Abstract
A conserved family of eukaryotic RNA-dependent RNA polymerases (RDRs) initiates or amplifies the production of small RNAs to provide sequence specificity for gene regulation by Argonaute/Piwi proteins. RDR-dependent silencing processes affect the genotype-phenotype relationship in many eukaryotes, but the principles that underlie the specificity of RDR template selection and product synthesis are largely unknown. Here, we characterize the initiation specificity of the Tetrahymena RDR, Rdr1, as a heterologously expressed single subunit and in the context of its biologically assembled multisubunit complexes (RDRCs). Truncation analysis of recombinant Rdr1 revealed domain requirements different from those of the only other similarly characterized RDR, suggesting that there are subfamilies of the RDR enzyme with distinct structural requirements for activity. We demonstrate an apparently obligate Rdr1 mechanism of initiation in which the template end is looped to provide the hydroxyl group priming the synthesis of dsRNA. RDRC subunits with poly(U) polymerase activity can act on the template end prior to looping to increase the duplex length of product, thus impacting the small RNA sequences generated by the RDRC-coupled Dicer. Overall, our findings give new perspective on mechanisms of RDR initiation and demonstrate that non-RDR subunits of an RDRC can affect the specificity of product synthesis.
Keywords: Dicer, RNA, RNA Polymerase, RNA Silencing, RNA Synthesis
Introduction
Argonaute/Piwi proteins loaded with small RNAs (sRNAs)3 direct diverse types of eukaryotic gene regulation (1, 2). Most extensively characterized are the post-transcriptional silencing roles of Argonaute subfamily proteins bound to sRNAs processed from annealed dsRNAs or snap-back hairpin structures (3, 4). In addition to transcript base pairing, dsRNA precursors of sRNAs can be generated by the activity of an RNA-dependent RNA polymerase (RDR). A family of eukaryotic RDRs is broadly represented in protists, plants, fungi, and animals (5, 6). Notably, eukaryotic RDRs have an origin independent of their viral counterparts, with a conserved active site and overall multidomain organization (6–8). Despite their apparently myriad essential biological functions, the biochemical properties of RDRs remain largely uncharacterized.
Only two RDRs have been assayed for catalytic activity following recombinant expression in a heterologous system. Neurospora crassa QDE-1 produced in Saccharomyces cerevisiae synthesizes predominantly 9–21-nucleotide (nt) short products with a 5′-triphosphate, as well as some longer products (9). In comparison, Arabidopsis thaliana RDR6 produced in Nicotiana benthamiana synthesizes long products from near full-length copying of input templates (10). RDR proteins from endogenous sources, which are purified as multisubunit RDR complexes (RDRCs), have also been found to catalyze distinct types of activity. The originally characterized RDR activity from tomato synthesizes products with a 5′-triphosphate (11), and triphosphate-capped 21–23-nt sRNAs are synthesized by the Caenorhabditis elegans RRF-1 RDRC (12). Wheat germ lysate, the Schizosaccharomyces pombe Rdp1 RDRC, and the Tetrahymena thermophila Rdr1 RDRCs instead synthesize long dsRNA products processed into sRNA duplexes by the coupled action of a Dicer enzyme (13–15).
It seems clear that at least two modes of dsRNA synthesis can be distinguished: constrained synthesis of sRNA-length or shorter products versus processive synthesis from initiation to the template 5′-end. Due to the nonparallel nature of previously employed assay conditions and product analysis methods, it is difficult to draw more detailed parallels from the cross-comparison of published studies. Importantly, in no case has the activity of a physiological RDRC been compared with the activity of its RDR subunit alone. Therefore, it remains unclear whether differences in the mode of product synthesis can arise from differences in the non-RDR subunits of an RDRC.
We have used the ciliated protozoan Tetrahymena as a model system for understanding the biochemical and biological specificity of RDR function. The only Tetrahymena RDR family member, Rdr1, assembles several RDRCs (15). Each RDRC harbors one of two related nucleotidyltransferase subunits, Rdn1 or Rdn2, with poly(U) polymerase activity. The Rdr1-Rdn1 complex additionally associates with one of two related novel subunits, Rdf1 or Rdf2. Tetrahymena RDRCs are stable to stringent wash conditions, but the use of gentle wash conditions preserves RDRC interaction with Dcr2, the Dicer that produces 23–24-nt sRNAs in vivo and in vitro (15, 16). Rdr1, Rdn1, and Dcr2 are essential, but the Rdn2, Rdf1, or Rdf2 subunit specific to a single RDRC can be depleted without loss of strain growth or viability (15–17). Cells lacking any one RDRC by RDN2, RDF1, or RDF2 gene knock-out have distinct cellular and sRNA accumulation phenotypes, indicating that each RDRC has a non-redundant function (17, 18).
Here, we investigated the template requirements and product structures of recombinant Tetrahymena Rdr1 and purified Tetrahymena RDRC complexes. We found that Rdr1 and its RDRCs produce dsRNA products that do not result from de novo initiation. Instead, Rdr1 initiates by looping the template 3′-end for intramolecular initiation of dsRNA synthesis. By comparing Rdr1 and RDRCs purified using the same tag, we show that RDRC context can influence dsRNA product structure. We demonstrate a novel role for RDRC-associated poly(U) polymerase activity, which, through uridylation of the template, increases the length of duplex product synthesis. Our findings suggest that template 3′-tailing prior to template looping allows dsRNA synthesis and Dicer cleavage of sRNA products from the original template 3′-end. Overall, this work characterizes a mode of RDR initiation distinct from the de novo initiation that produces triphosphate-capped sRNAs for amplification of gene silencing. We propose that at least a subset of RDR enzymes capable of long-product synthesis coupled to subsequent Dicer processing differ in their initiation mechanism from the short-product RDRs that generate triphosphate-capped sRNAs independent of Dicer. Our studies also provide the first evidence for an influence of RDR-associated subunits on the specificity of dsRNA product synthesis.
