Abstract
Evidence has recently accumulated suggesting that small noncoding RNAs, and particularly microRNAs, have the potential to strongly affect the replication and pathogenic potential of a range of human virus species. Here, we report the use of deep sequencing to comprehensively analyze small viral RNAs (18 to 27 nucleotides [nt]) produced during infection by influenza A virus. Although influenza A virus differs from most other RNA viruses in that it replicates its genome in the nucleus and is therefore exposed to the nuclear microRNA processing factors Drosha and DGCR8, we did not observe any microRNAs encoded by influenza virus genes. However, influenza virus infection did induce the expression of very high levels—over 100,000 copies per cell by 8 h postinfection—of a population of 18- to 27-nt small viral leader RNAs (leRNAs) that originated from the precise 5′ ends of all eight influenza virus genomic RNA (vRNA) segments. Like the vRNAs themselves, our data indicate that the leRNAs also bear a 5′-terminal triphosphate and are therefore not capable of functioning as microRNAs. Instead, the high-level production of leRNAs may imply a role in another aspect of the viral life cycle, such as regulation of the switch from viral mRNA transcription to genomic RNA synthesis.
IMPORTANCE
Influenza A virus is an important human pathogen that has the potential to give rise to serious pandemics. Here, we demonstrate that influenza A virus induces the expression of very high levels of small viral leader RNAs (leRNAs) within hours of infection. These RNAs are unusual in that they bear a 5′ triphosphate and originate from the very 5′ ends of the eight viral genomic RNA (vRNA) segments. Their high expression may imply an important role in the viral life cycle that could potentially serve as a novel target for antiviral drugs.
Introduction
MicroRNAs (miRNAs) are small regulatory RNAs of ~22 nucleotides (nt) in length that are expressed by all multicellular eukaryotes (1). The large majority of miRNAs are initially transcribed as part of a long capped, polyadenylated transcript referred to as a primary miRNA (pri-miRNA) precursor, where they form part of one arm of an internal ~80-nt stem-loop (2). This stem-loop is recognized by the nuclear RNase III enzyme Drosha, acting in concert with its cofactor DGCR8, leading to cleavage of the pri-miRNA stem. This cleavage liberates an ~60-nt RNA hairpin bearing an ~2-nt 3′ overhang, called the pre-miRNA intermediate. After nuclear export, the pre-miRNA encounters the cytoplasmic RNase III enzyme Dicer, which removes the terminal loop of the pre-miRNA, leaving a second ~2-nt 3′ overhang. One strand of the resultant miRNA duplex intermediate is then incorporated into the RNA-induced silencing complex (RISC), where it acts as a guide RNA to direct RISC to mRNA species bearing a complementary sequence. Binding of RISC to mRNA target sequences results in inhibition of protein synthesis and some degree of destabilization (1). It should be noted that both Drosha and Dicer leave a 5′ monophosphate after RNA cleavage and that this 5′ phosphate is, in fact, critical for miRNA incorporation into RISC (1, 3, 4).
It has recently become increasingly clear that miRNAs can exert a major positive or negative effect on viral replication (5). In particular, it has now been demonstrated that a number of nuclear DNA viruses carry genes that encode miRNAs that appear likely to play an important role in promoting viral pathogenesis (6–13). In contrast, analysis of a range of RNA viruses has failed to identify any miRNAs encoded by viral genes (9, 14, 42). At least two reasons for the current lack of RNA virus-derived miRNAs are apparent. On the one hand, excision of a viral miRNA from the genome or antigenome of an RNA virus would result in their cleavage and degradation, which is likely to be disadvantageous. On the other hand, it is also true that almost all the RNA viruses examined thus far replicate in the cytoplasm, away from the nuclear Drosha/DGCR8 heterodimer that initiates miRNA processing. If this is the key consideration, then RNA viruses that replicate in the nucleus, unlike RNA viruses that replicate in the cytoplasm, might express one or more viral miRNAs in infected cells. In the case of the retroviruses HIV-1 and human T-cell leukemia virus type 1 (HTLV-1), we and others have presented data arguing that these nuclear RNA viruses do not, in fact, contain genes that encode any miRNAs, although this issue has remained controversial (9, 14–16).
Another prominent RNA virus family that replicates its genome in the nucleus is the Orthomyxoviridae and, in particular, influenza A virus. Influenza A virus is a segmented, negative-strand RNA virus, and its polymerase complex (composed of the PB2, PB1, and PA proteins) generates not only mRNAs but also viral genomic RNAs (vRNAs) and plus-sense intermediates in vRNA biogenesis, called cRNAs, during infection (17). All these RNAs are derived from the eight vRNA segments that enter the cell in the infecting virus particle and that then rapidly migrate into the nucleus. We were therefore curious to determine whether influenza A virus might contain genes that encode miRNAs within one or more of its genomic vRNA segments or in the plus-sense viral mRNAs or cRNAs.
Using deep sequencing, we demonstrate that influenza virus does not express any viral miRNAs but instead generates high levels of ~18- to 27-nt small viral leader RNAs (leRNAs) that originate from the very 5′ end of each of the eight vRNA segments. These leRNAs accumulate to high levels in influenza virus-infected cells, reaching >100,000 copies per cell by 8 h postinfection (hpi), a level substantially higher than the total number of cellular miRNAs. Moreover, leRNAs differ from miRNAs both in their relatively heterogeneous lengths and, more importantly, in the fact that they retain a 5′ triphosphate, which precludes their incorporation into RISC (1, 3, 4). The high level of expression of the leRNAs is consistent with their playing an important role in the influenza virus life cycle.
