Abstract
Translation termination-coupled deadenylation is the first and often the rate-limiting step of eukaryotic mRNA decay in which two deadenylases, Ccr4-Caf1 and Pan2, play key roles. One of the deadenylases, Caf1, associates with Tob, which recruits Caf1 to the poly(A) tail through interactions with a cytoplasmic poly(A)-binding protein 1 (PABPC1). We previously proposed that the competition between Tob and eRF3 (a translation termination factor that interacts with PABPC1) is responsible for the regulation of deadenylase activity. However, the molecular mechanism of the regulation should be addressed by investigating the binding affinity and the cellular levels of these proteins. In this work, we characterized the human Tob interactions with Caf1 and a C-terminal domain of PABPC1 (PABC). Nuclear magnetic resonance (NMR) and Western blot analyses revealed that Tob consists of a structured N-terminal BTG-Tob domain and an unstructured C-terminal region with two conserved PAM2 (PABPC1-interacting motif 2) motifs. The BTG-TOB domain associates with Caf1, whereas the C-terminal PAM2 motif binds to PABC, with a Kd value of 20 μm. Furthermore, we demonstrated that the levels of eRF3 and Tob in HeLa cells are 4–5 μm and less than 0.2 μm, respectively. On the basis of these results, we propose a thermodynamic mechanism for the translation termination-coupled deadenylation mediated by the Tob-Caf1 complex.
Keywords: Enzyme Mechanisms, NMR, Protein Motifs, Protein-Protein Interactions, RNA-binding Protein, RNA Metabolism, Translation, ITC, PAM2, Poly(A)-binding Protein
Introduction
The degradation of mRNA (mRNA decay) is an important step in the down-regulation of gene expression. Most mRNA decay is initiated by deadenylation of the 3′-poly(A) tail (1–3) in which two major mRNA deadenylase complexes, Ccr4-Caf1 and Pan2-Pan3, play central roles in mammals (4).
Tob was identified as a protein that associates with Caf1, although it was originally found as a member of an antiproliferative protein family, which includes Tob2 and BTG1–4 (5–11). Among the proteins of this family, the 120 N-terminal residues are highly conserved and form a motif known as the BTG-Tob domain, which binds to Caf1 (12). The C-terminal region is not conserved, and thus, Tob (also known as Tob1) and Tob2 are classified as members of a subfamily distinct from the other BTG-Tob family members.
The C-terminal region of Tob contains the two PAM2 2 motifs PAM2-N and PAM2-C at residues 130–141 and 265–276, respectively (13, 14), and are known to bind to the PABC domain of cytosolic poly(A)-binding protein (PABPC1). PAM2-N was previously identified as the PABP-binding site (15). We recently reported that the mutation of Phe-274 of PAM2-C to alanine completely abolishes the binding affinity of Tob for PABPC1. This suggests that the interaction of PAM2-C and PABC is crucial for Tob-PABPC1 interaction (16). The translation termination-coupled mRNA decay rate was significantly slowed by mutations in the PAM2 region of eRF3, the PAM2 region of Tob, or the PAM2-binding site of PABC. Thus, we proposed that the coupling occurs with competition between eRF3 and Tob for PABC, which recruits Ccr4-Caf1 to the 3′-poly(A) tail (16). However, the mechanism of the competition remains elusive and should be quantitatively clarified on the basis of the binding affinities and intracellular levels of these proteins.
Here, we investigated the physicochemical properties of the interaction of Tob with Caf1 and PABPC1 using isothermal titration calorimetry (ITC) and nuclear magnetic resonance (NMR). The analyses revealed that Tob consists of a folded N-terminal BTG-Tob domain and an unstructured C-terminal region that contains two PAM2 motifs. The affinities of Tob for Caf1 and PABC were assessed using several Tob fragments. The results indicated that the C-terminal PAM2 sequence is the primary PABC-binding site and that Tob binds independently to Caf1 and PABC. The Caf1-Tob-PABPC1 ternary complex can thus be formed. Furthermore, we investigated the intracellular levels of eRF3, PABPC1, and Tob, all of which are involved in translation termination-coupled mRNA decay. The affinities and cellular levels of these proteins revealed the mechanism by which the deadenylase activity of Ccr4-Caf1 is regulated by eRF3 in a translation termination-coupled manner.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNAs encoding full-length human Caf1, Tob (full-length, residues 1–285, 1–148, 147–285, 121–148, and 260–285), and the PABC domain of PABPC1 (residues 541–623) were amplified by PCR using pCMVfrag-hCAF1, hTob, and PABPC1 constructs (16) as templates and cloned into the expression vector pGEX-6p-1 for Caf1 and Tob and pET42 for PABC. The constructs were expressed in Escherichia coli BL21(DE3) codon plus-RP, cultured in LB media, or in M9 medium containing [13C]glucose and/or 15[N]NH4Cl for uniformly 13C/15N- or 15N-labeled samples for NMR in minimal media. Recombinant GST-fused proteins were purified using a glutathione-SepharoseTM 4B affinity column. The GST tag was removed by PreScission protease (Amersham Biosciences). The TobN285,3 TobN148, and Tob(147–285) proteins were further purified by gel filtration chromatography, and the PABC, Tob(121–148), and Tob(260–285) proteins were purified by reversed-phase chromatography.