EXPERIMENTAL PROCEDURES
Rdr1 and RDRC Expression and Purification
Recombinant Rdr1 was expressed from a synthetic open reading frame (DNA2.0) in rabbit reticulocyte lysate (RRL) by coupled transcription and translation. For assays requiring analytical SDS-PAGE, recombinant protein was labeled with [35S]methionine. Most experiments used Rdr1 expressed with an N-terminal tag of tandem protein A domains (referred to as zz) joined to Rdr1 by an intervening cleavage site for tobacco etch virus (TEV) protease, identical to the tagged Rdr1 expressed in Tetrahymena (15, 17). For the Rdr1 dimerization assay, plasmids encoding Rdr1 tagged with zz and Rdr1 tagged with three tandem repeats of the FLAG peptide (referred to as F) were expressed separately or in combination.
RRL-produced zzRdr1 was recovered from extract prior to activity assays by binding to IgG-agarose and elution with TEV protease using conditions developed for RDRC purification from Tetrahymena extracts (15). T2MG binding and wash buffer (20 mm Tris-HCl (pH 7.5), 1 mm MgCl2, and 10% glycerol) was supplemented with 50 mm NaCl for all recombinant Rdr1 purifications. For dimerization assays, resin binding time was reduced from the standard 1.5 h to 30 min. Tetrahymena RDRC complexes assembled in vivo on zzRdr1 or a zzF-tagged Rdn subunit were purified by binding to IgG-agarose and elution with TEV protease as described previously (15, 17). T2MG with 50 mm NaCl was used for gentle washes, whereas T2MG with 200 mm NaCl was used for high-stringency washes.
Template Preparation and Activity Assays
With the exceptions noted, RNA templates for activity assays were transcribed from PCR products using T7 RNA polymerase and purified by denaturing PAGE using gels of 9% acrylamide/bisacrylamide (19:1), 7 m urea, and 0.6× Tris borate/EDTA. RNA concentration was determined by spectrophotometry. RNA32, Aless, and DNA-containing templates were synthesized chemically (Integrated DNA Technologies). RNA end modification to produce a 3′-phosphate group was performed as described (19). The purity, integrity, and concentration of RNA template stocks were confirmed by denaturing PAGE with SYBR Gold staining. Assays of dsRNA synthesis, dsRNA processing to sRNA, and poly(U) polymerase activity were carried out largely as described (15, 17) using [α-32P]CTP unless indicated otherwise. Reactions contained a final concentration of 50 nm template.
Assay products were extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and then precipitated in ethanol with linear polyacrylamide carrier in the presence of 300 mm NaCl. Products were resuspended in 2 μl of 10 mm Tris-HCl (pH 7.5) with 50 mm NaCl. S1 nuclease (Fermentas) treatment was performed for 10–20 min at 37 °C in the manufacturer's buffer. RNase H (U. S. Biochemical Corp.) digestion was performed according to the manufacturer's recommendation at 37 °C for 1 h. Reactions with dT18 or dA18 included ∼20 μm 18-nt DNA oligonucleotide (∼500-fold molar excess over template). S1 and RNase H reactions were quenched by the addition of a 20-fold excess volume of 10 mm Tris-HCl (pH 7.5), 1 mm EDTA, and 500 mm NaCl, followed by another round of product extraction and precipitation.
Samples were resuspended in loading dye containing 94% (v/v) formamide and 30 mm EDTA, denatured at 98 °C for 2 min, and then iced prior to analysis by formamide-PAGE (12% acrylamide/bisacrylamide (19:1), 45% formamide, 7 m urea, and 1× Tris borate/EDTA). The lengths of products were estimated by their migration relative to 5′-end-labeled RNA templates and other oligonucleotide markers. For sRNA analysis by blot hybridization, scaled-up (40 μl) RDRC reactions were performed with a 0.025 mm concentration of each non-radiolabeled NTP. Products were separated by formamide-PAGE and transferred to Hybond-N+ membrane (GE Healthcare) using electrophoretic transfer (GENIE). Blots were hybridized at room temperature for at least 4 h with 5′-end-labeled oligonucleotide probes antisense to the Rdr1 product strand. The sequence of the 3′-region probe was 5′-TGGATTCTGAAATGCTTTCTTACAACC; the sequence of the mid-region probe was 5′-GATGACGATAAATAAATACAACAATTGA.
RESULTS
Rdr1 Domain Requirements Differ from Previous Findings for QDE-1
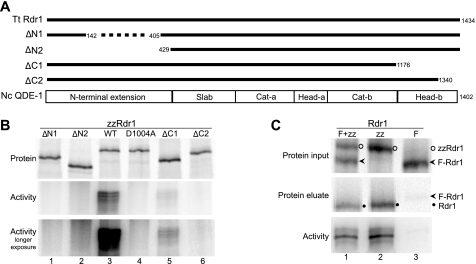
RDRs share regions of sequence conservation beyond the catalytic core (6). The only published study of RDR domain requirements examined recombinant N. crassa QDE-1, which is soluble and active without the RDR N-terminal extension (9). High-resolution structure determination for the remaining domains of QDE-1, designated slab, catalytic, and head (Fig. 1A), revealed that the head domain mediates subunit dimerization (8). Because recombinant QDE-1 appears to have unique biochemical features compared with subsequently characterized RDRCs, we analyzed the domain requirements of Tetrahymena Rdr1 for comparison. We used RRL to express a tagged version of Rdr1 (zzRdr1) known to be biologically functional (15). Recombinant Rdr1 was recovered from RRL using IgG-agarose to bind the N-terminal tag of protein A domains, followed by elution with TEV protease. Full-length Rdr1 and several Rdr1 domain truncation variants were examined for dsRNA synthesis activity using the same conditions optimized for the physiologically assembled Tetrahymena RDRCs (15, 17).
