Abstract
Agrin isoforms with different bioactivities are synthesized by the nerve and the muscle. Neural agrin containing an 8-amino acid insert (z8) introduced by alternative splicing is the active form that induces synaptic differentiation at the neuromuscular junction. In addition to alternative splicing, extracellular calcium is also required for the activity of neural agrin. To understand better how the activity of agrin is regulated by alternative splicing, we have applied alanine substitution mutagenesis to the z8 insert and the calcium binding site in the minimally functional AgG3z8 fragment. Single alanine substitutions in the 4th through the 7th amino acid of the z8 splice insert significantly reduced the function of agrin, in terms of acetylcholine receptor clustering activity and the affinity for binding to the muscle surface. Mutation of the asparagine at the 4th position drastically reduces bioactivity such that it is equivalent to that of muscle form AgG3z0. These reduced activity mutants also show reduced magnitudes of the calcium-induced CD spectrum change from that observed in AgG3z8 fragments, indicating that cross-talk between calcium and the z8 insert is critical for the normal activity of agrin. However, removal of Ca2+ binding via mutation of both aspartic acids in the calcium binding site did not totally eliminate the activity of AgG3z8. These results suggest a model wherein the z8 insert is a Ca2+-responsive allosteric element that is essential in forming an active conformation in neuronal agrin.
Keywords: Circular Dichroism, Nicotinic Acetylcholine Receptors, Site-directed Mutagenesis, Skeletal Muscle, Synapses, Agrin, Neuromuscular Junction, Receptor Cluster
Introduction
Agrin is a heparan sulfate proteoglycan first isolated from the electric organ of the marine ray Torpedo californica (1). Studies in the recent decades have established that agrin plays a critical role in directing the formation and stabilization of the mammalian neuromuscular junction. Agrin is secreted by the axonal terminals of motor neurons of the spinal cord and is highly concentrated in basal lamina of the synaptic cleft (2). Incubation of agrin with cultured myotubes or ectopic expression of agrin in adult skeletal muscles induces virtually all major aspects of postsynaptic specialization in the endplates, which include clustering of the nicotinic acetylcholine receptors (AChRs)2 and recruitment of other major postsynaptically expressed proteins such as acetylcholine esterase, utrophin, and rapsyn (3–5). Mutant mice deficient in agrin are born with severe defects of the neuromuscular synapse and die shortly after birth due to respiratory failure (6). In response to agrin released from the motor neurons, muscle produces retrograde signals to immobilize growth cones and to induce accumulation of synaptic vesicles and voltage-gated calcium channels in the presynaptic nerve terminals (6–9).
Recent study indicates that the downstream signaling in the neuromuscular junction is mediated by a receptor complex in myotubes containing the muscle-specific receptor tyrosine kinase, MuSK, and LRP4, a member of the LDL receptor family. LRP4 knock-out mice exhibit severe neuromuscular defects similar to those found in mutant animals deficient in MuSK or neural agrin (10). LRP4 serves as a co-receptor for the binding of agrin to MuSK and is essential for agrin-induced tyrosine phosphorylation of MuSK and AChR clustering (11, 12).
Multiple isoforms of agrin are expressed under spatial and temporal control by cells in the brain and in non-neural tissues including skeletal muscle (13, 14). The mammalian agrin gene contains three alternative splicing sites called X, Y, and Z sites. Alternative splicing at the Z site introducing an 8-amino acid insert (ELTNEIPA, z8 insert) is required for the synapse-forming activity of agrin, and this z8 isoform is only produced by neurons (13, 15, 16). The C-terminal 21-kDa fragment of neural agrin, which consists of the third globular domain with the z8 insert (AgG3z8), is necessary and sufficient for inducing the formation of AChR clusters in vitro under standard physiological conditions (15, 17).
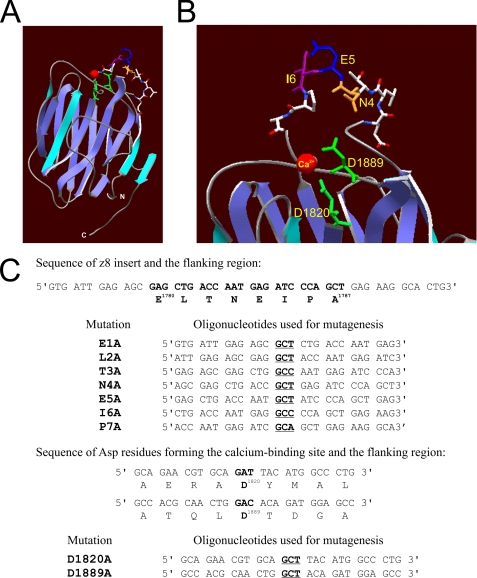
In addition to z8 insertion, extracellular calcium is also critical for the activity of agrin. It is required for the activation of MuSK as well as the induction and maintenance of AChR clustering (18–20). The AgG3 domain is homologous to the globular (LG5) domain of α2 laminin, which binds calcium through the side chains of two aspartic acids and two main chain carbonyls (21, 22). These aspartate residues are conserved in the AgG3 domain, and structural studies indicate that these Asp residues similarly mediate calcium binding in agrin (23). The AgG3 also belongs to the LNS (laminin/neurexin/sex hormone-binding globulin) domain family that typically displays a β-sandwich folding motif. The z8 insert is part of a surface loop on the same edge of the β-sandwich as the Ca2+ binding site (Fig. 1, A and B) (23). Intriguingly, Ca2+ binding induces a significant change in the circular dichroism (CD) spectrum of AgG3z8 but not AgG3z0 (24).
FIGURE 1.
Alanine substitution mutagenesis of AgG3z8. A, β-sandwich folding motif of the calcium-bound AgG3z8 shown in a ribbon diagram. The z8 insert forms a loop between two β-strands on the top edge of the protein (modeled by using laminin globular domain 5 of the laminin α2 chain (αL2LG5; Protein Data Bank code 1DYK, A chain; see supplemental materials). B, enlarged view showing the side chains of the z8 insert and the aspartic acids (Asp1820 and Asp1889) involved in calcium binding. Red, calcium; green, side chains of Asp1820 and Asp1889; orange, side chain of asparagine Asn4. C, synthetic oligonucleotides used for mutagenesis in this study. Only the sense strands of a pair of complementary primers for each mutation are shown. The codons for alanine are shown in bold and underlined. The first and last amino acids of the z8 insert as well as the two calcium binding site aspartates are numbered according to the published rat agrin protein sequence (Protein Identifier G3990241).
The mechanism by which the z8 insert mediates the signaling bioactivity of this alternatively spliced isoform of neural agrin is not clear. Alanine scanning studies are a widely used approach to examine structural and functional effects of regions of proteins and have been utilized in this study to examine the role of Ca+2 binding sites and the z8 insert on agrin function (25). We have investigated the correlation of the receptor-clustering activity of agrin with the ability of the AgG3 domain to be allosterically responsive to Ca2+ in CD studies of z8 mutants. In the alanine scanning studies conducted in this study, a single point mutation in the z8 insert, an N4A substitution, resulted in elimination of AChR clustering activity, marked reduction in binding affinity to myotubes, and a failure to undergo Ca2+-induced conformational changes. Importantly, the effect of N4A mutation is more severe than abrogating Ca2+ binding via mutagenesis of the Ca2+ binding site, suggesting a distinct accessory role for Ca2+ from that of the z8 insert. Our findings support a model wherein the z8 insert is required for agrin signaling as it is an allosteric responsive element necessary for neural signaling activity and its active conformation is favored by calcium binding.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Restriction and modification enzymes for DNA cloning were purchased from either New England Biolabs or Invitrogen. Synthetic oligonucleotide primers and synthetic z8 peptide were synthesized by Integrated DNA Technology (Coralville, IA) and Genemed Synthesis (San Francisco, CA), respectively. Nickel-nitrilotriacetic acid metal-affinity resin was the product of Qiagen (Valencia, CA). Rhodamine-α-bungarotoxin was purchased from Molecular Probes. General chemicals were purchased from Sigma.
