Abstract
Tristetraprolin (TTP) directs its target AU-rich element (ARE)-containing mRNAs for degradation by promoting removal of the poly(A) tail. The p38 MAPK pathway regulates mRNA stability via the downstream kinase MAPK-activated protein kinase 2 (MAPKAP kinase 2 or MK2), which phosphorylates and prevents the mRNA-destabilizing function of TTP. We show that deadenylation of endogenous ARE-containing tumor necrosis factor mRNA is inhibited by p38 MAPK. To investigate whether phosphorylation of TTP by MK2 regulates TTP-directed deadenylation of ARE-containing mRNAs, we used a cell-free assay that reconstitutes the mechanism in vitro. We find that phosphorylation of Ser-52 and Ser-178 of TTP by MK2 results in inhibition of TTP-directed deadenylation of ARE-containing RNA. The use of 14-3-3 protein antagonists showed that regulation of TTP-directed deadenylation by MK2 is independent of 14-3-3 binding to TTP. To investigate the mechanism whereby TTP promotes deadenylation, it was necessary to identify the deadenylases involved. The carbon catabolite repressor protein (CCR)4·CCR4-associated factor (CAF)1 complex was identified as the major source of deadenylase activity in HeLa cells responsible for TTP-directed deadenylation. CAF1a and CAF1b were found to interact with TTP in an RNA-independent fashion. We find that MK2 phosphorylation reduces the ability of TTP to promote deadenylation by inhibiting the recruitment of CAF1 deadenylase in a mechanism that does not involve sequestration of TTP by 14-3-3. Cyclooxygenase-2 mRNA stability is increased in CAF1-depleted cells in which it is no longer p38 MAPK/MK2-regulated.
Keywords: Inflammation, p38 MAPK, RNA Binding Protein, RNA Turnover, Signal Transduction, Deadenylation, MAPKAPK-2, Phosphorylation, Post-transcriptional Regulation, Tristetraprolin
Introduction
Many mammalian mRNAs contain adenosine/uridine-rich elements (AREs)3 in their 3′-untranslated regions (UTRs) that target mRNAs for rapid degradation. The importance of AREs in regulation of mRNA stability has been demonstrated particularly for mRNAs of the inflammatory response. The p38 MAPK pathway inhibits ARE-mediated decay allowing for dynamic control of the expression of these mRNAs (1–3). p38 MAPK regulates mRNA stability via the downstream kinase MAPKAP kinase 2 (MK2), which phosphorylates (4, 5) and prevents the function (6, 7) of the mRNA-destabilizing ARE-binding protein, tristetraprolin (TTP). We recently showed that dual control of mRNA stability by TTP and the p38 MAPK pathway is a general mechanism, which operates for many mRNAs of the inflammatory response (8). The importance of TTP in the control of inflammatory gene expression is demonstrated by spontaneous inflammatory arthritis in TTP−/− mice arising mainly from increased tumor necrosis factor (TNF) production (9). p38 MAPK also regulates mRNA stability by direct phosphorylation and inactivation of another ARE-binding protein, KH domain-splicing regulatory protein (10, 11). p38 MAPK inhibitors fail to destabilize mRNAs of the inflammatory response in macrophages from TTP−/− mice (8, 12). Thus, it appears that in these cells, at least, TTP is entirely responsible for the effects on mRNA stability mediated by the p38 MAPK pathway.
In addition to controlling inflammatory mediator mRNA stability, the p38 MAPK pathway stabilizes both TTP mRNA (13) and protein (14), thereby increasing TTP expression. The current understanding is that the mRNA destabilizing protein, TTP, is induced and held in a latent state by the p38 MAPK pathway and is poised to direct mRNA destabilization when activity in the pathway dissipates. Thus a proinflammatory stimulus triggers the induction of inflammatory genes and the subsequent resolution of their expression, the latter being crucial to prevent chronic inflammation. It has long been known that treatment of cells with p38 MAPK inhibitor following an inflammatory stimulus destabilizes inflammatory mediator mRNAs (1). Recently, we showed that treatment of cells with p38 MAPK inhibitor prior to an inflammatory stimulus fails to destabilize TNF mRNA (8), presumably because under these conditions, TTP protein is not induced (14). Thus, prolonged treatment of cells with p38 MAPK inhibitors in vivo may not take advantage of blocking this important post-transcriptional mechanism of gene regulation. This is one potential reason why p38 MAPK inhibitors have failed recently in clinical trials for rheumatoid arthritis (15). Elucidation of the post-transcriptional mechanism downstream of p38 MAPK may offer future promise for the therapy of such diseases.
It is known that TTP directs its target mRNAs for degradation by promoting removal of the poly(A) tail or deadenylation (16), the first step in mRNA decay. The p38 MAPK pathway stabilizes mRNAs by inhibiting deadenylation (17, 18) but the precise mechanism whereby phosphorylation of TTP by MK2 inhibits poly(A) tail shortening is not known. Phosphorylation of TTP by MK2 at Ser-52 and Ser-178 results in binding of 14-3-3 to TTP (6, 19), and the formation of this complex has been suggested to prevent TTP from interacting with mRNA decay factors (6).
Two distinct deadenylase complexes, poly(A) nuclease (PAN)2-PAN3, and carbon catabolite repressor protein (CCR)4-CCR4-associated factor (CAF)1, originally were discovered in yeast (20, 21). Human orthologues of both complexes exist (22). In humans, the CCR4·CAF1 complex comprises two subunits with deadenylase activity (CCR4 and CAF1) together with seven other CNOT proteins (23). Human CCR4 and CAF1 each have two different paralogues: CCR4a (CNOT6) and CCR4b (CNOT6L); and CAF1a (CNOT7) and CAF1b (CNOT8). In general, for mRNA decay in mammalian cells, PAN2-PAN3 is thought to catalyze initial poly(A) shortening, and CCR4-CAF1 then removes the remaining ∼110 nucleotides (nt) of the poly(A) tail (24).
CAF1 deadenylase has been implicated in ARE-mediated deadenylation. Knockdown of CAF1 by RNA interference (RNAi) has been shown to impair the deadenylation and decay of an ARE-containing β-globin mRNA (25, 26). In contrast, CCR4 depletion has been reported to have no effect on deadenylation of an ARE reporter mRNA (26). Mammalian cells also contain another, predominantly nuclear enzyme, poly(A) ribonuclease (PARN) (27). It has been suggested to be involved in ARE-mediated deadenylation (28) and to promote TTP-directed deadenylation in vitro (29). TTP has been reported to interact with mRNA decay factors including the exosome (30), Dcp1a, Dcp2, Xrn1, and also CCR4 (31) but not PARN (29). It is thus not clear which deadenylase is involved in TTP-directed deadenylation in cells.
To elucidate the mechanism whereby MK2 inactivates TTP, it was necessary to first identify which deadenylase is involved in TTP-directed deadenylation. To investigate this, we modified an in vitro ARE-dependent and TTP-directed deadenylation assay described by Lai et al. (29) to use bacterially expressed recombinant TTP. This allowed the involvement of deadenylases to be determined by assaying extracts from cells depleted of different deadenylases by RNAi in the presence of a constant amount of TTP. The use of recombinant TTP in the in vitro system also allowed us to investigate the role of MK2 in the absence of changes in TTP protein expression, which occurs in cells following activation or inhibition of this kinase (7, 14). The assay uses TNF and granulocyte/macrophage-colony stimulating factor (GM-CSF) ARE RNA substrates with 100-nt poly(A) tails. Deadenylation of both of these mRNAs has been shown previously to be regulated by TTP (16, 32). Both mRNAs also are stabilized by the p38 MAPK/MK2 pathway (33, 34). R18 and difopein (dimeric fourteen-three-three peptide inhibitor) are high affinity 14-3-3 antagonists that allow for essentially complete inhibition of 14-3-3 binding to target proteins (35). The in vitro deadenylation assay enabled us to use R18 and difopein to test the function of 14-3-3 in deadenylation and to determine a novel mechanism whereby MK2 inhibits TTP-directed deadenylation.
