Abstract
Localization of protein kinase A (PKA) via A-kinase-anchoring proteins (AKAPs) is important for cAMP responsiveness in many cellular systems, and evidence suggests that AKAPs play an important role in cardiac signaling. To test the importance of AKAP-mediated targeting of PKA on cardiac function, we designed a cell-permeable peptide, which we termed trans-activator of transcription (TAT)-AKAD for TAT-conjugated A-kinase-anchoring disruptor, using the PKA binding region of AKAP10 and tested the effects of this peptide in isolated cardiac myocytes and in Langendorff-perfused mouse hearts. We initially validated TAT-AKAD as a PKA localization inhibitor in cardiac myocytes by the use of confocal microscopy and cellular fractionation to show that treatment with the peptide disrupts type I and type II PKA regulatory subunits. Knockdown of PKA activity was demonstrated by decrease in phosphorylation of phospholamban and troponin I after β-adrenergic stimulation in isolated myocytes. Treatment with TAT-AKAD reduced myocyte shortening and rates of contraction and relaxation. Injection of TAT-AKAD (1 μm), but not scrambled control peptide, into the coronary circulation of isolated perfused hearts rapidly (<1 min) and reversibly decreased heart rate and peak left ventricular developed pressure. TAT-AKAD also had a pronounced effect on developed pressure (−dP/dt), consistent with a delayed relaxation of the heart. The effects of TAT-AKAD on heart rate and contractility persisted in hearts pretreated with isoproterenol. Disruption of PKA localization with TAT-AKAD thus had negative effects on chronotropy, inotropy, and lusitropy, thereby indicating a key role for AKAP-targeted PKA in control of heart rate and contractile function.
Keywords: Cardiac Muscle, G Protein-coupled Receptors (GPCR), Peptide Chemical Synthesis, Peptides, Protein Kinase A (PKA), A-Kinase Anchoring Protein
Introduction
Activation of protein kinase A (PKA)3 by second messenger cAMP enhances the phosphorylation of many downstream protein targets in the heart. A key concept that has emerged in recent years is the importance of localization of PKA through A-kinase-anchoring proteins (AKAPs) in maintaining cAMP-regulated activity in many cellular systems, including those found in cardiac cells (1–4). However, definitive evidence for a contribution of PKA localization in cardiac tissue or in the intact heart has been lacking. Most experiments that evaluate the role of PKA in cardiac signaling at the organ or whole animal level have used cyclic nucleotide analogs (e.g. (Rp)-cAMP) or inhibitors of the kinase (i.e. H89, PKA inhibitor), which may target kinase that is either localized or nonlocalized with substrates, thus leaving the following question unanswered; “To what extent does AKAP-mediated targeting contribute to cAMP/PKA-promoted effects?”
PKA is a tetramer that contains two regulatory (R) subunits and two catalytic (C) subunits. The R subunits not only bind to the active site cleft of the C subunit but also provide the binding surface for interaction with AKAPs. At least 13 different AKAPs have been identified in cardiac tissue, some of which have been characterized with respect to their mode of action and protein assemblies (4). Functionally, AKAP-mediated targeting of PKA has been implicated in calcium mobilization (AKAP18/15 and muscle-specific AKAP) (5–8), contractility (9), potassium channel function (yotiao) (10), β-adrenergic resensitization (AKAP79) (11, 12), inhibition of adenylyl cyclase V/VI (AKAP79) (13), and Ca2+ reuptake into the sarcoplasmic reticulum (AKAP18δ) (14). In addition, AKAP10 (d-AKAP2), a dual specificity AKAP that binds both type I (RI) and type II R (RII) subunits of PKA, has been implicated in regulating heart rate in mice and humans (15).
A well established approach to disrupt AKAP-mediated PKA anchoring is the use of competing peptides of the PKA binding region of AKAPs (16–18). Cellular uptake of peptides usually is accomplished by attaching a fatty acid moiety to the sequence or by using cell penetrating peptide sequences such as TAT or antennapedia (19, 20). Peptides are taken up by cells and compete with full-length AKAP for binding to the R subunit of PKA, thereby shifting the equilibrium toward unanchored (free) PKA, removing PKA from localized cAMP gradients and preventing its activation (Fig. 1). Disruption is not selective with respect to the AKAP, and the extent to which each AKAP-anchored PKA pool is affected by the disruptor depends on the dynamic equilibrium of the interaction. Tightly anchored PKA would be less likely to be competed away from the targeted site than would weakly anchored PKA. AKAPs anchor PKA through the R subunits; in general, the PKA-RI isoform binds AKAPs weaker than the RII isoforms (21). The RI isoform appears to be more dynamic, more sensitive to cyclic nucleotides and localized under conditions of stimulation (22–24). RII primarily localizes to substructures or organelles and usually is associated with particulate fractions (23). Therefore, PKA-RI mediated functions may be modulated more easily with exogenously added disrupting peptides.
FIGURE 1.
A model for AKAP-targeted PKA disruption. The disruptor peptide competes with full-length, localized AKAP for binding to PKA R subunit, resulting in displacement of inactive kinase (and its C subunit) from intracellular cAMP gradients, which prevents PKA activation but still allows hydrolysis of cAMP by cyclic nucleotide phosphodiesterase (PDE). Additional regulators involved may include protein phosphatases (PP).
The current studies tested the ability of a cell-permeable competing AKAP peptide (TAT-AKAD) to disrupt localization of PKA-RI and PKA-RII regulatory subunits in cardiac myocytes and to alter physiology in the isolated perfused heart. We validated the use of TAT-AKAD as an inhibitor of PKA action and demonstrated that AKAP-mediated targeting of PKA is important for maintaining heart rate and myocardial contractility in isolated perfused mouse hearts.
