Abstract
Antibiotics of the thiocillin, GE2270A, and thiostrepton class, which block steps in bacterial protein synthesis, contain a trithiazolyl (tetrahydro)pyridine core that provides the architectural constraints for high affinity binding to either the 50 S ribosomal subunit or elongation factor Tu. These mature antibiotic scaffolds arise from a cascade of post-translational modifications on 50–60-residue prepeptide precursors that trim away the N-terminal leader sequences (∼40 residues) while the C-terminal 14–18 residues are converted into the mature scaffold. In the producing microbes, the genes encoding the prepeptide open reading frames are flanked in biosynthetic clusters by genes encoding post-translational modification enzymes that carry out lantibiotic-type dehydrations of Ser and Thr residues to dehydroamino acid side chains, cyclodehydration and oxidation of cysteines to thiazoles, and condensation of two dehydroalanine residues en route to the (tetrahydro)pyridine core. The trithiazolyl pyridine framework thus arises from post-translational modification of the peptide backbone of three Cys and two Ser residues of the prepeptide.
Keywords: Antibiotics, Antimicrobial Peptides, Post-translational Modification, Ribosome Function, Translation Elongation Factors, Natural Products, Thiocillin, Thiopeptide
Context for Peptide Antibiotic Biosynthesis
Antibiotic-producing bacteria and fungi show remarkable versatility in fashioning simple building blocks from primary metabolism into complex scaffold architectures with specifically arrayed functional groups to interact with particular targets in susceptible neighboring microbes (1, 2). This is particularly notable in the economic utilization of malonyl- and methylmalonyl-CoA monomer units to make remarkably diverse polyketide frameworks as noted in two of the minireviews in this thematic series, covering the macrocyclic erythromycin family and the polycyclic aromatic tetracyclines (3, 4).
Parallel logic and machinery have evolved to use amino acids to elaborate peptide-based antibiotics (5–7). The nonribosomal peptide synthetase (NRPS)4 assembly lines are famous for turning out clinically important antibiotics, including the β-lactams (penems, cephems, and carbapenems) (8) and the glycopeptides of the vancomycin family (9, 10). More recently, the nonribosomally derived lipodepsipeptide daptomycin (11) has achieved notable clinical success in treatment of bacterial infections, as noted in the third minireview in this series (12).
A counterpoint strategy to these RNA-independent biosyntheses of peptide-like compounds involves ribosomal proteins that are processed post-translationally to antibiotic peptides (6, 7). Although perhaps less well known than the NRPS-assembled peptide antibiotics, the ribosomal antibiotic peptides are widely distributed throughout prokaryotes; multicellular eukaryotes, including humans, also turn out large quantities of ribosomally encoded defensins for innate immunity (13). The focus of this minireview is ribosomal peptide antibiotics that have undergone more extensive processing than just the proteolytic removal of leader peptides (14). These are peptide antibiotics that have undergone post-translational modification of side chains and backbones to turn hydrolytically labile and acyclic floppy peptides into conformationally rigid architectures that are protease-resistant. In this class fall the lantibiotics (15, 16) such as nisin, long used in food preservatives; the thiazole- and oxazole-containing Escherichia coli antibiotic peptide microcin B17, which targets DNA gyrase (17, 18); and the recently codified cyanobactin family (19, 20). But the pièces de résistance for iterative post-translational sculpting of peptide frameworks are the thiazolyl peptide antibiotics, produced by many Gram-positive bacteria.
Thiazolyl Peptide Antibiotic Family
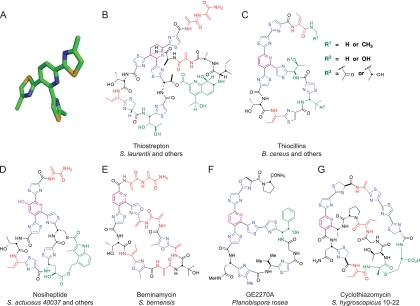
The characteristic features of the thiazolyl peptide antibiotic class are a central pyridine/tetrahydropyridine ring, with up to three thiazolyl substituents at the 2-, 3-, and 6-positions of the central heterocycle to yield a rigid tetracyclic core with propeller-shaped architecture (Fig. 1A) (for review, see Ref. 21). In members with the central nitrogen ring in the pyridine oxidation state (e.g. thiocillins) (Fig. 1B), the peptide chain runs as a macrocyclic loop between the thiazole rings from the 3- to the 2-position of the pyridine. The chain then exits from the thiazole at the 6-position and terminates two to four residues downstream (22–28). When the central ring is in the tetrahydropyridine oxidation state (e.g. thiostrepton) (Fig. 1C), the upstream peptide chain (the presequence or leader sequence) of the nascent peptide precursor is still attached to the tetrahydropyridine at the same carbon as thiazole 2. Some members of the thiostrepton subfamily have a second macrocyclic loop, utilizing this upstream peptide for modifications including macrocyclization and additional rigidification (29–32).
FIGURE 1.
Molecular structures of thiopeptide antibiotics. A, three-dimensional rendering of the thiocillin core depicting the propeller-like architecture resulting from macrocyclization. B, molecular structure of the eight members of the thiocillin family produced by B. cereus containing a trithiazolyl pyridine core. C, molecular structure of thiostrepton produced by Streptomyces laurentii and containing a trithiazolyl tetrahydropyridine core. Note the presence of the upstream sequence, attached to the tetrahydropyridine at the same carbon as thiazole 2, which constitutes part of the second macrocyclic loop. D, nosiheptide contains a second macrocycle consisting of an indolic acid attached between the side chains of Cys-10 and Glu-8 of the primary macrocycle. E, the macrocycle of berninamycin is extended by nine bonds between connections with the central pyridine compared with thiocillin and nosiheptide (35 bonds versus 26 bonds, respectively). F, GE2270A also has an elongated macrocycle compared with thiocillin, with 29 bonds between pyridine connections. G, the second macrocycle of cyclothiazomycin is generated by conjugation of the C-terminal segment to the side chain of a primary macrocycle amino acid.