RESULTS
To determine whether influenza A virus expresses miRNAs or other small viral RNAs in infected cells, we infected the highly permissive Madin-Darby canine kidney (MDCK) cell line at a multiplicity of infection (MOI) of 2 per cell with the human H3N2 influenza virus isolate A/Wuhan/359/95 (18). To minimize cytopathic effects, which could induce the production of cellular RNA breakdown products, we chose to harvest total cellular RNA at 8 hpi, by which point the viral titer has plateaued, yet no cytopathic effect was apparent (18). As our initial focus was on miRNAs potentially encoded by viral genes, we used a previously described cDNA synthesis protocol that directly ligates 3′ and then 5′ adapters onto small (~18- to 30-nt) RNAs purified from the total RNA pool (9, 13). As a result, this cDNA synthesis protocol is dependent on the presence of a 5′ phosphate, which is typical of miRNAs, but excludes RNAs bearing other 5′ modifications, such as a triphosphate or a cap.
This initial effort resulted in 135,408 usable Solexa sequence reads of 18 to 27 nt in length, of which the majority (~79%) were known cellular miRNAs that are conserved in humans and other mammalian species, including canines (Table 1). This number likely underestimates the number of cellular miRNAs, as canine miRNAs have not yet been well characterized. In addition, we also recovered 654 sequence reads (~0.5%) that were of influenza virus origin. Interestingly, the large majority of these reads (550 of 654, or ~0.4% of the total reads obtained) originated from the exact 5′ end, or very near the 5′ end, of each of the eight influenza virus vRNA segments. To further examine whether this represented a discrete population of viral small RNAs, we next performed a Northern blot analysis using a mixture of four oligonucleotide probes specific for the 5′ ends of the PB1, M, NA, and NP vRNA segments. This experiment (Fig. 1) resulted in the detection of a very strong RNA signal in the 20- to 30-nt range in influenza virus-infected MDCK cells that was not present in uninfected cells.
TABLE 1 .
Characterization of two influenza virus-infected small RNA cDNA libraries
| Characteristic | Value (%) for characteristic |
|
|---|---|---|
| Library 1a | Library 2b | |
| Total no. of reads | 135,408 | 5,365,655 |
| No. of influenza readsc | 654 (~0.5) | 1,792,020 (~33) |
| No. of cellular miRNAsd | 107,106 (~79) | 1,029,813 (~19) |
| No. of other readse | 27,648 (~20) | 2,543,822 (~47) |
Library 1 utilized a cDNA synthesis protocol that captures only small (18- to 27-nt) RNAs bearing a 5′ monophosphate.
Library 2 used the same total RNA preparation but utilized a cDNA synthesis protocol that captures small RNAs regardless of their 5′ phosphorylation status.
Influenza reads are reads that map anywhere on the influenza virus genome.
Cellular miRNAs refers to reads that are homologous to known human miRNAs (miRBase version 15.0).
“Other reads” refers to reads that represent mRNA, rRNA, etc., breakdown products or that could not be assigned.
FIG 1 .
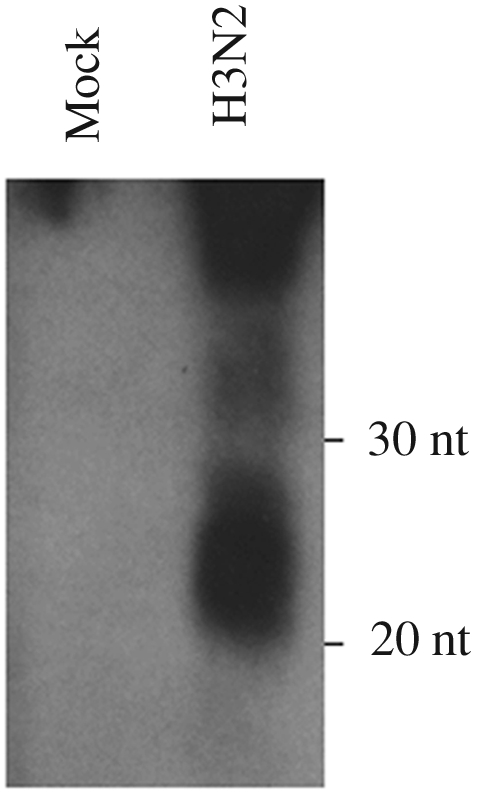
Northern blot analysis of influenza virus leader RNA expression. Total RNA was isolated 8 hpi from MDCK cells infected with the H3N2 influenza virus isolate A/Wuhan/359/95 and analyzed using a pool of oligonucleotide probes designed to recognize the 5′ 18 nt of the PB1, NA, M, and NP vRNAs.
A highly expressed population of influenza virus leader RNAs.
The cDNA synthesis protocol used to generate the first small RNA sequence library from influenza virus-infected cells is not able to capture RNAs bearing 5′ triphosphates, so the fact that the majority of the influenza virus RNAs recovered were derived from the 5′ ends of the influenza vRNA segments, which bear a terminal triphosphate (17), was unexpected. Moreover, given the low level of recovery of these leRNAs, <0.5% of the total number of small RNAs sequenced, the intensity of the signal detected by Northern analysis for these same leRNAs was also not anticipated (Fig. 1). We therefore reasoned that there might be a high level of expression of viral small RNAs originating from the 5′ ends of the eight vRNAs in influenza virus-infected cells that was not captured in our initial deep sequencing effort, as the library used would contain only rare 5′-terminal influenza vRNA fragments that had undergone partial hydrolysis in the cells, or during RNA isolation, to leave a 5′ monophosphate.