Construction of Expression Plasmids and Site-directed Mutagenesis
cDNA sequences encoding the rat AgG3 domain of neural (amino acids 1756–1948, AgG3z8) or muscle agrin (amino acids 1756–1940, AgG3z0) were amplified by PCR and cloned into pPICZαA as described before (24). Point mutations were introduced into agrin cDNA in the pPICZαA expression plasmid using the QuikChange® Site-Directed Mutagenesis kit (Stratagene) following the protocol provided by the manufacturer. Briefly, the mutation was introduced into AgG3z8 by 18-cycle PCR using 50 ng of pPICZαA-AgG3z8 plasmid as the template and a pair of complimentary mutagenesis primers (125 ng each). After degradation of original template by DpnI digestion, the reaction mixture was electroporated into TOP 10 cells. Plasmids with the mutation were purified from Zeocin-resistant colonies using a minipreparation kit (Bio-Rad) and confirmed by dideoxynucleotide DNA sequencing. The oligonucleotide primers used for the mutagenesis purpose are shown in Fig. 1C.
Yeast Expression of AgG3 Proteins
The agrin expression construct was linearized by PmeI digestion and transformed into Pichia pastoris KM71 strain by electroporation. Positive clones were selected on YPD plates with Zeocin (0.1 mg/ml). A single colony was used to inoculate a 50-ml overnight culture in buffered glycerol-complex medium (1% yeast extract, 2% peptone, 3.4 g/liter yeast nitrogen base, 0.1 m potassium phosphate (pH 6.0), 0.4 mg/liter biotin, and 1% glycerol). The initial culture was then expanded to 1 liter in buffered methanol-complex medium (BMMY, 1% yeast extract, 2% peptone, 3.4 g/liter yeast nitrogen base, 0.1 m potassium phosphate (pH 6.0), 0.4 mg/liter biotin). Methanol was added daily to the BMMY medium at a final concentration of 0.75% to induce and maintain protein expression. On the 4th day of induction, the culture supernatant was collected, and proteins were salted out in 70% ammonium sulfate by centrifugation at 6,000 × g for 30 min at 4 °C. The precipitates were dissolved in 40 ml of binding buffer (250 mm NaCl, 10 mm imidazole, 50 mm sodium phosphate (pH 7.4)), loaded onto a 10-ml nickel-nitrilotriacetic acid column, washed consecutively with 40 ml of binding buffer and 40 ml of washing buffer (250 mm NaCl, 25 mm imidazole, 50 mm sodium phosphate, (pH 7.4)), and finally eluted in 40 ml of PBS containing 250 mm imidazole. After the imidazole was removed by dialysis against 2 liters of 5 mm phosphate buffer (pH 7.4), the eluent was concentrated by gentle dehydration using Aquacide II (Calbiochem-Novabiochem).
AChR Clustering Assay
C2C12 myoblast cultures were maintained in the growth medium (DMEM containing 20% fetal calf serum, 2 mm glutamine, 0.5% chick embryo extract (Invitrogen), and penicillin/streptomycin). Differentiation was induced by switching myoblasts with 70% confluences to the fusion medium (DMEM containing 5% horse serum, 2 mm glutamine), and the medium was refreshed daily. AChR clusters were induced by adding recombinant agrin fragments to fully differentiated myotube cultures. Five h later, the cells were fixed in 2% paraformaldehyde (5 min), stained with rhodamine-conjugated α-bungarotoxin, rinsed in PBS, and viewed under a 400 × lens of an Olympus IX-70 fluorescent microscope. Ten random fields were photographed with a SPOT-2e camera in each experiment. The digital photographs of the stained myotubes from each field were converted to grayscale mode and analyzed by using the public domain Image program (National Institutes of Health). Background signals were eliminated by adjusting the threshold. The mean gray value of each photograph was used to represent the degree of AChR clustering on the cell surface.
Ligand Binding Assays
Recombinant agrin proteins were iodinated using the chloramine T method (26). The labeling reaction was carried out by adding 1 mCi of 125I-Na (Amersham Biosciences) to 0.5 mg of protein in potassium phosphate buffer (pH 7.0). The reaction was started with 20 μl of a chloramine T solution (2 mg/ml). After 45 s, 20 μl of 2 mg/ml Na2S2O5 was added to terminate the labeling reaction. Free 125I was separated from labeled proteins by gel filtration on Sepharose G-25 columns. Approximately 85% of the proteins were recovered after gel filtration. The biological activity of neural agrin was retained after iodination as shown by their ability to induce AChR clustering in cultured C2C12 myotubes. Iodinated agrin proteins were added at concentrations indicated to C2C12 myotubes grown on 35-mm dishes in the fusion medium. After incubation for 60 min at room temperature, cells were rinsed four times with 50 mm phosphate buffer (pH 7.4) and lysed with 0.2 m NaOH. Bound radioactivity was counted using a gamma counter. Nonspecific binding was measured by including an excess amount (500×) of each unlabeled ligand in the reaction and was subtracted from the total binding. In competition studies with agrin mutant proteins, the concentration of 125I-AgG3z8 was kept constant at 10 nm. Ligand binding activity in the absence of unlabeled competitors was normalized to 100%, and the amount of bound ligands was calculated from the reduction in 125I-AgG3z8 binding. KI values were calculated from the IC50 concentration of competing ligands by the equation of Cheng and Prusoff (46).
Circular Dichroism Spectroscopy
Circular dichroism spectra were collected at 25 °C on an Aviv 202 spectrometer. Protein was diluted in 6 mm Tris (pH 7.4), 15 mm NaCl to ∼0.2 mg/ml to reduce low UV absorbance. At least six scans/sample were recorded at 1-nm intervals from 280 to 195 nm with 1-s integration time. Protein concentration was determined using BCA Protein Assay kit (Pierce). Mean residue ellipticity values [θ] were calculated based on the protein sequences and concentration determinations. The composition of protein secondary structures was analyzed by using CDPro program (47, 48). Calcium titration curves were conducted by measuring [θ]215 at constant protein concentrations with varying concentrations of Ca2+ introduced using a syringe driven pump assembly.
