Abstract
Oxytetracycline (OTC) is a broad-spectrum antibiotic that acts by inhibiting protein synthesis in bacteria. It is an important member of the bacterial aromatic polyketide family, which is a structurally diverse class of natural products. OTC is synthesized by a type II polyketide synthase that generates the poly-β-ketone backbone through successive decarboxylative condensation of malonyl-CoA extender units, followed by modifications by cyclases, oxygenases, transferases, and additional tailoring enzymes. Genetic and biochemical studies have illuminated most of the steps involved in the biosynthesis of OTC, which is detailed here as a representative case study in type II polyketide biosynthesis.
Keywords: Antibiotics, Enzyme Catalysis, Enzyme Kinetics, Enzyme Mechanisms, Ribosome Function
Introduction
Oxytetracycline (OTC)3 is a well studied polyketide natural product and is an important example of type II polyketide biosynthesis. Type II polyketides, also known as bacterial aromatic polyketides, are a group of compounds produced naturally in bacteria from a poly-β-ketone intermediate that is tailored to form a polycyclic product containing at least one aromatic ring (1, 2). This class of metabolites includes many important bioactive compounds such as anticancer agents doxorubicin (3) and mithramycin (4), the antiviral and antifungal pradimicin (5), and the antibiotic tetracyclines (6). The biosynthetic machinery responsible for the synthesis of these compounds is structurally and biochemically similar to that for type II fatty acid synthases, leading to the classification type II polyketide synthases.
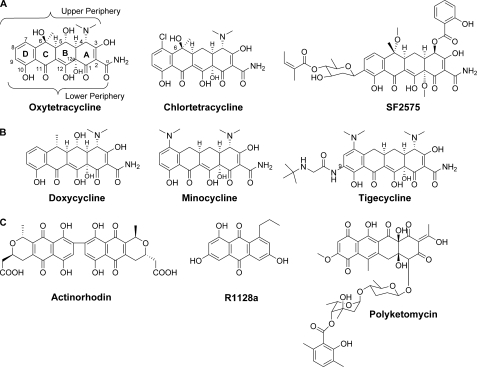
The discovery of chlortetracycline (CTC) in 1948 by Benjamin Duggar (7) marks the beginning of the tetracycline family history. Initially termed Aureomycin for its yellow hue, CTC was immediately recognized for its remarkable antibiotic properties and patented by American Cyanamid (7, 8). In 1950, A. C. Finlay with Pfizer published the discovery of a similar antibiotic produced by Streptomyces rimosus that they named Terramycin (later renamed OTC) (9). Both compounds are broad-spectrum antibiotics and inhibit protein synthesis in bacteria by binding to the 30 S ribosomal subunit (10, 11). Although the value of these compounds was readily apparent, it was several more years until the chemical structures were solved by the Woodward group (12, 13). The tetracyclines are characterized by a unique C2 amide functionality and the linearly fused tetracyclic backbone, which is heavily decorated to yield the 2-naphthacene carboxamide aglycon (Fig. 1). The oxidized lower periphery of the molecule includes a keto-enol configuration across the C11, C11a, and C12 positions, which is responsible for chelation of magnesium ion. This invariant feature and hydroxyl groups at C10 and C12a, which are involved in hydrogen bonding and the conformation of tetracyclines, are important for interaction with the 30 S ribosomal subunit (11). Modifications to the lower periphery, such as those at C1 and C10–C12a, are detrimental to antibiotic activity (14). In contrast, parts of the upper periphery of tetracycline are tolerant of chemical changes and thus have been the target of semisynthetic modifications (15).
FIGURE 1.
A, naturally produced tetracyclines; B, semisynthetic second and third generation tetracyclines; C, additional naturally occurring type II polyketides referenced in text.
The widespread production and use of natural tetracyclines in both human and animal medicine in the decades following their discovery led to emergence of resistance mechanisms and decreased effectiveness as front-line antibiotics (16, 17). The severity of bacterial resistance created an urgent need to develop new tetracycline derivatives capable of evading these resistance mechanisms. The most clinically valuable second generation tetracyclines are minocycline and doxycycline. Doxycycline is produced from OTC by a process that first converts OTC into methacycline, which is further reacted to form doxycycline (18, 19). These analogs are more lipophilic than the natural products and were shown to be more readily absorbed (20). The glycylcyclines or so-called third generation tetracycline analogs were developed recently, with tigecycline receiving Food and Drug Administration approval in 2005. Tigecycline is a minocycline derivative and contains a tert-butylglycylamido group at C9 (21, 22). The modification results in evasion of both efflux and ribosomal protection mechanisms of resistance (23), making it a viable choice against tetracycline-resistant infections.
Since the discovery of OTC in 1950, the biosynthetic pathway has been probed with the tools available to elucidate the mechanism of biosynthesis. Early feeding studies demonstrated the polyketide origin of OTC (24, 25), and blocked mutant studies determined much of the biosynthetic sequence and led to isolation of intermediates and shunt products (26–31). With advances in molecular biology and genetic techniques and the sequencing of the OTC gene cluster (6, 32, 33), roles of individual enzymes in the pathway have been examined. Combining genetic studies using the natural host S. rimosus and systematic reconstitution studies in heterologous Streptomyces hosts has led to the functional assignment of nearly all of the genes in the oxy cluster (33–35). Additional details of genetic and metabolic engineering aspects of tetracyclines can be found in earlier reviews (36, 37). Here, we review the current knowledge of OTC biosynthesis.