EXPERIMENTAL PROCEDURES
Animal Care
Animals were treated according to the Guide for the Care and Use of Laboratory Animals (25). Animal protocols were approved by the Department of Veterans Affairs, San Diego Healthcare System, Institutional Animal Care and Use Committee (San Diego, CA). C57BL/6 male mice (8–10-weeks-old and 24–26 g weight) were purchased from The Jackson Laboratory (Bar Harbor, ME) and kept on a 12-h light-dark cycle in a temperature controlled facility until the day of the experiment.
Synthesis of Cell-permeable Peptides
Peptides were synthesized by standard fluorenylmethoxycarbonyl solid-phase peptide synthesis followed by cleavage of the side chain protection groups using trifluoroacetic acid (TFA) (26, 27). AKAD (Ac-QEELAWKIAKMIVSDVMQQ C-CONH2) and a scrambled control (AKADscr) (Ac-QMADISALEVIKWKMVEQQC-CONH2) were synthesized with an N-terminal fluorescein isothiocynate (5-FITC, Invitrogen) containing a 6-aminohexanoic acid linker for cellular imaging studies or acetylated. The C terminus was amide-protected. A sequence from HIV-1 TAT protein TAT(47–57) (YGRKKRRQRRR-CONH2) was synthesized with a free amino terminus, and prior to side chain deprotection, a thiol-containing, heterobifunctional cross-linker: succinimidyl-3-(2-pyridyldithio)propionate (Invitrogen) was conjugated to the free N-terminal amine as described previously (28). Briefly, TAT:succinimidyl-3-(2-pyridyldithio)propionate:diisopropylethylamine (DIPEA) (molar ratios 1:3.2:1.5) were dissolved in anhydrous dichloromethane and reacted for 3 h at room temperature. The product was verified by mass spectrometry, and the conjugate was cleaved from the resin and purified by HPLC. To make disulfide-bonded, TAT-conjugated peptides (i.e. TAT-AKAD or TAT-AKADscr), succinimidyl-3-(2-pyridyldithio)propionate-TAT was added in equal amounts by weight to AKAD or AKADscr peptides containing a free thiol. The sample was dissolved in a small volume of dimethylformamide and diluted to 1 ml with 0.1 m triethylammonium acetate. After 1 h at room temperature, the product was verified and purified by HPLC. All peptides were HPLC-purified to >95% purity and lyophilized. Given the large number of positive charges in the peptide, the molecular mass was corrected for TFA salt by multiplying the number of positive charges in the peptide by the mass of TFA (114 dalton) and adding this to the peptide molecular mass. This corresponds to the corrected molecular mass and was used to calculate concentrations.
In Vitro Binding Assay
Direct binding of FITC-labeled AKAD and AKADscr to type I (RIα) and type II (RIIα) R subunits was determined by fluorescence polarization. Expression and purification of bovine RIα and mouse RIIα were described previously (29). RI and RII were diluted in buffer (10 mm Hepes, 150 mm NaCl, 3 mm EDTA, 10 mm DTT, pH 7.4) containing either 5 or 10 nm AKAD and AKADscr, respectively. The samples were incubated for 24 h at 25 °C and read on a Tecan Ultra plate reader with polarizing filters with an excitation 485 (20) and emission of 535 (25).
Preparation of Adult Rat Ventricular Myocytes
Cardiac myocytes (CM) were isolated from adult Sprague-Dawley rats (250–300 g, male) as described previously (30). Briefly, animals were heparinized (1,000 units i.p.) 5 min prior to being anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg), and their hearts were removed and placed in ice-cold cardioplegic (20 mm KCl) heart media solution (112 mmol/liter NaCl, 5.4 mmol/liter KCl, 1 mmol/liter MgCl2, 9 mmol/liter NaH2PO4. 11.1 mmol/liter d-glucose; supplemented with 10 mmol/liter Hepes, 30 mmol/liter taurine, 2 mmol/liter dl-carnitine, 2 mmol/liter creatine, pH. 7.4). The hearts were retrograde-perfused on a Langendorff apparatus with Ca2+-free heart medium for 5 min at 5 ml/min at 37 °C, followed by perfusion with a Ca2+-free heart medium containing collagenase II (250 units/mg; Worthington) for 20 min. Following perfusion, both ventricles were removed from the heart and minced in collagenase II containing heart medium for 10–15 min. The cell solution was washed several times to remove collagenase II and reacclimated to 1.2 mm Ca2+ for 25 min to produce calcium-tolerant CM. Myocytes were plated in heart media plus 4% fetal bovine serum on laminin (2 μg/cm2)-coated plates for 1 h. Plating media was changed to heart media supplemented with 1% bovine serum albumin (BSA) to remove all nonmyocytes, and CMs were placed in an incubator set at 37 °C and 5% CO2 and incubated for 1–24 h prior to experiments.