There are close to 100 members of the thiazolyl peptide antibiotic family isolated to date (21), beginning with micrococcin, isolated from Oxford sewage in 1948 (33), with new isolates reported as recently as 2009 (34). Structure determination was historically challenging until sophisticated NMR methodology arrived. Total syntheses of a small number of this class reflect heroic efforts on the part of the synthetic community (35–43) but are not likely to be practical routes if large quantities would be needed, leaving fermentation as the practical manufacturing option.
Two Classes of Bacterial Targets
As might be expected for antibiotics in the 1100–1200-Da range, the thiazolyl peptides do not get past the outer membrane barriers of Gram-negative bacteria but typically have submicromolar potencies against Gram-positive bacteria. They block bacterial protein synthesis selectively, as will be reviewed below, but have not achieved human clinical success due to twin failings of poor solubility and unfavorable pharmacokinetics.
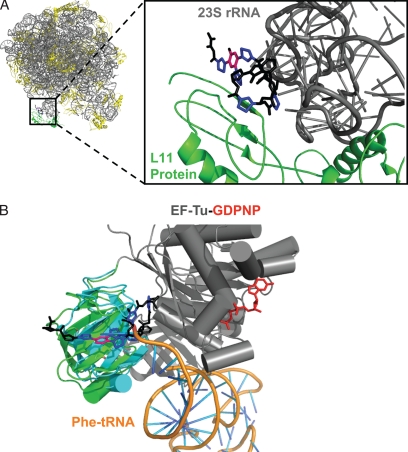
Thiazolyl peptide compounds with antibiotic activity target consecutive steps of bacterial translation elongation (for a review of translation elongation, see Ref. 44). Molecules such as thiostrepton, thiocillins, and nosiheptide (Fig. 1, B–D) have been known for decades to bind to the 50 S ribosomal subunit; resistance-generating mutations in particular loops of 23 S rRNA and residues in large ribosomal protein subunit L11 allowed initial target site mapping (45–56). The recently determined x-ray structures of the 50 S ribosomal subunit with thiostrepton and nosiheptide bound show the precise interaction of the trithiazolyl pyridine core and macrocyclic loops with the 23 S rRNA/L11 protein interface (57). This region of the ribosome constitutes part of the GTPase-activating center, which is responsible for the GTP/GDP cycling required to coordinate the events of bacterial translation. Binding of the antibiotics in this region blocks productive docking of two GTPase elongation factors, EF-G and EF-Tu, responsible for translocation of the A-site tRNA into the ribosomal P-site and subsequent aminoacylated tRNA delivery to the vacated A-site, respectively (Fig. 2A) (46, 58–60).
FIGURE 2.
Thiopeptide antibiotics bound to their targets in translation elongation. A, crystal structure of nosiheptide bound to the 50 S ribosomal subunit in a cleft formed by protein L11 (green) and the 23 S rRNA (gray). The central pyridine (pink) and thiazoles (blue) of nosiheptide are highlighted (Protein Data Bank code 2ZJP (57)). B, overlay of crystal structures of EF-Tu binding Phe-tRNA and GE2270A independently. This figure was adapted from Ref. 65. For clarity, only domain 2 has been aligned; domains 1 and 3 (gray), domain 2 (green), and guanosine 5-(β,γ-imido)triphosphate (GDPNP; red) correspond to EF-Tu bound to GE2270A (Protein Data Bank code 2C77 (68)). Domain 2 (blue) corresponds to EF-Tu bound to Phe-tRNA (orange) (Protein Data Bank code 1TTT (66)).
On the other hand, the subfamily of thiazolyl peptides represented by GE2270A (Fig. 1F) (61–63) and thiomuracin (64) congeners blocks protein synthesis at a prior step by tight binding to the EF-Tu conditional GTPase/chaperone that ferries aminoacylated tRNAs to the ribosome. X-ray analyses (Fig. 2B) show that the binding site of GE2270A overlaps with that of Phe-tRNA and thereby blocks its binding (65–68). One of the unsolved questions in this area of science involves determining which factors direct a thiazolyl peptide antibiotic to the 50 S subunit versus EF-Tu and whether one could engineer a hybrid scaffold that could hit both targets. Because the structures have been so complex and the total syntheses so long and difficult, essentially no structure-activity chemical relationships have been explored regarding this question.
Biosynthesis: Ribosomal or Nonribosomal
For many years, the logic and machinery of the biosynthesis of this antibiotic class remained obscure. The key features to be explained include 1) the timing and mechanism of dehydroalanine (Dha) and dehydrobutyrine (Dhb) residue formation, 2) the timing and mechanism of thiazole and oxazole heterocycles, and 3) the construction of the central (tetrahydro)pyridine ring. On the one hand, it is known that thiazoles can be formed by NRPS assembly lines (69–73), and a publication in 2001 argued for NRPS machinery in production of micrococcin P1 (in the thiocillin class) by Staphylococcus equorum (74). On the other hand, dehydration of Ser side chains to Dha and Thr to Dhb has been elucidated in detail for the post-translational processing of nisin and other lantibiotics that are ribosomal peptides (15, 75–78). Analogously, microcin B17 arises from a ribosomally generated precursor peptide (hereafter, prepeptide) in which the leader sequence is a crucial recognition element for a set of three enzymes that convert four Cys residues to thiazoles and four Ser residues to oxazoles (69, 79–82). More recently, the work of Schmidt and co-workers (19, 20) showed that the 100 or so cyanobactins, cyclic peptides with post-translationally generated heterocyclic rings, were similarly ribosomal in origin.