Biosynthesis of the Amidated Polyketide Backbone
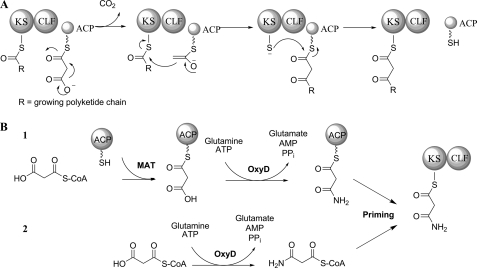
Biosynthesis of the polyketide backbone is catalyzed by the minimal polyketide synthase (PKS), which includes three components, the ketosynthase (KS) or KSα, the chain length factor (CLF) or KSβ, and the acyl carrier protein (ACP). In the OTC gene cluster, these are encoded by oxyA, oxyB, and oxyC, respectively (33). Details on the enzymology of the minimal PKS components have been covered in several recent reviews (1, 2, 38). Briefly, the KS and CLF, which share high sequence similarity, associate to form a heterodimer complex responsible for chain elongation as shown in Fig. 2A. The extender unit malonyl-CoA is transferred to the ACP by a malonyl-CoA:ACP acyltransferase, which can be shared with fatty acid biosynthesis (39), to form malonyl-ACP. The KS-CLF catalyzes C–C bond formation by Claisen-like decarboxylative condensation of incoming malonyl-ACP with the nascent polyketide chain. The net result of one such iteration is the addition of one ketide unit to the growing polyketide. Chain length is thought to be controlled by the CLF subunit, which determines the size of the polyketide binding cavity (40, 41).
FIGURE 2.
A, polyketide elongation by iterative Claisen-like condensation; B, two proposed pathways for biosynthesis of the malonamate starter unit by OxyD. MAT, malonyl-CoA:ACP acyltransferase.
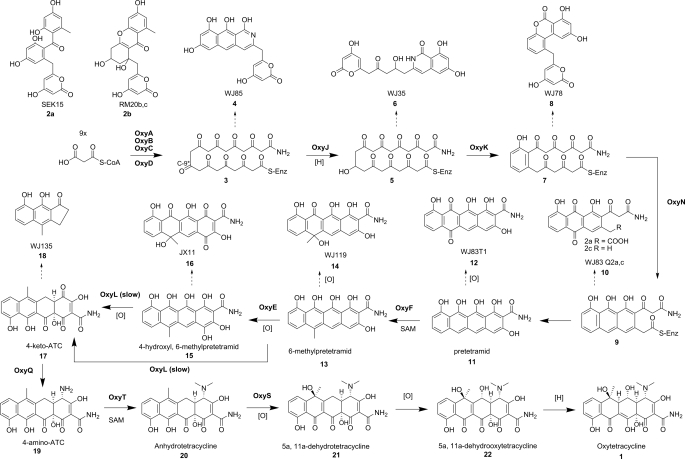
Starter unit selection is an important source of chemical diversity among aromatic polyketides (42), and the inclusion of an amide starter unit is one of the hallmarks of tetracycline biosynthesis. The most common starter unit for minimal PKS is acetate; however, the tetracycline family utilizes a malonamate starter unit, which is the origin of the C2 amide. The enzyme responsible for the biosynthesis of this unusual starter unit is the amidotransferase OxyD. OxyD shares high homology with ATP-dependent class II asparagines synthases, which catalyze the conversion of aspartate to asparagine using glutamine as the amine donor (43). OxyD similarly contains an N-terminal nucleophilic cysteine that has been shown to be responsible for the hydrolysis of glutamine amide in Escherichia coli AsnB (43). OxyD was therefore proposed to catalyze the conversion of a malonate equivalent to the corresponding malonamate in an ATP-dependent fashion. The exact substrate of OxyD, which can be either malonyl-S-CoA or malonyl-S-ACP, has not been determined due to the inability to reconstitute OxyD activities in vitro (33). The two proposed pathways are shown in Fig. 2B. The role of OxyD in starter unit biosynthesis was confirmed by coexpression with the minimal PKS OxyABC in a heterologous host and the isolation of WJ85 (4) (Fig. 3) (33). The presence of the amide in 4 confirms the role of OxyD in priming the minimal PKS. Coexpression of the C9 ketoreductase OxyJ similarly resulted in the biosynthesis of a reduced amidated polyketide backbone, which spontaneously cyclized to afford the isoquinolone WJ35 (6) (33). Therefore, the oxy minimal PKS and OxyD can be collectively termed the “oxy extended minimal PKS.”
FIGURE 3.
Biosynthesis of OTC and reported shunt products.
