adolescents. Alcohol Clin Exp Res. 2001c;25:672–679. [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Garcia-Andrade C, Phillips E. Auditory P3 findings in mission Indian youth. J Stud Alcohol. 2001d;62:562–570. doi: 10.15288/jsa.2001.62.562. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Carr L, Betancourt M, Montane-Jaime K. Association of the ADH2*3 allele with greater alcohol expectancies in African-American young adults. J Stud Alcohol. 2003;64:176–181. doi: 10.15288/jsa.2003.64.176. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Betancourt M, Gilder DA. The clinical course of alcoholism in 243 Mission Indians. Am J Psychiatry. 2004a;161:1204–1210. doi: 10.1176/appi.ajp.161.7.1204. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004b;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatr Genet. 2005;15:71–75. doi: 10.1097/00041444-200503000-00012. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Slutske WS, Gilder DA, Lau P, Wilhelmsen KC. Age at first intoxication and alcohol use disorders in Southwest California Indians. Alcohol Clin Exp Res. 2006;30:1856–1865. doi: 10.1111/j.1530-0277.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Slutske WS, Gilder DA, Lau P. Age of first marijuana use and the occurrence of marijuana use disorders in Southwest California Indians. Pharmacol Biochem Behav. 2007a;86:290–296. doi: 10.1016/j.pbb.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Phillips E, Finnerman G, Gilder D, Lau P, Criado J. P3 components and adolescent binge drinking in Southwest California Indians. Neurotoxicol Teratol. 2007b;29:153–163. doi: 10.1016/j.ntt.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Corey L, Lau P, Gilder DA, Wilhelmsen K. Heritability of illicit drug use and transition to dependence in Southwest California Indians. Psychiatr Genet. 2007c;17:171–176. doi: 10.1097/01.ypg.0000242201.56342.1a. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic screen for substance dependence and body mass index in Southwest California Indians. Genes Brain Behav. 2007;6:184–191. doi: 10.1111/j.1601-183X.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Slutske WS, Lind PA, Wilhelmsen KC. Externalizing disorders in American Indians: Comorbidity and a genome wide linkage analysis. Am J Med Genet B Neuropsychiatr Genet. 2008a;147B:690–698. doi: 10.1002/ajmg.b.30666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Phillips E. P3 components of the event-related potential and marijuana dependence in Southwest California Indians. Addict Biol. 2008c;13:130–142. doi: 10.1111/j.1369-1600.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Lind PA, Wilhelmsen KC. Association between single nucleotide polymorphisms in the mu opioid receptor gene (OPRM1) and self-reported responses to alcohol in American Indians. BMC Med Genet. 2008b;9:35. doi: 10.1186/1471-2350-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Heritability and a genome-wide linkage analysis of a Type II/B cluster construct for cannabis dependence in an American Indian community. Addict Biol. 2009a;14:338–348. doi: 10.1111/j.1369-1600.2009.00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Vieten C, Wilhelmsen KC. Linkage analyses of cannabis dependence, craving, and withdrawal in the San Francisco Family Study. Am J Med Genet. 2009b Nov 24; doi: 10.1002/ajmg.b.31050. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Phillips E, Gizer IR, Gilder DA, Wilhelmsen KC. EEG spectral phenotypes: Heritability and association with marijuana and alcohol dependence in an American Indian community study. Drug Alcohol Depend. 2010a;106:101–110. doi: 10.1016/j.drugalcdep.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Phillips E, Wilhelmsen KC. EEG alpha phenotypes: linkage analyses and relation to alcohol dependence in an American Indian community study. BMC Med Genet. 2010b doi: 10.1186/1471-2350-11-43. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Andrade C, Wall TL, Ehlers CL. Alcohol expectancies in a Native American population. Alcohol Clin Exp Res. 1996;20:1438–1442. doi: 10.1111/j.1530-0277.1996.tb01146.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Andrade C, Wall TL, Ehlers CL. The firewater myth and response to alcohol in Mission Indians. Am J Psychiatry. 1997;154:983–988. doi: 10.1176/ajp.154.7.983. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, et al. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry. 2009;65:111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilder DA, Wall TL, Ehlers CL. Co-morbidity of select anxiety and effective disorders in Southwest California Indians. Alcohol Clin Exp Res. 2004;28:1805–1813. doi: 10.1097/01.alc.0000148116.27875.b0. [DOI] [PubMed] [Google Scholar]