Thus, the stage was set, in the era of a thousand sequenced bacterial genomes, for both genome mining and directed cloning approaches to definitively rule in the ribosomal paradigm for this antibiotic class. Four groups, publishing within a 1-month period early in 2009 (64, 83–85), reported genes encoding short prepeptides and adjacent post-translational processing enzyme candidates for thiomuracin, GE2270A (Nonomuraea strains), thiostrepton, siomycin A (Streptomyces strains), and several thiocillin variants (Bacillus cereus). Subsequently, two additional gene clusters for nosiheptide (86) and cyclothiazomycin (87) have since been reported, validating the general organization of the clustering of the gene for a prepeptide and genes for tailoring enzymes to convert the prepeptide to the mature antibiotic.
The approach we took (83) with thiocillin was to assume that the Dhb residues at positions 4 and 13 came from threonines, the six thiazoles from cysteines, and the central pyridine from two Dha residues, the last a hypothesis put forth by Bycroft and Gowland (22) and then refined by Floss et al. (88, 89). As shown in Fig. 3A, this rational deconvolution would turn thiocillin back into a 14-residue peptide precursor. We then scanned the bacterial genome data base and found an exact match of these 14 residues at the C terminus of a 52-residue ORF in a B. cereus genome (ATCC 14579). We validated that this strain was a thiocillin producer that generates a set of eight variants.
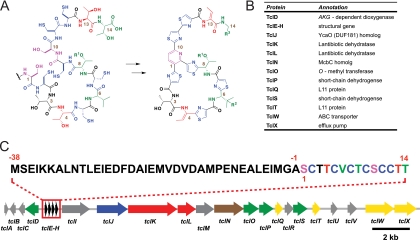
FIGURE 3.
Gene cluster responsible for biosynthesis of the thiocillin family of thiopeptides. A, predicted deconvolution of the mature thiocillin scaffold to generate a 14-amino acid precursor that could yield thiocillin through post-translational modifications. The resulting amino acid sequence is SCTTCVCTCSCCTT. B, list of genes and their purported functions responsible for thiocillin biosynthesis. C, graphical depiction of the thiocillin biosynthetic gene cluster. Four identical copies of a gene were identified that encode a 52-amino acid peptide containing the predicted 14-amino acid precursor of thiocillin.
A complementary approach, design of degenerate PCR primers to match the anticipated prepeptide-coding sequence or one of the cyclodehydration enzymes, allowed cloning of the thiostrepton (84, 85), thiomuracin (64), and nosiheptide (86) clusters. The gene cluster organization for all of these thiazolyl peptides is remarkably conserved: a prepeptide gene, followed by a suite of genes encoding post-translational maturation enzymes and export pumps and, in some clusters, predicted self-resistance genes (64, 83–87).
Cascade of Post-translational Modifications
Analysis of the thiocillin biosynthetic gene cluster from B. cereus ATCC 14579 revealed four identical copies of the prepeptide-coding gene (tclE–H), followed by seven/eight genes (Fig. 3, B and C) that are predicted to have the desired features for a post-translational cascade, including a pair of genes homologous to lantibiotic-processing enzymes (tclK/tclL) that could make Dha at positions 1 and 10 and Dhb at positions 4 and 13 (Fig. 3A). Two microcin B17 operon homologs (tclJ/tclN) could turn six Cys side chains at positions 2, 5, 7, 9, 11, and 12 into thiazolines by cyclodehydration, followed by oxidation to the heteroaromatic thiazoles (Fig. 4, A and B). The remaining ORFs encode likely candidates for catalyzing hydroxylation of Val-6, O-methylation of Thr-8, and oxidative decarboxylation of Thr-14. These three enzymes appear to act stochastically, yielding 23 = 8 thiocillin variants, differing in the modification state of Val-6, Thr-8, and Thr-14 but otherwise containing the same trithiazolyl pyridine core.
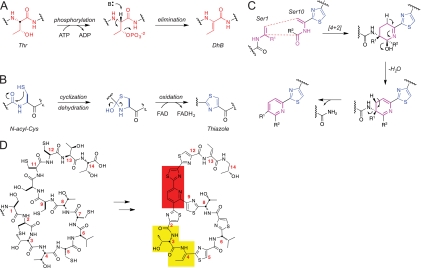
FIGURE 4.
Mechanisms and essential residues involved in maturation of thiopeptide compounds. A, Dha and Dhb are formed by a two-step mechanism involving phosphorylation of the Ser or Thr side chain. Base abstraction of the β-carbon proton results in formation of the sp2 center and elimination of the phosphate. B, thiazole formation begins with attack of a Cys thiol on the carbonyl of the previous amino acid residue. Rearrangement results in dehydration producing thiazoline, which is then oxidized to thiazole in an FAD-dependent mechanism. C, formation of the central pyridine ring of thiocillin is thought to proceed by [4 + 2] cycloaddition of the Dha residues derived from Ser-1 and Ser-10. Subsequent dehydration and elimination of the leader peptide by scission of the C–N bond complete pyridine formation. D, amino acids Ser-1 and Ser-10, responsible for formation of the central pyridine of thiocillin (red), are essential for production of thiocillin. Substitutions preventing thiazole formation from Cys-11 (red) completely inhibit production of thiocillin, suggesting that Cys-11 is crucial in the maturation of the thiocillin scaffold. Although substitutions at Thr-3 and Dhb-4 (yellow) should not alter the overall architecture of the macrocycle, alanine substitutions at these positions result in loss of antibiotic activity, consistent with their purported role in ribosome recognition and binding.