EXPERIMENTAL PROCEDURES
Materials
General laboratory reagents were from Sigma. 4-(4-Fluorophenyl)-2-(4-hydroxyphenyl)-5-(4-pyridyl)1H-imidazole (SB202190) was from Calbiochem-Novabiochem. pSP73-GM-CSF ARE-p(A)+ and -GM-CSF ARE mut-p(A)+ plasmids were described previously (36). The GM-CSF ARE sequence was removed by digestion with XbaI and replaced with a SpeI XbaI fragment from pBluescript-hTNF 75 (37) to create pSP73-TNF ARE-p(A)+ and -TNF ARE antisense-p(A)+ plasmids. Glutathione S-transferase (GST)-TTP expression plasmids were constructed by subcloning the previously described wild-type and a S52A/S178A mutated form of murine TTP cDNA (13) into pGEX-6P-3 (GE Healthcare) at the EcoRI site. GM-CSF ARE and GM-CSF ARE mut oligoribonucleotides were synthesized commercially (Dharmacon). The sequences were as follows: GM-CSF ARE, AGUAAUAUUUAUAUAUUUAUAUUUUUAAAAUAUUUAUUUAUUUAUUUAUUUAA; and GM-CSF ARE mut, GUAAUAUGAAUACAUCUGAAUGUCUGGAAUAUUGAUUGAUAAGCUUAGUCGAC. The antibodies used were as follows: anti-α-tubulin (Sigma), anti-MK2 (Upstate), anti-CAF1a (Abnova), anti-CAF1b (LifeSpan BioSciences), anti-pan-14-3-3 (Chemicon), and anti-TTP (4). Anti-PARN, anti-PAN2, and anti-PABP1 antibodies were gifts from Professor Anders Virtanen (Uppsala University), Professor Shin-ichi Hoshino (University of Tokyo), and Dr. Matt Brook (University of Edinburgh), respectively.
Cell Culture
HeLa (gift from Yamanouchi) and RAW 264.7 (ATCC) cells were cultured in Dulbecco's modified Eagle's medium (PAA Laboratories) supplemented with 10% fetal calf serum (PAA Laboratories). Penicillin/streptomycin (PAA Laboratories) was included in HeLa cell culture medium. Cells were maintained at 37 °C in the presence of 5% CO2.
Preparation of Recombinant TTP
GST-TTP expression plasmids were used to transform Escherichia coli TOP10 (Invitrogen). Bacteria were grown in LB containing 100 μg/ml ampicillin, and 1 mm isopropyl 1-thio-β-d-galactopyranoside was added at mid-exponential phase to induce expression for 12 h at 28 °C. Cells were harvested and suspended in 20 mm HEPES, pH 7.9, with 10% (v/v) glycerol, 0.5 m KCl, 2 mm DTT, 1 mm PMSF, 1 μg/ml pepstatin A, 13.5 μg/ml aprotinin, and 10 μm E-64. Cells were lysed by four passages through a French pressure cell at 15,000 psi. Cell debris was removed by centrifugation at 30,000 × g for 20 min, and the supernatant was incubated with glutathione-Sepharose 4B (GE Healthcare) at 4 °C for 30 min with shaking. The resin was washed with 15 column volumes of PBS, and bound protein was eluted with 50 mm Tris-HCl, pH 8.0, 10 mm reduced glutathione. On-column cleavage of the GST tag was performed with PreScission protease (GE Healthcare) treatment following the manufacturer's instructions. Glycerol was added to a final concentration of 10% (v/v), and the protein was stored at −80 °C until use. TTP protein concentration was determined by Bradford assay.
In Vitro Deadenylation Assay
This was performed according to Lai et al. (29) using HeLa cells lysed by Dounce homogenization. Briefly, RNA substrates with 100 nt poly(A) tails were prepared by in vitro transcription in the presence of [α-32P]UTP (PerkinElmer) as described previously (37). A radiolabeled poly(A) tail was added to the GM-CSF ARE or GM-CSF ARE mut oligoribonucleotides using a poly(A) tailing kit (Epicentre) and [α-32P]ATP (PerkinElmer) according to the manufacturer's instructions. RNA species with poly(A) tails of ∼100 nt were obtained by gel purification. HeLa S100, GST-TTP, and radiolabeled substrate RNA were mixed and incubated at 37 °C for the times indicated, and EDTA (final concentration, 20 mm) was added to terminate the reaction. RNA was isolated by phenol-chloroform extraction, electrophoresed on polyacrylamide-urea gels, and visualized and quantified by phosphorimaging (FLA-5100 imager, Fuji, Japan; AIDA 1D quantification software, Raytest, Germany).
In Vitro Phosphorylation of GST-TTP by MK2
In vitro phosphorylation of TTP by MK2 was performed with immunoprecipitated MK2 or with recombinant MK2 (Millipore). 1 μg of GST-TTP was combined with immunoprecipitated MK2 or with recombinant activated MK2 (0.1 unit) in a final volume of 30 μl containing 20 μm ATP and incubated at 30 °C. To assess phosphorylation of GST-TTP, 4 μCi of [γ-32P]ATP (PerkinElmer Life Sciences) was included in the reaction, and it was stopped by the addition of 10 μl of 4× SDS-PAGE sample buffer and subjected to electrophoresis. 32P incorporation was visualized and quantified by phosphorimaging. The stoichiometry of phosphorylation was measured by excising the radioactive bands and scintillation counting.
GST Pulldown Assay
2 μg of GST-TTP was incubated with 30 μl of a 50% slurry of glutathione-Sepharose 4B and 200 μg of cytoplasmic extract in 1 ml of binding buffer (10 mm HEPES, pH 7.6, 100 mm KCl, 6 mm MgCl2, 1 mm DTT, 1% (v/v) Igepal CA-360) for 60 min at 4 °C in the presence or absence of 150 units of benzonase (Sigma). The beads were washed with binding buffer and boiled in SDS-PAGE sample buffer for 5 min, and eluted proteins were analyzed by Western blotting as described previously (38).
RNase H Mapping and Northern Blot
RNase H mapping was performed as described previously (17) using an antisense murine TNF 3′-UTR oligodeoxynucleotide (TNF 1246) spanning nt 1246–1276 of the TNF mRNA (5′-GCTGGCTCTGTGAGGAAGGCTGTGCATTGC-3′). RNA was detected by Northern blotting using a murine antisense riboprobe that hybridizes with TNF mRNA between the cleavage site and the poly(A) tail. This was generated by PCR amplification of a 250-bp sequence spanning nt 1359–1609 of the 3′-UTR of TNF mRNA and in vitro transcribed as described previously (37). Full-length TNF and GAPDH mRNAs were detected by Northern blotting as described previously (17).
Electrophoretic Mobility Shift Assay (EMSA)
A GM-CSF ARE oligonucleotide was end-labeled with [γ-32P]ATP (3000 Ci/mmol) using T4 polynucleotide kinase. A 32P-labeled RNA probe (∼0.1 pmol) was incubated in the presence or absence of cold competitors for 15 min at room temperature with GST, unphosphorylated TTP, or TTP in vitro phosphorylated by MK2 in 20 μl of bandshift buffer (20 mm HEPES, pH 7.2, 100 μm ZnCl2, 50 mm KCl, 1 mm DTT, 5% glycerol) containing heparin sulfate (5 mg/ml). Two μl of loading buffer (80% glycerol, 0.1% bromphenol blue) was then added, and RNA·protein complexes were resolved by electrophoresis on a 4% (w/v) acrylamide/Tris borate gel using Tris borate running buffer. Complexes were visualized using a phosphorimaging device. Quantified data were plotted as fraction bound versus protein concentration and fit with the Hill equation using Prism 5 (GraphPad Software).