Like many type II PKSs that utilize nonacetate starter units, the oxy PKS can also be initiated by an acetate primer in the absence of OxyD. As illustrated first by Khosla and co-workers (44), the oxy minimal PKS alone synthesizes exclusively acetate-primed decaketides. In other pathways in which nonacetate starter units are employed, such as daunorubicin (45), frenolicin (46), R1128 (46, 47), and hedamycin (48, 49), an additional initiation module has been implicated in starter unit selection. In the case of frenolicin and R1128, it was further shown that a dedicated ACP is required for the chain initiation steps and plays a different role than the ACP utilized by the minimal PKS for chain elongation (46). The oxy pathway lacks these additional components, and a complete amidated backbone can be produced by the extended minimal PKS without additional enzymes. An enzyme that ensures starter unit fidelity in the R1128 pathway is the malonyl-CoA:ACP acyltransferase homolog ZhuC. ZhuC was shown to have potent thioesterase activity toward acetyl-ACP. This led to the proposal that the role of ZhuC was to selectively hydrolyze the competing acetyl-ACP in favor of the longer chain acyl-ACP primer units (50). The oxy cluster contains a ZhuC homolog, OxyP, and there are indications that it may serve a similar role in suppressing acetate priming and reducing the level of 2-acetyl-2-decarboxamido-OTC (ADOTC).4 Reduction of the ADOTC:OTC ratio is desirable, as ADOTC has greatly reduced antibiotic activity compared with OTC and is difficult to remove from fermentation broths (51). Unlike the R1128 pathway, which produces only acetate-primed polyketides in the absence of the proofreading activities of ZhuC, the oxy PKS does not require OxyP to incorporate the malonamate starter unit. Interestingly, an OxyP homolog is not found in the CTC gene cluster (52), which further indicates that such an enzyme is not essential for tetracycline biosynthesis but rather functions as an accessory enzyme.
Cyclization of the Polyketide Backbone
Because of the reactivity of the nascent poly-β-ketone backbone, cyclases are required to suppress spontaneous cyclizations and to promote regioselectivity of intramolecular aldol condensations. In the absence of cyclases, the KS-CLF and C9 ketoreductase have also been suggested to play key roles in influencing the folding of the polyketide chain (40, 53, 54). Most minimal PKSs produce C7–C12 cyclized polyketides as major products in the absence of additional tailoring enzymes, indicating that the KS-CLF may promote the regioselectivity of the first cyclization event (55). Analysis of the crystal structure of the actinorhodin (act) KS-CLF showed that buckling of the polyketide chain must occur within the KS-CLF tunnel to accommodate the full-length polyketide chain. This requirement may position the chain in a configuration that is favorable for C7–C12 cyclization (40). For many type II polyketides, C9 ketoreduction is the first tailoring step following the completion of the polyketide chain by the minimal PKS. Nearly all the C9 reduced polyketide backbones undergo C7–C12 cyclization to form the first ring, including that of tetracyclines. The crystal structure of act ketoreductase (KR) with a bound inhibitor indicated a preference for cyclized substrates (54).
However, these proposed roles for minimal PKS- and C9 KR-controlled C7–C12 cyclization do not apply to the amidated polyketide backbone synthesized by the oxy extended minimal PKS. As shown in Fig. 3, the previously mentioned 4 produced by OxyABCD adopts C11–C16 first ring cyclization, whereas 6 produced by OxyABCDJ is cyclized via C13–C18 aldol condensation (33). The regioselectivities of the first cyclization steps in 4 and 6 were especially surprising considering that acetate-primed decaketide synthesized by the oxy minimal PKS does indeed cyclize preferentially via C7–C12. For example, the oxy minimal PKS alone produces SEK15 (2a) (33, 55), whereas in the presence of act KR, it produces RM20b/c (2b) (44). Therefore, the presence of the amidated starter unit may significantly influence the orientation of the polyketide chain in the active site of the KS-CLF, as well as that of the C9 KR, to result in the unexpected cyclization regioselectivities of 4 and 6. Alternatively, whereas the backbone may be oriented in similar configurations, the presence of the amide starter unit somehow suppressed C7–C12 aldol condensation, leading to release of uncyclized products that rapidly undergo spontaneous cyclization to yield 4 and 6. Therefore, in the biosynthesis of OTC, first ring cyclase/aromatase is absolutely required for the correct C7–C12 cyclization and formation of the D-ring.
The OTC first ring cyclase OxyK was first identified by Petkovic et al. (56) as a cyclase/aromatase. Disruption of this gene resulted in the elimination of all recognizable tetracycline intermediates in the mutant culture (56). Zhang et al. (35) later demonstrated the role of OxyK. Heterologous expression of OxyK with OxyABCDJ in Streptomyces coelicolor CH999 led to the isolation of WJ78 (8), which contains the aromatized D-ring with the correct C7–C12 regioselectivity. OxyN was then identified as a second ring cyclase based on high sequence similarity to DpsY in the daunorubicin pathway (57) and MtmY in the mithramycin pathway (58). Addition of oxyN to the aforementioned cassette resulted in the production of the linear tetracyclic product WJ83T1 (12), which is a spontaneous oxidized version of pretetramid (11), as the major metabolite (35). The oxy gene cluster also encodes an additional putative cyclase, OxyI, which has sequence homology to MtmX, a putative fourth ring cyclase in the mithramycin pathway (59). Because the fully cyclized product 12 was produced upon coexpression with OxyN, the role of OxyI in the oxy pathway has remained unclear. Coexpression of oxyI in the heterologous host producing 12 had no effect on the product profile (35). To further investigate the basis of the fourth ring cyclization, Zhang et al. (35) created another construct in which the amidotransferase OxyD was removed. CH999 transformed with a cassette encoding OxyABCJKN produced only the tricyclic desmethylaklanonic acid, confirming that the terminal amide may influence the cyclization of the A-ring en route to 12.