The eighth ORF, tclM in the thiocillin cluster, is a candidate for the enzyme that may catalyze cyclization of Dha-1 and Dha-10 (derived from Ser-1 and Ser-10) to the pyridine ring. Nothing is known yet about timing or intermediates, although this is probably the central mechanistic question in assembly of the scaffold of this antibiotic class. If the process were to be concerted, it would constitute a [4 + 2] type of aza-Diels-Alder cyclization (Fig. 4C), an as yet undetected class of biochemical transformation (90). By the end of the modification process for enzymatic conversion of the 52-mer prepeptide into the mature scaffold of thiocillin, 13 of the 14 amino acids have undergone modification, and 14 post-translational enzymatic steps have occurred.
This is typical for the other subfamilies of the thiazolyl peptide antibiotics. In the thiostrepton case, additional modifications are required to build the second conformation-restraining macrocyclic loop, including fashioning a bicyclic quinaldic acid spacer from tryptophan and inserting it into the peptide chain (Fig. 1C) (84, 85). Likewise, the secondary macrocycle of nosiheptide also contains a tryptophan-derived bicyclic ring system, in this case, indolic acid (Fig. 1D) (21, 86).
The cyclothiazomycin gene cluster (87) reveals a 60-residue prepeptide whose 18 C-terminal amino acids are modified to a macrobicyclic scaffold (Fig. 1G) with two notable variant features: first, the central pyridine has only one thiazole ring attached, whereas the other two attached residues are Asn-2 and Ala-10, indicating that the pyridine ring formation does not require the maturing peptide chain to have been preconstrained by three thiazoles; second, there is an unusual tertiary thioether bridge between what had been Ser-5 and the C-terminal Cys-18, perhaps arising from dehydrative conversion of Ser-5 to Dha-5, Michael addition by the thiol side chain of Cys-18, and rearrangement to the observed α-thiol Ala-5–Cys-18 linkage. Cyclothiazomycin is not a protein synthesis inhibitor but rather an inhibitor of plasma renin (91–93), presumably reflecting a distinct architecture around the pyridine core, not recognized by EF-Tu or the large ribosomal subunits.
Genetic Approaches to Structure and Activity of Thiocillins
Because of the tractability of B. cereus to genetic manipulations, we have undertaken initial structure-activity studies in vivo by manipulating the sequence of the 52-mer prepeptide at the chromosomal level. To that end, a clean knock-out of the four identical genes encoding the prepeptide substrate (tclE–H) was generated by double-crossover homologous recombination, abolishing thiocillin production (94). Then, antibiotic production was restored by reintroduction of a single copy of the wild-type tclE gene into the chromosome, setting the stage for introduction of mutant forms of the tclE gene. Only limited studies have been conducted on changes in the leader peptide region (residues −1 to −38) because initial truncations led to loss of thiocillin production5; instead, our efforts have focused on residues 1–14, retained in the mature antibiotic scaffold.
Of the 14 residues, only Thr-3 is unmodified in the mature antibiotic, suggesting that all other sites may be important in maturation. As an initial probe of the importance of side chains, alanine scanning was effected by replacement of each of the 14 residues individually. Production and post-translational processing of 11 of the 14 alanine mutants were observed; no products were detected when Ser-1, Ser-10, or Cys-11 was replaced with Ala (Fig. 4D) (94, 95). It was anticipated that the Ala-1 and Ala-10 mutants would be incompetent to form the central pyridine core. Lack of product from Ala-11 could imply that formation of a thiazole from Cys-11 is an essential step in generation of the trithiazolyl pyridine framework of the mature antibiotics. All 11 of the single Ala mutants still had the central pyridine intact. The C2A and C9A mutants had bithiazolyl pyridine cores rather than the trithiazolyl pyridine core and had lost antibiotic activity, suggesting that the propeller architecture of the central core and the directing effect on the macrocyclic peptide loop (residues 3–8) are essential for recognition by the 50 S ribosomal subunit. The alanine scan also revealed that activity was attenuated when the thiazoles arising from Cys-5 and Cys-7 were replaced with Ala-5 and Ala-7, presumably a result of reducing the conformational rigidity in the macrocyclic loop. Also Ala-3 and Ala-4 replacements were inactive, indicating the Thr-3–Dhb-4 sector of the mature thiazolyl peptide is important in binding to the ribosome target (Fig. 4D) (57, 94, 95).
Because one of the limitations of the thiazolyl peptide antibiotic class in treating bacterial infections in humans is poor solubility, we examined whether the tcl post-translational machinery could handle Lys-3, Lys-6, and Lys-8 as replacements for Thr, Val, and Thr, respectively; the trithiazolyl pyridine scaffolds were produced, in lesser quantities, but antibiotic activity was not maintained (94). The same was true when negatively charged Asp side chains were used at positions 3 and 6: the trithiazolyl pyridine core was formed, but the antibiotic activity was reduced at least 2 logs in potency.
One other set of mutants in the tclE prepeptide gene involved replacement of each of the six Cys residues with Ser. Three of the Cys residues (at positions 2, 9, and 11) flank the central pyridine ring in the propeller array. Cys-5 and Cys-7 are in the macrocyclic loop, providing rigidifying conformational restraint, whereas Cys-12 becomes part of the bithiazole moiety in mature thiocillin scaffolds.
The corresponding Ser replacements test the partition of each Ser residue between two post-translational processing alternatives. One is the lantibiotic-type pathway (tclK/tclL), involving transient O-phosphorylation and then elimination of Pi to yield the olefinic Dha, as occurs in the processing of Ser-1 and Ser-10. The other (tclJ/tclN) is the microcin-type cyclodehydration to oxazoline and aromatizing oxidation to the oxazole ring. Therefore, a Cys-to-Ser replacement could generate four possible alternatives: 1) no modification of the CH2OH serine side chain, 2) Dha formation, 3) cyclodehydration and accumulation of the oxazoline ring, and 4) dehydrogenation of the oxazoline intermediate to the stable oxazole ring. For example, the C2S mutant yielded the unmodified Ser and the oxazoline, whereas oxazoline and oxazole derivatives were detected for the C12S mutant. Notably absent were any products from the C11S mutant, reminiscent of the absence of maturation of the C11A mutant, consistent with a special role for the thiazole at residue 11 in the trithiazolyl pyridine core construction.