To address this issue, we therefore generated a second small RNA library, using the same influenza virus-infected cell total RNA preparation, that included a step whereby all 5′ phosphates were first removed by incubation with alkaline phosphatase, followed by addition of a single 5′ phosphate using polynucleotide kinase (PNK). The advantage of this modified technique is that it would allow us to recover any viral RNAs bearing a 5′ triphosphate in this second cDNA library. The disadvantage is that we would also recover common RNA breakdown products lacking any 5′ phosphate moieties.
Solexa deep sequencing of this second small RNA library yielded 5,365,655 usable reads of 18 to 27 nt in length (Table 1). Of these, 2,543,822 (~47%) represented RNA breakdown products (cellular mRNA, rRNA, tRNA, snRNA, etc.) or were not assignable, a substantially higher percentage than seen with library 1 (Table 1). On the other hand, we recovered only 1,029,813 identifiable canine cellular miRNA reads in library 2, which represented ~19% of the total number of reads, far less than the ~79% seen in library 1. Most strikingly, the number of influenza virus reads increased from ~0.5% (654) to ~33% (1,792,020) of the total number of reads. Analysis of the genomic origin of these viral small RNAs revealed that, overall, 98.4% coincided with the precise 5′ end of each of the eight vRNA segments, varying from a low of 97.3% in the case of the PB2 vRNA to a high of 99.6% for the NA segment (Table 2). In terms of segment origin, we recovered a low of 63,435 leRNAs derived from the NS segment to a high of 562,102 derived from PB2 (Table 2). By analogy to the small, 5′-triphosphorylated viral leader RNAs that have been reported in cells infected by other negative-strand RNA viruses (19–23), we refer to influenza virus small RNAs that originate from the 5′ ends of the vRNA segments as influenza virus leader RNAs (leRNAs).
TABLE 2 .
Percentage of leader RNAs compared to all influenza virus-derived sequencesa
| Gene | No. of influenza virus RNAs | No. of leRNAs | % of leRNAs |
|---|---|---|---|
| PB2 | 577,849 | 562,102 | 97.3 |
| PB1 | 195,040 | 192,073 | 98.5 |
| PA | 378,204 | 373,593 | 98.8 |
| HA | 72,844 | 72,085 | 99.0 |
| NP | 283,744 | 282,006 | 99.4 |
| NA | 113,579 | 113,167 | 99.6 |
| M | 106,629 | 105,270 | 98.7 |
| NS | 64,134 | 63,435 | 98.9 |
| Overall | 1,792,020 | 1,763,730 | 98.4 |
This table presents the number and percentage of small influenza virus RNAs that start at position 1 of each vRNA segment, defined as leader RNAs (leRNAs).
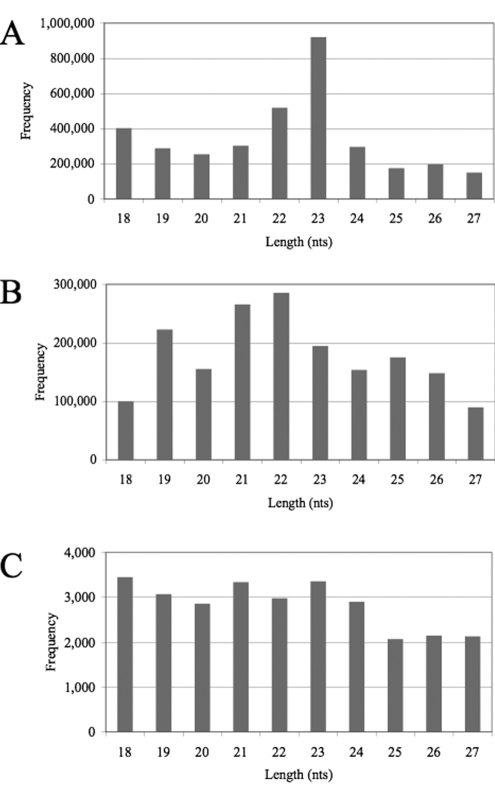
Analysis of the size distribution of the 18- to 27-nt cellular small RNAs in library 2 (Fig. 2A) revealed that the total cellular small RNA population, which represents a mixture of RNA breakdown products and cellular miRNAs, was heterogeneous in length but with a clear peak at the predicted 22- or 23-nt size expected for miRNAs (1), even though miRNAs appear to represent only a small proportion of these cellular RNA sequences (Table 1). (The peak at 23 nt reflects the very high level of expression of a 23-nt isoform of miR-21 in MDCK cells.) In contrast, the influenza virus leRNAs showed no clear size preference, although 19-nt, 21-nt, and 22-nt species were slightly predominant (Fig. 2B). Further breakdown of the leRNA size distribution by viral segment of origin (Table 3) again showed no particular size preference, with the most common size varying from 19 nt (PB2, M, NS, and HA) to 25 nt (PB1), with substantial numbers recovered at all sizes between the 18-nt and 27-nt cutoffs. (Due to technical limitations intrinsic to the Solexa sequencing protocol, RNAs of ≥28 nt in size could not be unequivocally identified.) The absence of tight clustering of the leRNAs at the 22/23-nt size characteristic of miRNAs is expected, as our data strongly suggest that the leRNAs contain a 5′-terminal triphosphate, which would preclude incorporation into RISC.
FIG 2 .