Immunofluorescence
FITC-labeled TAT-AKAD and TAT were dissolved in dimethyl sulfoxide and diluted in heart media plus 1% BSA (see above) at 37 °C. Final dimethyl sulfoxide concentrations did not exceed 0.85%. Peptides (300 μl) were incubated at 37 °C with freshly cultured myocytes adhered to laminin-coated coverslips in a 24-well plate for 16 h. Cells were washed with 500 μl of phosphate-buffered saline (PBS), fixed in 4% formalin, and prepared for immunofluorescence. Cells were washed with 100 mm glycine in PBS (pH to 7.4 with NaOH) and permeabilized in 0.1% Triton X-100/PBS for 10 min. Next, cells were washed twice in PBS/0.1% Tween 20 and blocked for 20 min in 1% BSA/PBS/0.05% Tween 20. Primary antibodies to RIα (monoclonal, BD Transduction Laboratories) (20 μg/ml final concentration) and RIIα (rabbit-polyclonal, Santa Cruz Biotechnology) (2 μg/ml final concentration) were mixed together in BSA block solution and incubated with cells for 3 h at room temperature. Cells were washed 3× at 5-min intervals with PBS/0.1% Tween. Donkey anti-mouse Cy5 (Jackson ImmunoResearch Laboratories) for RI and donkey anti-rabbit RRX (Jackson ImmunoResearch Laboratories) for RII were diluted 1:200 in BSA block solution plus 0.1% Tween and incubated for 1 h at room temperature. Cells were washed 6×, 5-min each in PBS/0.1% Tween, 2 × 5 min each with PBS, and 1× in distilled water. Coverslips were mounted onto slides with gelvatol and dried overnight. Fixed cells were imaged with a Bio-Rad Radiance 2000, laser scanning confocal microscope at the National Center for Microscopy and Imaging Research (University of California, San Diego). A 60× objective lens was used, and, for each cell image, three channels were scanned sequentially for Cy5 (long pass filter (excitation, 637; emission, 660)) RRX (excitation, 568; emission, 600/40) and FITC (excitation, 488; emission, 515/30) using LaserSharp software (Bio-Rad). Three data sets were acquired at 50 lines per second and averaged for each image. The data were imported into Adobe Photoshop, and each channel was saved as a JPEG file. JPEGs were imported into PowerPoint and aligned. Contrast and lighting was increased in Photoshop to the same extent for each image of the same color to allow for intensity comparisons.
Myocyte Fractionation Studies
TAT-AKAD and TAT-AKADscr peptides were diluted into heart media with 1% BSA to 5 μm and incubated with freshly isolated CM adherent to 10-cm, laminin-coated culture plates at 37 °C. Cells were incubated for 30 min, resuspended in ice-cold PBS (plus protease inhibitors), lysed by syringing through a 21-gauge needle, and fractionated using the FractionPREPTM cell fractionation kit (BioVision) into cytosolic, membrane, and nuclear fractions using the manufacturer's protocol. Western blots were run on the fractions. Experiments were performed four times using separate cell preparations. For quantitation, fractionated bands were normalized to the total amount of PKA-RI and PKA-RII in the cell lysate.
Western Blots of PKA Phosphosubstrates
TAT-AKAD (0.1–10 μm), TAT-AKADscr (0.1–10 μm) or H89 (200 μm) were diluted into heart media with 1% BSA and incubated with freshly isolated CM adherent to 6-well, 30-mm laminin-coated culture plates for 10 min for TAT-AKAD and TAT-AKADscr and 30 min for H89 at 37 °C. The medium was aspirated, and fresh, prewarmed medium or medium plus 10 nm of isoproterenol (Sigma) was added and incubated with cells for 2 min. Additional, studies were performed with phenylephrine (100 μm) as a negative control. Ice-cold lysis buffer (150 mm Na2CO3, 1 mm EDTA, pH 11), was added to each well, and the cells were resuspended and lysed by sonication. Lysed cells were stored at −80 °C until analyzed. Samples were probed with the following primary antibodies: troponin I (anti-rabbit poly IgG, Cell Signaling Technology), phosphotroponin I (anti-rabbit poly IgG, Cell Signaling Technology), phospholamban (anti-mouse monoclonal, Abcam), phospho-phospholamban (anti-rabbit poly IgG, Abcam) and phospho(Ser/Thr) PKA substrate (anti-rabbit IgG, Cell Signaling Technology). For quantitation, unstimulated and inhibitor-treated cells were normalized to the total protein.
Cell-shortening Studies
Healthy ventricular myocytes from adult rats were identified based on cells being quiescent, rod-shaped (∼10–12 microns by 100–120 microns), and having uniformly spaced cross striations. Unloaded cell shortening data were recorded from single ventricular myocytes using a video edge-detection system (Crescent Electronics, Sandy, UT). An aliquot of myocytes was placed in a small superfusion chamber in which the bottom was coated with a thin layer of laminin (Sigma) on the stage of a Nikon Diaphot microscope. Myocytes attached to the coverslip were continuously superfused with Krebs buffer (gassed with 5% CO2 and 95% O2, pH 7.4) at room temperature. Healthy, rod-shaped myocytes were electrically stimulated at 1 Hz with pulses of 5-ms duration at threshold voltage plus 10% (Grass SD9 Stimulator, Quincy, MA); shortening was detected at both ends. A custom designed electrode was used for field stimulation of each ventricular myocyte. The maximal rates of contraction and relaxation were calculated after the primary data had been “smoothed” with 3 point averaging to reduce high frequency noise (31).
Langendorff-perfused Hearts
Cardiac function was determined in isolated perfused hearts using an intraventricular balloon catheter to measure isovolumic left ventricular (LV) pressure as described previously (32). Briefly, aortas of isolated mouse hearts were cannulated for retrograde perfusion with warmed Krebs-Henseleit solution oxygenated with 95% O2 and 5%CO2. A fluid-filled balloon attached to a high fidelity pressure transducer (Millar Instruments, Houston, Texas) was placed through the mitral valve into the LV cavity. The balloon was inflated to 10 mmHg end diastolic pressure, and the hearts were allowed to equilibrate for 15 min, when baseline measurements were made. TAT-AKAD or TAT-AKADscr (0.01–10 μm) was infused into the retrograde perfusion line. The hearts were allowed to equilibrate to baseline levels of cardiac function between doses. Changes in heart rate and left ventricular pressure were recorded, and dP/dt was calculated using Windaq acquisition software (Dataq, Akron, Ohio). For isoproterenol experiments, hearts were allowed to stabilize and were then given a constant infusion of isoproterenol at 0.1 μm with the use of a syringe infusion pump. TAT-AKAD or TAT-AKADscr (1 μm) was then infused as above.