FIGURE 1.
Tetrahymena Rdr1 requires its N-terminal extension for activity and purifies as a monomer. A, RDR domains and expressed Rdr1 polypeptides. T. thermophila (Tt) Rdr1 truncations were designed based on alignment with N. crassa (Nc) QDE-1. Amino acid end points for Rdr1 truncations are indicated using the terminal amino acid included in the expressed protein. QDE-1 Head-a and Head-b or Cat-a and Cat-b are regions that fold together to form the head or catalytic domains, respectively. B, Rdr1 domain analysis. Recombinant proteins were radiolabeled by methionine incorporation, resolved by SDS-PAGE, and visualized by Typhoon PhosphorImager analysis (upper panel). Purified WT Rdr1 and variant and truncated proteins were assayed for catalytic activity using the RNA1 template without nuclease digestion of products (middle and lower panels). C, Rdr1 dimerization analysis. Differentially tagged Rdr1 proteins and their coexpressed combination were used as inputs for binding to IgG-agarose, followed by elution with TEV protease to cleave the tag from zzRdr1. Input and eluted samples were analyzed by SDS-PAGE (upper and middle panels), and eluted samples were analyzed for activity as described above (lower panel). The labeled Rdr1 polypeptide identities were confirmed by migration standards and by immunoblotting for the F tag (not shown).
RNA products of Rdr1 were radiolabeled by incorporation of [32P]CTP and resolved using highly denaturing formamide-PAGE, which we have found to be essential for complete suppression of product secondary structure formation (15). For comparison with previous studies of Tetrahymena RDRC activity, we used the 79-nt single-stranded RNA template RNA1 derived from a genomic locus subject to Rdr1-mediated silencing in vivo (16, 18). We compared the activity of WT Rdr1, the catalytic dead variant Rdr1-D1004A (with an alanine substitution for aspartic acid in the active site) (15), an internal deletion within the N-terminal extension, and N- and C-terminal truncations designed based on primary sequence conservation with QDE-1 to have end points at putative subdomain boundaries (Fig. 1A). All of the proteins were expressed at comparable levels (Fig. 1B, upper panel). Full-length Rdr1 (but not the catalytic dead variant) catalyzed robust product synthesis (Fig. 1B, middle and lower panels). Curiously, the N-terminal truncation of Tetrahymena Rdr1 analogous to that used for recombinant expression of QDE-1 eliminated enzyme activity, as did a smaller internal deletion of the Rdr1 N-terminal extension (Fig. 1B, lanes 1 and 2). In contrast, C-terminal truncation reduced but did not eliminate activity (Fig. 1B, lane 5; in some experiments, weak activity was also detectable for the protein assayed in lane 6).
Recombinant QDE-1 forms a homodimer (8). A similar interaction seems unlikely for Tetrahymena Rdr1 in RDRC context because RDRCs with distinct subunit composition are readily isolated from each other without the cross-purification that would be expected from Rdr1 multimerization (17). To test whether the isolated Tetrahymena Rdr1 subunit forms a multimer, we coexpressed differentially tagged versions of Rdr1 in RRL. In addition to zzRdr1 used above, we expressed Rdr1 with an N-terminal tag of three tandem FLAG epitopes (F-Rdr1). Either tagged Rdr1 or the combination was expressed in RRL, followed by binding to and elution from IgG-agarose (Fig. 1C). Elution with TEV protease removes the tag from zzRdr1, resulting in the isolation of untagged Rdr1 (Fig. 1C, lanes 1 and 2). A low-background binding of F-Rdr1 to IgG-agarose was detectable (Fig. 1C, lane 3), but there was no co-enrichment of F-Rdr1 with zzRdr1. Furthermore, no enrichment of zzRdr1 by copurification with F-Rdr1 was detected in the reciprocal purification on FLAG antibody resin (data not shown). Rdr1 isolated by zzRdr1 purification remained functional when coexpressed with F-Rdr1 (Fig. 1C, lower panel), as did F-Rdr1 when coexpressed with zzRdr1 (data not shown). These findings suggest that recombinant Tetrahymena Rdr1 functions as a subunit monomer. Overall, we conclude that there are likely to be subclasses of RDR with distinct domain requirements for function.
Rdr1 and RDRCs Can Produce Similar dsRNA Products of Less than Full Template Length
We next assayed product synthesis by recombinant Rdr1 compared with Rdr1 assembled in RDRC context. We purified the pool of physiologically assembled RDRCs from an extract of a Tetrahymena strain expressing zzRdr1 using stringent wash conditions to deplete the associated Dcr2 dsRNA cleavage activity (15). To our surprise, in assays using a variety of template lengths and sequences including the standard template RNA1 (Fig. 2A), recombinant Rdr1 and stringently washed RDRCs produced a similar profile of product synthesis. Notably, despite the absence of RDRC subunits with poly(U) polymerase activity in the RRL-expressed Rdr1 enzyme preparation, recombinant Rdr1 synthesized products that, like RDRC products, were longer than the input 79-nt template length (Fig. 2B, lanes 1 and 2). If Rdr1 or RDRC products were treated with S1 nuclease to remove any single-stranded regions, dsRNA product lengths were resolved to be near full template length and somewhat shorter (Fig. 2B, lanes 3 and 4). No bias toward the synthesis of sRNA-sized products was observed, consistent with the Dcr2 dependence of 23–24-nt sRNA production in vivo.