Outlook
A major goal for engineering of the post-translational modification of prepeptides into the various subgroups of thiazolyl peptide antibiotics is to improve the aqueous solubility and unfavorable pharmacokinetic properties that have to date obviated their clinical development for human infections by Gram-positive bacterial pathogens. Because of the structural complexity and lengthy total synthetic routes to the fully mature scaffolds, very few medicinal chemistry efforts have been reported, mostly involving reconstruction of the C terminus to make more soluble variants (96–99). Also, it appears that fragments of the core have lost orders of magnitude of antibiotic potency (100), consistent with requirement for the full architectural array for potent bacterial protein synthesis target recognition.
Thus, reprogramming both the prepeptide sequence and the tailoring specificity and capacity of the cascade of affiliated post-translational modification enzymes offers promise to explore what resculpting of the scaffold can be engineered and then which modifications retain full antibiotic potency. Although initial in vivo efforts to engineer more hydrophilic thiocillins have been achieved, the Lys and Asp side chains so far introduced do not retain antibiotic activity (94). However, producer organisms can hydroxylate and/or glycosylate selected thiazolyl peptide antibiotics (25, 32, 101–103), so it will be of interest to move those tailoring enzymes to heterologous thiazolyl peptide producers and examine the outcomes for increased aqueous solubility and potency retention.
In turn, reconstitution of the maturation steps by in vitro studies with purified enzymes will help to establish the timing of multiple dehydroamino acid formations (four in thiocillins), of heterocyclization of Cys to thiazoles (six in thiocillins), and of the dimerization of two Ser-derived Dha residues to a dihydropyridine ring, with subsequent redox adjustment, up to the heteroaromatic pyridine and down to the tetrahydropyridine found in thiostrepton. Which, if any, of the thiazole rings must form first (perhaps to induce a kinked conformation in the prepeptide backbone) to bring Dha-1 and Dha-10 in sufficient proximity to undergo the proposed aza-Diels-Alder cyclization is yet unknown, but we have cited evidence that Cys-11 cannot be replaced in thiocillin maturation.
Of particular interest will be deciphering the parameters that control regiochemistry of pyridine ring formation. In both the thiocillin and thiostrepton subclasses, Ser-1 and Ser-10 end up, after dehydration, as the framework of the pyridine/tetrahydropyridine ring with an intervening 26-atom peptide loop. In GE2270A, the core pyridine is instead formed from Ser-1 and Ser-11, whereas in berninamycin, the pyridine most likely derives from Ser-1 and Ser-13. Whether a single producer such as B. cereus ATCC 14579 can carry out such distinct regiochemistries of cyclization based solely on different prepeptide sequences with altered Ser spacing or whether putative “aza-Diels-Alderases” in each organism are specific for 26-, 29-, or 35-membered rings or other size variants remains to be explored in “mix and match” studies. In this context, the 29-membered GE2270A does not hit the 50 S ribosome but rather EF-Tu as its target, so it may be possible to redirect target selectivity by control of macrocyclic loop size. Given the growing number of gene clusters for the prepeptides and accompanying modification enzymes, evaluation of various gene combinations for in vivo engineering of thiazolyl peptide scaffolds should reveal the scope of the post-translational maturation cascades.
Supplementary Material
Acknowledgments
We thank Sarah Mahlstedt and Dr. Travis Young for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GIM 20011 and New England Regional Center of Excellence Grant U54 AI057 159 from NIAID (to C. T. W.). This is the fourth article in the Thematic Minireview Series on Antibacterial Natural Products. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
M. G. Acker and C. T. Walsh, unpublished data.
- NRPS
- nonribosomal peptide synthetase
- EF
- elongation factor
- Dha
- dehydroalanine
- Dhb
- dehydrobutyrine.