Statistical Analysis
Statistical analyses were performed by one-way analysis of variance followed by Bonferroni post hoc test or unpaired Student's t test. All data are expressed as mean ± S.E. Statistical significance was defined as p < 0.05.
RESULTS
Disruption of Endogenous Type I and Type II Regulatory Subunit Isoforms of PKA
A peptide containing 19 amino acids that corresponded to the PKA-regulatory subunit binding region of AKAP10 was synthesized with an N-terminal fluorescein and a C-terminal cysteine (see “Experimental Procedures”). The resulting peptide was tested for binding to the type I (RIα) and type II (RIIα) isoforms in vitro by fluorescence polarization. The peptide designated as AKAD bound similarly to RI and RII with Kd values of 3.5 nm ± 0.24 and 2.7 nm ± 0.08, respectively, demonstrating that this core peptide sequence does not discriminate between RI and RII binding (Fig. 2). Previously, we determined the dissociation constants for binding of a longer 27-residue sequence of AKAP10 to RI and RII to be 48 and 2.2 nm, respectively, indicating that in the context of a larger AKAP fragment, binding can be reduced to the RI isoform (33). Recent NMR studies revealed that a larger 40-amino acid fragment of AKAP10 bound to RI with a shifted helical register relative to binding of the same sequence to RII, such that the more N-terminal residues of the core AKAP10 sequence make contacts with the residues on the RI surface (34, 35). Truncating the more distal non-contacting N-terminal residues in the 19-residue peptide described above, may increase binding to RI by increasing accessibility of the core contacting AKAP10 residues with the binding pockets on RI. Irrespective of the mechanism, the smaller AKAP10 fragment designated as AKAD can serve as a dual-specific PKA disruptor to test in vivo. No significant binding was detected for AKADscr (tested up to 5 μm, (Fig. 2)).
FIGURE 2.
In vitro fluorescence polarization binding assay showing AKAD (filled symbols) and AKADscr (open symbols) binding to RI (●) and RII (■).
The peptides were disulfide-conjugated to HIV-TAT(47–57) to facilitate cellular uptake and conjugated to a FITC probe to monitor uptake using fluorescence microscopy. The advantage of this approach is that disulfide conjugation is labile in the reducing environment of the cell, resulting in release of the cargo (AKAD) from the carrier peptide (TAT) and thus preventing TAT from interfering with binding to the downstream target. Cellular uptake was evaluated in freshly isolated CM that were pretreated with FITC-labeled TAT-AKAD or FITC-TAT control. The cells were fixed and stained for endogenous RI and RII (Fig. 3). TAT-AKAD and TAT were visualized easily with the FITC label; ∼ 80–90% of cells were stained green, indicating efficient uptake. TAT-AKAD densely stained the transverse tubule network and the nucleus. Because the FITC label was on the N terminus of the peptide, the localization was presumably attributable to AKAD and not TAT (assuming disulfide bond reduction). The TAT carrier peptide was more diffuse with limited staining in the nucleus.
FIGURE 3.
Localization of R1 and RII in adult cardiac myocytes. Confocal fluorescence microscopy (60× objective) showing RI and RII localization in fixed cardiac myocytes (from a rat) and the effect of FITC-labeled TAT-AKAD (5 μm; green) and FITC-labeled TAT alone 5 μm (green) on endogenous RII (red) and RI (blue) localization. TAT-AKAD reduced the T-tubule network and perinuclear staining of RII (arrows) and reduced nuclear staining of RI (arrows) compared with TAT alone.
In cells exposed to carrier peptide (TAT) or no peptide, endogenous RII aligned with the transverse tubule network, the outer membrane and was concentrated in perinuclear locations (Fig. 3). RI was more diffuse with subtle cross-striated staining throughout the cell. Unlike endogenous RII, RI was more concentrated throughout the nucleus; in the space immediately adjacent to the nucleus, there was an absence of RI staining (Fig. 3). In the presence of TAT-AKAD, RII staining was reduced at the transverse tubule network, and there was less intense perinuclear staining, indicating some disruption of localized signal by the peptide. For RI, because the staining pattern was more subtle throughout the cytosol, the effect of TAT-AKAD was difficult to assess by confocal microscopy. However, there was an absence of nuclear staining, suggesting that the inhibitor peptide prevents nuclear localization of RI. There also was no defined area adjacent to the nucleus that was free of RI staining, suggesting that TAT-AKAD disrupts RI localization around the nucleus (Fig. 3).
Next, we used cellular fractionation to evaluate RI and RII disruption by the TAT peptides. Myocytes exposed to TAT-AKAD or TAT-AKADscr were lysed and fractionated into cytosolic, membrane, and nuclear fractions, and the lysates were probed by immunoblotting for RI and RII. In the absence of peptide (medium alone) or with TAT-AKADscr, the majority of RI was in the cytoplasmic fraction with a minor fraction in the membrane and a smaller amount in the nuclear fraction, consistent with the results obtained by assessing immunofluorescence (Fig. 4A). RII was present at similar levels in all fractions with more localized to the nuclear fraction compared with RI (Fig. 4B). TAT-AKAD reduced RI and RII staining in the membrane and nuclear fractions consistent with the idea that the peptide alters membrane localization (Fig. 4, C and D).
FIGURE 4.