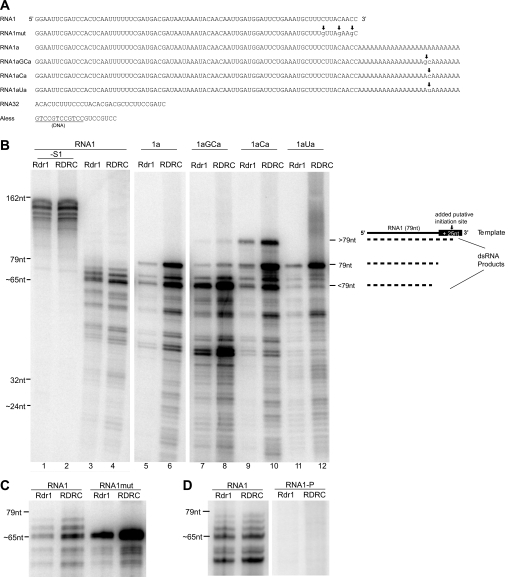
FIGURE 2.
Rdr1 alone and Rdr1 in RDRC context have similar template specificities. A, template sequences. Sequence substitutions made in the 3′-region of variants of RNA1 or RNA1a are indicated with lowercase letters and arrows. The region of the Aless template composed of DNA is underlined. B and C, template dependence of dsRNA synthesis. Recombinant Rdr1 purified following zzRdr1 expression in RRL (Rdr1) and the mixed RDRC population purified from Tetrahymena expressing zzRdr1 (RDRC) were assayed for activity using the templates indicated. Unless indicated otherwise (B, lanes 1 and 2), reaction products were digested with S1 nuclease to remove the single-stranded region(s) of dsRNA products and the single-stranded products of RDRC poly(U) polymerase activity prior to formamide-PAGE. End-labeled template RNAs and smaller synthetic oligonucleotides were run in lanes not shown as markers, with the approximate size of some of the products estimated by relative migration. At right in B, a diagram indicates the relative lengths of the RNA1 template (solid line) and its 3′-A tract extension in RNA1a (black box labeled + 25nt) along with three representative product strands (dashed lines) that would remain base-paired with portions of the template to be protected from digestion by S1 nuclease (dsRNA products). The addition of an initiation site within the A tract allows the synthesis of an additional product (lanes 7–10) of the expected length for complete template copying to its 5′-end. D, influence of template 3′-end structure. Shown are nuclease-treated reaction products of RNA1 with 3′-end hydroxyl groups (RNA1) or a 3′-phosphate group (RNA1-P). Lanes are cropped from the same exposure of the same gel.
A likely cause of heterogeneity in dsRNA product lengths is the potential use of heterogeneous sites within the template for dsRNA synthesis initiation. Extending the RNA1 template by the addition of a poly(A) tail (RNA1a) (Fig. 2A) favored the synthesis of a dsRNA product with the approximate length of RNA1, suggesting that the homopolymer A tract did not support dsRNA synthesis initiation (Fig. 2B, lanes 5 and 6). Insertion of a GC or C within the A tract (RNA1aGCa and or RNA1aCa templates) (Fig. 2A) provided an additional initiation site, resulting in the synthesis of an extra product with the length expected for processive synthesis from the introduced initiation site to the template 5′-end (Fig. 2B, lanes 7–10). Activity assays using numerous additional templates did not suggest any specific sequence requirement for dsRNA synthesis initiation by Rdr1 or RDRC, although some sequence preference was evident. For example, both Rdr1 and RDRC greatly preferred a template with an introduced cytidine initiation site rather than a uridine initiation site (Fig. 2B, lanes 9–12). Likewise, substitution of the three cytidine residues within the 3′ 15 nt of the RNA1 template for guanosines (RNA1mut) (Fig. 2A) decreased the yield of the longest products and increased the yield of the slightly shorter ∼64–65-nt products (Fig. 2C). These findings suggest that Tetrahymena Rdr1 can initiate dsRNA synthesis from variable positions near the template 3′-end but then copies the remainder of the template processively. Curiously, in contrast to the lack of requirement for any specific template sequence, Rdr1 showed an absolute dependence on template 3′-end structure. If the 3′-terminal template nucleotide was converted to a 3′-phosphate group, no activity was observed in reactions with Rdr1 or RDRC (Fig. 2D).