RESULTS
Synthetic z8 Peptide Is Not Biologically Active
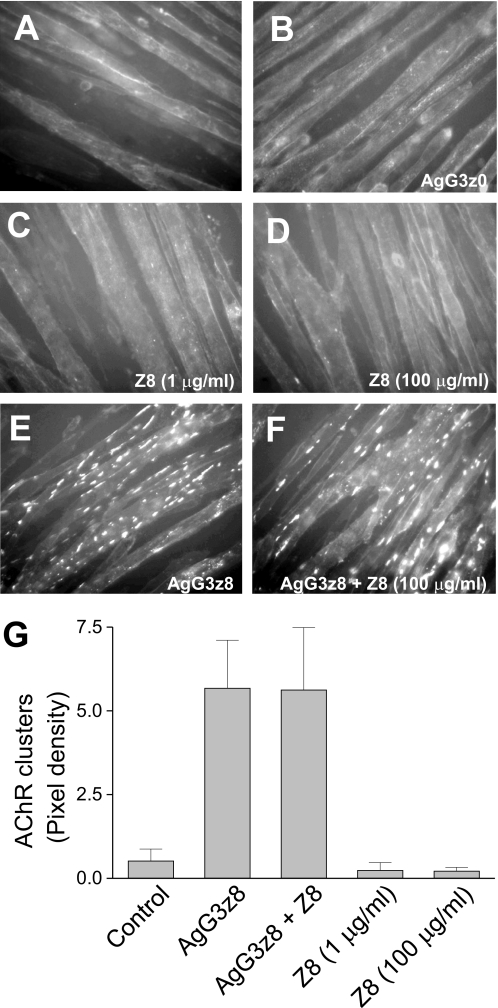
The recombinant C-terminal ∼21-kDa fragment of the AgG3 domain containing the z8 splice insert is the minimal agrin domain necessary and sufficient to induce the formation of AChR clusters in cultured myotubes. The same domain of muscle agrin, which lacks the z8 insert, binds poorly to muscle membrane, and has no AChR clustering activity (27–29). To test whether the z8 insert itself is able to bind directly to the agrin receptor and to regulate AChR clustering, synthetic z8 peptide (ELTNEIPA) was applied to differentiated C2C12 myotubes, and the extent of AChR clustering was examined by staining with rhodamine-α-bungarotoxin. Fluorescent microscopy revealed that few spontaneous AChR clusters formed on the plasma membrane in control myotubes or myotubes treated with muscle form agrin AgG3z0 (Fig. 2, A and B). Incubation of myotubes with the z8 peptide at a concentration of either 1 or 100 μg/ml (∼1.2 mm and 120 mm, respectively) did not induce formation of more receptor aggregates (Fig. 2, C, D, and G). In contrast, application of neural form agrin (AgG3z8, 1 μg/ml, ∼50 nm) induced significant aggregation of AChR on the cell surface (Fig. 2, E and G). Thus, the z8 peptide alone is insufficient to activate signaling through the agrin receptor. However, it might bind the receptor but not activate it. If this was the case, the peptide should act as a competitive antagonist of neural agrin. Therefore we treated myotube culture with excess z8 peptide (100 μg/ml) 1 h before adding the recombinant AgG3z8 protein (1 μg/ml) and incubated it for an additional 5 h. The z8 peptide had no apparent effect on the AChR clustering activity of neural agrin (Fig. 2, F and G). Therefore, in agreement with previous studies using a synthetic chick ortholog of the z8 sequence (17), the z8 insert alone does not bind the agrin receptor with any measureable affinity in the absence of other parts of AgG3 domain.
FIGURE 2.
Activity of synthetic Z8 peptide and recombinant agrin G3 domains. AChR clustering was not detected in C2C12 myotubes treated with (A) no agrin protein, (B) 1 μg/ml AgG3z0, (C) 1 μg/ml or (D) 100 μg/ml synthetic z8 peptide. Incubation with 1 μg/ml AgG3z8 induced patch-like AChR cluster formation (E), which persisted in the presence of 100 μg/ml synthetic z8 peptide (F). The AChRs on muscle cell surface were labeled with rhodamine-conjugated α-bungarotoxin and examined under a fluorescent microscope. G, fluorescent labeled receptor clusters in each visual field photographed and measured using the ImageJ software. The data represent the mean ± S.E. (error bars) of results from four independent experiments.
Effect of Alanine Substitution Mutagenesis in the z8 Insert of AgG3z8 on AChR Clustering Activity
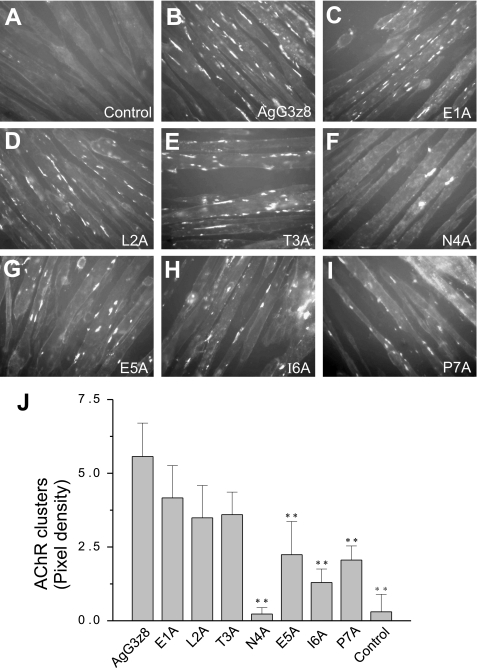
To determine the functional significance of each amino acid in the z8 insert, we substituted each of the first seven residues (ELTNEIP) with alanine by site-directed mutagenesis (Fig. 1C). Alanine is tolerated in both hydrophilic and hydrophobic environments, and it does not alter the main chain conformation, nor does it impose extreme electrostatic or steric effects (30). Introducing alanine into this region is not likely to affect the correct folding of the protein. Mutant recombinant AgG3z8 proteins were expressed as secretory proteins using yeast P. pastoris and purified to homogeneity as described previously (24). Each mutant protein was added at 1 μg/ml concentration (∼50 nm) to C2C12 myotubes, and 5 h later the levels of AChR clustering were measured. Mutations in the first three amino acids (ELT) weakly, but not significantly, reduced the AChR clustering activity of AgG3z8 (Fig. 3, B–D and J). Importantly, substitution of the 4th residue in z8, an asparagine, appeared to eliminate the activity of neural agrin, reducing the degree of receptor clustering to that of control myotubes (Fig. 3, F and J; p > 0.05, n = 4). Mutation of the 5th, 6th, and 7th amino acids (EIP) significantly reduced the AChR clustering activity of AgG3z8 (p < 0.01, n = 4), but to a lesser extent than the N4A mutation (Fig. 3, G–J). These results show that the 4th to 7th amino acids of the z8 insert are functionally critical in neural agrin signaling, with the asparagine at position 4 playing a particularly critical role.
FIGURE 3.
Activity of alanine substitution mutants of the AgG3z8 protein. A–I, 1st to 7th amino acids of the z8 peptide in AgG3z8 replaced with alanine by site-directed mutagenesis. The AChR clustering activities of the wild-type AgG3z8 and each of the mutant proteins at 1 μg/ml were measured in C2C12 myotube culture. J, statistic analysis of the degrees of AChR clustering. The data represent the mean ± S.E. (error bars) of results from four separate experiments. **, p < 0.01 compared with AgG3z8.