Disruption of subcellular localization of RI and RII with TAT-AKAD. Cellular myocyte fractionation studies show that TAT-AKAD (5 μm), but not TAT-AKADscr (5 μm), can disrupt membrane anchoring of endogenous RI (A) and RII (B). 100 μg of protein was loaded per lane. Data are mean ± S.E.
Addition of TAT-AKAD to calcium-tolerant myocytes plated on laminin-coated coverslips produced an unexpected effect; within 1min of adding TAT-AKAD (1 μm) to cells, they began to contract with ripples of contraction along their length (data not shown). This response was not observed for TAT-AKADscr or TAT alone. At >5 μm TAT-AKAD, the cells rounded up, and the majority came off of the laminin-coated plates, making experiments difficult with adherent cells. The observed changes likely indicate that TAT-AKAD affects the contractile machinery (data not shown).
Inhibition of PKA-dependent Phosphorylation after β-Adrenergic Stimulation
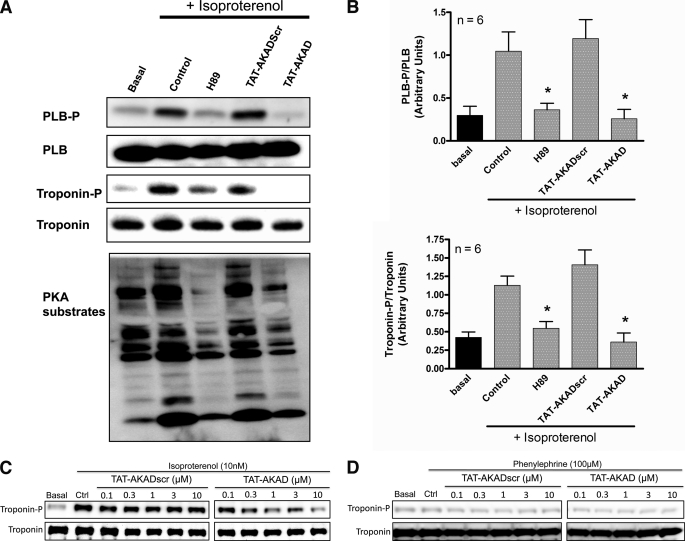
To test whether disruption of PKA resulted in decreased kinase activity, we stimulated myocytes with isoproterenol in the presence and absence of TAT-AKAD and probed for phosphorylation of known PKA substrates by Western blot. Treatment with TAT-AKAD significantly reduced phosphorylation of troponin I and phospholamban. H89 was used as a control. Using a general serine/threonine phospho-antibody, a reduction in PKA substrate phosphorylation was observed, indicating the peptide has a global effect on PKA activity (Fig. 5, A and B). A concentration-response effect was observed for TAT-AKAD in the presence of isoproterenol (Fig. 5C). Phenylephrine, an α-adrenergic agonist, did not alter phosphorylation of troponin and TAT-AKAD did not affect the phosphorylation of troponin in the presence of phenylephrine (Fig. 5D).
FIGURE 5.
Disruption of PKA activity with TAT-AKAD. A, immunoblots of phospho-phospholamban (PLB-P), phosphotroponin, and a general PKA substrate antibody showing that H89 (200 μm) and TAT-AKAD (5 μm) decrease PKA substrate phosphorylation. B, quantitation by densitometry of different cell preparations *, p < 0.05 compared with control (Ctrl) with isoproterenol. C, concentration response for TAT-AKADscr and TAT-AKAD in the presence of isoproterenol. D, concentration response for TAT-AKADscr and TAT-AKAD in the presence of phenylephrine. PLB, phospholamban. Data are mean ± S.E.
Decreased Contraction in Isolated Cardiac Myocytes
In field-stimulated isolated CM, incubation with TAT-AKAD reduced shortening amplitude relative to baseline (Fig. 6, A and B). In addition, maximal rates of relaxation and contraction were also reduced (Fig. 6, C and D).
FIGURE 6.
Effect of TAT-AKAD on cardiac myocyte shortening, contraction, and relaxation in vitro. A, representative cell shortening traces are superimposed to compare baseline results, with data obtained after application of the peptide. Contractions of single myocytes were elicited by field stimulation at 1 Hz at room temperature. B, cell shortening amplitude was first measured under baseline conditions and then after TAT-AKAD peptide (1 μm, n = 5) for 10 min. Myocyte-shortening amplitude was significantly reduced during TAT-AKAD peptide treatment relative to baseline (*, p < 0.01). TAT-AKAD peptide effects on rates of relaxation (C) and contraction (D) also are shown. Maximal rates of relaxation and contraction in field-stimulated myocytes were measured at baseline and following TAT-AKAD treatment (1 μm, n = 5). TAT-AKAD peptide treatment significantly reduced maximal rates of relaxation and contraction relative to baseline (*, p < 0.001). Data are mean ± S.E.
Decreased Heart Rate and Contractile Function in Langendorff-perfused Mouse Hearts
To determine whether TAT-AKAD affects cardiac contraction ex vivo, we injected TAT-AKAD or TAT-AKADscr peptide into the coronary circulation of isolated perfused hearts. TAT-AKAD injected at <1 μm produced no significant changes in heart rate or LV-developed pressure compared with baseline (data not shown). Fig. 7A shows representative LV pressure tracings at baseline and during TAT-AKAD injection. By contrast, injection of 1 μm TAT-AKAD decreased heart rate and LV-developed pressure (Fig. 7, B and C). The rates of LV pressure development (+dP/dtmax) and relaxation (−dP/dtmin) also were reduced with a more pronounced effect on −dP/dt (Fig. 7E). The effects on cardiac function were reversible within 10 min of washout of the peptide. Administration of 10 μm TAT-AKAD produced a profound reduction of LV-developed pressure; this effect was irreversible (data not shown). The TAT-AKADscr peptide had no significant effect on heart rate or LV-developed pressure. The effects of the TAT-AKAD persisted in hearts of animals treated with reserpine, which depletes endogenous catecholamines (supplemental Fig. 1). In the context of isoproterenol stimulation, TAT-AKAD (1 μm) injected into the coronary circulation significantly decreased heart rate (13% ± 13), developed pressure (27% ± 10), +dP/dtmax (30% ± 10), and −dP/dtmin (27% ± 10) (Fig. 8, A–D).