RNAi
The following siRNAs were used: scramble, 5′-CAGUCGCGUUUGCGACUGGdTdT-3′ and PARN, described previously by Lin et al. (28) were obtained from Eurofins MWG Operon; and CCR4a (s33100), CCR4b (s48341), CAF1a (s26638), CAF1b (s17849), PAN2 (s19252), and PAN3 (s48721) were purchased from Ambion (Applied Biosystems).
HeLa cells were transfected twice using Oligofectamine (Invitrogen), 24 h apart, as described previously (38), except that 15 nm siRNA (final concentration) was used. The efficiency of protein depletion was determined by Western blotting (38) and quantitative reverse-transcription PCR (Q-RT-PCR).
Q-RT-PCR
RNA was isolated using the RNeasy Kit (Qiagen), and cDNA was generated using the Reverse Transcription system kit and oligo(dT) (Promega). This cDNA was analyzed by Q-RT-PCR using TaqMan technology and primer-probe sets for CCR4a (Hs01019492_m1), CCR4b (Hs00375913_m1), CAF1a (Hs01020564_m1), CAF1b (Hs00231841_m1), PARN (Hs00377733_m1), PAN2 (Hs00208356_m1), PAN3 (Hs00107000_m1), COX-2 (Hs00153133_m1), and GAPDH (Hs99999905_m1) from Applied Biosystems. A Rotor-Gene 6000 thermal cycler and software (Corbett Research) were used. The ΔΔCt method and relative quantitation (with standard curves) were used for mRNA quantitation using GAPDH as internal control (39).
RESULTS
The p38 MAPK Pathway Inhibits Deadenylation of Endogenous TNF mRNA
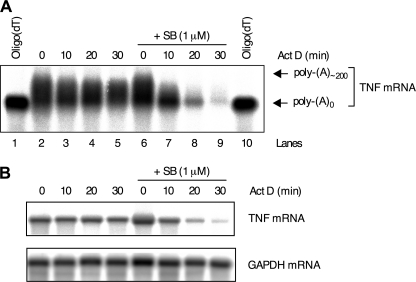
We showed previously that p38 MAPK stabilizes ARE-containing reporter mRNAs by inhibiting deadenylation (17). To investigate whether p38 MAPK regulates deadenylation of an endogenous mRNA, the poly(A) tail length of TNF mRNA induced by lipopolysaccharide (LPS) in the macrophage-like cell line RAW 264.7 in the presence or absence of the p38 MAPK inhibitor SB202190 was assessed. For this, RNase H mapping was used because full-length TNF mRNA is too long for accurate resolution of mRNAs with different poly(A) tail lengths. RNase H cleaves RNA/DNA heteroduplexes allowing mRNAs to be shortened in a specific fashion in the presence of oligodeoxyribonucleotides. Cells were treated with LPS for 2 h, and then actinomycin D was added to block transcription together with dimethyl sulfoxide vehicle or 1 μm SB202190. RNA was isolated from the cells and treated with RNase H and an oligodeoxyribonucleotide against the TNF mRNA 3′-UTR to cleave TNF mRNA. Some samples were additionally treated with oligo(dT) to completely remove poly(A) tails. Cleaved TNF mRNA was detected by Northern blot with a probe against the 3′-end of the mRNA. In cells treated with LPS and vehicle, TNF mRNA was relatively stable and little poly(A) shortening occurred (Fig. 1A). The mRNA underwent rapid simultaneous decay and deadenylation in the presence of SB202190 (Fig. 1A), confirming an important role for p38 MAPK in blocking deadenylation of an endogenous transcript. It was not possible to quantify GAPDH mRNA on the same membranes as TNF mRNA owing to nonspecific degradation of GAPDH mRNA during RNase H treatment. Therefore, an identical actinomycin D chase experiment was performed in parallel, and full-length TNF and GAPDH mRNAs were detected by Northern blotting. The even GAPDH signals on the gel (Fig. 1B) confirm that the identical TNF mRNA decay profiles in Fig. 1 (A and B) are an accurate assessment of TNF mRNA decay.
FIGURE 1.
p38 MAPK regulates the poly(A) tail length of endogenous TNF mRNA. A and B, RAW 264.7 cells were treated with LPS (10 ng ml−1) for 2 h, and then actinomycin D (Act D; 10 μg ml−1) was added together with vehicle (0.1% dimethyl sulfoxide) or 1 μm SB202190 (SB). Cells were harvested at the time intervals shown, and RNA was extracted. A, 10 μg RNA was incubated with TNF 1246 (lanes 2–9) or TNF 1246 oligodeoxynucleotide and oligo(dT) (lanes 1 and 10) and cleaved with RNase H. The samples were Northern blotted with an antisense riboprobe against the TNF 3′-UTR. B, Northern blot of full-length TNF mRNA. GAPDH is shown as a loading control.
TTP-directed in Vitro Deadenylation of ARE-containing RNA
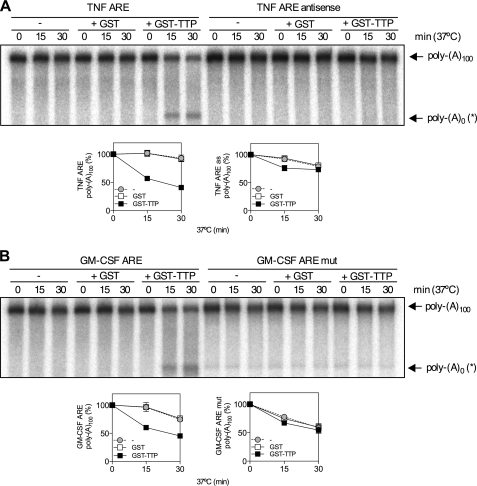
p38 MAPK stabilizes inflammatory mRNAs via activation of the downstream kinase MK2, which phosphorylates and blocks the function of the mRNA destabilizing factor, TTP. To investigate the mechanism whereby MK2 inhibits poly(A) shortening, it was necessary to modify an ARE-dependent and TTP-directed in vitro deadenylation assay developed by Lai et al. (29) to use recombinant bacterially expressed GST-TTP rather than TTP overexpressed in 293 cells. In vitro deadenylation reactions contained a 100,000 × g supernatant of HeLa cytoplasmic extract (S100), which served as a source of deadenylases, GST-TTP or GST, and a radiolabeled RNA substrate comprising ARE-containing RNA linked to a 100 nt poly(A) tail. Reaction mixtures were incubated for different times in the presence of GST-TTP, GST or S100 alone. This showed time- and TTP-dependent disappearance of full-length TNF RNA substrate (poly(A)100) and the accumulation of an intermediate of higher mobility on the gel (Fig. 2A). The increase in the deadenylated product did not fully correspond to the reduction of full-length substrate (Fig. 2A), suggesting that it is an intermediate that undergoes further decay. No TTP-directed deadenylation was seen in in vitro deadenylation assay with antisense TNF ARE (Fig. 2A). Similar results were obtained for a GM-CSF ARE RNA and mutant form in which the ARE was disrupted with multiple substitutions of A and U to G and C (Fig. 2B), consistent with ARE binding by TTP being a mandatory event for TTP-directed deadenylation. Titration of GST-TTP confirmed that 100 ng is the optimum amount of TTP to use in the assay and oligo(dT) and RNase H treatment showed that the GM-CSF ARE RNA intermediate represents an RNA species that lacks the poly(A) tail (poly(A)0(*)) and an additional ∼5 nt (supplemental Fig. S1).
FIGURE 2.
TTP promotes ARE-dependent deadenylation in vitro. A, in vitro deadenylation of 32P-labeled TNF ARE or TNF ARE antisense RNA substrates were incubated in the presence of HeLa S100 (5 μg) and 100 ng GST or GST-TTP at 37 °C for the times indicated. Representative phosphorimage of urea-PAGE of 32P-labeled reaction products is shown. Graphs show mean poly(A)100 expressed as a percentage of t = 0 ± S.E. from three independent experiments. Where not shown, error bars are smaller than the symbols. The positions of polyadenylated substrate (poly(A)100) and deadenylated product (poly(A)0(*)) are indicated. B, as for A but with GM-CSF ARE or GM-CSF ARE mut RNA substrates.