Rdr1 Can Copy DNA if Ribonucleotides Are Present at the Template 3′-End
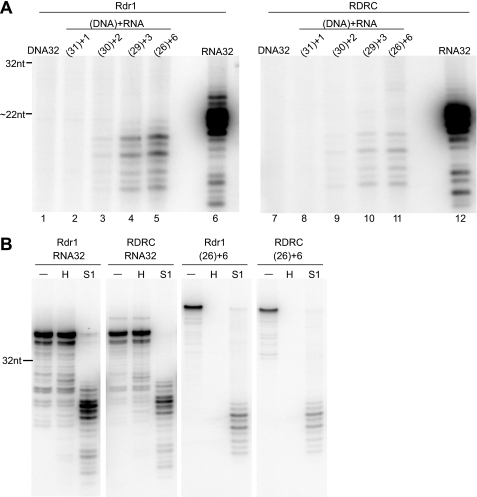
An endogenous tomato RDR activity, tagged N. crassa QDE-1 expressed in vivo, and recombinant A. thaliana RDR6 each have been shown to synthesize RNA using DNA as template (10, 11, 20). We therefore tested Tetrahymena Rdr1 and RDRC activity on DNA and RNA oligonucleotides of the same 32-nt sequence (RNA32) (Fig. 2A). Rdr1 had robust activity on the synthetic RNA, yielding heterogeneously sized dsRNA products shorter than the full-length template following treatment with S1 nuclease (Fig. 3A, lane 6). No products were observed in Rdr1 reactions with the DNA template, but chimeric templates with a DNA 5′-end and two to six residues of RNA at the 3′-end supported the synthesis of RNA products resistant to treatment with S1 nuclease (Fig. 3A, lanes 1–5). Nearly identical results were observed for Rdr1 assayed in RDRC context (Fig. 3A, lanes 7–12). Products from reactions with the template containing 26 nt of DNA were degraded by RNase H, confirming the presence of product DNA-RNA hybrid, whereas RNA32 template products were resistant to RNase H (Fig. 3B). We conclude that Tetrahymena Rdr1 can act as a DNA-dependent RNA polymerase in vitro, albeit with relatively low product yield compared with its activity as an RNA-dependent RNA polymerase. However, even when copying DNA, Rdr1 has an absolute requirement for RNA structure at the template 3′-end. We note that in previous studies it is possible that DNA templates were extended by ribonucleotide tailing prior to dsRNA synthesis, in particular in the case of recombinant A. thaliana RDR6, which, unlike Tetrahymena Rdr1, has an inherent nucleotidyltransferase activity (10, 17).
FIGURE 3.
Rdr1 can copy a DNA template if the 3′-end is RNA. A, Rdr1 and RDRC assay products from reactions containing 32-nt templates of entirely DNA (lanes 1 and 7), 5′-DNA (with the number of DNA nucleotides in parentheses) with 1, 2, 3, or 6 nt of RNA at the 3′-end (lanes 2–5 and 8–11, respectively), or entirely RNA (lanes 6 and 12) were treated with S1 nuclease before formamide-PAGE. B, Rdr1 and RDRC assay products from reactions with RNA32 and template (26)+6 (26 nt of DNA followed by 6 nt of RNA at the 3′-end) were treated with RNase H (H), S1 nuclease, or no nuclease (−) before formamide-PAGE.
The Duplex Product Contains an Internal Loop
Some of the single-stranded RNA content of RDRC products could derive from the poly(U) polymerase activity of the Rdn subunits, but even recombinant Rdr1 lacking an associated nucleotidyltransferase produced partially single-stranded products (Fig. 2B, lanes 1–4). The migration of these partially single-stranded products was not affected by single-stranded RNA exonuclease treatments (data not shown). Also, the specific activity of products from reactions with radiolabeled CTP, UTP, or ATP was not greatly altered by digestion of the single-stranded product regions with S1 nuclease, as would be expected if a nucleotide-selective transferase activity was adding a 3′-tail to the dsRNA products (data not shown). We therefore tested a model of product structure in which the template and product strands are joined by an internal loop of single-stranded RNA. In Rdr1 products, this loop would be derived from template residues only, whereas the loop of RDRC products could include both template residues and nucleotides added to the 3′-end of the template by Rdn activity (see below). A requirement for priming from a “looped” rather than base-paired template 3′-end would explain why no product synthesis was observed from an annealed primer (data not shown). The required use of the template 3′-end as primer would preclude copying of templates with a blocked 3′-end, as we observed (Fig. 2D), and would also explain why we have been unable to detect a product 5′-triphosphate group by direct or indirect labeling approaches (data not shown).
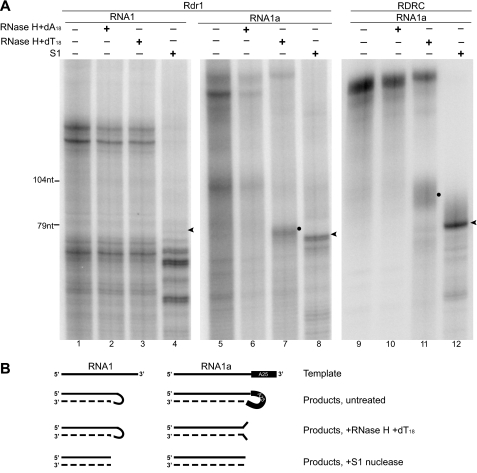
To investigate template looping as a mechanism of initiation, we exploited the observation that templates with a poly(A) 3′-region did not initiate dsRNA synthesis within the homopolymer A tract (Fig. 2B). We compared products of the 79-nt RNA1 and 104-nt RNA1a templates for their susceptibility to digestion with RNase H in the presence of dT18 DNA oligonucleotide. For the RNA1 template, Rdr1 products were insensitive to digestion with RNase H in the presence of dA18 or dT18 DNA oligonucleotide; as a control, products were shifted to faster migration by treatment with S1 nuclease (Fig. 4A, lanes 1–4). For the RNA1a template, Rdr1 products were insensitive to digestion with RNase H in the presence of dA18 (Fig. 4A, lanes 5 and 6). On the other hand, digestion with RNase H in the presence of dT18 produced discrete products not much larger than products digested by S1 nuclease (Fig. 4A, lanes 7 and 8). Similar results were observed for RDRC products, with slightly slower product mobility following RNase H treatment in the presence of dT18 (Fig. 4A, lanes 9–12). These results support the template-looping mechanism for Rdr1 initiation of dsRNA synthesis (Fig. 4B). We note that the largest Rdr1 and RDRC products were more efficiently digested by S1 nuclease than by RNase H and dT18 under our reaction conditions (Fig. 4A, compare the longest products in lane 7 versus lane 8 or lane 11 versus lane 12). This could derive from the need for dT18 hybridization to compete with structure formation in the looped poly(A) region of RNA1a products.