The recently sequenced gene cluster responsible for biosynthesis of the tetracycline-like compound SF2575 allowed reconstitution of the cyclization events in this pathway as well (61). The second ring cyclase in the ssf pathway, SsfY2, has low homology to OxyN while bearing strong homology to second ring cyclases from benzoisochromanequinone pathways such as Gra-ORF33 (60). Also present in the ssf gene cluster is an OxyI homolog, SsfY4, which was also determined to be unnecessary for cyclization of the tetracyclic scaffold, confirming the result seen with OxyI. More importantly, a putative acyl-CoA ligase, SsfL2, was required to produce the tetracyclic intermediate 12 in a heterologous host (61). A possible role of SsfL2 may be to catalyze the Claisen-like cyclization between C1 and C18 of hydrolyzed tricyclic anthracene carboxylic acids in a CoA-dependent fashion. SsfL2 has homologs in other pathways that produce tetracycline-like compounds, including OTC (33), CTC (52), mithramycin (58), and the recently sequenced polyketomycin (62) gene clusters. The oxy homolog OxyH was shown, however, to be dispensable by knock-out studies.4 Therefore, the exact mechanism for fourth ring cyclization in this system is not yet completely understood.
Formation of Anhydrotetracycline
The heavily decorated cyclohexenone A-ring of tetracyclines possesses unique structural features not observed among other aromatic polyketides. These functional groups, which are essential for the antibiotic properties of tetracyclines, include the C2 primary amide, C4 dimethylamine, and the C12a tertiary alcohol. One of the most important intermediates in the transformation of pretetramid to OTC is anhydrotetracycline (ATC; 20). ATC is the first intermediate in the biosynthetic pathway to contain the fully functionalized A-ring. Recent studies by Zhang et al. (34) have identified and characterized the minimal set of enzymes required for the formation of 20.
The first tailoring step following cyclization is C6 methylation. OxyF was identified in the gene cluster as one of two S-adenosylmethionine-dependent methyltransferases and was shown to be responsible for C6 methylation by reconstitution in a heterologous host. Inclusion of the gene encoding OxyF in the aforementioned construct producing pretetramid resulted in the production of the known intermediate 6-methylpretetramid (13), which was spontaneously oxidized to WJ119 (14) (35). The methylation of C6, which is located in the lower periphery of OTC, has been known to be dispensable for the subsequent transformations by downstream enzymes (28). Accordingly, removal of OxyF from the heterologous host that produced 20 led to the biosynthesis of 6-demethyl-ATC (34).
Following C6 methylation, 6-methylpretetramid is converted to 4-keto-ATC (17) via double hydroxylation of the A-ring. A total of five putative oxygenases were present in the oxy gene cluster, and systematic reconstitution studies identified OxyL as the NADPH-dependent oxygenase required to produce 17 via hydroxylation of the C4 and C12a positions. Under in vivo conditions, 17 readily rearranged through intramolecular rearrangement of the B-ring and two tandem retro-Claisen cleavages to yield the observed degradation product WJ135 (18) (34, 63). The hydroxylation activities of OxyL were confirmed in vitro, as addition of purified OxyL to 13 produced a short-lived product identified as 17. A subsequent report by Wang et al. (64) indicated that OxyE, another FAD-dependent oxygenase, is a C4 hydroxylase and plays an ancillary role in this key dihydroxylation step that converts 13 to 17. Although OxyL can perform the dihydroxylation of C12a and C4 alone, the presence of OxyE significantly increases the titer of OTC, likely by increasing the rate of this key tailoring step (Fig. 3). Coexpression of OxyE with enzymes that produced 13 in a heterologous host led to the biosynthesis of JX11 (16), whereas inactivation of oxyE in S. rimosus led to the accumulation of shunt products that arose via spontaneous oxidation of 13. The ancillary role of OxyE to OxyL is similar to the activities of homologs observed in the polyketomycin pathway. In this pathway, Daum et al. (62) proposed the C4 hydroxylation catalyzed by the OxyE homolog PokO1 can also be catalyzed by the OxyL homolog PokO2, which also performs the C4 and C12a hydroxylations.
OxyQ has high sequence homology to the pyridoxal 5′-phosphate-dependent aspartate aminotransferases (65). As a member of this family, OxyQ was predicted to catalyze the transamination of the α-amino group from a donor amino acid to the C4 ketone of 17 to form 19. Indeed, upon expression of a cassette encoding OxyABCDJKNLQ in CH999, 19 was shown to accumulate in the fermentation broth (34). The amino acid donor of OxyQ has not been established because a soluble version of recombinant OxyQ has not been obtained so far.
Addition of the gene encoding OxyT completed the ATC-producing cassette, and these 10 enzymes were shown to be the minimal set of enzymes required for the biosynthesis of 20 in a heterologous host (34). OxyT was further characterized by in vitro assays using 19 as a substrate and S-adenosylmethionine as a methyl donor. The in vitro assay revealed that a monomethylated intermediate can be detected after short reaction times. Full conversion of 19 to 20 was observed after prolonged reaction times (34).
Formation of OTC
The remaining steps in OTC biosynthesis have not been heterologously reconstituted in vivo; however, in vitro assays and blocked mutant studies have shed light on the tailoring of 20 to OTC. The next step following formation of 20 is hydroxylation of C6. An ATC oxygenase has been purified and studied in vitro for both the CTC (66) and OTC (67, 68) pathways. In both cases, the ATC oxygenase was identified as a monooxygenase catalyzing the conversion of 20 to 21 in the presence of NADPH and atmospheric oxygen. Point mutation studies by Peric-Concha et al. (68) showed that mutation of the essential glycine residues of the NADPH-binding site eliminated activity of OtcC (also named OxyS) when expressed heterologously in E. coli. Interestingly, when this putatively inactive mutant gene was transformed into an oxyS null mutant strain, the C6 hydroxylation activity was restored, resulting in production of OTC (68). The authors proposed that this surprising result may be caused by an additional hydroxylase that may also have some activity toward the C6 position, similar to the ancillary role of OxyE to OxyL. However, in this case, the OxyS mutant is needed for hydroxylation, perhaps serving as a structural partner to the unidentified hydroxylase.