- Gilder DA, Lau P, Dixon M, Corey L, Phillips E, Ehlers CL. Co-morbidity of select anxiety, affective, and psychotic disorders with cannabis dependence in Southwest California Indians. J Addict Dis. 2006;25:67–79. doi: 10.1300/J069v25n04_07. [DOI] [PubMed] [Google Scholar]
- Gilder DA, Lau P, Corey L, Ehlers CL. Factors associated with remission from cannabis dependence in Southwest California Indians. J Addict Dis. 2007;26:23–30. doi: 10.1300/J069v26n04_04. [DOI] [PubMed] [Google Scholar]
- Gilder DA, Lau P, Ehlers CL. Item response theory analysis of age of first use of cannabis and lifetime cannabis use disorder criteria in an American Indian community sample. JSAD. 2009;70:839–849. doi: 10.15288/jsad.2009.70.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Heckathorn DD. Respondent-driven sampling: a new approach to the study of hidden populations. Soc Problems. 1997;44:174–199. [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Hesselbrock MN, Hesselbrock VM, Segal B, Schuckit MA, Bucholz K. Ethnicity and psychiatric comorbidity among alcohol-dependent persons who receive inpatient treatment: African Americans, Alaska Natives, Caucasians, and Hispanics. Alcohol Clin Exp Res. 2003;27:1368–1373. doi: 10.1097/01.ALC.0000080164.21934.F9. [DOI] [PubMed] [Google Scholar]
- Hesselbrock VM, Segal B, Hesselbrock MN. Alcohol dependence among Alaska Natives entering alcoholism treatment: a gender comparison. J Stud Alcohol. 2000;61:150–156. doi: 10.15288/jsa.2000.61.150. [DOI] [PubMed] [Google Scholar]
- Kalton G, Anderson DW. Sampling rare populations. J Royal Stat Society. 1986;149:65–82. [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, et al. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol Clin Exp Res. 2006;30:1807–1816. doi: 10.1111/j.1530-0277.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Lange K, Elston RC. Extensions to pedigree analysis. I. Likelihood calculations for simple and complex pedigrees. Hum Hered. 1975;25:95–105. doi: 10.1159/000152714. [DOI] [PubMed] [Google Scholar]
- Leland J. Firewater myths: North American Indian drinking and alcohol addiction. New Brunswick, N.J: Publications Division, Rutgers Center of Alcohol Studies; 1976. [Google Scholar]
- Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, et al. A genomewide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, et al. Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry. 2008;13:407–416. doi: 10.1038/sj.mp.4002038. [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Loukola A, Broms U, Maunu H, Widen E, Heikkila K, Siivola M, et al. Linkage of nicotine dependence and smoking behavior on 10q, 7q and 11p in twins with homogeneous genetic background. Pharmacogenomics J. 2008;8:209–219. doi: 10.1038/sj.tpj.6500464. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morean ME, Corbin WR. Subjective Response to Alcohol: A Critical Review of the Literature. Alcohol Clin Exp Res. 2009 Dec 17; doi: 10.1111/j.1530-0277.2009.01103.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Monteiro MG, Klein JL, Schuckit MA. High levels of sensitivity to alcohol in young adult Jewish men: a pilot study. J Stud Alcohol. 1991;52:464–469. doi: 10.15288/jsa.1991.52.464. [DOI] [PubMed] [Google Scholar]
- Moss HB, Yao JK, Maddock JM. Responses by sons of alcoholic fathers to alcoholic and placebo drinks: perceived mood, intoxication, and plasma prolactin. Alcohol Clin Exp Res. 1989;13:252–257. doi: 10.1111/j.1530-0277.1989.tb00322.x. [DOI] [PubMed] [Google Scholar]
- Muhib FB, Lin LS, Stueve A, Miller RL, Ford WL, Johnson WD, et al. A venue-based method for sampling hard-to-reach populations. Public Health Rep. 2001;116(Suppl 1):216–222. doi: 10.1093/phr/116.S1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlin DB, Thomson JB. Alcohol challenge with sons of alcoholics: a critical review and analysis. Psychol Bull. 1990;108:383–402. doi: 10.1037/0033-2909.108.3.383. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley SS, Maisto SA. Effects of family drinking history and expectancies on responses to alcohol in men. J Stud Alcohol. 1985;46:289–297. doi: 10.15288/jsa.1985.46.289. [DOI] [PubMed] [Google Scholar]
- Pedersen SL, McCarthy DM. An examination of subjective response to alcohol in African Americans. J Stud Alcohol Drugs. 2009;70:288–295. doi: 10.15288/jsad.2009.70.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock VE. Meta-analysis of subjective sensitivity to alcohol in sons of alcoholics. Am J Psychiatry. 1992;149:1534–1538. doi: 10.1176/ajp.149.11.1534. [DOI] [PubMed] [Google Scholar]
- S.F.B.R. [Last accessed on 2/18/10.];Sequential Oligogenic Linkage Analysis Routines. 2009 Available at: http://solar.sfbrgenetics.org/