Size distribution of small RNAs returned by Solexa/Illumina deep sequencing. (A) Size distribution of all non-influenza virus small RNAs recovered in library 2 (Table 1). (B) Size distribution of all influenza virus small RNAs. (C) Size distribution of all non-leRNA influenza virus small RNAs.
TABLE 3 .
Size distribution of H3N2 leader RNAsa
| Length (nt) | No. of leRNAs of the indicated length derived from the following influenza vRNA segment: |
|||||||
|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | |
| 18 | 33,567 | 8,512 | 15,097 | 6,858b | 4,498 | 14,251 | 7,049 | 6,858b |
| 19 | 115,343 | 10,093 | 15,888 | 15,330b | 13,470 | 6,462 | 28,056 | 15,330b |
| 20 | 36,539 | 18,962 | 34,487 | 3,742b | 24,710 | 21,361 | 9,064 | 3,742b |
| 21 | 70,309 | 28,898 | 70,226 | 7,308b | 42,140 | 9,728 | 26,498 | 7,308b |
| 22 | 82,197 | 12,056 | 105,011 | 7,526 | 46,306 | 8,184 | 16,255 | 5,247 |
| 23 | 22,272 | 20,949 | 63,814 | 2,878 | 48,082 | 15,643 | 11,174 | 6,664 |
| 24 | 42,376 | 16,307 | 26,101 | 5,006 | 42,786 | 14,607 | 1,500 | 2,296 |
| 25 | 32,327 | 34,429 | 24,996 | 14,640 | 42,608 | 15,033 | 2,002 | 7,152 |
| 26 | 75,262 | 32,694 | 8,399 | 5,673 | 12,353 | 3,100 | 3,346 | 5,094 |
| 27 | 51,910 | 9,173 | 9,574 | 3,125 | 5,053 | 4,798 | 326 | 3,745 |
| Total | 562,102 | 192,073 | 373,593 | 72,085 | 282,006 | 113,167 | 105,270 | 63,435 |
This table presents the size distribution, in nucleotides, of the leRNAs derived from each of the indicated influenza vRNA segments, with the most prevalent size in boldface type.
Because the HA and NS vRNAs are identical in sequence through nucleotide 21, we could not assign reads derived from these sequences unequivocally. We have therefore arbitrarily assigned 50% of the reads to each of these two segments.
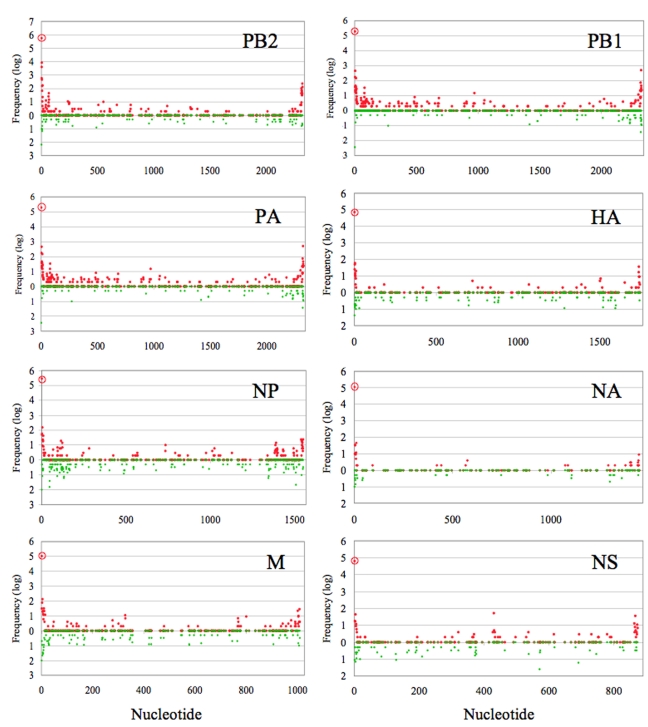
We also recovered a small number of influenza virus RNA fragments (28,292) that originate from regions of the influenza virus genome other than the 5′ ends of vRNAs. Could these represent viral miRNAs? In fact, analysis of their size distribution again revealed no preference for the 22/23-nt size characteristic of miRNAs (Fig. 2C). Analysis of the precise origin of all small influenza virus RNAs, including the leRNAs, within the eight genomic segments of influenza virus (Fig. 3) revealed the expected high level of origin from the 5′ ends of the vRNA segments that is the defining characteristic of influenza virus leRNAs and also showed a modest enrichment of influenza virus small RNAs from near the 3′ end of the vRNA, as well as from the 3′ end of the mRNA/cRNA population, although these are all present at a level that is >1,000-fold lower than the level of leRNAs. Other influenza virus-derived small RNAs were derived from random locations within both the vRNA and mRNA/cRNA transcripts, and not from one or a small number of discrete sites, as would be predicted for miRNAs (Fig. 3). Instead, this distribution suggests that these are random viral RNA breakdown products.
FIG 3 .
Genomic origin of all influenza virus small RNAs. Small red circles indicate the frequency and genomic locations of the starting nucleotide of vRNA-derived sequences recovered by deep sequencing, while small green circles represent the frequency and genomic locations of mRNA/cRNA-derived sequences for all eight influenza virus genome segments. The viral leRNAs, which start at nucleotide 1 of the vRNA, are indicated by a large red circle containing a small red circle. The data used were derived from cDNA library 2 (Table 1). All sequences are aligned on the basis of their 5′ ends, which are aligned to the left for the vRNAs and to the right for the viral cRNAs/mRNAs. The frequency is the number of viral sequence reads obtained that bear identical 5′ ends.