The enzymatic basis of the C5 hydroxylation step unique to the OTC pathway has yet to be resolved. Of the five putative oxygenases found in the OTC gene cluster, OxyR and OxyG have not been assigned biosynthetic functions. Sequence homology places OxyR within the pyridoxine 5′-phosphate oxidase-like protein family, which also includes ActVA-ORF2, an unassigned protein from the actinorhodin gene cluster (69). On the other hand, OxyG is predicted to be a small quinone-forming monooxygenase with homology to enzymes found in several other polyketide clusters such as mithramycin (70), aklavinone (71), and polyketomycin (62). AknX was shown in vitro to catalyze the quinone-forming reaction converting emodin anthrone to emodin, leading the authors to believe that it catalyzes the parallel reaction in the conversion of aklanonic acid anthrone to aklanonic acid (71). In the case of mithramycin, however, inactivation of the OxyG homolog MtmOIII was shown to have no effect on mithramycin production (72). The potential quinone-forming activities of OxyG homologs led to initial speculation that it may be involved in the formation of 17; however, as described above, OxyL (with the help of OxyE) has been shown to be sufficient for catalysis of this step. This does not eliminate, however, the possibility of an auxiliary role of OxyG such as that shown for OxyE (64). An interesting aspect of the C5 hydroxylation is that because it is a step unique to OTC biosynthesis, one would expect the gene responsible to be absent from the CTC gene cluster, much as the CTC C7 halogenase (73) is absent from the OTC gene cluster. A comparison of the two gene clusters reveals, however, that each of the five oxygenases in the oxy cluster has a homolog in the CTC gene cluster (33, 52). It is possible that the enzyme responsible for C5 hydroxylation is present in both gene clusters but is inactive toward a chlorinated intermediate in the CTC pathway. Alternatively, the enzyme responsible for this step in the CTC gene cluster contains an inactivating mutation. For additional comparison, the ssf gene cluster contains only two oxygenases, an OxyS homolog, SsfO1, and an OxyL homolog, SsfO2 (33, 61). Therefore, the role of the two additional oxygenases and the enzymatic basis of the C5 hydroxyl remain an unsolved mystery of OTC biosynthesis.
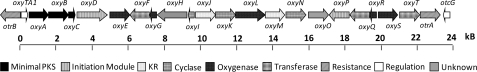
The final step in OTC biosynthesis is the reduction of the C5a–C11a double bond in 22. Nakano et al. (74) identified a gene from Streptomyces aureofaciens that was shown to be responsible for this last step in the biosynthesis of CTC. This gene, encoding TchA, was mapped outside of the previously sequenced CTC gene cluster. TchA requires 7,8-dedimethyl-8-hydroxy-5-deazariboflavin as a cofactor (74), which explains why mutants blocked in 7,8-dedimethyl-8-hydroxy-5-deazariboflavin biosynthesis, previously referred to as cosynthetic factor I, were also blocked in this final reduction step (31, 75). In the oxy pathway, there is no clear enzyme candidate that may be associated with this step. Therefore, the reductase may similarly be encoded elsewhere on the genome. In a study by Binnie et al. (75), the 34-kb region containing the oxy gene cluster and the immediate flanking regions were transformed into Streptomyces lividans and Streptomyces albus. Both recombinant strains were shown to be positive in OTC production, which suggests that the gene cluster contained all the enzymes required for OTC biosynthesis. If any genes involved in OTC production are indeed located outside the known gene cluster, they are perhaps widely found in Streptomyces strains and not specific to tetracycline biosynthesis. Most recently, Boddy and co-workers (76) demonstrated heterologous production of OTC in Myxococcus xanthus by inserting a construct containing the gene cluster shown in Fig. 4 into the host genome. This further reinforces the proposal that all the genes required to produce OTC in a prokaryote host are indeed located on this cluster.
FIGURE 4.
OTC gene cluster from S. rimosus.
Regulatory and Resistance Genes
Apart from biosynthetic genes, antibiotic producers must carry self-resistance genes to protect the host strain. The most common mechanisms of providing tetracycline resistance are through efflux proteins and ribosomal protection (17). The oxy gene cluster contains two proteins involved in tetracycline resistance, OtrA and OtrB, which are 34 kb apart and flank the biosynthetic gene cluster (77, 78). Cloning and characterization of TetA (analogous to OtrA) by Ohnuki et al. (79) indicated that it was involved in a ribosomal protection mechanism of resistance. This was further confirmed by Doyle et al. (80), who also demonstrated sequence similarity between OtrA and TetM. TetM has been shown to bind noncovalently to the ribosome and promote removal of tetracycline (81, 82). On the other hand, OtrB was shown to be a membrane-bound protein responsible for reduced accumulation of tetracycline in the cell (79, 83). The gene encoding OtrB lies with opposite polarity to the gene encoding the regulatory protein OxyTA1 in a similar fashion as the tetR/tetA pair, in which TetR negatively regulates expression of the efflux protein TetA (84). Sequence comparison places OxyTA1 closer to the MarR family of transcriptional regulators, which are known to repress expression of resistance genes in the absence of the inducer (85). Interestingly, the oxy cluster does not encode a transcriptional activator such as the SARP (Streptomyces antibiotic regulatory protein) family proteins found in many antibiotic gene clusters, including the CTC cluster (86). Recently, however, a LuxR family transcriptional activator, OtcG, has been identified by Lešnik et al. (87) to be encoded just outside of otrA. Inactivation of otcG resulted in a decrease in production of OTC, indicating that it is a positive regulator of OTC biosynthesis (87).