FIGURE 4.
Rdr1 links the template to product by looping the template 3′-end. A, Rdr1 and RDRC assay products from reactions containing RNA1 or RNA1a were reduced in length by S1 nuclease treatment. RNA1a assay products were also reduced in length by product treatment with RNase H in the presence of dT18 (but not dA18) oligonucleotide. Arrowheads mark the largest product strands from the duplex region protected by S1 nuclease treatment, whereas circles indicate slightly longer product strands likely to contain some single-stranded region not fully removed by RNase H. B, shown is a model for Rdr1 and RDRC initiation by template looping. The 3′-end of RNA1 and RNA1a templates (solid lines) is extended by product synthesis (dashed lines). Initiation does not occur within the 25-nt 3′-A tract of RNA1a, so this region of the template is contained within the internal loop of the product and is therefore accessible for degradation either by S1 nuclease or by RNase H in the presence of dT18.
RDRC Context and Rdr1-associated Rdn Activity Can Influence Template Looping
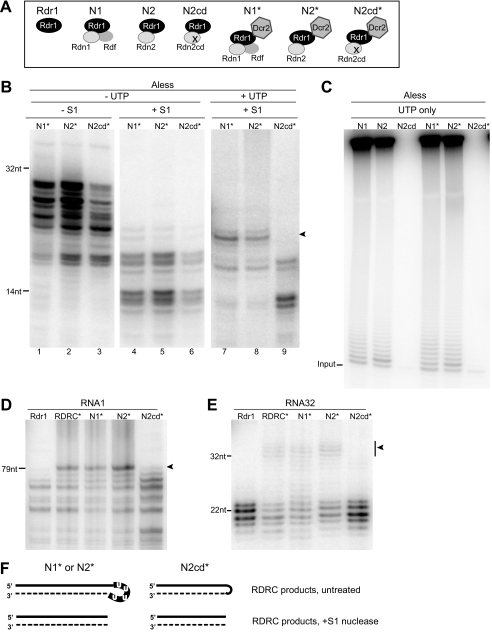
The RDRC activity assays described above used the cellular pool of Rdr1 complexes. The related RDRC poly(U) polymerase subunits, Rdn1 and Rdn2, assemble with Rdr1 in a mutually exclusive manner. Both enzymes can extend single-stranded RNA by mono-, oligo-, or polyuridine addition in vitro, but their in vivo depletion phenotypes are distinct: RDN1 knockdown or expression of a catalytic dead Rdn1 variant is lethal, whereas RDN2 knock-out or expression of a catalytic dead Rdn2 variant (referred to as Rdn2cd) is not (17). When assayed as stringently washed complexes, RDRCs purified from cell extracts by tagged Rdn1, Rdn2, or Rdn2cd had similar specificities of dsRNA product synthesis (17). Curiously, we found that gently washed RDRCs with associated Dcr2, designated RDRC*, showed differences in product synthesis dependent on the presence of catalytically active Rdn.
RDRC purifications from extracts of cells expressing tagged Rdn1, Rdn2, or Rdn2cd (17) were conducted using gentle wash conditions to purify RDRC complexes designated N1*, N2*, and N2cd* according to their Rdn subunit identity (Fig. 5A). These complexes were assayed for dsRNA product synthesis using a 20-nt chimeric template with a DNA 5′-end and RNA 3′-end (Aless) (Fig. 2A), the short products of which are unsuitable for Dcr2 processing. Because the Aless template lacks adenosines, UTP should not be required for dsRNA synthesis. Indeed, N1*, N2*, and N2cd* RDRC reactions lacking UTP produced predominant products longer than the template that were converted to dsRNA products of less than template length by S1 nuclease treatment (Fig. 5B, lanes 1–6). The addition of UTP to the reaction did not affect the profile of dsRNA products produced by the N2cd* RDRC, which lacks an active poly(U) polymerase (Fig. 5B, lane 9). In contrast, in assays of the N1* and N2* RDRCs harboring a catalytically active Rdn subunit, the presence of UTP allowed the synthesis of longer dsRNA products (Fig. 5B, lanes 7 and 8). The poly(U) polymerase activity of N1* and N2* (but not N2cd*) was confirmed in reactions with radiolabeled UTP, with other NTPs absent to prevent dsRNA synthesis on the Aless template (Fig. 5C). Reactions using other templates also showed a change in the product profile of dsRNA synthesis dependent on the presence of catalytically active Rdn: N1*, N2*, and the pooled RDRC* produced longer products in assays with RNA1 (Fig. 5D) or RNA32 (Fig. 5E), whereas Rdr1 alone or N2cd* did not.
FIGURE 5.