Influenza virus leader RNAs are expressed early after infection.
After infection by influenza virus, the eight vRNA segments are initially copied to yield viral mRNA species that are then translated to yield viral proteins (17). These newly synthesized influenza virus proteins, in particular the viral nucleoprotein NP, are then thought to induce a partial switch from mRNA transcription to genome replication, i.e., to cRNA synthesis followed by the generation of new vRNAs, beginning at 1 or 2 hours after infection (24–26). It has been proposed that these progeny vRNAs can make a significant contribution to viral mRNA, and hence protein, synthesis, although this has remained controversial (25, 27, 28).
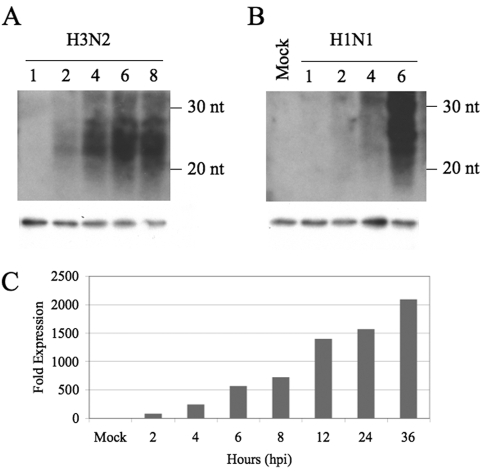
The leRNAs we observe must be derived from transcripts copied from viral cRNAs and are likely to result from nonprocessive transcription of the 3′ ends of the cRNAs or, less probably, from endonucleolytic cleavage of vRNA transcripts. In either case, one might expect leRNA expression to occur somewhat later in the viral life cycle. To address this issue, we performed a time course analysis of leRNA production in MDCK cells infected with either the H3N2 virus isolate A/Wuhan/359/95 (18) or an H1N1 clinical isolate, A/HK/54/98 (29), at a multiplicity of infection (MOI) of ~2/cell (Fig. 4A and B). Interestingly, we observed that the H3N2 virus gave rise to detectable leRNA expression as soon as 2 hpi, which increased rapidly up to 8 hpi. The pattern observed with the more cytopathic H1N1 isolate was slightly different, with leRNA expression being first detected at ~4 hpi and then increasing rapidly to 6 hpi. We also noted some differences in leRNA size, with the H3N2 isolate giving rise to leRNAs that were essentially all between 20 and 30 nt in size (Fig. 4A), while the H1N1 isolate gave rise to some leRNAs that appeared to be slightly over 30 nt, although leRNAs 20 to 30 nt in size again predominated (Fig. 4B). To extend these data to a third influenza virus isolate and to a second cell line, we harvested total RNA samples from human 293T cells infected with the H3N2 influenza virus isolate A/Udorn/72 (30) at an MOI of ~1. The assay used to quantify leRNA expression in this instance was quantitative reverse transcriptase PCR (qRT-PCR), using a stem-loop probe specific for the PB2 leRNA, which was expected to allow detection of the leRNAs over a wide dynamic range. As shown in Fig. 4C, we were again able to detect leRNAs as soon as 2 hpi, but we now observed that the leRNAs continued to accumulate up to 36 hpi. Overall, these data suggest that leRNAs and vRNAs are likely to arise simultaneously and quite early in the influenza virus life cycle and also suggest that the leRNAs, like the full-length vRNAs, accumulate in infected cells and can reach high levels at late times after infection. We note that previous reports have demonstrated that vRNA synthesis can initiate as soon as 1 hpi (25).
FIG 4 .
Time course analysis of influenza virus leader RNA expression. (A) Northern analysis of RNA samples isolated from MDCK cells infected with H3N2 isolate A/Wuhan/359/95 and harvested at the indicated time points, given as hours postinfection (hpi) above the lanes. This Northern blot was analyzed using the same leRNA-specific probes used in Fig. 1. (B) Expression of H1N1 isolate A/HK/54/98-derived leRNAs in infected MDCK cells at the indicated times postinfection. In panels A and B, the migration positions of small RNA size markers are indicated to the right of the blots, and U6 RNA served as a loading control. (C) qRT-PCR analysis of influenza virus leRNA expression in 293T cells infected with the influenza virus isolate A/Udorn/72. Samples were harvested at the indicated time points, given as hpi. The qRT-PCR probe used is specific for leRNAs derived from the PB2 RNA segment. Data are given as a multiple of the qRT-PCR signal obtained using mock-infected cells, which was set at 1.
DISCUSSION
The initial goal of this research project was to determine whether influenza A virus, which copies its RNA genome in the nucleus, might give rise to viral miRNAs. Unexpectedly, we instead observed that influenza virus generates very high levels of short (18- to 27-nt-long) viral leader RNAs (leRNAs) that originate from the very 5′ ends of all eight vRNA segments. Our data strongly suggest that these leRNAs, like the vRNAs themselves, contain a 5′-terminal triphosphate. As a result, the leRNAs were recovered very inefficiently when a standard small RNA sequencing protocol, which requires a 5′ phosphate, was used. However, leRNAs became the dominant small RNA species when a modified protocol was used; the modified protocol allows the generation of cDNAs from small RNAs regardless of the 5′ phosphorylation status (Table 1). Remarkably, under these conditions, we recovered substantially more leRNAs from cells infected 8 h previously with influenza A virus than total cellular miRNAs. As individual cells generally express from 80,000 to over 100,000 miRNAs (1), this means that influenza leRNAs were expressed at well over 100,000 copies per cell by 8 hpi.