REFERENCES
- 1.Sattely E. S., Fischbach M. A., Walsh C. T. (2008) Nat. Prod. Rep. 25, 757–793 [DOI] [PubMed] [Google Scholar]
- 2.Walsh C. T., Fischbach M. A. (2010) J. Am. Chem. Soc. 132, 2469–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cane D. E. (2010) J. Biol. Chem. 285, 27517–27523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickens L. B., Tang Y. (2010) J. Biol. Chem. 285, 27509–27515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischbach M. A., Walsh C. T. (2006) Chem. Rev. 106, 3468–3496 [DOI] [PubMed] [Google Scholar]
- 6.Nolan E. M., Walsh C. T. (2009) ChemBioChem 10, 34–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIntosh J. A., Donia M. S., Schmidt E. W. (2009) Nat. Prod. Rep. 26, 537–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schofield C. J., Baldwin J. E., Byford M. F., Clifton I., Hajdu J., Hensgens C., Roach P. (1997) Curr. Opin. Struct. Biol. 7, 857–864 [DOI] [PubMed] [Google Scholar]
- 9.Hubbard B. K., Walsh C. T. (2003) Angew. Chem. Int. Ed. Engl. 42, 730–765 [DOI] [PubMed] [Google Scholar]
- 10.Li T. L., Huang F., Haydock S. F., Mironenko T., Leadlay P. F., Spencer J. B. (2004) Chem. Biol. 11, 107–119 [DOI] [PubMed] [Google Scholar]
- 11.Miao V., Coëffet-Legal M. F., Brian P., Brost R., Penn J., Whiting A., Martin S., Ford R., Parr I., Bouchard M., Silva C. J., Wrigley S. K., Baltz R. H. (2005) Microbiology 151, 1507–1523 [DOI] [PubMed] [Google Scholar]
- 12.Robbel L., Marahiel M. A. (2010) J. Biol. Chem. 285, 27501–27508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganz T. (2003) Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 14.Oman T. J., van der Donk W. A. (2010) Nat. Chem. Biol. 6, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee C., Paul M., Xie L., van der Donk W. A. (2005) Chem. Rev. 105, 633–684 [DOI] [PubMed] [Google Scholar]
- 16.Patton G. C., van der Donk W. A. (2005) Curr. Opin. Microbiol. 8, 543–551 [DOI] [PubMed] [Google Scholar]
- 17.Severinov K., Semenova E., Kazakov A., Kazakov T., Gelfand M. S. (2007) Mol. Microbiol. 65, 1380–1394 [DOI] [PubMed] [Google Scholar]
- 18.Heddle J. G., Blance S. J., Zamble D. B., Hollfelder F., Miller D. A., Wentzell L. M., Walsh C. T., Maxwell A. (2001) J. Mol. Biol. 307, 1223–1234 [DOI] [PubMed] [Google Scholar]
- 19.Donia M. S., Hathaway B. J., Sudek S., Haygood M. G., Rosovitz M. J., Ravel J., Schmidt E. W. (2006) Nat. Chem. Biol. 2, 729–735 [DOI] [PubMed] [Google Scholar]
- 20.Donia M. S., Ravel J., Schmidt E. W. (2008) Nat. Chem. Biol. 4, 341–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagley M. C., Dale J. W., Merritt E. A., Xiong X. (2005) Chem. Rev. 105, 685–714 [DOI] [PubMed] [Google Scholar]
- 22.Bycroft B. W., Gowland M. S. (1978) J. Chem. Soc. Chem. Commun. 256–258 [Google Scholar]
- 23.Shoji J., Kato T., Yoshimura Y., Tori K. (1981) J. Antibiot. 34, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 24.Pascard C., Ducruix A., Lunel J., Prangé T. (1977) J. Am. Chem. Soc. 99, 6418–6423 [DOI] [PubMed] [Google Scholar]
- 25.Prange T., Ducruix A., Pascard C., Lunel J. (1977) Nature 265, 189–190 [DOI] [PubMed] [Google Scholar]
- 26.Liesch J. M., Rinehart K. L., Jr. (1977) J. Am. Chem. Soc. 99, 1645–1646 [DOI] [PubMed] [Google Scholar]
- 27.Shimanaka K., Takahashi Y., Iinuma H., Naganawa H., Takeuchi T. (1994) J. Antibiot. 47, 1153–1159 [DOI] [PubMed] [Google Scholar]
- 28.Shimanaka K., Takahashi Y., Iinuma H., Naganawa H., Takeuchi T. (1994) J. Antibiot. 47, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 29.Ebata M., Miyazaki K., Otsuka H. (1969) J. Antibiot. 22, 434–441 [DOI] [PubMed] [Google Scholar]
- 30.Anderson B., Hodgkin D. C., Viswamitra M. A. (1970) Nature 225, 233–235 [DOI] [PubMed] [Google Scholar]
- 31.Puar M. S., Ganguly A. K., Afonso A., Brambilla R., Mangiaracina P., Sarre O., MacFarlane R. D. (1981) J. Am. Chem. Soc. 103, 5231–5233 [Google Scholar]
- 32.Puar M. S., Chan T. M., Hegde V., Patel M., Bartner P., Ng K. J., Pramanik B. N., MacFarlane R. D. (1998) J. Antibiot. 51, 221–224 [DOI] [PubMed] [Google Scholar]
- 33.Su T. L. (1948) Br. J. Exp. Pathol. 29, 473–481 [PMC free article] [PubMed] [Google Scholar]
- 34.Mukai A., Fukai T., Hoshino Y., Yazawa K., Harada K., Mikami Y. (2009) J. Antibiot. 62, 613–619 [DOI] [PubMed] [Google Scholar]
- 35.Moody C. J., Bagley M. C. (1998) Chem. Commun. 2049–2050 [Google Scholar]
- 36.Bagley M. C., Bashford K. E., Hesketh C. L., Moody C. J. (2000) J. Am. Chem. Soc. 122, 3301–3313 [Google Scholar]
- 37.Nicolaou K. C., Safina B. S., Zak M., Estrada A. A., Lee S. H. (2004) Angew. Chem. Int. Ed. Engl. 43, 5087–5092 [DOI] [PubMed] [Google Scholar]