Conclusions
Elucidation of the biosynthetic pathway of OTC has contributed to our knowledge of type II polyketide biosynthesis and illuminated aspects unique to this pathway, such as the biosynthesis of the unusual malonamate starter unit and its effect on downstream enzymatic steps. The tetracycline scaffold has been a valuable inspiration for medicinal chemists, and tetracycline derivatives still play an important role in treating bacterial infections. Identification of the role of individual enzymes in the pathway is another step forward toward rational engineering of new compounds. Characterization of additional tetracycline and polyketide pathways will further aid our understanding of the remaining questions regarding this pathway and enhance our ability to engineer new compounds.
This work was supported in part by National Science Foundation Grant CBET 0545860. This is the second article in the Thematic Minireview Series on Antibacterial Natural Products. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
P. Wang and Y. Tang, unpublished data.
- OTC
- oxytetracycline
- CTC
- chlortetracycline
- PKS
- polyketide synthase
- KS
- ketosynthase
- CLF
- chain length factor
- ACP
- acyl carrier protein
- ADOTC
- 2-acetyl-2-decarboxamido-OTC
- KR
- ketoreductase
- ATC
- anhydrotetracycline.
REFERENCES
- 1.Hertweck C., Luzhetskyy A., Rebets Y., Bechthold A. (2007) Nat. Prod. Rep. 24, 162–190 [DOI] [PubMed] [Google Scholar]
- 2.Das A., Khosla C. (2009) Acc. Chem. Res. 42, 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutchinson C. R. (1997) Chem. Rev. 97, 2525–2536 [DOI] [PubMed] [Google Scholar]
- 4.Lombó F., Menéndez N., Salas J. A., Méndez C. (2006) Appl. Environ. Microbiol. 73, 1–14 [DOI] [PubMed] [Google Scholar]
- 5.Tsunakawa M., Nishio M., Ohkuma H., Tsuno T., Konishi M., Naito T., Oki T., Kawaguchi H. (1989) J. Org. Chem. 54, 2532–2536 [Google Scholar]
- 6.Hunter I. S., Hill R. A. (1997) in Biotechnology of Antibiotics (Strohl W. R. ed.) pp 659–682, 2 Ed., Marcel Dekker, Inc., New York [Google Scholar]
- 7.Duggar B. M. (1948) Ann. N.Y. Acad. Sci. 51, 177–181 [DOI] [PubMed] [Google Scholar]
- 8.Duggar B. M. (September13, 1949) U. S. Patent 2,482,055
- 9.Finlay A. C., Hobby G. L., P'an S. Y., Regna P. P., Routein J. B., Seeley D. B., Shull G. M., Sobin B. A., Solomons I. A., Vinson J. W., Kane J. H. (1950) Science 111, 8515400447 [Google Scholar]
- 10.Chopra I., Roberts M. (2001) Microbiol. Mol. Biol. Rev. 65, 232–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodersen D. E., Clemons W. M., Jr., Carter A. P., Morgan-Warren R. J., Wimberly B. T., Ramakrishnan V. (2000) Cell 103, 1143–1154 [DOI] [PubMed] [Google Scholar]
- 12.Hochstein F. A., Stephens C. R., Conover L. H., Regna P. P., Pasternack R., Gordon P. N., Pilgrim F. J., Brunings K. J., Woodward R. B. (1953) J. Am. Chem. Soc. 75, 5455–5475 [Google Scholar]
- 13.Stephens C. R., Conover L. H., Pasternack R., Hochstein F. A., Moreland W. T., Regna P. P., Pilgrim F. J., Brunings K. J., Woodward R. B. (1954) J. Am. Chem. Soc. 76, 3568–3575 [Google Scholar]
- 14.Nelson M. L., Park B. H., Levy S. B. (1994) J. Med. Chem. 37, 1355–1361 [DOI] [PubMed] [Google Scholar]
- 15.Nelson M. L. (2002) Annu. Rep. Med. Chem. 37, 105–114 [Google Scholar]
- 16.Levy S. B., McMurry L. M., Burdett V., Courvalin P., Hillen W., Roberts M. C., Taylor D. E. (1989) Antimicrob. Agents Chemother. 33, 1373–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy S. B., McMurry L. M., Barbosa T. M., Burdett V., Courvalin P., Hillen W., Roberts M. C., Rood J. I., Taylor D. E. (1999) Antimicrob. Agents Chemother. 43, 1523–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens C. R., Beereboom J. J., Rennhard H. H., Gordon P. N., Murai K., Blackwood R. K., von Wittenau M. S. (1963) J. Am. Chem. Soc. 85, 2643–2652 [Google Scholar]
- 19.Villax I., Page P. (February19, 1985) U. S. Patent 4,500,458
- 20.Agwuh K. N., MacGowan A. (2006) J. Antimicrob. Chemother. 58, 256–265 [DOI] [PubMed] [Google Scholar]
- 21.Chopra I. (2001) Curr. Opin. Pharmacol. 1, 464–469 [DOI] [PubMed] [Google Scholar]
- 22.Sum P. E., Petersen P. (1999) Bioorg. Med. Chem. Lett. 9, 1459–1462 [DOI] [PubMed] [Google Scholar]