RDRCs can produce a larger size of dsRNA product than Rdr1 alone, dependent on a catalytically active poly(U) polymerase subunit and UTP. A, schematic of different enzyme preparations assayed. Active-site substitution is indicated by ×. B, products from activity on the Aless template in the absence (lanes 1–6) or presence (lanes 7–9) of UTP without (lanes 1–3) or with (lanes 4–9) subsequent removal of single-stranded RNA regions by S1 nuclease. The largest Rdn-dependent product strands are indicated by an arrowhead. C, Rdn-mediated tailing of Aless template with radiolabeled UTP. D and E, assay products from additional templates, with subsequent removal of single-stranded RNA regions by S1 nuclease. The Rdn-dependent product strands are indicated by arrowheads. F, model for Rdn influence on RDRC product synthesis. Rdr1 product synthesis is shown as a dashed line. Uridine(s) added to the initial template 3′-end by Rdn activity (illustrated as a thick line with four Us, although the number of uridines is uncertain) would contribute loop nucleotides and thereby allow a longer length of the template to be copied by Rdr1.
The Rdn-dependent increase in the length of template that is copied into dsRNA provides the first biochemical demonstration of an RDRC subunit influence on dsRNA product synthesis by an RDR. We suggest that Rdn-mediated template U tailing provides an optimal structure for 3′-loop formation and/or priming of second-strand synthesis, favoring dsRNA synthesis across the maximal extent of the original template (Fig. 5F). Only a limited length of template U tailing is likely to occur prior to second-strand synthesis because the internal loop region of RDRC* products was not digested by RNase H in the presence of dA18 (data not shown). Unfortunately, our attempts to define loop sequences by cloning template-looped products were unsuccessful, even for the relatively short products of the Aless template. Template use was not stimulated by the preaddition of various lengths of U tail (data not shown), suggesting the need for coordination of the Rdn and Rdr1 phases of template extension. Indeed, gently purified RDRCs showed a greatly enhanced Rdn dependence of dsRNA product synthesis, suggesting that an RDRC* conformation correlated with Dcr2 association promotes productive coordination of Rdn and Rdr1 activities.
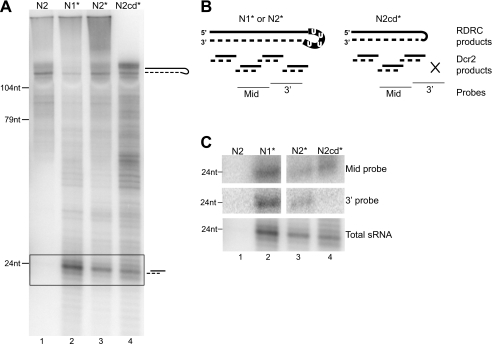
Impact of Rdn Activity on sRNA Production by Dcr2
Because Rdn activity affects the profile of dsRNA product synthesis by Rdr1, it could alter the sequence of sRNA products generated by Dcr2. We used the RNA1 template in assays of the stringently washed N2 RDRC lacking associated Dcr2 or the gently washed N1*, N2*, and N2cd* RDRCs with copurified Dcr2 (Fig. 5A). All of these Rdr1-containing RDRCs generated products that, without S1 nuclease treatment, were longer than input template length (Fig. 6A). In addition, as expected, the N1*, N2*, and N2cd* RDRCs generated 23–24-nt sRNA products from dsRNA cleavage by Dcr2 (Fig. 6A, lanes 2–4). We note that not all RDRC* complexes retain Dcr2, with generally higher Dcr2 stoichiometry in N1* compared with N2* purifications (17). As expected, the N2 RDRC depleted of Dcr2 did not generate sRNA-sized products (Fig. 6A, lane 1), and the N2cd* RDRC lacking nucleotidyltransferase activity did not generate the background of long single-stranded products extending to the top of the gel (lane 4).
FIGURE 6.
Rdr1-associated poly(U) polymerase activity extends the length of template represented in the Dicer cleavage products. A, assays of various RDRC preparations indicated were performed using RNA1 template. Products were not treated with S1 nuclease prior to formamide-PAGE. In addition to RDRC products, Dcr2 present in RDRC* purifications generated 23–24-nt sRNAs. B, shown is a schematic of proposed Rdn influence on RDRC product synthesis and subsequent cleavage by Dcr2. Rdr1 product synthesis is shown as a dashed line. Oligonucleotide probes (mid- and 3′-probes) were designed complementary to the indicated regions of the product strand. C, assays were performed as described for A except for the use of unlabeled NTPs. Products of specific sequence were subsequently detected by blot hybridization with the mid- or 3′-probe (upper and middle panels). The total amount of Dcr2 product was gauged by the parallel reaction containing radiolabeled CTP (the boxed region of the gel in A; shown again as the lower panel in C).
Parallel reactions were performed with unlabeled NTPs to evaluate sRNA sequences by blot hybridization. End-labeled oligonucleotide probes complementary to the product strand were used to detect putative sRNAs derived from RDRC synthesis across different regions of the template (Fig. 6B). No sRNA sequences were detected in reactions with the stringently washed N2 RDRC lacking Dcr2 (Fig. 6C, lane 1). Both N1* and N2* RDRC reactions generated sRNAs complementary to the mid- and 3′-regions of template, but sRNAs complementary to the template 3′-region were not detected in the N2cd* reaction (Fig. 6C, lanes 2–4). These results extend the demonstration of Rdn-dependent copying of the template 3′-end into dsRNA. Without Rdn-mediated U tailing of the template, more of the template 3′-region must be incorporated into the internal loop. Our attempts to determine whether RDRC-coupled Dcr2 cleavage occurred preferentially on the loop side of a dsRNA product were not successful due to the apparent lack of partially Dcr2-cleaved dsRNA intermediates (data not shown). Dcr2 may be most active when cotranscriptionally cleaving nascent dsRNA, prior to RDRC completion of dsRNA synthesis on a long RNA template.