NMR Spectroscopy
For the backbone assignments of TobN148, uniformly 13C/15N-lableled TobN148 were prepared at a concentration of 0.3 mm in a buffer containing 10 mm phosphate (pH 6.0), 400 mm NaCl, 1 mm dithiothreitol, 0.5 m choline-O-sulfate, and 8% (v/v) D2O. A set of the triple resonance experiments, HNCO, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH, in conjunction with 15N-edited NOESY heteronuclear single quantum coherence (HSQC) with a mixing time of 120 ms, was performed at 35 °C on a Bruker Avance 500 spectrometer equipped with a cryogenic probe, and the spectra were processed by using Sparky software (26).
The backbone assignments of Tob(260–285) were established by analyzing homonuclear NOESY and total correlation spectroscopy, 15N-edited NOESY-HSQC, and 15N-edited total correlation spectroscopy-HSQC, which were transferred to the signals in the 1H-15N HSQC spectrum of [15N]Tob(147–285). The PABC titrations (0–1.6 mm) were carried out for [15N]Tob(260–285) and [15N]Tob(147–285) with Tob fragment concentrations of 0.4 mm at 30 °C.
The previously reported backbone assignments of PABC were confirmed by HNCACB and CBCA(CO)NH. The titrations of Tob(147–285) at 0–0.2 mm were carried out for [15N]PABC at 0.1–0.2 mm, 30 °C.
ITC
The binding of TobN285 and TobN148 to Caf1 was measured by ITC (17) using a MicroCal VP-ITC microcalorimeter (MicroCal Inc., Northampton, MA). The protein samples were dialyzed against a buffer containing 10 mm NaH2PO4, 500 mm NaCl, and 1 mm dithiothreitol (pH 6.0). A total of 290 μl of 79 μm TobN285 and 161 μm TobN148 was injected into the 7.6 and 12 μm Caf1 solutions, respectively, at 25 °C.
To analyze the interactions between Tob fragments and PABC, aliquots of 408–550 μm PABC solutions were injected into solutions of 29–39 μm Tob fragments using the ITC buffer containing 10 mm NaH2PO4 and 50 mm NaCl (pH 7.0). In addition, the ITC experiments were carried out in salt-free buffer for the binding of TobN148, Tob(121–147), Tob(147–285), and Tob(260–285) to PABC.
Heats of dilution were determined by titrating the injectant solution into the dialysis buffer, after which the heats were subtracted from the raw titration data before data analysis. The titration data were analyzed using the MicroCal Origin software version 5.0, provided by the manufacturer. The thermodynamic parameters with the error values calculated from the fittings are listed in Tables 1 and 2.
TABLE 1.
Thermodynamic parameters for the interactions of Tob fragments with Caf1
| Tob | Stoichiometry | Kd | ΔH | ΔS |
|---|---|---|---|---|
| μm | kcal/mol | cal/mol K | ||
| TobN285 | 1.23 ± 0.01 | 0.648 ± 0.009 | −4.73 ± 0.06 | 12.5 |
| TobN148 | 0.901 ± 0.005 | 0.59 ± 0.03 | −5.18 ± 0.04 | 11.1 |
TABLE 2.
Thermodynamic parameters for the interactions of Tob fragments with PABC
| Tob | NaCl | Stoichiometry (PABC/Tob) | Kd | ΔH | ΔS |
|---|---|---|---|---|---|
| mm | μm | kcal/mol | cal/mol K | ||
| Tob(147–285) | 50 | 0.813 ± 0.009 | 20.8 ± 0.4 | −21.6 ± 0.3 | −50.9 |
| Tob(147–285) | 0 | 0.80 ± 0.04 | 8.03 ± 0.01 | −11.7 ± 0.7 | −17.2 |
| Tob(260–285)a | 0 | 0.938 ± 0.006 | 9.3 ± 0.3 | −35.3 ± 0.3 | −95.3 |
| TobN148 | 0 | Not Determined | |||
| Tob(120–147)a | 0 | 1.16 ± 0.04 | 57 ± 4 | −1.20 ± 0.06 | 15 |
a Tob fragments were injected into the PABC solution.