Effect of Alanine Substitution Mutagenesis in the z8 Insert of AgG3z8 on Myotube Surface Affinity
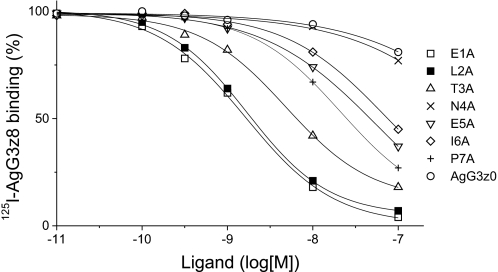
To determine whether mutations within the z8 sequence changes the binding affinity of AgG3z8 to myotubes, competition binding assays using C2C12 myotubes were conducted. The binding of iodinated AgG3z8 protein (5 nm) to myotubes cultured on 35-mm dishes was competed by unlabeled mutant proteins containing single alanine substitutions in the z8 insert. Alanine substitutions in the z8 sequence reduced the binding affinity of AgG3z8 to different degrees (Fig. 4). Similar to the effects of z8 mutations to the AChR clustering activities seen in Fig. 3, the affinity of AgG3z8 was slightly reduced by mutations of the first three z8 sequences, moderately affected by mutations of the 5th to 7th amino acids, and severely abolished by N4A substitution (Fig. 3). The dissociation constant (KI) was 4.6, 6.2, and 39 nm, for E1A, L2A, and T3A mutant proteins, respectively (Table 1). In contrast, the N4A mutant showed a strikingly low affinity (KI = 2.1 μm), similar to that of muscle agrin (AgG3z0, KI = 1.8 μm). Substitution at the 5th, 6th, or 7th residue reduced the protein binding affinity, but the effect was much less severe (KI = 85, 110, 74 nm for E5A, I6A, and P6A mutants, respectively). Thus, mutations of z8 residues resulted in parallel reduction in the signaling activity and surface affinity of AgG3z8 protein.
FIGURE 4.
Competition binding assay of agrin mutants. Myotubes grown in fusion medium on 35-mm dishes were incubated with 5 nm 125I-AgG3z8 plus a mutant protein. Ligand binding in the absence of the competitor was normalized to 100%. The amount of bound competing ligand was calculated from the reduction in 125I-AgG3z8. KI values were calculated from the IC50 concentration of competing ligands by the equation of Cheng and Prusoff (46) and presented in Table 1.
TABLE 1.
KI of AgG3z8 mutant proteins obtained by competition binding assays
The concentration of wild-type 125I-AgG3z8 was kept constant at 10 nm. Ligand binding activity in the absence of competitors was normalized to 100%, and the amount of bound ligands was calculated from the reduction in 125I-AgG3z8. KI values were calculated from the IC50 concentration of competing ligands by the equation of Cheng and Prusoff (46). For comparison, the KD values estimated by Scatchard plot were 9.3 nm, 367 nm, 1.1 μm, and 1.2 μm for AgG3z8, AgG3z8D1820/1899A, AgG3z8N4A, and AgG3z0, respectively (Fig. 9B).
| Ligand | KII |
|---|---|
| nm | |
| E1A | 4.6 |
| L2A | 6.2 |
| T3A | 39 |
| N4A | 2,100 |
| E5A | 85 |
| I6A | 110 |
| P7A | 74 |
| AgG3z0 | 1,800 |
AgG3z8N4A Is a Loss-of-function Mutant That Still Retains Its Secondary Structure
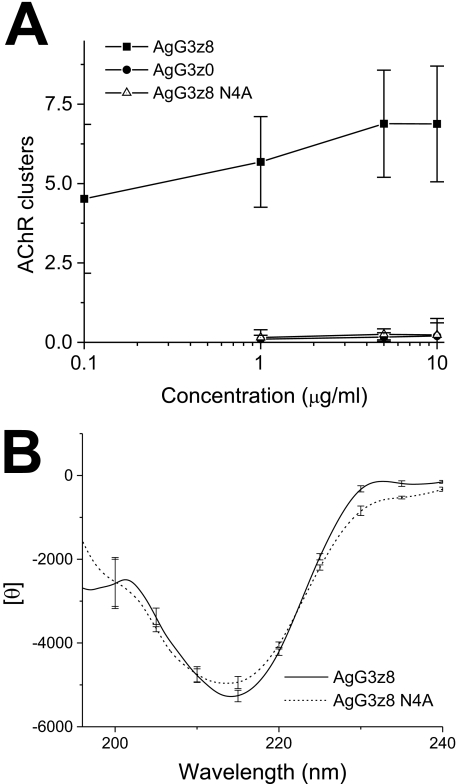
The AChR clusters readily appeared on C2C12 myotube treated with wild-type AgG3z8 at concentration as low as 0.1 μg/ml and reached plateau at 5 μg/ml (∼25 nm; Fig. 5A). This efficacy is comparable with that of a chick AgG3 fragment expressed in COS and HeLa cells (EC50 ∼ 13 nm) (17, 31). Both muscle agrin AgG3z0 and the N4A AgG3z8 mutant failed to induce receptor aggregation at concentrations up to 10 μg/ml (Fig. 5A). To test whether the N4A mutation inactivates AgG3z8 by disrupting the structure of this domain, comparative CD studies were conducted to analyze the net secondary structure of AgG3z8 and AgG3z8N4A protein. The CD spectrum of the N4A mutant was very similar to that of the wild-type AgG3z8 (Fig. 5B), indicating the two proteins had similar net secondary structures. Deconvolution of the spectra indicated that they both had ∼30% β-sheet, 25% β-turn, and 2% α-helix. These results suggest that the reduced activity of the N4A mutation is not attributable to any gross changes in protein structure.
FIGURE 5.
AgG3z8 N4A is a loss-of-function mutant that retains wild-type net secondary structure. A, dose-response curves of the AChR clustering activity of neural agrin (AgG3z8), N4A mutant, and muscle agrin (AgG3z0) plotted as log of concentration versus clustering activity. The wild-type and mutant proteins were each incubated at concentrations indicated with C2C12 myotubes grown in fusion medium for 5 h. The cells were then stained with rhodamine-α-bungarotoxin. AChR clusters (pixel density/microscopic field) in cultures exposed to 0.5 μm AgG3z8 were normalized to 100%. Each data point represents mean ± S.E. (error bars) of results from four independent experiments. B, superimposition of CD spectra of AgG3z8 and the N4A mutant in TBS. The data represent the mean of results from four separate experiments, each with six scans, and the S.E. is shown at every 10 nm of the wavelength.
N4A Mutation Eliminates Calcium-induced Conformational Change
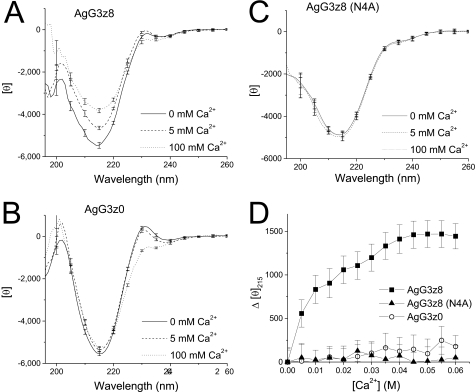
Extracellular calcium is critically required for the induction of AChR clustering by motoneurons or by agrin in cultured myotubes (18–20). AgG3z8 is homologous to the globular (LG5) domain of α2 laminin, which binds calcium through the side chains of two aspartic acids and two main chain carbonyls (21, 22). AgG3 domain also contains this conserved, low affinity Ca2+ binding site formed by the side chains of Asp1820 and Asp1889 (Fig. 1A) (23). In a previous study (24), we found Ca2+-induced conformational changes in AgG3z8, but not in AgG3z0, suggesting that the z8 insert imparts conformational sensitivity to the binding of Ca2+ at its distinct binding site and implicates a functional allosteric role for this neural-specific splice insert. Therefore, we tested whether this conformational sensitivity to Ca2+ is abrogated in the N4A mutant by comparing the effect of titrated Ca2+ on the CD spectra of AgG3z8, AgG3z0, and the N4A mutant. In the absence of added Ca2+, all three AgG3 proteins show similar typical β-rich CD spectra with the deep minima at 215 nm (Fig. 6, A–C). As reported before, calcium induced prominent changes in the CD spectrum of AgG3z8 (Fig. 6A). A change in the magnitude of the mean residue ellipticity at 215 nm ([θ]215) was readily observed in the presence of 5 mm Ca2+. A further reduction in the ellipticity at this wavelength was observed when the calcium concentration was raised to 100 mm to saturate the Ca2+ binding site. Plotting the net change of [θ]215 (Δ[θ]215) against calcium concentration gave an apparent KD of ∼10 mm for the Ca2+-induced allosteric effect (Fig. 6D). Deconvolution analysis of the averaged spectrum for AgG3z8 determined that Ca2+ binding resulted in an ∼11% increase in the content of β-sheet (p < 0.01, n = 4) with a concomitant reduction in the unordered structure of AgG3z8. Calcium caused a different effect on the CD spectra of AgG3z0, resulting in noticeable reductions around 230 nm. In contrast, the CD spectra of AgG3z8N4A exhibited virtually no change in response to increasing Ca2+ concentrations (Fig. 6, C and D). These results indicate that the N4A mutation results in loss of a conformational sensitivity to added Ca2+ and suggest that the Asn4 residue is critical for Ca2+-induced conformational changes in neural agrin.