FIGURE 7.
TAT-AKAD regulation of cardiac function in isolated, perfused hearts. A, representative images of left ventricular pressure tracings for isolated hearts perfused under basal conditions and after infusion of 1 μm TAT-AKADscr or TAT-AKAD peptide. B–E show heart rate (B), developed pressure (C), +dP/dt (D), and −dP/dt (E) after perfusion with 0.01, 0.1, and 1 μm TAT-AKADscr and TAT-AKAD peptide (n = 5/group). *, p < 0.01.
FIGURE 8.
Effect of TAT-AKAD on cardiac function in isolated, perfused hearts exposed to isoproterenol. A–D show heart rate (A), developed pressure (B), +dP/dt (C), and −dP/dt (D) in hearts stimulated with 0.1 μm isoproterenol and then infused with either 1 μm TAT-AKADscr or TAT-AKAD peptide (n = 8 in TAT-AKAD group and n = 6 in TAT-AKADscr group). *, p < 0.01.
DISCUSSION
We defined a 19-residue peptide within the PKA binding region of AKAP10/d-AKAP2 that had similar, high affinity binding to both RI and RII subunits of PKA. We found that a TAT-conjugated form of this peptide (TAT-AKAD) is taken up by isolated myocytes and uptake corresponds to altered cellular distribution of PKA-RI and PKA-RII subunits. TAT-AKAD significantly reduced isoproterenol-induced PKA phosphorylation, consistent with inhibition of PKA catalytic activity. TAT-AKAD also altered contractile function in isolated cardiac myocytes and decreased heart rate and developed pressure in intact mouse hearts, indicating a critical role for PKA-AKAP interactions in cardiac signaling.
PKA regulates many aspects of cardiac cell function, including metabolic processes, calcium handling, ion channel function, and contractility. PKA-dependent phosphorylation is regulated through binding of the R subunits of PKA to AKAPs that target PKA to its substrate (36). It is not known to what extent each R subunit regulates different aspects of molecular signaling or cardiac function, but it seems likely that distinct roles could be prescribed for each subunit. Both RI and RII regulatory subunits are expressed in the heart at similar levels (37). However, RII-mediated PKA regulation in the heart has been characterized more extensively (36). The RII isoform normally is associated with the plasma membrane, the sarcoplasmic reticulum, or subcellular organelles through targeting of various AKAPs (23). The RI isoform resides primarily in the cytosol and becomes localized under stimulated conditions (4, 22–24). RI has been shown to associate with the sodium calcium exchanger (NCX1), which is involved in calcium efflux, implicating the RI isoform in PKA regulation of this exchanger (38). We confirmed the localization pattern for the RI and RII isoforms described above in adult cardiac myocytes using immunofluorescence and cellular fractionation. Nuclear localization of RI in cardiac myocytes is consistent with prior results (39, 40). TAT-AKAD disrupted RI nuclear localization and is consistent with the idea that RI can be targeted to the nucleus via an AKAP (39, 40).
AKAPs are diverse but functionally related proteins linked by their ability to bind the R subunits of PKA (36). In general, AKAPs bind RII isoforms with higher affinity than RI isoforms (21). Thus, PKA-RI-mediated functions may be more susceptible to competition and modulation with disrupting peptides such as TAT-AKAD. AKAP10/d-AKAP2 is a dual specificity AKAP that binds both RI and RII regulatory subunits of PKA. AKAP10 is involved in heart rate regulation in mice and humans (15). The importance of the AKAP10 gene in humans is underscored by the presence of single nucleotide polymorphs (SNPs), one of which is present in the PKA binding region (33). This SNP codes for an amino acid change of isoleucine 646 to valine (I646V), and the only ascribed function is an increased binding affinity to the type I regulatory subunit of PKA (33). I646V can predispose patients to an increased mean heart rate and low heart rate variability, both indicators of decreased vagus nerve sensitivity and predictors of ventricular arrhythmias and increased risk for sudden cardiac death (15, 41). This suggests that antagonizing the RI-AKAP10 interaction may reduce heart rate and potentially provide a therapeutic benefit in the subset of patients with the deleterious allele. Consistent with this idea, we demonstrated that TAT-AKAD effectively binds RI and reduces heart rate and contractile function. It is important to note that our findings are in perfused hearts isolated from neurohumoral stimulation, suggesting that mechanisms in addition to altered parasympathetic drive and vagal tone are involved in the TAT-AKAD induced modulation of chronotropy and inotropy in the heart at baseline. Although the mechanism for this effect is unknown, both RI- and RII-mediated AKAP targeting may be involved. McConnell et al. (42) showed that treatment of rats hearts with Ht31, an RII subunit binding peptide, reduced left ventricular-positive and -negative dP/dt at baseline in vivo. The drop in dP/dt was not as large as that observed for TAT-AKAD in this study, which may indicate that RI-mediated events dominate in the regulation of cardiac relaxation. AKAD-mediated effects on contractility in the absence of isoproterenol stimulation were pronounced, indicating that PKA function may be important for basal levels of cardiac contraction. Basal levels of cAMP are implicated in calcium mediated pacemaker activity via PKA-regulated processes, both at the plasma membrane via the sodium-calcium exchanger (type I PKA subunit) and at the sarcoplasmic reticulum via the ryanodine receptor (type II PKA subunit) (43). Future studies using isoform-selective disruptors will be important to define the contribution of each isoform in cardiac signaling.