Because miRNA incorporation into RISC requires a 5′ monophosphate (1, 3, 4), the leRNAs we detected cannot represent miRNAs. Moreover, because both Dicer and Drosha cleavage leaves behind a 5′ monophosphate (1), these RNase III enzymes are clearly not involved in leRNA biogenesis. Most probably, the leRNAs result from premature termination by the viral RNA-dependent RNA polymerase (RdRP) while utilizing a cRNA template. While this has been previously reported to occur at high levels in vitro (31), it also remains formally possible that the leRNAs instead arise from endonucleolytic cleavage of nascent or mature vRNA molecules. The heterogeneous 3′ ends of the leRNAs (Table 3) would appear to make the latter hypothesis less likely.
While leRNAs account for 98.4% of all small influenza viral RNAs, we did detect small RNAs that were derived from regions other than the precise 5′ ends of the vRNAs. However, these influenza virus small RNAs did not show a discrete size of ~22 nt, as predicted for both miRNAs and small interfering RNAs (siRNAs) (Fig. 2C) (1), and are therefore likely RNA breakdown products. Consistent with this hypothesis, these small viral RNAs were not derived from discrete locations within the influenza virus genomic segments, as would be expected for viral miRNAs (Fig. 3).
It is interesting to compare our observations with previously published work documenting the expression of short leader RNAs, derived from the 5′ ends of the genomic RNA, in cells infected with a range of other negative-strand RNA viruses, including vesicular stomatitis virus (VSV) (21, 23), rabies virus (22), and respiratory syncytial virus (19). The most studied of these are the VSV leRNAs, which contain a 5′-terminal triphosphate, range from ~44 to ~54 nt in length, and have been reported to be expressed at ~300 copies per infected cell (20, 32, 33). Despite their very low level of expression, the VSV leRNAs have been proposed to play a key role in the VSV life cycle by regulating the switch from mRNA transcription to genomic RNA replication by binding to the VSV nucleocapsid protein (20). The VSV leRNAs have also been shown to bind the cellular protein La (33, 34), which may prevent activation of the cellular innate immunity factor RIG-I by these 5′-triphosphorylated RNA molecules (19).
While this article was being prepared, Perez et al. (35) reported the expression of analogous influenza virus leader RNAs, which they termed small viral RNAs, in a range of influenza virus-infected cell types, including human lung epithelial cells and embryonated chicken eggs. These small viral RNAs were initially detected by conventional deep sequencing, using a protocol that captures only small RNAs bearing a 5′ monophosphate, and as a result, the small viral RNAs comprised only 1,524 (0.04%) of the total of 4,217,121 short RNA sequence reads reported by this group. This is comparable to, but even lower than, the ~0.4% of total sequence reads contributed by the leRNAs when we used a comparable sequencing protocol and contrasts with the very high level of leRNAs that were detected when a protocol that captures 5′-triphosphorylated short RNAs was used (Table 1).
Perez et al. (35) reported that their influenza virus small RNAs were between 22 and 27 nt in length and were derived from the 5′ ends of all eight influenza virus genome segments, which is similar to what we have observed for the leRNAs (Fig. 2B and Table 3), although it is also clear that the leRNAs bear fairly heterogeneous 3′ ends. Our data do differ somewhat from Perez et al. (35) in that these authors reported that their small viral RNAs were detectable starting at ~12 hpi, while we could clearly see leRNA expression at 6 hpi and sometimes even earlier (Fig. 4). Indeed, by 8 hpi, leRNAs were found to be expressed at >105 copies per cell, as noted above. This discrepancy may result from the use of different cell types in these analyses, as Perez et al. (35) primarily used A549 cells, while we primarily used MDCK cells.
An important observation reported by Perez et al. (35) is that introduction of an antisense locked nucleic acid (LNA) oligonucleotide (36) complementary to the HA segment leRNA inhibits HA vRNA synthesis yet had little or no effect on HA mRNA or cRNA transcription, thus suggesting that leRNAs might play a critical role in virus genome replication, perhaps by triggering the switch from mRNA transcription to genome replication. This result is perhaps unexpected, as it has previously been proposed that production of optimal levels of influenza virus mRNAs, and hence proteins, requires mRNAs generated by transcription of newly generated vRNAs, not just mRNAs transcribed from the input virus vRNA genomes (25, 28, 31). If this hypothesis is indeed correct, then loss of leRNA function should inhibit not only vRNA, but also mRNA, synthesis, which was not observed (35). We note that the LNA used in this experiment has the potential to anneal to not only the HA leRNAs but also to the identical vRNA 5′ ends present on both the input and newly generated HA vRNA segments. Given data showing that the 5′ 13 nt or so of the vRNAs plays a critical role in influenza virus transcription (17, 37), it may be difficult to segregate phenotypic effects resulting from LNA annealing to the viral leRNAs from effects resulting from LNA annealing to the vRNAs themselves. Nevertheless, the hypothesis that the influenza virus leRNAs play a critical role in the temporal regulation of influenza virus RNA synthesis (35) is an important one and certainly consistent with their very high level of expression. Moreover, the proposed role for leRNAs in the influenza virus replication cycle is similar to that proposed for the viral leader RNAs encoded by VSV, which may play an equivalent role in activating VSV genomic RNA replication (20). On the other hand, it is also possible that the leRNAs simply represent prematurely terminated vRNA transcripts and, therefore, do not regulate vRNA synthesis. On the basis of previous data obtained using other negative-strand RNA viruses, one could also envision leRNAs playing a role in vRNA encapsidation into newly generated progeny virions (38) or in regulating innate immune responses (19, 22, 33, 34). In any case, the high level of expression of viral leRNAs in influenza A virus-infected cells does suggest an important role(s) in the virus replication cycle that could represent a novel target for the treatment of influenza virus-induced disease.