Relative Abundance in HeLa Cells
HeLa cells were transiently transfected as specified in the legends for Figs. 1 and 6 using Lipofectamine 2000. Total cellular protein was isolated using buffer A (50 mm Tris-HCl (pH 6.8), 8% glycerol, 2% SDS, 2% 2-mercaptoethanol) and analyzed by Western blotting using anti-FLAG (Sigma), anti-PABPC1 (Abcam), anti-Tob (raised against His-tagged TobN285 protein), anti-Pan3 (raised against GST-Pan3(488–687) protein), or anti-eRF3 (18). The levels of cellular Tob, Pan3, and eRF3 were normalized to FLAG-tagged Tob, Pan3, and eRF3, respectively. The levels of FLAG-tagged Tob, Pan3, and eRF3 were normalized to FLAG-tagged PABPC1. The numbers specified in Fig. 6C represent the levels of cellular Tob, Pan3, and eRF3, where the level of cellular PAPBC1 is defined as 100%.
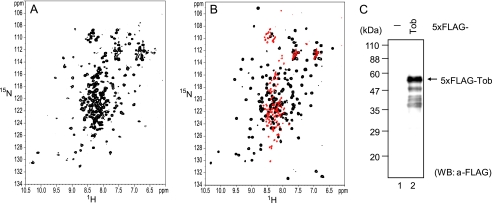
FIGURE 1.
Tob consists of the folded N-terminal domain and the unstructured C-terminal region. A, 1H-15N HSQC spectrum of [15N]TobN285. B, an overlay of the 1H-15N HSQC spectra of [15N]TobN148 (black) and [15N]Tob(147–285) (red). C, degradation of Tob in HeLa cells. HeLa cells were transiently transfected with pCMV-5×FLAG (−) or pCMV-5×FLAG-Tob (Tob). Total cell protein was isolated and analyzed by Western blotting (WB) using anti-FLAG. A relatively darker exposure was required to detect the degradation of 5×FLAG Tob. Note that intact Tob migrated as a 58-kDa protein in the SDS-PAGE gel, whereas its theoretical molecular mass is 44 kDa.
FIGURE 6.
Expression levels of cellular PABPC1, Tob, Pan3, and eRF3 in HeLa cells. A, HeLa cells were transiently transfected with pFLAG-CMV5 (−), pFLAG-PABPC1 (PABPC1), or pFLAG-eRF3 (eRF3). Total cell proteins were analyzed by Western blotting using anti-PABPC or anti-eRF3. The three left-most lanes, which represent the analysis of 4-fold dilutions of total cell proteins, indicate that the analysis is semiquantitative. B, as in A, except that cells were transiently transfected with pCMV-5 × FLAG (−), pCMV-5 × FLAG-PABPC1 (PABPC1), pCMV-5 × FLAG-Pan3 (Pan3), or pCMV-5 × FLAG-Tob (Tob). Total cell proteins were analyzed by Western blotting using anti-FLAG, or anti-PABPC1, anti-Pan3, or anti-Tob. The three or five left-most lanes represent the analysis of 4-fold dilutions of total cell proteins. C, quantification of cellular PABP, Tob, Pan3, and eRF3 in HeLa cells. The levels of Tob, Pan3, and eRF3 were normalized to the level of PABPC1 and were subsequently expressed as a percentage of the level of PABP.
RESULTS
Tob Consists of the Folded BTG-Tob Domain and the Unstructured C-terminal Region
Structural data for Tob are currently limited to the BTG-Tob domain, which consists of the N-terminal 120 residues. However, the rest of the protein, which is less conserved among the BTG family proteins, provides distinguishing characteristics among the family members. We attempted to investigate the structure of full-length Tob (345 residues) and its interactions with Caf1 and PABC. Unfortunately, only a trace amount of the full-length Tob was expressed in E. coli, and therefore, we used a C-terminally truncated mutant (residues 1–285, hereafter referred to as TobN285), which was previously shown to form a ternary complex with Caf1 and PABPC1 (16).
Fig. 1A shows the 1H-15N HSQC spectrum of uniformly 15N-labled TobN285. A number of signals are well dispersed in the 1H dimension, and strong signals are located at 7.8–8.5 ppm. The former well dispersed signals are characteristic of folded proteins, whereas the latter strong and narrowly dispersed signals presumably arise from unstructured regions. We obtained two spectra of the uniformly 15N-labeled TobN148 and the C-terminal fragment of Tob (residues 147–285, hereafter referred to as Tob(147–285)). The overlay of these spectra (Fig. 1B) is essentially identical to the spectrum of TobN285 (Fig. 1A), where the well dispersed signals and narrowly dispersed signals correspond to those of TobN148 and Tob(147–285), respectively.