FIGURE 6.
Calcium-induced CD spectrum change in AgG3z8, AgG3z0, and N4A mutant protein. A–C, CD spectra of AgG3z8 (A), AgG3z0 (B), and the N4A mutant (C), measured in TBS with 0, 5, and 100 mm of calcium. D, calcium titration curves of AgG3z8, AgG3z8N4A, and AgG3z0 for the ellipticity shift at 215 nm. The concentration of calcium that causes the half-maximum change in [θ]215 of AgG3z8 is ∼10 mm. In contrast, calcium has little effect on the CD spectrum of N4A mutant and muscle agrin. Each data point represents the mean of four independent measurements, and the S.E. (error bars) are shown at every 10 nm of the wavelength.
Calcium-induced Conformational Change Is Correlated with the Bioactivity of Neural Agrin
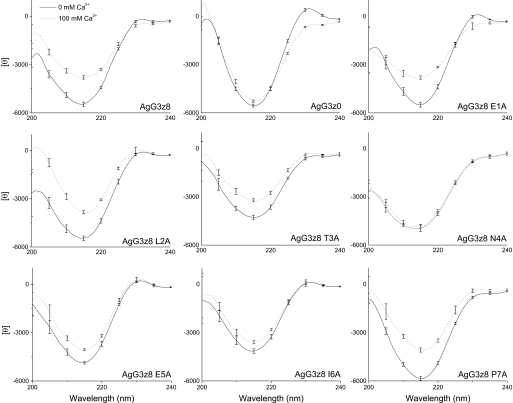
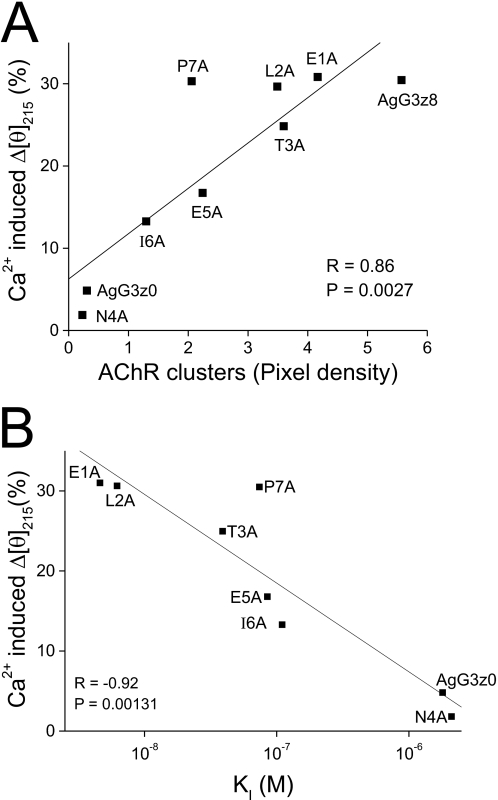
The finding that Ca2+-induced conformational changes (Δ[θ]215) only occur in active neural AgG3 implies that this conformational change might be critical in its AChR clustering activity. To test this hypothesis, we determined the Ca2+-induced conformational changes of all z8 Ala-substituted mutants using CD spectroscopy. These mutant proteins showed different degrees of CD spectrum change in response to 100 mm calcium (Fig. 7). The Ca2+-induced net change in the magnitude of the mean residue ellipticity at 215 nm (Δ[θ]215) for each mutant protein was plotted against its AChR clustering activity (from Fig. 3J) and the dissociation constant, KI (from Table 1), respectively. As shown in Fig. 8, a strong correlation exists between the Ca2+-induced conformational change and bioactivity of neural AgG3 (Fig. 8A; r = 0.86, p = 0.00027) and between the Ca2+-induced conformational change and AgG3 binding affinity (Fig. 8B; r = −0.92, p = 0.00131). The only exception is the P7A mutant, probably because replacing proline with alanine increased the flexibility of the z8 insert. The resulting greater conformational change is likely random; therefore it does not contribute to the affinity and activity of the protein. These results suggest that the decreased activity and affinity of these mutants are associated by their reduced ability to form an active conformation in the presence of calcium.
FIGURE 7.
Analysis of AgG3z8 mutant proteins by CD spectrophotometry. The CD spectrum of each protein was measured in TBS with 0 or 100 mm calcium, respectively, and superimposed to show the Ca2+-induced conformational change. The data represent the mean of results from three separate experiments, each with six scans, and the S.E. (error bars) is shown at every 10 nm of the wavelength.
FIGURE 8.
Ca2+-induced protein conformational change is correlated with the AChR clustering activity and the binding affinity of agrin. A, Ca2+-induced reduction in the magnitude of the mean residue ellipticity at 215 nm (Δ[θ]215) of each protein shown in Fig. 5 plotted against the AChR clustering activity of the protein in Fig. 2B. Linear regression analysis indicated a positive correlation (r = 0.86, p = 0.0027). B, linear regression analysis of the correlation between calcium-induced conformational change (Δ[θ]215) and the log KI value of AgG3 proteins shown in Table 1. r = −0.92, p = 0.00131.