- 23.Bergeron J., Ammirati M., Danley D., James L., Norcia M., Retsema J., Strick C. A., Su W. G., Sutcliffe J., Wondrack L. (1996) Antimicrob. Agents Chemother. 40, 2226–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas R., Williams D. J. (1983) J. Chem. Soc. Chem. Commun. 12, 128–130 [Google Scholar]
- 25.Thomas R., Williams D. J. (1983) J. Chem. Soc. Chem. Commun. 12, 677–679 [Google Scholar]
- 26.McCormick J. R., Jensen E. R. (1965) J. Am. Chem. Soc. 87, 1794–1795 [DOI] [PubMed] [Google Scholar]
- 27.McCormick J. R., Jensen E. R., Johnson S., Sjolander N. O. (1968) J. Am. Chem. Soc. 90, 2201–2202 [DOI] [PubMed] [Google Scholar]
- 28.McCormick J. R., Johnson S., Sjolander N. O. (1963) J. Am. Chem. Soc. 85, 1692–1694 [Google Scholar]
- 29.McCormick J. R., Miller P. A., Johnson S., Arnold N., Sjolander N. O. (1962) J. Am. Chem. Soc. 84, 3023–3025 [Google Scholar]
- 30.McCormick J. R., Reichenthal J., Johnson S., Sjolander N. O. (1963) J. Am. Chem. Soc. 85, 1694–1695 [Google Scholar]
- 31.Rhodes P. M., Winskill N., Friend E. J., Warren M. (1981) J. Gen. Microbiol. 124, 329–338 [Google Scholar]
- 32.Kim E. S., Bibb M. J., Butler M. J., Hopwood D. A., Sherman D. H. (1994) Gene 141, 141–142 [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Ames B. D., Tsai S. C., Tang Y. (2006) Appl. Environ. Microbiol. 72, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W., Watanabe K., Cai X., Jung M. E., Tang Y., Zhan J. (2008) J. Am. Chem. Soc. 130, 6068–6069 [DOI] [PubMed] [Google Scholar]
- 35.Zhang W., Watanabe K., Wang C. C., Tang Y. (2007) J. Biol. Chem. 282, 25717–25725 [DOI] [PubMed] [Google Scholar]
- 36.Petković H., Cullum J., Hranueli D., Hunter I. S., Perić-Concha N., Pigac J., Thamchaipenet A., Vujaklija D., Long P. F. (2006) Microbiol. Mol. Biol. Rev. 70, 704–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickens L. B., Tang Y. (2009) Metab. Eng. 11, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hertweck C. (2009) Angew. Chem. Int. Ed. Engl. 48, 4688–4716 [DOI] [PubMed] [Google Scholar]
- 39.Revill W. P., Bibb M. J., Hopwood D. A. (1995) J. Bacteriol. 177, 3946–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keatinge-Clay A. T., Maltby D. A., Medzihradszky K. F., Khosla C., Stroud R. M. (2004) Nat. Struct. Mol. Biol. 11, 888–893 [DOI] [PubMed] [Google Scholar]
- 41.Tang Y., Tsai S. C., Khosla C. (2003) J. Am. Chem. Soc. 125, 12708–12709 [DOI] [PubMed] [Google Scholar]
- 42.Moore B. S., Hertweck C. (2002) Nat. Prod. Rep. 19, 70–99 [DOI] [PubMed] [Google Scholar]
- 43.Boehlein S. K., Richards N. G., Schuster S. M. (1994) J. Biol. Chem. 269, 7450–7457 [PubMed] [Google Scholar]
- 44.Fu H., Ebert-Khosla S., Hopwood D. A., Khosla C. (1994) J. Am. Chem. Soc. 116, 6443–6444 [Google Scholar]
- 45.Bao W., Sheldon P. J., Hutchinson C. R. (1999) Biochemistry 38, 9752–9757 [DOI] [PubMed] [Google Scholar]
- 46.Tang Y., Lee T. S., Kobayashi S., Khosla C. (2003) Biochemistry 42, 6588–6595 [DOI] [PubMed] [Google Scholar]
- 47.Meadows E. S., Khosla C. (2001) Biochemistry 40, 14855–14861 [DOI] [PubMed] [Google Scholar]
- 48.Bililign T., Hyun C. G., Williams J. S., Czisny A. M., Thorson J. S. (2004) Chem. Biol. 11, 959–969 [DOI] [PubMed] [Google Scholar]
- 49.Das A., Khosla C. (2009) Chem. Biol. 16, 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Y., Koppisch A. T., Khosla C. (2004) Biochemistry 43, 9546–9555 [DOI] [PubMed] [Google Scholar]
- 51.Lykkeberg A. K., Sengeløv G., Cornett C., Tjørnelund J., Hansen S. H., Halling-Sørensen B. (2004) J. Pharm. Biomed. Anal. 34, 559–567 [DOI] [PubMed] [Google Scholar]
- 52.Ryan M. J. (December31, 1996) U. S. Patent 5,589,385
- 53.Fritzsche K., Ishida K., Hertweck C. (2008) J. Am. Chem. Soc. 130, 8307–8316 [DOI] [PubMed] [Google Scholar]
- 54.Korman T. P., Tan Y. H., Wong J., Luo R., Tsai S. C. (2008) Biochemistry 47, 1837–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu H., Ebert-Khosla S., Hopwood D. A., Khosla C. (1994) J. Am. Chem. Soc. 116, 4166–4170 [Google Scholar]
- 56.Petkovic H., Thamchaipenet A., Zhou L. H., Hranueli D., Raspor P., Waterman P. G., Hunter I. S. (1999) J. Biol. Chem. 274, 32829–32834 [PubMed] [Google Scholar]