The backbone assignments of 93 residues of TobN148 out of the 130 detectable signals were established by a series of triple resonance experiments, using uniformly 13C/15N-labeled TobN148 (supplemental Fig. 1 and supplemental Table 1). A chemical shift index analysis (19) provided an indication that the secondary structure of the BTG-Tob domain is consistent with the previously reported crystal structure (20, Protein Data Bank (PDB) code: 2d5r) and that the segment defined by residues 120–148 is unstructured. Thus, TobN285 consists of the N-terminal folded BTG-Tob domain followed by the C-terminal unstructured region.
To investigate whether the C-terminal region of intact Tob is also unstructured, we carried out Western blotting using the N-terminally FLAG-tagged full-length protein that was expressed in HeLa cells (Fig. 1C). In addition to the main band at 58 kDa corresponding to the full-length Tob, there is smearing of bands ranging from 37 to 58 kDa, indicating that the C-terminal region of the intact protein is degraded by proteases from the cells. The high sensitivity to protease degradation suggests that most of the C-terminal region of the intact Tob protein is unstructured. We concluded that the C-terminal region of intact Tob is also unstructured.
Binding Affinity of Tob for Caf1
The binding affinity of TobN285 for Caf1 was investigated by ITC. Fig. 2 shows the isotherms obtained upon titration of the Caf1 solution with TobN285. The best fit of the integrated isotherms, using a single-site model, indicates a dissociation constant (Kd) value of 0.65 μm. It should be noted that a NaCl concentration of 500 mm was required to maintain the stability of Caf1.
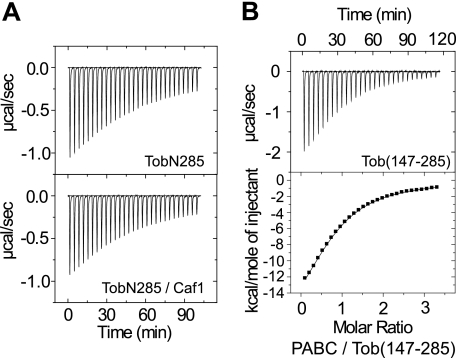
FIGURE 2.
ITC analyses of Tob-Caf1 interaction. Upper panel, trace of the calorimetric titrations of 29 × 10-μl aliquots of 79 μm TobN285 into the cell containing the 7.6 μm Caf1 solution; lower panel, the integrated binding isotherm obtained from the experiments. The buffer used in the ITC experiments comprised 10 mm NaH2PO4, 500 mm NaCl, and 1 mm dithiothreitol (pH 6.0).
In addition, we analyzed the Caf1 binding affinity of a further truncated mutant containing the N-terminal 148 residues (TobN148), which includes the BTG-Tob domain and the N-terminal PAM2 sequence. The interaction between TobN148 and Caf1 yielded comparable thermodynamic parameters, indicating that the Caf1-binding site is within the segment defined by residues 1–148 (Table 1).
Interaction of Tob and PABC
ITC analysis of the interaction between TobN285 and PABC indicated the occurrence of a significant amount of heat exchange. However, the heat exchange did not become saturated at the end of the titrations (Fig. 3A, upper), and the fitting of the integrated isotherm after correcting for the heats of dilution using a one-site binding model did not converge. Thus, we fixed the binding stoichiometry at 1:1, which generated an apparent Kd value of 55 μm. An identical isotherm was obtained in the presence of Caf1 (Fig. 3A, lower), indicating that the binding of Tob to PABC is not affected by Caf1.
FIGURE 3.
ITC analyses of Tob-PABC interaction. A, trace of the calorimetric titrations of 29 × 10-μl aliquots of 408 μm PABC into the cell containing 39 μm TobN285 (upper panel) and the 1:1 mixture of TobN285 and Caf1 (lower panel, 29 μm each). B, upper panel, trace of the calorimetric titrations of 29 × 10-μl aliquots of 550 μm PABC into the cell containing 37 μm Tob(147–285); lower panel, the integrated binding isotherm obtained from the experiments. The buffer used in the ITC experiments is comprised of 10 mm NaH2PO4 and 50 mm NaCl (pH 7.0). The thermodynamic parameters are summarized in Table 2.