- 38.Nicolaou K. C., Zak M., Safina B. S., Lee S. H., Estrada A. A. (2004) Angew. Chem. Int. Ed. Engl. 43, 5092–5097 [DOI] [PubMed] [Google Scholar]
- 39.Müller H. M., Delgado O., Bach T. (2007) Angew. Chem. Int. Ed. Engl. 46, 4771–4774 [DOI] [PubMed] [Google Scholar]
- 40.Delgado O., Müller H. M., Bach T. (2008) Chemistry 14, 2322–2339 [DOI] [PubMed] [Google Scholar]
- 41.Nicolaou K. C., Dethe D. H., Chen D. Y. (2008) Chem. Commun. 2632–2634 [DOI] [PubMed] [Google Scholar]
- 42.Nicolaou K. C., Dethe D. H., Leung G. Y., Zou B., Chen D. Y. (2008) Chem. Asian J. 3, 413–429 [DOI] [PubMed] [Google Scholar]
- 43.Lefranc D., Ciufolini M. A. (2009) Angew. Chem. Int. Ed. Engl. 48, 4198–4201 [DOI] [PubMed] [Google Scholar]
- 44.Agirrezabala X., Frank J. (2009) Q. Rev. Biophys. 42, 159–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cameron D. M., Thompson J., Gregory S. T., March P. E., Dahlberg A. E. (2004) Nucleic Acids Res. 32, 3220–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cameron D. M., Thompson J., March P. E., Dahlberg A. E. (2002) J. Mol. Biol. 319, 27–35 [DOI] [PubMed] [Google Scholar]
- 47.Cundliffe E. (1971) Biochem. Biophys. Res. Commun. 44, 912–917 [DOI] [PubMed] [Google Scholar]
- 48.Cundliffe E., Dixon P., Stark M., Stöffler G., Ehrlich R., Stöffler-Meilicke M., Cannon M. (1979) J. Mol. Biol. 132, 235–252 [DOI] [PubMed] [Google Scholar]
- 49.Cundliffe E., Thompson J. (1979) Nature 278, 859–861 [DOI] [PubMed] [Google Scholar]
- 50.Cundliffe E., Thompson J. (1981) J. Gen. Microbiol. 126, 185–192 [DOI] [PubMed] [Google Scholar]
- 51.Hummel H., Böck A. (1987) Biochimie 69, 857–861 [DOI] [PubMed] [Google Scholar]
- 52.Mankin A. S., Leviev I., Garrett R. A. (1994) J. Mol. Biol. 244, 151–157 [DOI] [PubMed] [Google Scholar]
- 53.Porse B. T., Cundliffe E., Garrett R. A. (1999) J. Mol. Biol. 287, 33–45 [DOI] [PubMed] [Google Scholar]
- 54.Rosendahl G., Douthwaite S. (1993) J. Mol. Biol. 234, 1013–1020 [DOI] [PubMed] [Google Scholar]
- 55.Rosendahl G., Douthwaite S. (1994) Nucleic Acids Res. 22, 357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wienen B., Ehrlich R., Stöffler-Meilicke M., Stöffler G., Smith I., Weiss D., Vince R., Pestka S. (1979) J. Biol. Chem. 254, 8031–8041 [PubMed] [Google Scholar]
- 57.Harms J. M., Wilson D. N., Schluenzen F., Connell S. R., Stachelhaus T., Zaborowska Z., Spahn C. M., Fucini P. (2008) Mol. Cell 30, 26–38 [DOI] [PubMed] [Google Scholar]
- 58.Tanaka T., Sakaguchi K., Yonehara H. (1971) J. Biochem. 69, 1127–1130 [DOI] [PubMed] [Google Scholar]
- 59.Modolell J., Cabrer B., Parmeggiani A., Vazquez D. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 1796–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez R. L., Jr., Chu S., Puglisi J. D. (2007) RNA 13, 2091–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selva E., Beretta G., Montanini N., Saddler G. S., Gastaldo L., Ferrari P., Lorenzetti R., Landini P., Ripamonti F., Goldstein B. P. (1991) J. Antibiot. 44, 693–701 [DOI] [PubMed] [Google Scholar]
- 62.Kettenring J., Colombo L., Ferrari P., Tavecchia P., Nebuloni M., Vékey K., Gallo G. G., Selva E. (1991) J. Antibiot. 44, 702–715 [DOI] [PubMed] [Google Scholar]
- 63.Anborgh P. H., Parmeggiani A. (1991) EMBO J. 10, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris R. P., Leeds J. A., Naegeli H. U., Oberer L., Memmert K., Weber E., LaMarche M. J., Parker C. N., Burrer N., Esterow S., Hein A. E., Schmitt E. K., Krastel P. (2009) J. Am. Chem. Soc. 131, 5946–5955 [DOI] [PubMed] [Google Scholar]
- 65.Parmeggiani A., Nissen P. (2006) FEBS Lett. 580, 4576–4581 [DOI] [PubMed] [Google Scholar]
- 66.Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B. F., Nyborg J. (1995) Science 270, 1464–1472 [DOI] [PubMed] [Google Scholar]
- 67.Heffron S. E., Jurnak F. (2000) Biochemistry 39, 37–45 [DOI] [PubMed] [Google Scholar]
- 68.Parmeggiani A., Krab I. M., Okamura S., Nielsen R. C., Nyborg J., Nissen P. (2006) Biochemistry 45, 6846–6857 [DOI] [PubMed] [Google Scholar]
- 69.Roy R. S., Gehring A. M., Milne J. C., Belshaw P. J., Walsh C. T. (1999) Nat. Prod. Rep. 16, 249–263 [DOI] [PubMed] [Google Scholar]
- 70.Duerfahrt T., Eppelmann K., Müller R., Marahiel M. A. (2004) Chem. Biol. 11, 261–271 [DOI] [PubMed] [Google Scholar]
- 71.Konz D., Klens A., Schörgendorfer K., Marahiel M. A. (1997) Chem. Biol. 4, 927–937 [DOI] [PubMed] [Google Scholar]
- 72.Walsh C. T., Chen H., Keating T. A., Hubbard B. K., Losey H. C., Luo L., Marshall C. G., Miller D. A., Patel H. M. (2001) Curr. Opin. Chem. Biol. 5, 525–534 [DOI] [PubMed] [Google Scholar]