AKAP involvement in cardiac physiology and pathophysiology is of increasing interest (36). AKAPs are involved in excitation-contraction coupling (44) and cardiac hypertrophy (45), two areas critical to the pathophysiology of heart failure. We find that TAT-AKAD acutely reduces isoproterenol-induced phosphorylation of phospholamban and troponin I in vitro. Phospholamban and troponin I phosphorylation are altered in the failing heart (46, 47). Chronic isoproterenol infusion produces cardiomyopathy (48), and PKA activation is involved in the pathophysiology of heart failure (49). Peptide inhibitors of PKA and AKAP interaction, such as TAT-AKAD could potentially prove to be useful tools in heart failure research and therapeutics for heart failure.
In conclusion, we designed a cell-penetrating peptide using TAT bound to a truncated form of AKAP10 to disrupt interactions of RI and RII PKA subunits with AKAPs in the heart in vitro and ex vivo. We found that disruption of RI and RII subcellular localization inhibits cellular PKA activity and has negative chronotropic and inotropic effects in cardiac myocytes and the isolated heart. Our results thus demonstrate the efficacy of small peptide regulation of PKA-AKAP interactions to modulate cardiac function and suggest potential applications of disruption of AKAP-mediated PKA anchoring in the study and treatment of cardiovascular disease.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL081400 (to D. M. R.), HL066941 (to P. A. I. and D. M. R.), HL091071 (to H. H. P.), DK054441 (to S. S. T.) and HL079788 (to L. L. H.) from the United States Public Health Service. This work was also supported by Scientist Development Grant 060039N (to H. H. P.) from the American Heart Association and a Veterans Affairs Merit grant (to D. M. R.) from the Department of Veterans Affairs.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PKA
- protein kinase A
- AKAP
- A-kinase-anchoring proteins
- TAT
- trans-activator of transcription
- TAT-AKAD
- TAT-conjugated A-kinase-anchoring disruptor
- CM
- cardiac myocyte
- R
- regulatory
- C
- catalytic.
REFERENCES
- 1.Dodge-Kafka K. L., Langeberg L., Scott J. D. (2006) Circ. Res. 98, 993–1001 [DOI] [PubMed] [Google Scholar]
- 2.Kapiloff M. S. (2002) Mol. Pharmacol. 62, 193–199 [DOI] [PubMed] [Google Scholar]
- 3.McConnachie G., Langeberg L. K., Scott J. D. (2006) Trends Mol. Med. 12, 317–323 [DOI] [PubMed] [Google Scholar]
- 4.Ruehr M. L., Russell M. A., Bond M. (2004) J. Mol. Cell Cardiol. 37, 653–665 [DOI] [PubMed] [Google Scholar]
- 5.Dodge K. L., Khouangsathiene S., Kapiloff M. S., Mouton R., Hill E. V., Houslay M. D., Langeberg L. K., Scott J. D. (2001) EMBO J. 20, 1921–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulme J. T., Lin T. W., Westenbroek R. E., Scheuer T., Catterall W. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao T., Yatani A., Dell'Acqua M. L., Sako H., Green S. A., Dascal N., Scott J. D., Hosey M. M. (1997) Neuron 19, 185–196 [DOI] [PubMed] [Google Scholar]
- 8.Kapiloff M. S., Jackson N., Airhart N. (2001) J. Cell Sci. 114, 3167–3176 [DOI] [PubMed] [Google Scholar]
- 9.Fink M. A., Zakhary D. R., Mackey J. A., Desnoyer R. W., Apperson-Hansen C., Damron D. S., Bond M. (2001) Circ. Res. 88, 291–297 [DOI] [PubMed] [Google Scholar]
- 10.Marx S. O., Kurokawa J., Reiken S., Motoike H., D'Armiento J., Marks A. R., Kass R. S. (2002) Science 295, 496–499 [DOI] [PubMed] [Google Scholar]
- 11.Gardner L. A., Naren A. P., Bahouth S. W. (2007) J. Biol. Chem. 282, 5085–5099 [DOI] [PubMed] [Google Scholar]
- 12.Gardner L. A., Tavalin S. J., Goehring A. S., Scott J. D., Bahouth S. W. (2006) J. Biol. Chem. 281, 33537–33553 [DOI] [PubMed] [Google Scholar]
- 13.Bauman A. L., Soughayer J., Nguyen B. T., Willoughby D., Carnegie G. K., Wong W., Hoshi N., Langeberg L. K., Cooper D. M., Dessauer C. W., Scott J. D. (2006) Mol. Cell 23, 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lygren B., Carlson C. R., Santamaria K., Lissandron V., McSorley T., Litzenberg J., Lorenz D., Wiesner B., Rosenthal W., Zaccolo M., Taskén K., Klussmann E. (2007) EMBO Rep. 8, 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tingley W. G., Pawlikowska L., Zaroff J. G., Kim T., Nguyen T., Young S. G., Vranizan K., Kwok P. Y., Whooley M. A., Conklin B. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8461–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr D. W., Hausken Z. E., Fraser I. D., Stofko-Hahn R. E., Scott J. D. (1992) J. Biol. Chem. 267, 13376–13382 [PubMed] [Google Scholar]