We next carried out an ITC analysis for PABC binding, using TobN148 and Tob(147–285). Fig. 3B shows the binding isotherm obtained upon titration of PABC into a solution of Tob(147–285). The integrated isotherm was fitted using the one-site model. The fitting produced a Kd value of 20 μm with a binding stoichiometry of 1. On the other hand, essentially constant heat exchange was observed for the injections of PABC into the TobN148 solution under the same experimental conditions (including the same sample concentrations). This indicates that the interaction between TobN148 and PABC is weak (data not shown). These results demonstrate that the primary PABC-binding site is located within residues 147–285 of Tob. It should be noted that the unsaturated titration curve for the TobN285-PABC interaction is presumably caused by the weak interaction of the N-terminal 148 residues of Tob with PABC.
PAM2-C Is the Primary PABC-binding Site in Tob
To identify the PABC-binding site in Tob, a chemical shift perturbation (CSP) analysis was carried out. The perturbation caused by the addition of PABC to [15N]Tob(147–285) was analyzed.
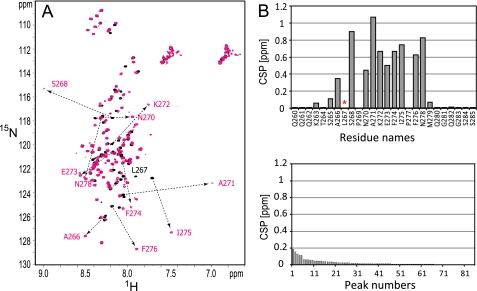
The HSQC spectra of [15N]Tob(147–285) revealed large spectral changes upon the addition of PABC (Fig. 4A). As expected, the spectral changes were identical to those of the 15N-labeled peptide corresponding to Tob residues 260–285 (hereafter referred to as Tob(260–285)), which includes PAM2-C (residues 265–276). This observation indicates that PAM2-C acts as the Tob-binding site within residues 147–285. This was confirmed by the ITC analyses of the binding of PABC to Tob(147–285) and Tob(260–285). These experiments produce essentially identical thermodynamic parameters (Table 2). It should be noted that another Tob construct comprising residues 121–148 (referred to as Tob(121–148)), containing PAM2-N (residues 130–141), exhibits a 6-fold lower affinity for PABC than Tob(260–285) and Tob(147–285).
FIGURE 4.
PABC-binding site of Tob. A, overlay of the 1H-15N HSQC spectra of free (black) and PABC-bound Tob(147–285) (magenta). Signals with CSPs larger than 0.20 ppm upon the addition of PABC are labeled and connected by dotted lines. B, plot of the amount of CSPs of Tob(147–285) upon the addition of the PABC domain. The CSPs were calculated as Δδ = {ΔδHN2 + (Δδ15N/6.5)2}½. The asterisk indicates that CSP was not obtained due to disappearing of the NMR signals upon the addition of PABC. The unassigned 84 peaks of Tob(147–285) are plotted in the bottom panel in order of decreasing amplitude in the CSPs.
Tob-binding Site of PABC
To investigate the Tob-binding site of PABC, a CSP analysis was carried out with perturbation produced by the addition of TobN285 to [15N]PABC (Fig. 5). The backbone assignments of free PABC (Biological Magnetic Resonance Data Bank entry 4915) were confirmed by a series of triple resonance experiments using uniformly 13C/15N-labeled PABC. Because the fast exchange regime was observed upon the addition of 0.5 and 1.0 equivalents of TobN285 to [15N]PABC (with the exception of three signals of Lys-580, Met-584, and Val-613 that disappeared), most of the resonance assignments were transferred to the Tob-bound state. Fig. 5B shows a plot of CSPs, which indicates that the residues exhibiting CSPs larger than 0.08 ppm are Met-561, Glu-564, His-574, Thr-576, Thr-582, L585, Ile-588, L593, Lys-606, Ala-610, Ala-612, Gln-615, His-617, and Ala-619. The three signals that disappeared presumably underwent line broadening in the intermediate exchange regime. This suggests that the CSPs for Lys-580, Met-584, and Val-613 are larger than those of the other affected residues listed above. These 17 residues include 11 residues out of 12 that exhibited large CSPs upon the addition of Paip2 (21). This indicates that Tob binds to PABPC1 through the PAM2-PABC interaction, which is shared by other PAM2-containing proteins.
FIGURE 5.
CSP analysis of TobN285 binding to [15N]PABC. A, NMR spectral change of [15N]PABC upon the addition of TobN285. 1H-15N HSQC spectra of [15N]PABC are shown in the absence (black) and presence (magenta) of TobN285. The signals with CSPs larger than 0.08 ppm are labeled. B, CSPs caused by the addition of TobN285 are plotted. The asterisks indicate residues that were previously reported to exhibit CSPs larger than 0.4 ppm upon the addition of Paip2 (21).