Phosphorylation of TTP by MK2 Inhibits Deadenylation
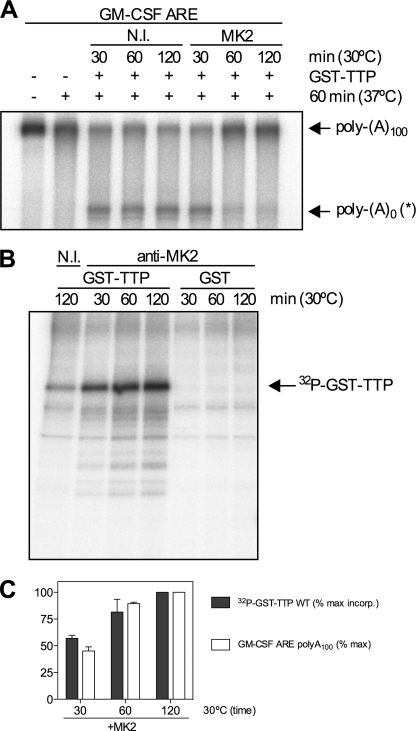
To elucidate how phosphorylation of TTP by MK2 regulates deadenylation of ARE-containing mRNAs, inhibition of TTP-directed deadenylation by MK2 was reconstituted in vitro. In initial experiments, MK2 was immunoprecipitated from lysates of HeLa cells stimulated with IL-1α, a potent activator of the p38 MAPK pathway, and used to phosphorylate recombinant GST-TTP. Phosphorylation reactions were performed for different times, and phosphorylated GST-TTP was assayed for its ability to promote deadenylation following a 1 h incubation at 37 °C. A dummy phosphorylation reaction with a nonimmune antibody immunoprecipitate served as a control. Phosphorylation of GST-TTP by MK2 for 1 or 2 h inhibited the ability of TTP to promote GM-CSF ARE substrate deadenylation (Fig. 3, A and C). Analysis of GST-TTP phosphorylated for different times in the presence of [γ-32P]ATP showed that phosphorylation was essentially complete at 2 h, and little phosphorylation of GST-TTP was seen with a nonimmune immunoprecipitate (Fig. 3B). Thus, the inhibition of deadenylation appeared to closely correlate with phosphorylation of GST-TTP by MK2 (Fig. 3C).
FIGURE 3.
Phosphorylation of TTP by MK2 inhibits deadenylation. MK2 was immunoprecipitated from lysates of HeLa cells stimulated with IL-1α and used to phosphorylate recombinant GST-TTP in vitro. Phosphorylation reactions were performed for different times (30, 60, and 120 min) at 30 °C, and a nonimmune antibody (N.I.) served as a control. A, phosphorylated GST-TTP assayed by in vitro deadenylation assay for 60 min at 37 °C. B, phosphorimage of GST-TTP or GST phosphorylation by immunoprecipitated MK2 or nonimmune control (N.I.) for different times in the presence of [γ-32P]ATP. C, graph showing correlation between GST-TTP phosphorylation by MK2 (% max incorp. of 32P) and inhibition of deadenylation.
Phosphorylation of TTP by MK2 Has No Effect on GM-CSF ARE RNA Binding
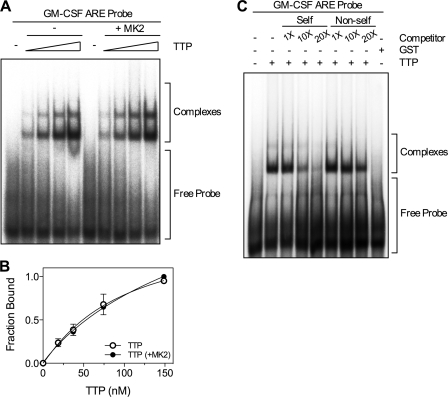
It has been suggested that phosphorylation of TTP by MK2 reduces its affinity for mRNA (7). To test this, EMSA was performed with a GM-CSF ARE RNA probe lacking a poly(A) tail and either unphosphorylated TTP or TTP phosphorylated by recombinant active MK2 in vitro. For this experiment, cleaved TTP lacking the GST tag was used to avoid formation of complexes that may be formed due to dimerization of GST. Bands corresponding to two distinct RNA·protein complexes were observed and there was no significant difference in binding between unphosphorylated and phosphorylated TTP (Fig. 4, A and B). ARE-dependent binding of TTP was confirmed by competition experiments using unlabeled GM-CSF ARE (self) and GM-CSF ARE mut (non-self) probes as competitors in the binding reaction. Competition was strongest with GM-CSF ARE RNA, consistent with specific ARE binding (Fig. 4C). Incubation of GM-CSF ARE RNA with a bacterially expressed GST preparation confirmed that complexes were formed by TTP and not E. coli contaminants (Fig. 4C). Phosphorylation of TTP by MK2 was confirmed to be complete by the incorporation of 32P into TTP (supplemental Fig. S2). The stoichiometry of phosphorylation was 0.48 mol/mol TTP. This value is approximate as TTP protein concentration was measured by Bradford assay and not all of the preparation was full-length (see Fig. 3B).
FIGURE 4.
Phosphorylation of TTP by MK2 does not affect ARE binding by TTP. A, 32P end-labeled GM-CSF ARE RNA probe (0.1 pmol) was incubated with different concentrations of either unphosphorylated TTP or TTP phosphorylated by recombinant active MK2 for 30 min at 30 °C. B, graph of bound RNA against [TTP] and best fit with the Hill equation from three independent experiments. Where not shown, error bars are smaller than the symbols. C, 32P end-labeled GM-CSF ARE was incubated with TTP (25 nm) or GST (25 nm) in the presence or absence of increasing amounts (1, 10, and 20×) of cold GM-CSF ARE (self) or GM-CSF ARE mut (non-self) RNA competitors. Results are representative of three experiments. RNA·protein complexes were resolved by electrophoresis and visualized using a phosphorimaging device. The free probe and the TTP·RNA complexes are indicated.
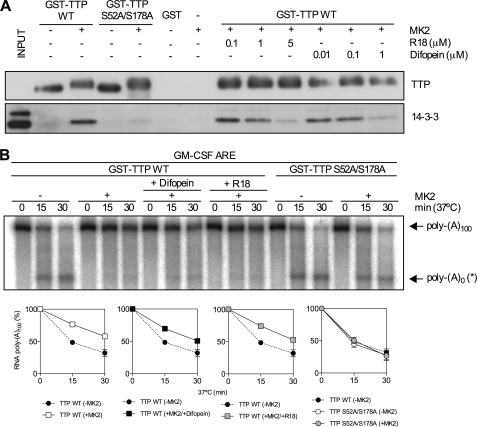
14-3-3 Binding Does Not Mediate the Effect of MK2 on TTP-directed Deadenylation
The involvement of 14-3-3 in sequestering TTP from deadenylases was tested using the antagonists R18 and difopein (35). GST pulldown was used to determine the concentrations of R18 and difopein needed to disrupt the 14-3-3·TTP complex. Wild-type and S52A/S178A GST-TTP were phosphorylated by recombinant MK2 as before or left unphosphorylated. Fusion proteins or GST alone were incubated with glutathione-Sepharose 4B beads and HeLa S100 in the presence or absence of 14-3-3 binding inhibitors. Western blots of pulled down material for 14-3-3 and TTP showed, as expected, that phosphorylation of TTP at Ser-52 and Ser-178 by MK2 gave rise to 14-3-3 binding (Fig. 5A). 5 μm R18 or 1 μm difopein efficiently inhibited 14-3-3 binding to phosphorylated TTP (Fig. 5A). Phosphorylation of GST-TTP by recombinant active MK2 inhibited deadenylation (Fig. 5B). Inactive MK2 had no effect (data not shown). Inclusion of R18 or difopein in in vitro deadenylation assay did not significantly impair the ability of MK2 to inhibit TTP-directed deadenylation, indicating that this process is not regulated by 14-3-3 (Fig. 5B). Moreover, GST-TTP S52A/S178A did not display MK2-dependent inhibition of deadenylation (Fig. 5B), indicating that Ser-52 and Ser-178 mediate the effect of MK2 on deadenylation directed by TTP. Both wild-type and S52A/S178A GST-TTP migrated with reduced mobility on SDS-PAGE (Fig. 5A), consistent with phosphorylation of residues other than Ser-52 and Ser-178 by MK2. This was confirmed by significant 32P incorporation into the mutant form of the protein in the presence of MK2 (supplemental Fig. S2).