Mutation of Calcium Binding Sites Reduces but Does Not Eliminate the Activity of Neural Agrin
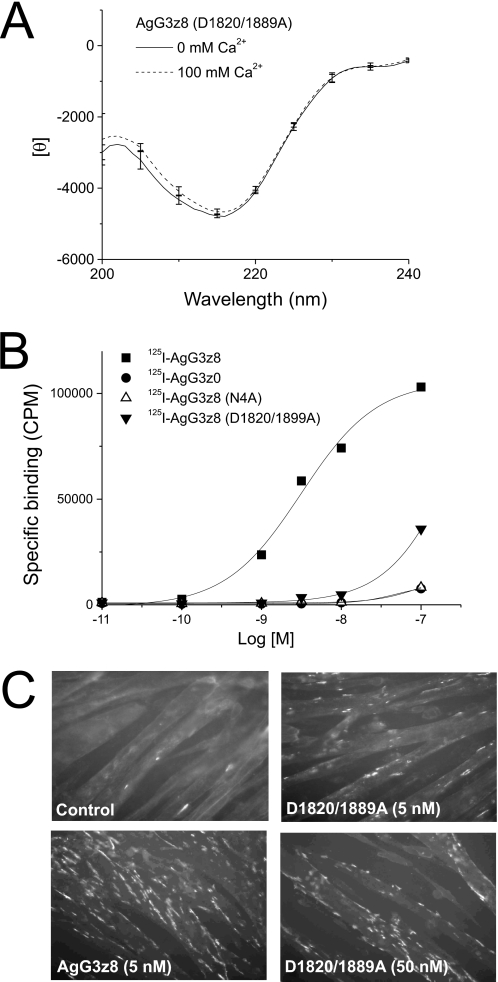
To test whether allowing the formation of active conformation in the presence of Ca2+ is the only function of the z8 insert, we mutated both aspartic acids that were shown to be directly involved in Ca2+ binding in AgG3z8 and compared the resulting activity of this double mutant with the activity of AgG3z8N4A. As expected, the CD spectrum of the Ca2+ binding site mutant AgG3z8D1820/1899A was insensitive to 100 mm calcium (Fig. 9A), similar to that of AgG3z8N4A. In direct binding assays with iodinated AgG3 proteins, AgG3z8 was shown to bind cultured myotubes with high affinity, whereas AgG3z8D1820/1899A, AgG3z8N4A, and AgG3z0 exhibited poor binding (Fig. 9B). The KD values estimated by Scatchard plot were 9.3 nm, 367 nm, 1.1 μm, and 1.2 μm for AgG3z8, AgG3z8D1820/1899A, AgG3z8N4A, and AgG3z0, respectively. Importantly, the nonbinding Ca2+ mutant containing the wild-type z8 insert still displayed residual affinity to myotubes. Finally, the effect of mutating the Ca2+ binding site of AgG3z8 on neural agrin signaling (i.e. the AChR clustering activity) was examined. The number of receptor clusters on myotubes treated with 5 nm AgG3z8D1820/1899A mutant protein was markedly lower than that of cells exposed to the same amount of AgG3z8 (Fig. 9C). However, unlike the N4A mutation, the D1820/1899A double mutant was able to induce AChR clustering at a higher concentration (50 nm; Fig. 9C). These results are in agreement with the finding that the binding of neuronal agrin to LRP4 is reduced but not totally abolished in Ca2+-free conditions (11). Therefore mutating the Ca2+ binding site in AgG3z8 has less severe consequence than the N4A mutation. This result indicates that first, calcium plays critical but accessory roles by enhancing the activity of agrin protein and second, besides being required for the Ca2+-induced conformational change to occur, the z8 insert (and Asn4 in particular) has an indispensible role as an allosteric response element that is modulated by Ca2+ and is necessary for effective signaling through the agrin receptor.
FIGURE 9.
Mutation of the calcium binding site reduces but does not eliminate the activity of AgG3z8. A, AgG3z8D1820/D1889A mutant protein displayed identical CD spectra in the presence or absence of calcium. B, specific binding of iodinated AgG3z8, AgG3z0, N4A mutant, and the D1820/D1889A mutant to C2C12 myotubes in 35-mm culture dishes is shown. Note that the D1820/D1889A mutant showed higher specific binding than AgG3z8N4A or AgG3z0. C, number of AChR clusters on myotube surface induced by the calcium-binding site mutant (at 0.1 μg/ml, ∼5 nm) is considerably lower than that induced by AgG3z8 at the same concentration. However, the mutant protein shows clear AChR clustering activity at a higher concentration (50 nm).
DISCUSSION
Alternative splicing at the Z site introducing an 8-amino acid insert (ELTNEIPA, z8 insert) is required for the synapse forming activity of agrin and is only produced by neurons (13, 15, 16). These critical sequences are also potential mutation sites in congenital neuromuscular diseases and are thus a focus of much interest. To understand how the structure and function of agrin may be regulated by alternative splicing, alanine scanning studies were conducted to identify key residues in the z8 insert in the expressed truncated AgG3 domain. In general, alanine substitutions of z8 sequences in the AgG3z8 fragment decrease neural agrin signaling, in terms of AChR clustering activity and its affinity for binding to the muscle surface. The reduction of activity is significantly larger when the mutation is introduced to the 4th to 7th amino acids of z8. These 4 amino acids, NEIP in single-letter code, are conserved in vertebrate agrin protein. Most strikingly, the N4A mutant loses all activities uniquely associated with neural agrin and is functionally indistinguishable from the muscle form, AgG3z0, in our assays. Interestingly, the z8 mutants all show correlated reductions in the magnitude of their Ca2+-induced CD spectral changes (Δ[θ]215).
NMR and crystallographic studies of the chick G3-B0 and G3-B8 domains (orthologs of mammalian AgG3z8 and AgG3z0) revealed that the core of AgG3 domain is an LNS/β-sandwich folding motif featuring two layers of β-sheets formed by 13 β-strands (23, 32). Structure models indicate that the B8 insert is part of a mobile loop overhanging from one edge of the AgG3 domain. A Ca2+ binding site was identified (and is distinct from the B8 loop), and the KD for Ca2+ is reported to be about 0.1 mm and 0.6 mm for neural and muscle form agrin, respectively (23). Ca2+ binding sharpens the NMR signals of amino acids near the binding site and around the B insertion loop, indicating that Ca2+ stabilizes the structure of AgG3 domain. However the core β-sandwich structure does not show major structural change in response to Ca2+ binding or to the inclusion of the B8 loop. Moreover, the B8 insert is still mobile in the Ca2+-bound G3-B8 (23). In contrast, pronounced differential effects of Ca2+ on the structures of the mammalian neural and muscle agrin G3 domains can be observed by using CD spectroscopy. Our previous study indicates that AgG3z8 and AgG3z0 both display CD spectra typical of proteins rich in β-structures, with a deep minimum at 215 nm. Binding of Ca2+ markedly reduces the magnitude of 215 nm peak in the spectra of AgG3z8 but not of AgG3z0 (24). Calcium causes different effects on the CD spectra of AgG3z0, resulting in minor but noticeable reductions around 230 nm (Figs. 6B and 7). It probably reflects the structural change on the core β-sandwich structure shared by both AgG3z0 and AgG3z8. However, the larger observed Ca2+-responsive allosteric effects of the z8 insert may mask this spectral change in AgG3z8.
Because the Ca2+-induced CD spectral change (Δ[θ]215) is only seen in the active, neural AgG3, this suggests that interaction between the z8 insert and Ca2+ transforms agrin into an active conformation capable of neural signaling. This notion is further supported by this study; all of the AgG3z8 functional mutants show reduced ability to undergo Ca2+-induced conformational change. However, data from NMR or x-ray crystallography studies do not support that an inactive AgG3z8 structure is simply transitioned to an active one by Ca2+ binding.
Structural study suggests that the z8 insert forms a flexible loop on the surface of AgG3z8 and does not possess a definitive structure either in Ca2+-bound or Ca2+-free form (23). Because Ca2+ does not drastically change the core β-sandwich structure of AgG3 domains, the difference of CD spectra between Ca2+-free and Ca2+-bound form of AgG3z8 may be due to the z8 loop being an allosteric responsive element that, together with Ca2+, stabilizes the functionally active form of neural agrin. It is possible that the flexible z8 loop adopts multiple structures with the CD spectrum representing an average net structure of a pool of heterogeneous conformations. We propose that the equilibrium between these active and inactive conformations is allosterically responsive to Ca2+ binding and that the active conformation(s) (i.e. one that is capable of binding and signaling via effective binding to its receptor on myotubes) may be stabilized upon Ca2+ binding, enhancing the activity of agrin. The calcium-induced CD spectral change is the result of such shift of equilibrium. Although Ca2+ binding stabilizes the formation of the active z8 agrin structure, it is not absolutely essential because mutation of the two aspartic acids of the Ca2+ binding site did not fully eliminate agrin activity. In this Ca2+-nonbinding mutant, the z8 insert may still be able to adopt an active conformation but with less frequency and, consequently, displayed reduced activity. In agreement with this finding, the binding of neuronal agrin to LRP4 is found to be reduced but not totally abolished in Ca2+-free conditions (11).