- Savoie TM, Emory EK, Moody-Thomas S. Acute alcohol intoxication in socially drinking female and male offspring of alcoholic fathers. J Stud Alcohol. 1988;49:430–435. doi: 10.15288/jsa.1988.49.430. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Subjective responses to alcohol in sons of alcoholics and control subjects. Arch Gen Psychiatry. 1984;41:879–884. doi: 10.1001/archpsyc.1984.01790200061008. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Ethanol-induced changes in body sway in men at high alcoholism risk. Arch Gen Psychiatry. 1985;42:375–379. doi: 10.1001/archpsyc.1985.01790270065007. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. A clinical model of genetic influences in alcohol dependence. J Stud Alcohol. 1994;55:5–17. doi: 10.15288/jsa.1994.55.5. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Drug and alcohol abuse: a clinical guide to diagnosis and treatment. New York: Plenum Medical Book Co; 1995. [Google Scholar]
- Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Arch Gen Psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Tsuang JW, Anthenelli RM, Tipp JE, Nurnberger JI., Jr Alcohol challenges in young men from alcoholic pedigrees and control families: a report from the COGA project. J Stud Alcohol. 1996;57:368–377. doi: 10.15288/jsa.1996.57.368. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Tipp JE, Smith TL, Wiesbeck GA, Kalmijn J. The relationship between Self-Rating of the Effects of alcohol and alcohol challenge results in ninety-eight young men. J Stud Alcohol. 1997b;58:397–404. doi: 10.15288/jsa.1997.58.397. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Tipp JE. The Self-Rating of the Effects of alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction. 1997a;92:979–988. [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, et al. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001a;25:323–329. [PubMed] [Google Scholar]
- Schuckit MA, Kraft HS, Hurtado SL, Tschinkel SA, Minagawa R, Shaffer RA. A measure of the intensity of response to alcohol in a military population. Am J Drug Alcohol Abuse. 2001b;27:749–757. doi: 10.1081/ada-100107666. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Danko GP, Isacescu V. Level of response to alcohol measured on the self-rating of the effects of alcohol questionnaire in a group of 40-year-old women. Am J Drug Alcohol Abuse. 2003;29:191–201. doi: 10.1081/ada-120018846. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Wilhelmsen K, Smith TL, Feiler HS, Lind P, Lange LA, et al. Autosomal linkage analysis for the level of response to alcohol. Alcohol Clin Exp Res. 2005;29:1976–1982. doi: 10.1097/01.alc.0000187598.82921.27. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim R, Fukukura T, Allen R. The overlap in predicting alcohol outcome for two measures of the level of response to alcohol. Alcohol Clin Exp Res. 2009;33:563–569. doi: 10.1111/j.1530-0277.2008.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall TL, Thomasson HR, Schuckit MA, Ehlers CL. Subjective feelings of alcohol intoxication in Asians with genetic variations of ALDH2 alleles. Alcohol Clin Exp Res. 1992;16:991–995. doi: 10.1111/j.1530-0277.1992.tb01907.x. [DOI] [PubMed] [Google Scholar]
- Wall TL, Ehlers CL. Acute effects of alcohol on P300 in Asians with different ALDH2 genotypes. Alcohol Clin Exp Res. 1995;19:617–622. doi: 10.1111/j.1530-0277.1995.tb01557.x. [DOI] [PubMed] [Google Scholar]
- Wall TL, Garcia-Andrade C, Thomasson HR, Carr LG, Ehlers CL. Alcohol dehydrogenase polymorphisms in Native Americans: identification of the ADH2*3 allele. Alcohol Alcohol. 1997;32:129–132. doi: 10.1093/oxfordjournals.alcalc.a008246. [DOI] [PubMed] [Google Scholar]
- Wall TL, Johnson ML, Horn SM, Carr LG, Smith TL, Schuckit MA. Evaluation of the self-rating of the effects of alcohol form in Asian Americans with aldehyde dehydrogenase polymorphisms. J Stud Alcohol. 1999;60:784–789. doi: 10.15288/jsa.1999.60.784. [DOI] [PubMed] [Google Scholar]
- Wall TL, Carr LG, Ehlers CL. Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American Mission Indians. Am J Psychiatry. 2003;160:41–46. doi: 10.1176/appi.ajp.160.1.41. [DOI] [PubMed] [Google Scholar]
- Wall TL, Shea SH, Luczak SE, Cook TA, Carr LG. Genetic associations of alcohol dehydrogenase with alcohol use disorders and endophenotypes in white college students. J Abnorm Psychol. 2005;114:456–465. doi: 10.1037/0021-843X.114.3.456. [DOI] [PubMed] [Google Scholar]
- Wang D, Ma JZ, Li MD. Mapping and verification of susceptibility loci for smoking quantity using permutation linkage analysis. Pharmacogenomics J. 2005;5:166–172. doi: 10.1038/sj.tpj.6500304. [DOI] [PubMed] [Google Scholar]
- Weber JL, May PE. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, et al. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen KC, Ehlers C. Heritability of substance dependence in a Native American population. Psychiatr Genet. 2005;15:101–107. doi: 10.1097/00041444-200506000-00006. [DOI] [PubMed] [Google Scholar]