FIGURE 5.
14-3-3 binding does not mediate the effect of MK2 on TTP-directed deadenylation. A, GST-TTP (WT or S52A/S178A) was phosphorylated with MK2 as in Fig. 4 or left unphosphorylated and used in a GST pulldown assay. S100 extracts from HeLa cells were incubated with recombinant GST-TTP or GST (negative control) immobilized on glutathione-Sepharose in the presence or absence of R18 or difopein. Pulled down proteins were resolved by SDS-PAGE and Western blotted for 14-3-3 and TTP. Results are representative of three experiments. B, in vitro deadenylation assay using unphosphorylated and phosphorylated GST-TTP (wild-type and S52A/S178A) in the presence or absence of R18 (5 μm) or difopein (1 μm). Graphs of mean (±S.E.) for three independent experiments are shown.
The CCR4·CAF1 Complex Is the Major TTP-directed Deadenylase in HeLa S100
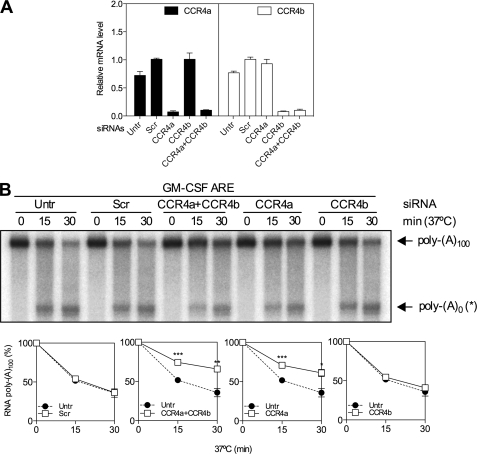
To further probe the mechanism it was necessary to identify the major deadenylase responsible for TTP-directed deadenylation. The known mammalian deadenylases (CCR4-CAF1, PAN2-PAN3 and PARN) were depleted in HeLa cells in two independent experiments by RNAi and high speed supernatants prepared and analyzed by duplicate in vitro deadenylation assays. First, HeLa cells were transfected with siRNAs against the two CCR4 paralogues (CCR4a and CCR4b alone or in combination), a scrambled double-stranded oligonucleotide as a control, or left untransfected. Cells were lysed, and lysates were either centrifuged at 100,000 × g for in vitro deadenylation assay or used for RNA isolation to assess deadenylase depletion by Q-RT-PCR. This showed a reduction of ∼85–90% of both CCR4a and CCR4b and no compensatory or off-target effects on reciprocal mRNAs (Fig. 6A). It was not possible to assess CCR4 knockdown by Western blotting as no suitable antibodies have yet been described. The effect of deadenylase depletion on deadenylation was investigated only in the presence of TTP, because in its absence, no appreciable deadenylation occurred. Knockdown of CCR4a alone resulted in reduced TTP-directed deadenylation compared with that observed with S100 from scramble-transfected or untransfected cells (Fig. 6B). Depletion of CCR4b was without effect (Fig. 6B). Simultaneous depletion of both CCR4a and CCR4b did not result in any further inhibition compared with CCR4a depletion alone (Fig. 6B).
FIGURE 6.
Depletion of CCR4a inhibits TTP-directed deadenylation. HeLa cells were left untransfected (Untr), transfected with a scramble (Scr) control double-stranded oligoribonucleotide or with siRNAs targeting CCR4a or CCR4b, separately or in combination. In vitro deadenylation assays were performed in the presence of GST-TTP as shown in Fig. 2. A, CCR4a and CCR4b mRNA knockdown efficiencies were determined by Q-RT-PCR. Graphs of mean (±S.D.) mRNA determined by Q-RT-PCR of RNA from knockdown cells. B, Graphs show mean poly(A)100 expressed as a percentage of t = 0 ± S.D. from two independent knockdown experiments performed in duplicate. Where not shown, error bars are smaller than the symbols. Significance was determined by two-tailed unpaired t test. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
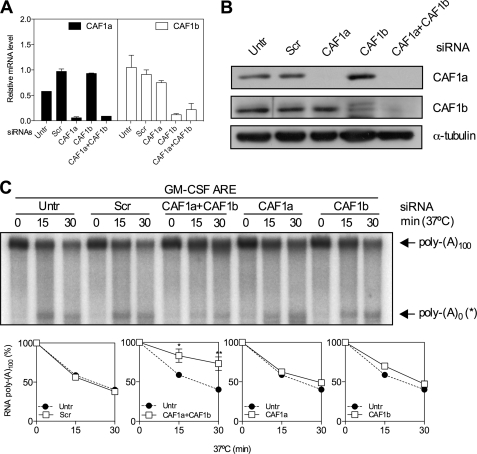
CAF1a and CAF1b were depleted by RNAi as before, and knockdown efficiency was evaluated by Q-RT-PCR and Western blotting. CAF1a and CAF1b mRNA (Fig. 7A) and protein (Fig. 7B) were strongly suppressed by their respective siRNAs. Depletion of either CAF1a or CAF1b alone had no effect on deadenylation directed by TTP (Fig. 7C). However, simultaneous depletion inhibited deadenylation (Fig. 7C).
FIGURE 7.
Depletion of CAF1a and CAF1b inhibits TTP-directed deadenylation. CAF1a or CAF1b expression was suppressed, separately or in combination (CAF1a+CAF1b) in HeLa cells by RNAi as shown in Fig. 6. A, CAF1a and CAF1b mRNA knockdown efficiencies were determined by Q-RT-PCR as in Fig. 6. B, CAF1a and CAF1b knockdown assessed by Western blot for CAF1a and CAF1b. C, in vitro deadenylation assay and graphs as in Fig. 6. Untr, untransfected; Scr, scramble.
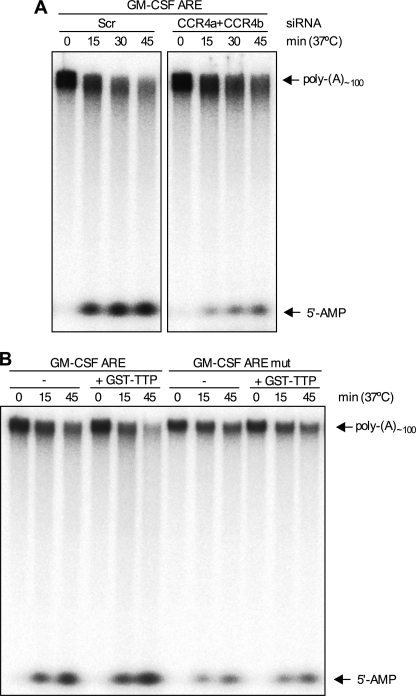
Depletion of CCR4 and CAF1 paralogues resulted in statistically significant inhibition of TTP-directed deadenylation, but the effects of knockdown on the formation of the deadenylated intermediate were variable. To confirm the involvement of the CCR4·CAF1 complex, CCR4 was depleted as before, and extracts were assayed in the presence of GST-TTP using an RNA substrate with a labeled poly(A) tail as opposed to a labeled body. CCR4 depletion caused some inhibition of the reduction in signal for full-length substrate as seen before and near complete inhibition of the production of a low molecular mass species (Fig. 8A). The identity of the high molecular mass product was confirmed as 5′-AMP by thin layer chromatography (data not shown). The modified assay was confirmed to display TTP and ARE dependence of deadenylation (Fig. 8B).