- 57.Lomovskaya N., Doi-Katayama Y., Filippini S., Nastro C., Fonstein L., Gallo M., Colombo A. L., Hutchinson C. R. (1998) J. Bacteriol. 180, 2379–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prado L., Lombó F., Braña A. F., Méndez C., Rohr J., Salas J. A. (1999) Mol. Gen. Genet. 261, 216–225 [DOI] [PubMed] [Google Scholar]
- 59.Künzel E., Wohlert S. E., Beninga C., Haag S., Decker H., Hutchinson R. C., Blanco G., Mendez C., Salas J. A., Rohr J. (1997) Chem. Eur. J. 3, 1675–1678 [Google Scholar]
- 60.Ichinose K., Bedford D. J., Tornus D., Bechthold A., Bibb M. J., Revill W. P., Floss H. G., Hopwood D. A. (1998) Chem. Biol. 5, 647–659 [DOI] [PubMed] [Google Scholar]
- 61.Pickens L. B., Kim W., Wang P., Zhou H., Watanabe K., Gomi S., Tang Y. (2009) J. Am. Chem. Soc. 131, 17677–17689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daum M., Peintner I., Linnenbrink A., Frerich A., Weber M., Paululat T., Bechthold A. (2009) ChemBioChem 10, 1073–1083 [DOI] [PubMed] [Google Scholar]
- 63.Scott A. I., Yamaguchi E., Chung S. K. (1975) Tetrahedron Lett. 16, 1369–1372 [Google Scholar]
- 64.Wang P., Zhang W., Zhan J., Tang Y. (2009) ChemBioChem 10, 1544–1550 [DOI] [PubMed] [Google Scholar]
- 65.Nakai T., Okada K., Akutsu S., Miyahara I., Kawaguchi S., Kato R., Kuramitsu S., Hirotsu K. (1999) Biochemistry 38, 2413–2424 [DOI] [PubMed] [Google Scholar]
- 66.Béhal V., Hoštálek Z., Vaněk Z. (1979) Biotechnol. Lett. 1, 177–182 [Google Scholar]
- 67.Butler M. J., Gedge B. N. (1989) Biotechnol. Tech. 4, 235–238 [Google Scholar]
- 68.Peric-Concha N., Borovicka B., Long P. F., Hranueli D., Waterman P. G., Hunter I. S. (2005) J. Biol. Chem. 280, 37455–37460 [DOI] [PubMed] [Google Scholar]
- 69.Caballero J. L., Martinez E., Malpartida F., Hopwood D. A. (1991) Mol. Gen. Genet. 230, 401–412 [DOI] [PubMed] [Google Scholar]
- 70.Lombó F., Blanco G., Fernández E., Méndez C., Salas J. A. (1996) Gene 172, 87–91 [DOI] [PubMed] [Google Scholar]
- 71.Chung J. Y., Fujii I., Harada S., Sankawa U., Ebizuka Y. (2002) J. Bacteriol. 184, 6115–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abdelfattah M. S., Rohr J. (2006) Angew. Chem. Int. Ed. Engl. 45, 5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dairi T., Nakano T., Aisaka K., Katsumata R., Hasegawa M. (1995) Biosci. Biotechnol. Biochem. 59, 1099–1106 [DOI] [PubMed] [Google Scholar]
- 74.Nakano T., Miyake K., Endo H., Dairi T., Mizukami T., Katsumata R. (2004) Biosci. Biotechnol. Biochem. 68, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 75.Binnie C., Warren M., Butler M. J. (1989) J. Bacteriol. 171, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stevens D. C., Henry M. R., Murphy K. A., Boddy C. N. (2010) Appl. Environ. Microbiol. 76, 2681–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Butler M. J., Friend E. J., Hunter I. S., Kaczmarek F. S., Sugden D. A., Warren M. (1989) Mol. Gen. Genet. 215, 231–238 [DOI] [PubMed] [Google Scholar]
- 78.Rhodes P. M., Hunter I. S., Friend E. J., Warren M. (1984) Biochem. Soc. Trans. 12, 586–587 [DOI] [PubMed] [Google Scholar]
- 79.Ohnuki T., Katoh T., Imanaka T., Aiba S. (1985) J. Bacteriol. 161, 1010–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doyle D., McDowall K. J., Butler M. J., Hunter I. S. (1991) Mol. Microbiol. 5, 2923–2933 [DOI] [PubMed] [Google Scholar]
- 81.Burdett V. (1996) J. Bacteriol. 178, 3246–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Connell S. R., Tracz D. M., Nierhaus K. H., Taylor D. E. (2003) Antimicrob. Agents Chemother. 47, 3675–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reynes J. P., Calmels T., Drocourt D., Tiraby G. (1988) J. Gen. Microbiol. 134, 585–598 [DOI] [PubMed] [Google Scholar]
- 84.Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., Gallegos M. T., Brennan R., Tobes R. (2005) Microbiol. Mol. Biol. Rev. 69, 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. (2001) Nat. Struct. Biol. 8, 710–714 [DOI] [PubMed] [Google Scholar]
- 86.Wietzorrek A., Bibb M. (1997) Mol. Microbiol. 25, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 87.Lešnik U., Gormand A., Magdevska V., Fujs Š, Raspor P., Hunter I., Petkovic H. (2009) Food Technol. Biotechnol. 47, 323–330 [Google Scholar]