Cellular Levels of Tob, PABPC1, and eRF3 in HeLa Cells
Previously, we proposed that Tob and Pan3 recruit deadenylases Caf1 and Pan2, respectively, to the 3′-poly(A) tail through the interaction of PAM2 in Tob-Pan3 with the PABC domain of PABPC1. The activity of the deadenylases is regulated in a translation termination-coupled manner through competition of the PAM2 motifs in eRF3 with Tob-Pan3 for PABC (16). To quantitatively address the relationship of these interactions, we performed Western blot analyses of HeLa cells to investigate expression levels of Tob, Pan3, and eRF3, relative to PABPC1, which has been reported to have a cellular level of 4 μm (22).
Fig. 6 shows the transient and cellular expression levels of the proteins as detected by antibodies for the FLAG tag and each protein, respectively. The cellular levels of Tob, Pan3, and eRF3 were quantified and normalized to the level of PABPC1 (Fig. 6C). Although the levels of Tob and Pan3 were evaluated as the upper limit, due to the detection limit of the analysis, the cellular level of Tob was less than 6.6% of that of PABPC1 and the cellular level of Pan3 was less than 2.3% of that of PABPC1. On the basis of the cellular level of PABPC1, the levels of Tob and Pan3 are estimated to be less than 260 and 92 nm, respectively. On the other hand, eRF3 is expressed at a level similar to that of PABPC1 (4–5 μm).
DISCUSSION
Structure of Tob and Binding of Tob to Caf1 and PABPC1
The NMR and Western blot analyses of Tob revealed that Tob consists of a well folded BTG-Tob N-terminal domain comprising 120 residues and the unstructured C-terminal region that contains two PAM2 motifs (Fig. 1). This observation is consistent with the report that PAM2 motifs are generally found in unstructured regions (14).
The ITC analyses indicated that the BTG-Tob domain of Tob is crucial for binding of Caf1. The C-terminal region of Tob is not involved in the binding of Caf1 (Fig. 2). Under high salt conditions (500 mm NaCl), the Kd value for the Tob-Caf1 interaction was found to be 0.6 μm (Table 1). The affinity might be much greater at a physiological salt concentration because the high concentration of NaCl appears to impair the electrostatic interactions between Tob and Caf1. The crystal structure of the TobN138-Caf1 complex (PDB code: 2d5r) contains at least three intermolecular ion pairs (Lys-51–Glu-247, Asp-65–Lys-254, and Glu-98–Lys-203) and a hydrogen bond (Glu-105–Tyr-255) at the relatively hydrophobic binding interface (20).
On the other hand, the NMR and ITC analyses revealed that the PAM2-C sequence of Tob is the primary PABC-binding site, which contributes to the binding affinity with a Kd value of 20 μm (Table 1). The PAM2-N sequence (residues, 130–141; sequence, NSFNPEAQVFMP) has binding affinity for PABC, which is 6-fold weaker than that of PAM2-C (residues, 265–276; sequence, SALSPNAKEFIF) under the salt-free conditions (Table 2). Thus, the Kd value appears to exceed 100 μm under the high salt conditions. Although the PAM2-N sequence of Tob contains conserved hydrophobic residues at positions 3, 5, 7, 10, and 12, its lower affinity for PABC is presumably a consequence of the absence of the glutamate residue at position 9, which forms hydrogen bonds with PABC via an ordered water molecule in the crystal structure of the PAM2 sequences of Paip2 complexed with PABC (23, PDB code: 1i2t). The Tob-binding site of PABC is a typical PAM2-binding site. Therefore, the interaction can be competitively suppressed by other PAM2-containing proteins, such as eRF3, which has a 20-fold higher affinity for PABC than Tob and a cellular level more than 15-fold higher than that of Tob (see below).
The affinity of Tob for PABC was not affected by Caf1 binding to Tob. This is consistent with the observation that the BTG-Tob domain (which binds to Caf1) and the PAM2-C sequence are separated by a 150-residue unstructured region. Thus, the binding of PABC to Tob and the binding of Caf1 to Tob are independent of each other. This indicates that the Caf1-Tob-PABPC1 ternary complex could be formed when the protein concentrations are sufficiently high with respect to their binding affinities.
Dual Interactions of the Tob-Caf1 Complex Enable Regulation of Ccr4-Caf1 Activity by eRF3 in a Translation Termination-coupled Manner
It was reported that purified mouse Caf1 has only 1 substituted residue with respect to human Caf1 (Asn-282 of human Caf1 is replaced by Ser), and this protein has a large Km value of 24 μm for a substrate, 35-residue RNA containing 10 adenine residues at the 3′-end (24). Therefore, the recruitment of Caf1 by Tob to the 3′-end of poly(A) through the Tob-PABPC1 interaction would be required to enhance the substrate accessibility of Caf1 and thus increase the deadenylase activity.