FIGURE 8.
Depletion of CCR4 inhibits TTP-directed deadenylation and formation of 5′-AMP. A, cells were transfected with scramble or CCR4a and CCR4b together, cells were lysed, and extracts were assayed in the presence of GST-TTP using an [α-32P]ATP-labeled RNA substrate. Two portions of the same gel are shown. B, same as A, but GM-CSF ARE or GM-CSF ARE mut RNA substrates were incubated in the presence or absence of GST-TTP (100 ng) and untransfected cell extracts for the times indicated. The position of the 5′-AMP product of the deadenylation reaction is indicated.
An siRNA targeting PARN strongly suppressed PARN mRNA and protein expression (supplemental Figs. S3, A and B). PARN depletion had no effect on TTP-directed deadenylation of GM-CSF ARE RNA (supplemental Fig. S3C). However, recombinant bacterially expressed PARN catalyzed TTP-directed deadenylation as previously reported (data not shown) (29). The involvement of both PAN2 and PAN3 in TTP-directed deadenylation was also tested as above. Despite strong depletion of both subunits, no effect on TTP-directed deadenylation in S100 supernatants was observed (supplemental Fig. S4).
To control for possible off-target effects, all siRNA-mediated depletions were repeated with a different set of siRNAs. Similar results were obtained in in vitro deadenylation assay (data not shown). These observations indicate that the CCR4·CAF1 complex is the major source of deadenylase activity in HeLa cells responsible for TTP-directed deadenylation. CAF1a and CAF1b may have redundant functions in HeLa cells as deadenylation was inhibited only when both CAF1a and CAF1b were knocked down together and not individually (Fig. 7C).
Recruitment of CAF1 to TTP Is Inhibited by MK2 Phosphorylation and 14-3-3 Independence
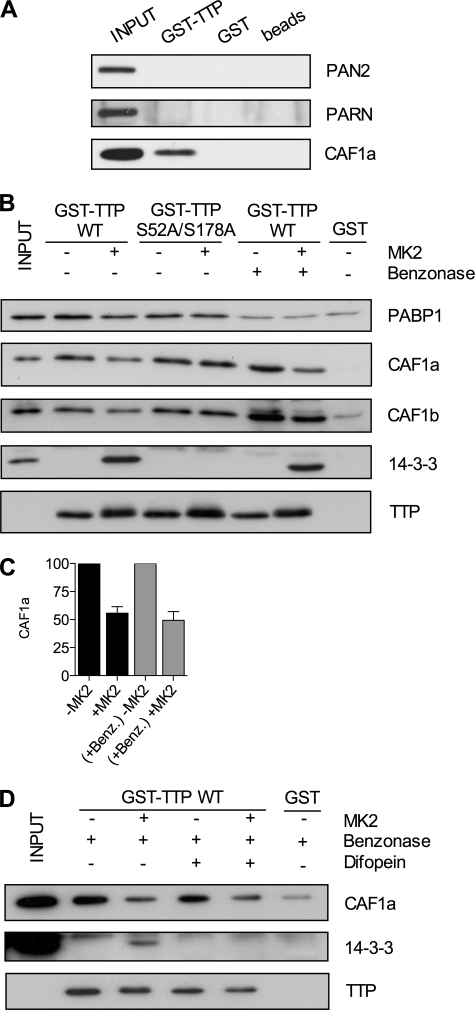
To investigate interactions between TTP and different deadenylases, GST pulldowns were performed with HeLa cell lysates using GST-TTP and GST or beads alone as controls. Neither PARN nor PAN2 were found to associate with GST-TTP (Fig. 9A). In the absence of antibodies to CCR4 proteins, CAF1a was probed for instead. As expected, and in agreement with results obtained by in vitro deadenylation assay, CAF1a interacted with GST-TTP (Fig. 9A).
FIGURE 9.
Recruitment of CAF1 to TTP is inhibited by MK2 phosphorylation and 14-3-3 independent. A–D, GST-TTP or GST alone was bound to glutathione -Sepharose 4B beads in presence of HeLa cell lysates. A, pulled down material probed for PAN2, PARN, or CAF1a. B, GST pulldown assay of GST-TTP (wild-type or S52A/S178A mutant) phosphorylated by MK2 in vitro or left unphosphorylated as in Fig. 4, in the presence or absence of 150 units of benzonase. C, a graph of mean (±S.E.) for three independent experiments of Caf1a protein quantified by densitometry. D, GST pulldown assay of wild-type GST-TTP phosphorylated by MK2 or left unphosphorylated, in the presence of 150 units of benzonase (Benz.) and in the presence or absence of 1 μm difopein.
To test whether the TTP-CAF1 interaction is modulated by MK2, pulldowns were repeated with GST-TTP that was phosphorylated by MK2 or left unphosphorylated. The nonspecific nuclease, benzonase, which cleaves poly(A) RNA in addition to other sequences was used to determine whether interactions are RNA-dependent. GST-TTP interacted with both CAF1a and CAF1b, and these interactions were inhibited by phosphorylation of the ARE-binding protein by MK2 (Fig. 9B). Phosphorylation reduced the mean binding in three independent experiments of CAF1a to GST-TTP both in the presence or absence of benzonase, by ∼50% (Fig. 9C). Thus, the interactions were RNA-independent, and benzonase treatment actually increased the amount of CAF1a and CAF1b interacting with unphosphorylated or phosphorylated GST-TTP (Fig. 9B).
In the case of miRNA-mediated deadenylation, PABP1 has been suggested to form a link between the CCR4·CAF1 and RNA-induced silencing complexes (40). PABP1 also has been suggested to interact with TTP, although it was not shown whether the interaction is RNA-independent (41). To investigate whether PABP1 mediates the interaction between TTP and the CCR4·CAF1 complex, membranes were reprobed for PABP1. This showed that PABP1 does bind GST-TTP, but the interaction is RNA-dependent (Fig. 9B). Detection of 14-3-3, and the shift in mobility of TTP, confirmed that GST-TTP was phosphorylated by MK2 (Fig. 9B).
Despite the lack of effect of difopein and R18 on deadenylation by phosphorylated TTP, it was important nevertheless to check that 14-3-3 binding did not impede binding of the CCR4·CAF1 complex. This was tested by the inclusion of difopein in a pulldown of unphosphorylated and phosphorylated TTP. Benzonase was included to block RNA-dependent effects. The addition of difopein to the GST-TTP binding reaction had no effect on CAF1a recruitment (Fig. 9D). 14-3-3 protein was not detectable in the presence of difopein (Fig. 9D), confirming the inhibition of 14-3-3 binding to phosphorylated GST-TTP. Phosphorylation of GST-TTP was confirmed by the shift in mobility (Fig. 9D).
CAF1 Is Required for Regulation of COX-2 mRNA Stability by p38 MAPK/MK2
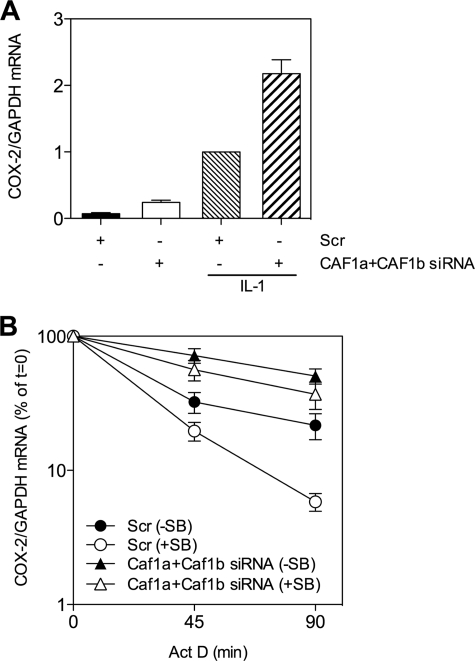
To confirm that CAF1 is needed for p38 MAPK-mediated regulation of ARE-mRNA decay, CAF1a and CAF1b were depleted from HeLa cells and then treated with or without IL-1 for 90 min to activate the p38 MAPK/MK2 pathway and induce COX-2 mRNA. This mRNA contains an ARE and is post-transcriptionally regulated by TTP and p38 MAPK/MK2 (8). Cells were treated with actinomycin D alone or together with p38 MAPK inhibitor. Depletion of CAF1a and CAF1b increased steady-state COX-2 mRNA in resting and IL-1-treated cells (Fig. 10A). COX-2 mRNA was more stable in CAF1-depleted than scramble-transfected cells (Fig. 10B). p38 MAPK inhibitor destabilized the mRNA in scramble-transfected cells but not in CAF1-depleted cells (Fig. 10B). Thus, CAF1 plays a key role in the regulation of inflammatory mRNA stability by the p38 MAPK pathway.