There could be a number of ways for the cross-talk between Ca2+ and z8 to occur. Calcium binding may stabilize the active z8 conformation by changing the nearby environment of z8, due to the close proximity of Ca2+ binding site to the z8 insert (Fig. 1, A and B). It is also possible that the slight change of the core AgG3z8 structure in its Ca2+-bound form may limit what possible structures z8 can adapt. In addition, previous structural study indicates that asparagine side chains participate in forming the calcium binding sites of AgG1 and AgG2 domains (21), transient direct interaction between Ca2+ and the Asn4 within the z8 insert is also possible. An interesting finding is that in both AgG3z8 and AgG3z0 calcium binding also results in different degrees of spectral change between 220 and 240 nm, whereas AgG3z8N4A displays virtually no calcium-induced spectral change in the whole wavelength range scanned (Fig. 4C), similar to AgG3z8D1820/1899A. This may also suggest that the N4A mutation could interfere with the calcium binding ability of the z8 insert.
An important finding of this study is the identification of Asn4 as the most critical amino acid in the z8 insert. The AgG3z8N4A mutant loses all activity, far more severe than the calcium binding site mutant. Meanwhile, it does not display calcium-induced CD spectrum change although its secondary structure remains intact (Figs. 5B and 6C). The precise role played by Asn4 in the binding and activation of the agrin receptor complex remains to be determined by further structural and mutagenesis studies. Here, we propose that the Asn4 residue is essentially required for the formation of an active z8 conformation(s). In the N4A mutant, the Ca2+ binding site of AgG3 domain should still be intact. The mutant protein is able to bind calcium but unable to form active conformation(s). As to why the N4A mutation results in total elimination of calcium-induced CD spectrum change, we suggest two possible explanations. First, the spectrum change may be the direct result of the emergence of calcium-induced active conformation. Because there is no active conformation to be stabilized by calcium, the CD spectrum of AgG3z8N4A mutant displays few changes upon Ca2+ binding. Second, Asn4 may play an additional role by interacting with calcium (although transiently), hence the conformation of AgG3z8N4A mutant is no longer sensitive to calcium.
The effect of calcium on the conformation of agrin reported in this study apparently occurs at concentrations higher than the general concentration of extracellular calcium (∼2.2 mm). Nevertheless, previous study indicated raising the concentration of extracellular calcium to 5 or 10 mm indeed increased the AChR clustering activity of AgG3z8 (24). This may suggest that under physiological conditions only a portion of agrin protein is active. In addition, considering that the synaptic cleft of neuromuscular junction is tightly surrounded by perisynaptic Schwann cells (33), it is tempting to speculate that the calcium concentration within this enclosed space may be modulated to enhance the synaptogenic activity of agrin. It should also be noted that the full-length agrin has much higher affinity to the muscle surface through multiple protein interactions (11, 34). The endogenous agrin may be allosterically induced into displaying its active conformation by the cooperative efforts of calcium and other members of the agrin receptor complex; therefore the observed response to Ca2+ may not require the elevated Ca2+ concentrations reported here.
Taken together, the above findings suggest the following model of z8-Ca2+ interplay. The z8 insert is an allosteric responsive element and is present on the surface of AgG3 domain as an intrinsically disordered loop that samples multiple conformations, a low proportion of which are capable of neural signaling via effective binding to its receptor complex. The 4th to the 7th amino acids of the z8 insert are critical for the formation of active conformation(s). The binding of Ca2+, at a distinct site separate from this loop region, further stabilizes the active conformation(s) thereby increasing the activity of AgG3z8 protein. A similar role for these z8 residues in muscle-specific receptor tyrosine kinase phosphorylation activity was observed by Scotton et al. (49). However, in that study using a longer chick agrin C45A4B8 fragment that contains two LG domains (LG2 and LG3), functional deficiency is apparent only when more than one B8 residue is mutated. While z8 insertion is the major determinant of activity, the LG2 domain enhances the activity of neural agrin by binding to α-dystroglycan on the muscle surface. Our finding indicates that affinity to the muscle surface also dictates the activity of mutants of the minimal functional AgG3z8 (Fig. 8). This suggests that in the absence of the Lg2 domain, the z8 insert is sufficient for an induced active conformation that initiates the downstream signaling pathway.
It is suggested that the z8 insert would interact with agrin receptor through an induced fit model (23). The actual contribution of Ca2+ and the z8 sequence will therefore require detailed structure analysis and protein interaction study of the complex formation by AgG3z8, MuSK, and LRP4. In addition, mutational studies that investigate the functional and structural effects of substituting other amino acids for Asn4 and structural comparisons between AgG3z8 and AgG3z8N4A using NMR or x-ray crystallography may further elucidate the molecular mechanisms underlying the role of the z8 insert on agrin function. A potential advantage of the unstructured z8 insert is that it may act as a flexible adaptor to allow agrin to bind to and select among multiple receptors. In addition to the typical AChR cluster formation in the neuromuscular junction, neural agrin activates multiple downstream events such as reorganization of cytoskeleton (35, 36) and formation of lipid raft (37–39) in muscle cells. Agrin is also involved in the formation of autonomic neuroeffector junctions (40–42) and immunological synapses (43). Testing whether the z8 mutations identified by this study also impair these functions of agrin may reveal whether a common signaling complex is involved. In addition, our study also provides insight into how locally flexible, intrinsically unstructured domains contribute to protein function and signaling. These domains comprise a significant portion of the proteome, yet their importance has only recently been appreciated (44, 45).
Supplementary Material
Acknowledgments
We thank Dr. Willi Halfter for helpful comments and Yun Yao for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant NS38301 and the Muscular Dystrophy Association (to Z.-Z. W.), National Science Council in Taiwan Grants NSC 95-2314-B-037-108 and NSC96-2320-B037-033-MY2, and Kaohsiung Medical University Faculty Development Grants Q095006 and Q097007 (to C.-N. T.). The CD studies were supported by National Institutes of Health/National Center for Research Resources Shared Instrumentation Grant 1S10RR11998 (to M. C.).
We dedicate this paper to the memory of Zuo-Zhong Wang, Ph.D., who died in a tragic hiking accident on June 15, 2008.

The on-line version of this article (available at http://www.jbc.org) contains supplemental material and additional references.
- AChR
- acetylcholine receptor
- AgG3
- the third laminin-like globular domain of agrin
- AgG3z0
- recombinant fragment of the third laminin-like globular domain of muscle agrin
- AgG3z8
- recombinant fragment of the third laminin-like globular domain of neural agrin
- MuSK
- muscle-specific receptor tyrosine kinase
- z8 insert
- 8-amino acid insert (ELTNEIPA) at the Z site.