- 17.Carr D. W., Stofko-Hahn R. E., Fraser I. D., Cone R. D., Scott J. D. (1992) J. Biol. Chem. 267, 16816–16823 [PubMed] [Google Scholar]
- 18.Colledge M., Scott J. D. (1999) Trends Cell Biol. 9, 216–221 [DOI] [PubMed] [Google Scholar]
- 19.Futaki S., Goto S., Sugiura Y. (2003) J. Mol. Recognit. 16, 260–264 [DOI] [PubMed] [Google Scholar]
- 20.Zhao M., Weissleder R. (2004) Med. Res. Rev. 24, 1–12 [DOI] [PubMed] [Google Scholar]
- 21.Feliciello A., Gottesman M. E., Avvedimento E. V. (2001) J. Mol. Biol. 308, 99–114 [DOI] [PubMed] [Google Scholar]
- 22.Cadd G. G., Uhler M. D., McKnight G. S. (1990) J. Biol. Chem. 265, 19502–19506 [PubMed] [Google Scholar]
- 23.Skalhegg B. S., Tasken K. (2000) Front Biosci. 5, D678–693 [DOI] [PubMed] [Google Scholar]
- 24.Skålhegg B. S., Taskén K., Hansson V., Huitfeldt H. S., Jahnsen T., Lea T. (1994) Science 263, 84–87 [DOI] [PubMed] [Google Scholar]
- 25.Clark D. (1996) Guide for the Care and Use of Laboratory Animals, National Academy of Science, Washington, D. C. [Google Scholar]
- 26.Guy C. A., Fields G. B. (1997) Methods Enzymol. 289, 67–83 [DOI] [PubMed] [Google Scholar]
- 27.Wellings D. A., Atherton E. (1997) Methods Enzymol. 289, 44–67 [DOI] [PubMed] [Google Scholar]
- 28.Soughayer J. S., Wang Y., Li H., Cheung S. H., Rossi F. M., Stanbridge E. J., Sims C. E., Allbritton N. L. (2004) Biochemistry 43, 8528–8540 [DOI] [PubMed] [Google Scholar]
- 29.Burns L. L., Canaves J. M., Pennypacker J. K., Blumenthal D. K., Taylor S. S. (2003) Biochemistry 42, 5754–5763 [DOI] [PubMed] [Google Scholar]
- 30.Communal C., Singh K., Pimentel D. R., Colucci W. S. (1998) Circulation 98, 1329–1334 [DOI] [PubMed] [Google Scholar]
- 31.Landeen L. K., Dederko D. A., Kondo C. S., Hu B. S., Aroonsakool N., Haga J. H., Giles W. R. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H736–749 [DOI] [PubMed] [Google Scholar]
- 32.Roth D. M., Lai N. C., Gao M. H., Drumm J. D., Jimenez J., Feramisco J. R., Hammond H. K. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H172–177 [DOI] [PubMed] [Google Scholar]
- 33.Kammerer S., Burns-Hamuro L. L., Ma Y., Hamon S. C., Canaves J. M., Shi M. M., Nelson M. R., Sing C. F., Cantor C. R., Taylor S. S., Braun A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4066–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinderman F. S., Kim C., von Daake S., Ma Y., Pham B. Q., Spraggon G., Xuong N. H., Jennings P. A., Taylor S. S. (2006) Mol. Cell 24, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarma G. N., Kinderman F. S., Kim C., von Daake S., Chen L., Wang B. C., Taylor S. S. (2010) Structure 18, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauban J. R., O'Donnell M., Warrier S., Manni S., Bond M. (2009) Physiology 24, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krall J., Taskén K., Staheli J., Jahnsen T., Movsesian M. A. (1999) J. Mol. Cell Cardiol. 31, 971–980 [DOI] [PubMed] [Google Scholar]
- 38.Schulze D. H., Muqhal M., Lederer W. J., Ruknudin A. M. (2003) J. Biol. Chem. 278, 28849–28855 [DOI] [PubMed] [Google Scholar]
- 39.Brown R. L., August S. L., Williams C. J., Moss S. B. (2003) Biochem. Biophys. Res. Commun. 306, 394–401 [DOI] [PubMed] [Google Scholar]
- 40.Reinitz C. A., Bianco R. A., Shabb J. B. (1997) Arch. Biochem. Biophys. 348, 391–402 [DOI] [PubMed] [Google Scholar]
- 41.Neumann S. A., Tingley W. G., Conklin B. R., Shrader C. J., Peet E., Muldoon M. F., Jennings J. R., Ferrell R. E., Manuck S. B. (2009) Psychophysiology 46, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McConnell B. K., Popovic Z., Mal N., Lee K., Bautista J., Forudi F., Schwartzman R., Jin J. P., Penn M., Bond M. (2009) J. Biol. Chem. 284, 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinogradova T. M., Lakatta E. G. (2009) J. Mol. Cell Cardiol. 47, 456–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manni S., Mauban J. H., Ward C. W., Bond M. (2008) J. Biol. Chem. 283, 24145–24154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Appert-Collin A., Cotecchia S., Nenniger-Tosato M., Pedrazzini T., Diviani D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10140–10145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bodor G. S., Oakeley A. E., Allen P. D., Crimmins D. L., Ladenson J. H., Anderson P. A. (1997) Circulation 96, 1495–1500 [DOI] [PubMed] [Google Scholar]
- 47.Waggoner J. R., Kranias E. G. (2005) Heart Fail Clin. 1, 207–218 [DOI] [PubMed] [Google Scholar]
- 48.Rona G. (1985) J. Mol. Cell Cardiol. 17, 291–306 [DOI] [PubMed] [Google Scholar]
- 49.Wehrens X. H., Lehnart S. E., Reiken S., Vest J. A., Wronska A., Marks A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.