FIGURE 10.
CAF1 is required for regulation of COX-2 mRNA stability by p38 MAPK/MK2. A and B, HeLa cells were transfected with scramble or CAF1a and CAF1b siRNAs as in Fig. 6. Cells were then treated with IL-1 (20 ng/ml) for 90 min (A) and then treated with actinomycin D (Act D) alone or together with 1 μm SB202190 (SB) for the times indicated (B). A, graph shows COX-2/GAPDH mRNA normalized to scramble (IL-1). B, plot of % of t = 0 COX-2/GAPDH mRNA. A and B show mean and S.E. from three independent experiments.
DISCUSSION
We have shown that phosphorylation of TTP by MK2 directly inactivates TTP by inhibiting the recruitment of the CCR4·CAF1 deadenylase complex. However, it is not clear precisely which subunit of the complex TTP interacts with. We find that 14-3-3 does not appear to sequester TTP away from the mRNA decay machinery as proposed previously (6) as R18 and difopein prevented 14-3-3 from binding TTP but had no effect on TTP-directed deadenylation or CAF1 recruitment. 14-3-3 binding to TTP has been reported to cause TTP to shuttle from the nucleus to the cytoplasm (19) and to prevent TTP from associating with stress granules (6). We cannot exclude that 14-3-3-mediated TTP subcellular localization might play a role in the regulation of ARE-mediated decay, but it does not appear to be the mechanism to explain the regulation of TTP-directed deadenylation by MK2 under the conditions used in this study.
Regulation of TTP-directed deadenylation by MK2 requires phosphorylation of Ser-52 and Ser-178 because no effect on deadenylation was observed for a mutant form in which these two serines were mutated to alanine. Moreover, no MK2-dependent difference in binding of CAF1a and CAF1b was observed with the mutant form of TTP. MK2 still caused significant phosphorylation of the S52A/S178A mutant form of TTP, consistent with phosphorylation of other residues, as reported previously (5). However, because regulation of deadenylation and CAF1 binding was blocked by mutation of Ser-52 and Ser-178 to alanine, the other MK2 phosphorylation sites may have a different function.
Several observations indicate that the effect of MK2 on deadenylation is caused by phosphorylation of TTP and not of other proteins in the S100 extract. First, the effect of MK2 on deadenylation was dependent on the time of incubation of TTP with MK2 (Fig. 3). Second, when the incubation of TTP with MK2 was limited to 30 min, deadenylation was not inhibited relative to TTP incubated with nonimmune controls (Fig. 3A), thus, carry-over of MK2 to the S100-containing reaction mixture was not responsible for the effect observed. Third, the effect of MK2 was reversed by mutation of Ser-52 and Ser-178 of TTP to alanine (Fig. 5B).
Phosphorylation of TTP by MK2 has been suggested to reduce the affinity of TTP for ARE-containing mRNA (7). However, others found that mutation of the two major MK2 phosphorylation sites to alanine had no effect on ARE binding (42). Using purified recombinant TTP and MK2, we also could not detect any difference in affinity of unphosphorylated or phosphorylated TTP for the GM-CSF ARE, showing that regulation of RNA binding does not account for the regulation of deadenylation observed in the in vitro system described here.
We find that the major deadenylase involved in TTP-directed deadenylation in HeLa cells is the CCR4·CAF1 complex. CAF1a and CAF1b appear to have important deadenylase activities that can substitute one another, whereas CCR4a also has a role. CCR4b appears not to be involved in TTP-directed deadenylation in HeLa cells as depletion of CCR4b in combination with CCR4a did not increase the inhibition of deadenylation seen for CCR4a depletion alone. HeLa cells may express a lower level of CCR4b than CCR4a or the two paralogues may have different activities. In contrast, depletion of PAN2-PAN3 had no effect on in vitro deadenylation directed by TTP. No interaction between PAN2-PAN3 and TTP could be detected either. PAN2-PAN3 has been suggested to catalyze initial deadenylation of mRNA (24), so it is possible that the 100-nt poly(A) tails of RNA substrates used in this study are not long enough to reconstitute this step in vitro. Alternatively, PAN2-PAN3 may play only a minor role in TTP-directed deadenylation. In agreement with Lai et al. (29), we found that TTP can direct deadenylation in the presence of recombinant PARN, but this enzyme does not appear to be responsible for deadenylation directed by TTP in HeLa S100. Moreover, it has been shown previously that deadenylation catalyzed by PARN is stimulated by the presence of an m7-guanosine cap on substrate RNAs (43). As reported previously (29), we found no difference in rates of deadenylation between capped and uncapped RNA substrates consistent with deadenylation in our system being independent of PARN. This deadenylase is a nuclear cytoplasmic shuttling protein with a predominantly nuclear localization (24). Because TTP also can shuttle between the nucleus and the cytoplasm, we speculate that PARN may catalyze TTP-directed deadenylation in the nucleus.
We recognize that our in vitro assay may not be entirely specific for deadenylation and exonuclease activities other than deadenylases are present in HeLa cell extracts. Therefore, we cannot exclude the possibility that other mRNA decay components are involved in TTP-directed decay, and they could be regulated by MK2. This does not detract from the main conclusion of this study that CCR4·CAF1 is the main deadenylase complex involved in TTP-directed deadenylation and that MK2-mediated regulation of its recruitment to TTP provides at least part of the mechanism for mRNA stabilization by the p38 MAPK pathway. The in vivo significance of the involvement of CAF1 in p38 MAPK-mediated regulation of mRNA decay is confirmed by our observation that COX-2 mRNA stability in CAF1-depleted cells is not regulated by the p38 MAPK/MK2 pathway.
PABP1 has been reported to act as a bridging factor between GW182, a protein essential for miRNA-mediated gene silencing, and the CCR4·CAF1 complex (40). We hypothesized a similar scenario for TTP-CCR4·CAF1 interaction, but PABP1 binding to TTP was RNA-dependent, excluding PABP1 as a bridging factor between TTP and the deadenylase complex. Other proteins might mediate the recruitment of CAF1 by TTP, and RNA-dependent interactions may also be involved. It would be interesting in the future to perform a more complete analysis of interactions with TTP and to determine how they are regulated by phosphorylation.
TTP has been shown to be a component of both stress granules and processing bodies (PBs) (44), and it has been suggested to deliver its mRNA cargo to PBs for degradation or sequestration from polysomes by interacting with other components of these particles (31). Moreover, deadenylation has been shown to be a prerequisite for PB formation and mRNAs in PBs appear to represent poly(A)-shortened species (25). Our finding that TTP-directed deadenylation is mediated by the CCR4·CAF1 complex, together with evidence that CCR4 and CAF1 are predominantly not PB-associated (25), suggests that TTP-directed deadenylation also may be largely PB-independent. We cannot, however, exclude the involvement of stress granules and PBs in TTP function, and it would be interesting to understand the role of these foci in the TTP-directed deadenylation mechanism.