However, the PABC domain of PABPC1, which includes the Tob-binding site, also binds to the translation termination factor eRF3. Although the Kd value for binding of PABC to Tob was found to be 20 μm, eRF3 was reported to possess 20-fold higher affinity for PABC (Kd = 1 μm) (25). Furthermore, we demonstrated that eRF3 exists in HeLa cells at a level comparable with that of the PABPC1, 4 μm (22), whereas the levels of Tob and Pan3 were estimated to be less than 260 and 92 nm, respectively (Fig. 6). Based on the Kd values and the cellular levels of these proteins, the concentrations of eRF3 and Tob that bind to PABPC1 are calculated to be 2.4 μm and less than 52 nm, respectively. This suggests that eRF3 dominates Tob in the interactions with PABPC1.
We propose here that the enhanced affinity of Caf1 for the substrate would be acquired by dual interactions, which occur upon association of Caf1 with Tob: (i) the recognition of the poly(A)-bound PABPC1 by the PAM2 sequence of Tob and (ii) the poly(A) recognition by Caf1 (Fig. 7A). Because the binding energy would be expected to represent the sum of the binding energy of each interaction, the apparent affinity of the Tob-Caf1 complex for the substrate could be theoretically increased to the product of the Kd (2.0 × 10−5 m) and Km (2.4 × 10−5 m) values, i.e. 0.5 nm, although the affinity is likely to be affected to some extent by other factors, such as the distance and flexibility of the linker regions between the BTG-Tob domain and the PAM2-C sequence. Considering the low cellular level of the Tob-Caf1 complex relative to the reported Km value, the deadenylase activity should be achieved by the dual interactions, which make the deadenylase complex accessible to the 3′-end of the PABPC1-bound poly(A).
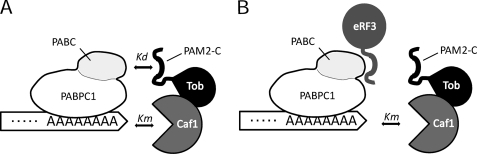
FIGURE 7.
Schematic drawing of the interactions between the Tob-Caf1 complex and the PABPC1-bound poly(A). A, dual interactions between PAM2-C of Tob and PABC and between Caf1 and the 3′-end of poly(A). The apparent Kd for the interaction of the Tob-Caf1 complex and the PABPC1-bound poly(A) can be calculated theoretically to be the product of Kd and Km. B, when eRF3 dominates the interaction with PABC, the apparent Kd of the Tob-Caf1 complex is decreased to the level of Km.
The dual interactions of the Tob-Caf1 complex adequately explain how the deadenylation occurs in a translation termination-coupled manner. The comparable Kd and Km values imply that each of the dual interactions contributes equally to the affinity of the Tob-Caf1 complex for the 3′-end of poly(A). As discussed above, eRF3 dominates Tob in the interactions with PABPC1, and thus, the affinity of the Tob-Caf1 complex for the substrate would be as low as the Km level (Fig. 7B). This precludes access of the deadenylase complex to the 3′-end of poly(A) at the cellular level. The deadenylase activity of Caf1 could be considerably enhanced only when the PABC domain of the poly(A)-bound PABPC1 and the exposed poly(A) tail are simultaneously available for the interactions. This scenario is likely to occur upon recruitment of eRF3 to the ribosome for translation termination. Taken together, the combination of the two weak interactions of the Tob-Caf1 complex may not only enable access of the complex to the 3′-end of the PABPC1-bound poly(A) (even at the low cellular level of the complex) but also enable eRF3 to regulate the deadenylase activity of the Tob-Caf1 complex. The affinities and the cellular levels of the proteins involved in deadenylation provide the thermodynamic basis for the translation termination-coupled mRNA decay.
Supplementary Material
Acknowledgment
We thank Prof. Kaori Wakamatsu for providing choline-O-sulfate.
This work was supported in part by grants from the Japan New Energy and Industrial Technology Development Organization (NEDO) and the Ministry of Economy, Trade, and Industry (METI) (to I. S.), a grant-in-aid for scientific research on priority areas from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to M. O. and I. S.), and a grant from the Takeda Science Foundation (to M. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
Throughout this report, the format TobNxxx indicates a Tob fragment containing residues 1–xxx. The format Tob(xxx–yyy) indicates a Tob fragment containing residues xxx–yyy.
- PAM2
- PABPC1-interacting motif 2
- PAM2-N
- N-terminal PAM2
- PAM2-C
- C-terminal PAM2
- PABPC1
- cytoplasmic poly(A)-binding protein 1
- PABC
- C-terminal domain of PABPC1
- ITC
- isothermal titration calorimetry
- HSQC
- heteronuclear single quantum coherence
- CSP
- chemical shift perturbation.