- 73.Schneider T. L., Shen B., Walsh C. T. (2003) Biochemistry 42, 9722–9730 [DOI] [PubMed] [Google Scholar]
- 74.Carnio M. C., Stachelhaus T., Francis K. P., Scherer S. (2001) Eur. J. Biochem. 268, 6390–6401 [DOI] [PubMed] [Google Scholar]
- 75.Xie L., Miller L. M., Chatterjee C., Averin O., Kelleher N. L., van der Donk W. A. (2004) Science 303, 679–681 [DOI] [PubMed] [Google Scholar]
- 76.Chatterjee C., Miller L. M., Leung Y. L., Xie L., Yi M., Kelleher N. L., van der Donk W. A. (2005) J. Am. Chem. Soc. 127, 15332–15333 [DOI] [PubMed] [Google Scholar]
- 77.Li B., Yu J. P., Brunzelle J. S., Moll G. N., van der Donk W. A., Nair S. K. (2006) Science 311, 1464–1467 [DOI] [PubMed] [Google Scholar]
- 78.Willey J. M., van der Donk W. A. (2007) Annu. Rev. Microbiol. 61, 477–501 [DOI] [PubMed] [Google Scholar]
- 79.Li Y. M., Milne J. C., Madison L. L., Kolter R., Walsh C. T. (1996) Science 274, 1188–1193 [DOI] [PubMed] [Google Scholar]
- 80.Milne J. C., Eliot A. C., Kelleher N. L., Walsh C. T. (1998) Biochemistry 37, 13250–13261 [DOI] [PubMed] [Google Scholar]
- 81.Belshaw P. J., Roy R. S., Kelleher N. L., Walsh C. T. (1998) Chem. Biol. 5, 373–384 [DOI] [PubMed] [Google Scholar]
- 82.Milne J. C., Roy R. S., Eliot A. C., Kelleher N. L., Wokhlu A., Nickels B., Walsh C. T. (1999) Biochemistry 38, 4768–4781 [DOI] [PubMed] [Google Scholar]
- 83.Wieland Brown L. C., Acker M. G., Clardy J., Walsh C. T., Fischbach M. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2549–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao R., Duan L., Lei C., Pan H., Ding Y., Zhang Q., Chen D., Shen B., Yu Y., Liu W. (2009) Chem. Biol. 16, 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelly W. L., Pan L., Li C. (2009) J. Am. Chem. Soc. 131, 4327–4334 [DOI] [PubMed] [Google Scholar]
- 86.Yu Y., Duan L., Zhang Q., Liao R., Ding Y., Pan H., Wendt-Pienkowski E., Tang G., Shen B., Liu W. (2009) ACS Chem. Biol. 4, 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J., Yu Y., Tang K., Liu W., He X., Huang X., Deng Z. (2010) Appl. Environ. Microbiol. 76, 2335–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mocek U., Knaggs A. R., Tsuchiya R., Nguyen T., Beale J. M., Floss H. G. (1993) J. Am. Chem. Soc. 115, 7557–7568 [Google Scholar]
- 89.Mocek U., Zeng Z., O'Hagan D., Zhou P., Fan L. D. G., Beale J. M., Floss H. G. (1993) J. Am. Chem. Soc. 115, 7992–8001 [Google Scholar]
- 90.Kelly W. L. (2008) Org. Biomol. Chem. 6, 4483–4493 [DOI] [PubMed] [Google Scholar]
- 91.Aoki M., Ohtsuka T., Yamada M., Ohba Y., Yoshizaki H., Yasuno H., Sano T., Watanabe J., Yokose K., Seto H. (1991) J. Antibiot. 44, 582–588 [DOI] [PubMed] [Google Scholar]
- 92.Aoki M., Ohtsuka T., Itezono Y., Yokose K., Furihata K., Seto H. (1991) Tetrahedron Lett. 32, 221–224 [Google Scholar]
- 93.Aoki M., Ohtsuka T., Itezono Y., Yokose K., Furihata K., Seto H. (1991) Tetrahedron Lett. 32, 217–220 [Google Scholar]
- 94.Acker M. G., Bowers A. A., Walsh C. T. (2009) J. Am. Chem. Soc. 131, 17563–17565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bowers A. B., Acker M. G., Koglin A., Walsh C. T. (2010) J. Am. Chem. Soc. 132, 7519–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lociuro S., Tavecchia P., Marzorati E., Landini P., Goldstein B. P., Denaro M., Ciabatti R. (1997) J. Antibiot 50, 344–349 [DOI] [PubMed] [Google Scholar]
- 97.Naidu B. N., Sorenson M. E., Zhang Y., Kim O. K., Matiskella J. D., Wichtowski J. A., Connolly T. P., Li W., Lam K. S., Bronson J. J., Pucci M. J., Clark J. M., Ueda Y. (2004) Bioorg. Med. Chem. Lett. 14, 5573–5577 [DOI] [PubMed] [Google Scholar]
- 98.Naidu B. N., Sorenson M. E., Bronson J. J., Pucci M. J., Clark J. M., Ueda Y. (2005) Bioorg. Med. Chem. Lett. 15, 2069–2072 [DOI] [PubMed] [Google Scholar]
- 99.Schoof S., Baumann S., Ellinger B., Arndt H. D. (2009) ChemBioChem 10, 242–245 [DOI] [PubMed] [Google Scholar]
- 100.Starosta A. L., Qin H., Mikolajka A., Leung G. Y., Schwinghammer K., Nicolaou K. C., Chen D. Y., Cooperman B. S., Wilson D. N. (2009) Chem. Biol. 16, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Constantine K. L., Mueller L., Huang S., Abid S., Lam K. S., Li W., Leet J. E. (2002) J. Am. Chem. Soc. 124, 7284–7285 [DOI] [PubMed] [Google Scholar]
- 102.Leet J. E., Li W., Ax H. A., Matson J. A., Huang S., Huang R., Cantone J. L., Drexler D., Dalterio R. A., Lam K. S. (2003) J. Antibiot. 56, 232–242 [DOI] [PubMed] [Google Scholar]
- 103.Northcote P. T., Siegel M., Borders D. B., Lee M. D. (1994) J. Antibiot. 47, 901–908 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.