DISCUSSION
Eukaryotic RDR proteins play important roles in numerous cellular processes. Despite a clear biological significance, little is known about the principles that govern RDR specificity for template recognition or synthesis of a dsRNA product. Here, we have provided new insight into these biochemical specificities. Also, for the first time, we compared the properties of a recombinant Rdr1 subunit alone and its physiologically assembled RDRCs.
The N. crassa QDE-1 C terminus mediates subunit dimerization, with an extensive interaction surface proposed to promote its biological function (8). Here, we did not detect dimerization of recombinant Tetrahymena Rdr1. Tetrahymena Rdr1 also does not appear to dimerize in RDRC context because distinct RDRCs harboring the same Rdr1 subunit purify separately from each other (17). Also unlike recombinant QDE-1 (9), we found that recombinant Tetrahymena Rdr1 requires its N-terminal extension for catalytic activity. These differences between N. crassa QDE-1 and Tetrahymena Rdr1 suggest that there may be biochemically distinct requirements for activity among subclasses of RDR enzyme. QDE-1 may be representative of an RDR class that preferentially generates short products using de novo initiation, as clearly demonstrated for the C. elegans RRF-1 RDRC (12). Tetrahymena Rdr1 may be representative of a distinct RDR class that initiates by template looping, with more processive synthesis of dsRNA coupled to Dicer processing. A template-looping requirement for RDR initiation has not previously been proposed, although products consistent with this mechanism have been detected in previous studies. QDE-1 generates some products of approximately twice the length of template when copying a subset of RNA templates (9), and some S. pombe RDRC products are longer than template length as well (14, 21). Even using highly purified RDR or RDRC preparations, trace contamination with a nuclease active on single-stranded RNA would be sufficient to prevent detection of a covalent linkage between the template and product strands.
Our comparisons of Tetrahymena Rdr1 and RDRCs uncovered evidence for an influence of RDRC subunits in dsRNA synthesis. The Rdn subunits of Tetrahymena RDRCs do not alter the requirement for a template-looping mechanism of dsRNA synthesis, but they do modulate the preferred site of dsRNA synthesis initiation. Likely through template 3′-tailing with a limited number of uridines, Rdn catalytic activity allows a primary transcript to be more extensively copied into dsRNA and more completely represented in the sRNA pool produced by Dcr2. In vitro, RDRC purification conditions affected the coupling of Rdn and Rdr1 activities. Like the Tetrahymena RDRCs, the S. pombe RDRC contains a predicted nucleotidyltransferase subunit, Cid12 (14). We speculate that S. pombe Cid12 and other non-canonical poly(A) polymerase superfamily subunits of RDRCs serve functions similar to those of the Tetrahymena Rdn subunits, which may or may not be possible to detect in vitro depending on purification and assay conditions.
Acknowledgment
We thank Suzanne R. Lee for initial studies.
This work was supported, in whole or in part, by National Institutes of Health Grant GM54198 (to K. C.).
- sRNA
- small RNA
- RDR
- RNA-dependent RNA polymerase
- nt
- nucleotide(s)
- RDRC
- RNA-dependent RNA polymerase complex
- RRL
- rabbit reticulocyte lysate
- TEV
- tobacco etch virus.
REFERENCES
- 1.Siomi H., Siomi M. C. (2009) Nature 457, 396–404 [DOI] [PubMed] [Google Scholar]
- 2.Ghildiyal M., Zamore P. D. (2009) Nat. Rev. Genet. 10, 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 4.Carthew R. W., Sontheimer E. J. (2009) Cell 136, 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerutti H., Casas-Mollano J. A. (2006) Curr. Genet. 50, 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zong J., Yao X., Yin J., Zhang D., Ma H. (2009) Gene 447, 29–39 [DOI] [PubMed] [Google Scholar]
- 7.Iyer L. M., Koonin E. V., Aravind L. (2003) BMC Struct. Biol. 28, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salgado P. S., Koivunen M. R., Makeyev E. V., Bamford D. H., Stuart D. I., Grimes J. M. (2006) PLoS Biol. 4, e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makeyev E. V., Bamford D. H. (2002) Mol. Cell 10, 1417–1427 [DOI] [PubMed] [Google Scholar]
- 10.Curaba J., Chen X. (2008) J. Biol. Chem. 283, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiebel W., Haas B., Marinković S., Klanner A., Sänger H. L. (1993) J. Biol. Chem. 268, 11858–11867 [PubMed] [Google Scholar]
- 12.Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H. (2007) EMBO J. 26, 5007–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang G., Reinhart B. J., Bartel D. P., Zamore P. D. (2003) Genes Dev. 17, 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., Moazed D. (2004) Cell 119, 789–802 [DOI] [PubMed] [Google Scholar]
- 15.Lee S. R., Collins K. (2007) Nat. Struct. Mol. Biol. 14, 604–610 [DOI] [PubMed] [Google Scholar]
- 16.Lee S. R., Collins K. (2006) Genes Dev. 20, 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S. R., Talsky K. B., Collins K. (2009) RNA 15, 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couvillion M. T., Lee S. R., Hogstad B., Malone C. D., Tonkin L. A., Sachidanandam R., Hannon G. J., Collins K. (2009) Genes Dev. 23, 2016–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akbergenov R., Si-Ammour A., Blevins T., Amin I., Kutter C., Vanderschuren H., Zhang P., Gruissem W., Meins F., Jr., Hohn T., Pooggin M. M. (2006) Nucleic Acids Res. 34, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H. C., Chang S. S., Choudhary S., Aalto A. P., Maiti M., Bamford D. H., Liu Y. (2009) Nature 459, 274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiyama T., Cam H., Verdel A., Moazed D., Grewal S. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]