MATERIALS AND METHODS
Cell culture, infections, and RNA isolation.
Madin-Darby canine kidney (MDCK) cells were maintained in Eagle’s minimum essential medium supplemented with 10% fetal bovine serum, nonessential amino acids, and sodium pyruvate. Cells were infected with influenza virus at an MOI of 2 and harvested at the indicated time points. Total RNA was harvested using TriReagent according to the manufacturer’s protocol.
Solexa/Illumina deep sequencing.
cDNA libraries for deep sequencing were prepared from total RNA samples as previously described (9, 13). Briefly, small RNAs 18 to 30 nt in length were isolated from ~30 µg of total RNA by gel fractionation and then sequentially ligated to 3′ and 5′ linkers. cDNA libraries for deep sequencing were then generated by standard reverse transcription and PCR amplification of the ligated small RNAs. Data were returned in FASTA format and analyzed using custom scripts in addition to the formatdb, megablast, blastoutparse, and filter alignment scripts of the miRDeep software package (39). To capture all small RNAs, regardless of their 5′ phosphorylation status, additional dephosphorylation and kinasing steps were included following the 3′ linker ligation, just prior to ligation of the 5′ linker. Specifically, gel-purified 3′ linker-ligated RNAs were dephosphorylated in a 20-µl reaction mixture with 20 U of alkaline phosphatase at 37°C for 1 h. Dephosphorylated RNAs were purified by precipitation, and 5′ monophosphates were added in a 20-µl reaction mixture with 10 U of T4 PNK and an excess of ATP at 37°C for 1 h. All other aspects of sample preparation were identical to the standard protocol (9, 13).
Northern blot analysis.
leRNA expression was analyzed by standard small RNA Northern blot analysis as previously described (6, 7). Briefly, 30 µg of total RNA was loaded onto 15% Tris-borate-EDTA (TBE)-urea gels. RNAs were transferred onto nitrocellulose and fixed by UV cross-linking. The top portion of the blots, which contain the large viral RNAs, were then removed to avoid titration of the pool of end-labeled probes recognizing the PB1 (5′-AAAATGCCTTGTTTCTACT-3′), NA (5′-AAACTCCTTGTTTCTACT-3′), M (5′-AACTACCTTGTTTCTACT-3′), and NP (5′-AAATACCCTTGTTTCTACT-3′) leRNAs.
Quantitation of leRNA synthesis using qRT-PCR.
Custom TaqMan stem-loop primers from Applied Biosystems were designed against the most common isoform of the PB2 leRNA recovered from deep sequencing (5′-AGUAGAAACAAGGUCGUUU-3′) and used to analyze RNA harvested from influenza virus-infected cells, as previously described (40). Briefly, 10 ng of total RNA was used as a template for reverse transcription. The sample was then diluted to 60 µl, of which 8 µl was used for PCR. Values were normalized to a cellular miRNA, miR-16, and displayed as fold expression relative to a mock-infected sample, as previously described (41).
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant R21-AI088327 to B.R.C. H.-L.Y. was supported in part by NIAID contract HHSN26620070005C, the American Lebanese Syrian Associated Charities (ALSAC), and the Area of Excellence Scheme of the University Grants Committee (grant AoE/M-12/06). J.L.U. was supported by NIH training grant T32-CA009111.
We thank Daniel Kolakofsky, Robert Webster, and Malik Peiris for helpful discussions and Peter Palese for the gift of the A/Udorn/72 virus.
Footnotes
Citation Umbach, J. L., H.-L. Yen, L. L. M. Poon, and B. R. Cullen. 2010. Influenza A virus expresses high levels of a novel class of small viral leader RNAs in infected cells. mBio 1(4):e00204-10. doi:10.1128/mBio.00204-10.