REFERENCES
- 1.Nitkin R. M., Smith M. A., Magill C., Fallon J. R., Yao Y. M., Wallace B. G., McMahan U. J. (1987) J. Cell Biol. 105, 2471–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escher G., Béchade C., Levi S., Triller A. (1996) J. Cell Sci. 109, 2959–2966 [DOI] [PubMed] [Google Scholar]
- 3.Nitkin R. M., Rothschild T. C. (1990) J. Cell Biol. 111, 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campanelli J. T., Hoch W., Rupp F., Kreiner T., Scheller R. H. (1991) Cell 67, 909–916 [DOI] [PubMed] [Google Scholar]
- 5.Reist N. E., Werle M. J., McMahan U. J. (1992) Neuron 8, 865–868 [DOI] [PubMed] [Google Scholar]
- 6.Gautam M., Noakes P. G., Moscoso L., Rupp F., Scheller R. H., Merlie J. P., Sanes J. R. (1996) Cell 85, 525–535 [DOI] [PubMed] [Google Scholar]
- 7.Gautam M., DeChiara T. M., Glass D. J., Yancopoulos G. D., Sanes J. R. (1999) Brain Res. Dev. Brain Res. 114, 171–178 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen Q. T., Son Y. J., Sanes J. R., Lichtman J. W. (2000) J. Neurosci. 20, 6077–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campagna J. A., Rüegg M. A., Bixby J. L. (1995) Neuron 15, 1365–1374 [DOI] [PubMed] [Google Scholar]
- 10.Weatherbee S. D., Anderson K. V., Niswander L. A. (2006) Development 133, 4993–5000 [DOI] [PubMed] [Google Scholar]
- 11.Kim N., Stiegler A. L., Cameron T. O., Hallock P. T., Gomez A. M., Huang J. H., Hubbard S. R., Dustin M. L., Burden S. J. (2008) Cell 135, 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B., Luo S., Wang Q., Suzuki T., Xiong W. C., Mei L. (2008) Neuron 60, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rüegg M. A., Tsim K. W., Horton S. E., Kröger S., Escher G., Gensch E. M., McMahan U. J. (1992) Neuron 8, 691–699 [DOI] [PubMed] [Google Scholar]
- 14.Hoch W., Ferns M., Campanelli J. T., Hall Z. W., Scheller R. H. (1993) Neuron 11, 479–490 [DOI] [PubMed] [Google Scholar]
- 15.Ferns M., Hoch W., Campanelli J. T., Rupp F., Hall Z. W., Scheller R. H. (1992) Neuron 8, 1079–1086 [DOI] [PubMed] [Google Scholar]
- 16.Rupp F., Ozçelik T., Linial M., Peterson K., Francke U., Scheller R. (1992) J. Neurosci. 12, 3535–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gesemann M., Denzer A. J., Rüegg M. A. (1995) J. Cell Biol. 128, 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloch R. J. (1983) J. Neurosci. 3, 2670–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace B. G. (1988) J. Cell Biol. 107, 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borges L. S., Lee Y., Ferns M. (2002) J. Neurobiol. 50, 69–79 [DOI] [PubMed] [Google Scholar]
- 21.Hohenester E., Tisi D., Talts J. F., Timpl R. (1999) Mol. Cell 4, 783–792 [DOI] [PubMed] [Google Scholar]
- 22.Timpl R., Tisi D., Talts J. F., Andac Z., Sasaki T., Hohenester E. (2000) Matrix Biol. 19, 309–317 [DOI] [PubMed] [Google Scholar]
- 23.Stetefeld J., Alexandrescu A. T., Maciejewski M. W., Jenny M., Rathgeb-Szabo K., Schulthess T., Landwehr R., Frank S., Rüegg M. A., Kammerer R. A. (2004) Structure 12, 503–515 [DOI] [PubMed] [Google Scholar]
- 24.Tseng C. N., Zhang L., Cascio M., Wang Z. Z. (2003) J. Biol. Chem. 278, 17236–17245 [DOI] [PubMed] [Google Scholar]
- 25.Sidhu S. S., Kossiakoff A. A. (2007) Curr. Opin. Chem. Biol. 11, 347–354 [DOI] [PubMed] [Google Scholar]
- 26.Hunter W. M., Greenwood F. C. (1964) Biochem. J. 91, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deyst K. A., McKechnie B. A., Fallon J. R. (1998) Brain Res. Dev. Brain Res. 110, 185–191 [DOI] [PubMed] [Google Scholar]
- 28.Gesemann M., Brancaccio A., Schumacher B., Rüegg M. A. (1998) J. Biol. Chem. 273, 600–605 [DOI] [PubMed] [Google Scholar]
- 29.Bowen D. C., Sugiyama J., Ferns M., Hall Z. W. (1996) J. Neurosci. 16, 3791–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham B. C., Wells J. A. (1989) Science 244, 1081–1085 [DOI] [PubMed] [Google Scholar]
- 31.Ferns M. J., Campanelli J. T., Hoch W., Scheller R. H., Hall Z. (1993) Neuron 11, 491–502 [DOI] [PubMed] [Google Scholar]
- 32.Alexandrescu A. T., Maciejewski M. W., Rüegg M. A., Engel J., Kammerer R. A. (2001) J. Biomol. NMR 20, 295–296 [DOI] [PubMed] [Google Scholar]
- 33.Koirala S., Reddy L. V., Ko C. P. (2003) J. Neurocytol. 32, 987–1002 [DOI] [PubMed] [Google Scholar]
- 34.Burgess R. W., Dickman D. K., Nunez L., Glass D. J., Sanes J. R. (2002) J. Neurochem. 83, 271–284 [DOI] [PubMed] [Google Scholar]
- 35.Dobbins G. C., Luo S., Yang Z., Xiong W. C., Mei L. (2008) Mol. Brain 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shadiack A. M., Nitkin R. M. (1991) J. Neurobiol. 22, 617–628 [DOI] [PubMed] [Google Scholar]
- 37.Ramseger R., White R., Kröger S. (2009) J. Biol. Chem. 284, 7697–7705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pato C., Stetzkowski-Marden F., Gaus K., Recouvreur M., Cartaud A., Cartaud J. (2008) Chem. Biol. Interact. 175, 64–67 [DOI] [PubMed] [Google Scholar]
- 39.Stetzkowski-Marden F., Gaus K., Recouvreur M., Cartaud A., Cartaud J. (2006) J. Lipid Res. 47, 2121–2133 [DOI] [PubMed] [Google Scholar]
- 40.Gingras J., Rassadi S., Cooper E., Ferns M. (2007) Dev. Neurobiol. 67, 521–534 [DOI] [PubMed] [Google Scholar]
- 41.Gingras J., Spicer J., Altares M., Zhu Q., Kuchel G. A., Ferns M. (2005) Cell Tissue Res. 320, 115–125 [DOI] [PubMed] [Google Scholar]
- 42.Thomas W. S., O'Dowd D. K., Smith M. A. (1993) Dev. Biol. 158, 523–535 [DOI] [PubMed] [Google Scholar]
- 43.Khan A. A., Bose C., Yam L. S., Soloski M. J., Rupp F. (2001) Science 292, 1681–1686 [DOI] [PubMed] [Google Scholar]
- 44.Dunker A. K., Silman I., Uversky V. N., Sussman J. L. (2008) Curr. Opin. Struct. Biol. 18, 756–764 [DOI] [PubMed] [Google Scholar]
- 45.Bhalla J., Storchan G. B., MacCarthy C. M., Uversky V. N., Tcherkasskaya O. (2006) Mol. Cell. Proteomics 5, 1212–1223 [DOI] [PubMed] [Google Scholar]
- 46.Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 47.Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 48.Sreerama N., Venyaminov S. Y., Woody R. W. (2001) Anal. Biochem. 299, 271–274 [DOI] [PubMed] [Google Scholar]
- 49.Scotton P., Bleckmann D., Stebler M., Sciandra F., Brancaccio A., Meier T., Stetefeld J., Ruegg M. A. (2006) J. Biol. Chem. 281, 36835–36845 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.