Supplementary Material
Acknowledgments
We are most grateful to J. Steitz, S. Hoshino, A. Virtanen, and M. Brook for provision of reagents. We also acknowledge Arthritis Research UK for support.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- ARE
- adenosine/uridine-rich element
- PAN
- poly(A) nuclease
- PARN
- poly(A) ribonuclease
- MK2
- MAPK-activated protein kinase 2
- nt
- nucleotide(s)
- Q-RT-PCR
- quantitative reverse-transcription PCR
- PB
- processing body.
REFERENCES
- 1.Dean J. L., Brook M., Clark A. R., Saklatvala J. (1999) J. Biol. Chem. 274, 264–269 [DOI] [PubMed] [Google Scholar]
- 2.Winzen R., Kracht M., Ritter B., Wilhelm A., Chen C. Y., Shyu A. B., Müller M., Gaestel M., Resch K., Holtmann H. (1999) EMBO J. 18, 4969–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasa M., Mahtani K. R., Finch A., Brewer G., Saklatvala J., Clark A. R. (2000) Mol. Cell. Biol. 20, 4265–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahtani K. R., Brook M., Dean J. L., Sully G., Saklatvala J., Clark A. R. (2001) Mol. Cell. Biol. 21, 6461–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chrestensen C. A., Schroeder M. J., Shabanowitz J., Hunt D. F., Pelo J. W., Worthington M. T., Sturgill T. W. (2004) J. Biol. Chem. 279, 10176–10184 [DOI] [PubMed] [Google Scholar]
- 6.Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F., Blackwell T. K., Anderson P. (2004) EMBO J. 23, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hitti E., Iakovleva T., Brook M., Deppenmeier S., Gruber A. D., Radzioch D., Clark A. R., Blackshear P. J., Kotlyarov A., Gaestel M. (2006) Mol. Cell. Biol. 26, 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tudor C., Marchese F. P., Hitti E., Aubareda A., Rawlinson L., Gaestel M., Blackshear P. J., Clark A. R., Saklatvala J., Dean J. L. (2009) FEBS Lett. 583, 1933–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., Schenkman D. I., Gilkeson G. S., Broxmeyer H. E., Haynes B. F., Blackshear P. J. (1996) Immunity 4, 445–454 [DOI] [PubMed] [Google Scholar]
- 10.Gherzi R., Lee K. Y., Briata P., Wegmüller D., Moroni C., Karin M., Chen C. Y. (2004) Mol. Cell 14, 571–583 [DOI] [PubMed] [Google Scholar]
- 11.Winzen R., Thakur B. K., Dittrich-Breiholz O., Shah M., Redich N., Dhamija S., Kracht M., Holtmann H. (2007) Mol. Cell. Biol. 27, 8388–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta S., Biswas R., Novotny M., Pavicic P. G., Jr., Herjan T., Mandal P., Hamilton T. A. (2008) J. Immunol. 180, 2545–2552 [DOI] [PubMed] [Google Scholar]
- 13.Tchen C. R., Brook M., Saklatvala J., Clark A. R. (2004) J. Biol. Chem. 279, 32393–32400 [DOI] [PubMed] [Google Scholar]
- 14.Brook M., Tchen C. R., Santalucia T., McIlrath J., Arthur J. S., Saklatvala J., Clark A. R. (2006) Mol. Cell. Biol. 26, 2408–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark A. R., Dean J. L., Saklatvala J. (2009) Arthritis Rheum. 60, 3513–3514 [DOI] [PubMed] [Google Scholar]
- 16.Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean J. L., Sarsfield S. J., Tsounakou E., Saklatvala J. (2003) J. Biol. Chem. 278, 39470–39476 [DOI] [PubMed] [Google Scholar]
- 18.Winzen R., Gowrishankar G., Bollig F., Redich N., Resch K., Holtmann H. (2004) Mol. Cell. Biol. 24, 4835–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson B. A., Stehn J. R., Yaffe M. B., Blackwell T. K. (2002) J. Biol. Chem. 277, 18029–18036 [DOI] [PubMed] [Google Scholar]
- 20.Boeck R., Tarun S., Jr., Rieger M., Deardorff J. A., Müller-Auer S., Sachs A. B. (1996) J. Biol. Chem. 271, 432–438 [DOI] [PubMed] [Google Scholar]
- 21.Tucker M., Valencia-Sanchez M. A., Staples R. R., Chen J., Denis C. L., Parker R. (2001) Cell 104, 377–386 [DOI] [PubMed] [Google Scholar]
- 22.Garneau N. L., Wilusz J., Wilusz C. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 113–126 [DOI] [PubMed] [Google Scholar]
- 23.Lau N. C., Kolkman A., van Schaik F. M., Mulder K. W., Pijnappel W. W., Heck A. J., Timmers H. T. (2009) Biochem. J. 422, 443–453 [DOI] [PubMed] [Google Scholar]
- 24.Yamashita A., Chang T. C., Yamashita Y., Zhu W., Zhong Z., Chen C. Y., Shyu A. B. (2005) Nat. Struct. Mol. Biol. 12, 1054–1063 [DOI] [PubMed] [Google Scholar]
- 25.Zheng D., Ezzeddine N., Chen C. Y., Zhu W., He X., Shyu A. B. (2008) J. Cell. Biol. 182, 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwede A., Ellis L., Luther J., Carrington M., Stoecklin G., Clayton C. (2008) Nucleic Acids Res. 36, 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Körner C. G., Wahle E. (1997) J. Biol. Chem. 272, 10448–10456 [DOI] [PubMed] [Google Scholar]
- 28.Lin W. J., Duffy A., Chen C. Y. (2007) J. Biol. Chem. 282, 19958–19968 [DOI] [PubMed] [Google Scholar]
- 29.Lai W. S., Kennington E. A., Blackshear P. J. (2003) Mol. Cell. Biol. 23, 3798–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., Pruijn G. J., Stoecklin G., Moroni C., Mann M., Karin M. (2001) Cell 107, 451–464 [DOI] [PubMed] [Google Scholar]
- 31.Lykke-Andersen J., Wagner E. (2005) Genes Dev. 19, 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carballo E., Lai W. S., Blackshear P. J. (2000) Blood 95, 1891–1899 [PubMed] [Google Scholar]
- 33.Brook M., Sully G., Clark A. R., Saklatvala J. (2000) FEBS Lett. 483, 57–61 [DOI] [PubMed] [Google Scholar]
- 34.Tebo J., Der S., Frevel M., Khabar K. S., Williams B. R., Hamilton T. A. (2003) J. Biol. Chem. 278, 12085–12093 [DOI] [PubMed] [Google Scholar]
- 35.Masters S. C., Fu H. (2001) J. Biol. Chem. 276, 45193–45200 [DOI] [PubMed] [Google Scholar]
- 36.Voeltz G. K., Ongkasuwan J., Standart N., Steitz J. A. (2001) Genes Dev. 15, 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean J. L., Wait R., Mahtani K. R., Sully G., Clark A. R., Saklatvala J. (2001) Mol. Cell. Biol. 21, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alford K. A., Glennie S., Turrell B. R., Rawlinson L., Saklatvala J., Dean J. L. E. (2007) J. Biol. Chem. 282, 6232–6241 [DOI] [PubMed] [Google Scholar]
- 39.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 40.Zekri L., Huntzinger E., Heimstädt S., Izaurralde E. (2009) Mol. Cell. Biol. 29, 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowlett R. M., Chrestensen C. A., Schroeder M. J., Harp M. G., Pelo J. W., Shabanowitz J., DeRose R., Hunt D. F., Sturgill T. W., Worthington M. T. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 295, G421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L., Stoecklin G., Van Way S., Hinkovska-Galcheva V., Guo R. F., Anderson P., Shanley T. P. (2007) J. Biol. Chem. 282, 3766–3777 [DOI] [PubMed] [Google Scholar]
- 43.Dehlin E., Wormington M., Körner C. G., Wahle E. (2000) EMBO J. 19, 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005) J. Cell. Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.