REFERENCES
- 1. Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 2. Cullen B. R. 2004. Transcription and processing of human microRNA precursors. Mol. Cell 16:861–865 [DOI] [PubMed] [Google Scholar]
- 3. Ma J. B., Ye K., Patel D. J. 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D. J. 2008. Structure of the guide-strand-containing argonaute silencing complex. Nature 456:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Umbach J. L., Cullen B. R. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cai X., Lu S., Zhang Z., Gonzalez C. M., Damania B., Cullen B. R. 2005. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai X., Schäfer A., Lu S., Bilello J. P., Desrosiers R. C., Edwards R., Raab-Traub N., Cullen B. R. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grey F., Antoniewicz A., Allen E., Saugstad J., McShea A., Carrington J. C., Nelson J. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F. A., van Dyk L. F., Ho C. K., Shuman S., Chien M., Russo J. J., Ju J., Randall G., Lindenbach B. D., Rice C. M., Simon V., Ho D. D., Zavolan M., Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 10. Pfeffer S., Zavolan M., Grasser F. A., Chien M., Russo J. J., Ju J., John B., Enright A. J., Marks D., Sander C., Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736 [DOI] [PubMed] [Google Scholar]
- 11. Samols M. A., Hu J., Skalsky R. L., Renne R. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 79:9301–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sullivan C. S., Grundhoff A. T., Tevethia S., Pipas J. M., Ganem D. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682–686 [DOI] [PubMed] [Google Scholar]
- 13. Umbach J. L., Kramer M. F., Jurak I., Karnowski H. W., Coen D. M., Cullen B. R. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin J ., Cullen B. R. 2007. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 81:12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Brisibe E. A., Saksena N. K., Fujii Y. R. 2004. HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ouellet D. L., Plante I., Landry P., Barat C., Janelle M. E., Flamand L., Tremblay M. J., Provost P. 2008. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 36:2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palese P., Shaw M. L. 2007. Orthomyxoviridae: the viruses and their replication. , p. 1647–1689 In Knipe D. M., Howley P. M. Fields virology 5th ed., vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 18. Yen H. L., Herlocher L. M., Hoffmann E., Matrosovich M. N., Monto A. S., Webster R. G., Govorkova E. A. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 49:4075–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bitko V., Musiyenko A., Bayfield M. A., Maraia R. J., Barik S. 2008. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J. Virol. 82:7977–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blumberg B. M., Leppert M., Kolakofsky D. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837–845 [DOI] [PubMed] [Google Scholar]
- 21. Colonno R. J., Banerjee A. K. 1978. Complete nucleotide sequence of the leader RNA synthesized in vitro by vesicular stomatitis virus. Cell 15:93–101 [DOI] [PubMed] [Google Scholar]
- 22. Kurilla M. G., Cabradilla C. D., Holloway B. P., Keene J. D. 1984. Nucleotide sequence and host La protein interactions of rabies virus leader RNA. J. Virol. 50:773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leppert M., Rittenhouse L., Perrault J., Summers D. F., Kolakofsky D. 1979. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell 18:735–747 [DOI] [PubMed] [Google Scholar]
- 24. Beaton A. R., Krug R. M. 1986. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc. Natl. Acad. Sci. U. S. A. 83:6282–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hatada E., Hasegawa M., Mukaigawa J., Shimizu K., Fukuda R. 1989. Control of influenza virus gene expression: quantitative analysis of each viral RNA species in infected cells. J. Biochem. 105:537–546 [DOI] [PubMed] [Google Scholar]
- 26. Medcalf L., Poole E., Elton D., Digard P. 1999. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J. Virol. 73:7349–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hay A. J., Lomniczi B., Bellamy A. R., Skehel J. J. 1977. Transcription of the influenza virus genome. Virology 83:337–355 [DOI] [PubMed] [Google Scholar]
- 28. Shapiro G. I., Gurney T., Jr, Krug R. M. 1987. Influenza virus gene expression: control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs. J. Virol. 61:764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheung C. Y., Poon L. L., Lau A. S., Luk W., Lau Y. L., Shortridge K. F., Gordon S., Guan Y., Peiris J. S. 2002. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease?. Lancet 360:1831–1837 [DOI] [PubMed] [Google Scholar]
- 30. Murphy B. R., Tierney E. L., Spring S. B., Chanock R. M. 1976. Temperature-sensitive mutants of influenza A virus. XI. Transfer of ts lesions in the Hong Kong/68-ts-1 [a] virus to the influenza A/Udorn/72 wild type. J. Infect. Dis. 134:577–584 [DOI] [PubMed] [Google Scholar]
- 31. Kawaguchi A., Nagata K. 2007. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 26:4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurilla M. G., Piwnica-Worms H., Keene J. D. 1982. Rapid and transient localization of the leader RNA of vesicular stomatitis virus in the nuclei of infected cells. Proc. Natl. Acad. Sci. U. S. A. 79:5240–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilusz J., Kurilla M. G., Keene J. D. 1983. A host protein (La) binds to a unique species of minus-sense leader RNA during replication of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 80:5827–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kurilla M. G., Keene J. D. 1983. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell 34:837–845 [DOI] [PubMed] [Google Scholar]
- 35. Perez J. T., Varble A., Sachidanandam R., Zlatev I., Manoharan M., Garcia-Sastre A., tenOever B. R. 2010. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U. S. A. 107:11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elmen J., Lindow M., Schutz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjarn M., Hansen H. F., Berger U., Gullans S., Kearney P., Sarnow P., Straarup E. M., Kauppinen S. 2008. LNA-mediated microRNA silencing in non-human primates. Nature 452:896–899 [DOI] [PubMed] [Google Scholar]
- 37. Fodor E., Palese P., Brownlee G. G., Garcia-Sastre A. 1998. Attenuation of influenza A virus mRNA levels by promoter mutations. J. Virol. 72:6283–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patton J. T., Davis N. L., Wertz G. W. 1984. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J. Virol. 49:303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Friedlander M. R., Chen W., Adamidi C., Maaskola J., Einspanier R., Knespel S., Rajewsky N. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26:407–415 [DOI] [PubMed] [Google Scholar]
- 40. Umbach J. L., Nagel M. A., Cohrs R. J., Gilden D. H., Cullen B. R. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J. Virol. 83:10677–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 42. Parameswaran P., Sklan E., Wilkins C., Burgon T., Samuel M. A., Lu R., Ansel K. M., Heissmeyer V., Einav S., Jackson W., Doukas T., Paranjape S., Polacek C., dos Santos F. B., Jalili R., Babrzadeh F., Gharizadeh B., Grimm D., Kay M., Koike S., Sarnow P., Ronaghi M., Ding S. W., Harris E., Chow M., Diamond M. S., Kirkegaard K., Glenn J. S., Fire A. Z. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6 e1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]