REFERENCES
- 1.Shyu A. B., Belasco J. G., Greenberg M. E. (1991) Genes Dev. 5, 221–231 [DOI] [PubMed] [Google Scholar]
- 2.Muhlrad D., Parker R. (1992) Genes Dev. 6, 2100–2111 [DOI] [PubMed] [Google Scholar]
- 3.Decker C. J., Parker R. (1993) Genes Dev. 7, 1632–1643 [DOI] [PubMed] [Google Scholar]
- 4.Yamashita A., Chang T. C., Yamashita Y., Zhu W., Zhong Z., Chen C. Y., Shyu A. B. (2005) Nat. Struct. Mol. Biol. 12, 1054–1063 [DOI] [PubMed] [Google Scholar]
- 5.Rouault J. P., Rimokh R., Tessa C., Paranhos G., Ffrench M., Duret L., Garoccio M., Germain D., Samarut J., Magaud J. P. (1992) EMBO J. 11, 1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda S., Kawamura-Tsuzuku J., Ohsugi M., Yoshida M., Emi M., Nakamura Y., Onda M., Yoshida Y., Nishiyama A., Yamamoto T. (1996) Oncogene 12, 705–713 [PubMed] [Google Scholar]
- 7.Montagnoli A., Guardavaccaro D., Starace G., Tirone F. (1996) Cell Growth Differ. 7, 1327–1336 [PubMed] [Google Scholar]
- 8.Yoshida Y., Matsuda S., Ikematsu N., Kawamura-Tsuzuku J., Inazawa J., Umemori H., Yamamoto T. (1998) Oncogene 16, 2687–2693 [DOI] [PubMed] [Google Scholar]
- 9.Ikematsu N., Yoshida Y., Kawamura-Tsuzuku J., Ohsugi M., Onda M., Hirai M., Fujimoto J., Yamamoto T. (1999) Oncogene 18, 7432–7441 [DOI] [PubMed] [Google Scholar]
- 10.Tzachanis D., Freeman G. J., Hirano N., van Puijenbroek A. A., Delfs M. W., Berezovskaya A., Nadler L. M., Boussiotis V. A. (2001) Nat. Immunol. 2, 1174–1182 [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T., K-Tsuzuku J., Ajima R., Nakamura T., Yoshida Y., Yamamoto T. (2002) Genes Dev. 16, 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mauxion F., Chen C. Y., Séraphin B., Shyu A. B. (2009) Trends Biochem. Sci. 34, 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda S., Rouault J., Magaud J., Berthet C. (2001) FEBS Lett. 497, 67–72 [DOI] [PubMed] [Google Scholar]
- 14.Albrecht M., Lengauer T. (2004) Biochem. Biophys. Res. Commun. 316, 129–138 [DOI] [PubMed] [Google Scholar]
- 15.Okochi K., Suzuki T., Inoue J., Matsuda S., Yamamoto T. (2005) Genes Cells 10, 151–163 [DOI] [PubMed] [Google Scholar]
- 16.Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., Shimada I., Tsujimoto M., Suzuki T., Katada T., Hoshino S. (2007) Genes Dev. 21, 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 18.Hoshino S., Imai M., Mizutani M., Kikuchi Y., Hanaoka F., Ui M., Katada T. (1998) J. Biol. Chem. 273, 22254–22259 [DOI] [PubMed] [Google Scholar]
- 19.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 20.Horiuchi M., Takeuchi K., Noda N., Muroya N., Suzuki T., Nakamura T., Kawamura-Tsuzuku J., Takahasi K., Yamamoto T., Inagaki F. (2009) J. Biol. Chem. 284, 13244–13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlov G., Trempe J. F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4409–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Görlach M., Burd C. G., Dreyfuss G. (1994) Exp. Cell Res. 211, 400–407 [DOI] [PubMed] [Google Scholar]
- 23.Kozlov G., Ménade M., Rosenauer A., Nguyen L., Gehring K. (2010) J. Mol. Biol. 397, 397–407 [DOI] [PubMed] [Google Scholar]
- 24.Viswanathan P., Ohn T., Chiang Y. C., Chen J., Denis C. L. (2004) J. Biol. Chem. 279, 23988–23995 [DOI] [PubMed] [Google Scholar]
- 25.Kozlov G., De Crescenzo G., Lim N. S., Siddiqui N., Fantus D., Kahvejian A., Trempe J. F., Elias D., Ekiel I., Sonenberg N., O'Connor-McCourt M., Gehring K. (2004) EMBO J. 23, 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goddard T. D., Kneller D. G. (2004) SPARKY 3, University of California, San Francisco [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.