Abstract
Herpes simplex virus-1 (HSV-1) is a large dsDNA virus that encodes its own DNA replication machinery and other enzymes involved in DNA transactions. We recently reported that the HSV-1 DNA polymerase catalytic subunit (UL30) exhibits apurinic/apyrimidinic and 5′-deoxyribose phosphate lyase activities. Moreover, UL30, in conjunction with the viral uracil DNA glycosylase (UL2), cellular apurinic/apyrimidinic endonuclease, and DNA ligase IIIα-XRCC1, performs uracil-initiated base excision repair. Base excision repair is required to maintain genome stability as a means to counter the accumulation of unusual bases and to protect from the loss of DNA bases. Here we show that the HSV-1 UL2 associates with the viral replisome. We identified UL2 as a protein that co-purifies with the DNA polymerase through numerous chromatographic steps, an interaction that was verified by co-immunoprecipitation and direct binding studies. The interaction between UL2 and the DNA polymerase is mediated through the UL30 subunit. Moreover, UL2 co-localizes with UL30 to nuclear viral prereplicative sites. The functional consequence of this interaction is that replication of uracil-containing templates stalls at positions −1 and −2 relative to the template uracil because of the fact that these are converted into non-instructional abasic sites. These findings support the existence of a viral repair complex that may be capable of replication-coupled base excision repair and further highlight the role of DNA repair in the maintenance of the HSV-1 genome.
Keywords: DNA Damage, DNA Polymerase, DNA Repair, DNA Replication, Herpesvirus, Base Excision Repair, Uracil DNA Glycosylase
Introduction
Herpes simplex virus-1 (HSV-1) is a large double-stranded DNA virus with a genome of ∼152 kbp (reviewed in Refs. 1 and 2). HSV-1 switches between a lytic replication cycle in epithelial cells that is tightly regulated by a temporal order of viral gene expression and a state of latency in sensory neurons during which the genome is proposed to be maintained in heterochromatin form with limited gene expression and no detectable DNA replication (3). Viral DNA replication is dependent on seven viral genes (4, 5). Six of these genes encode components of a typical DNA replication fork. The heterodimeric replicase (reviewed in Refs. 6 and 7) consists of a catalytic subunit encoded by UL30, with DNA polymerase (pol),2 3′-5′-proofreading exonuclease, and RNase H activities (8–11), and a tightly associated DNA-binding protein that confers a high degree of processivity on the pol, encoded by UL42 (11–17). The viral single-strand DNA-binding protein (ICP8) is encoded by UL29 (9), whereas the three subunits of the DNA helicase-primase are encoded by the UL5, UL8, and UL52 genes (18). Finally, UL9 encodes an origin-binding protein with associated helicase activity that targets the viral replication origins (oriS and oriL) to promote unwinding and thereby initiate DNA replication at those sites (19–24). A key step in HSV-1 DNA replication is the intricately orchestrated assembly of the essential viral replication proteins into prereplicative sites adjacent to nuclear domain 10 (ND10) structures that mature into viral replication compartments (25–29).
Because DNA damage may generate mutagenic lesions or physical obstacles that lead to replication fork collapse, efficient and unimpeded DNA replication is dependent on mechanisms that provide and maintain a robust DNA replication template. One mechanism that contributes to genome maintenance is homologous recombination, allowing replication to restart at sites of fork collapse (reviewed in Ref. 30). Indeed, HSV-1 DNA replication is accompanied by vigorous recombination that leads to the formation of large networks of viral DNA replication intermediates (31). ICP8 has been shown to play a major role in mediating recombination reactions including recombination-mediated replication (32–36). In addition, various DNA repair mechanisms may contribute to viral genome maintenance. In this regard, a recent study showed that the replication of HSV-1 containing UV-induced lesions is reduced to various levels in cells with defects in nucleotide excision repair (XP-A), translesion synthesis (pol η), transcription-coupled nucleotide excision repair (CS-A and CS-B), and homologous recombination (Rad 51, Rad 52 and Rad 54), with the most pronounced effect contributed by a deficiency in XP-A, decreasing virus production 106-fold (37). Base excision repair (BER) is a further mechanism that contributes to genome maintenance by removing unusual bases from the DNA and to repair apurinic/apyrimidinic (AP) sites resulting from spontaneous base loss (reviewed in Ref. 38). With respect to HSV-1, a previous study showed that viral DNA from infected cultured fibroblasts contains a steady state of 2.8–5.9 AP sites per viral genome equivalent (39). Because AP sites are non-instructional, the failure to repair such sites would terminate viral replication. Indeed, UL30 cannot replicate beyond a model AP site (tetrahydrofuran residue) (39), indicating a requirement for BER to process such lesions. HSV-1 encodes several enzymes that would safeguard from the accumulation of unusual bases, namely a uracil DNA glycosylase (UDG), encoded by UL2, as well as a dUTPase, encoded by UL50, to reduce the pool of dUTP and prevent misincorporation by the viral pol (40, 41). Moreover, we recently showed that UL30 possesses both AP and 5′-deoxyribose phosphate lyase activities (42). Furthermore, UL2 and UL30 in combination with cellular AP endonuclease and DNA ligase IIIα-XRCC1 are capable of mediating uracil-initiated BER in vitro (43). Therefore, HSV-1 has a means of surveying and repairing DNA damage, specifically for the removal of uracil and the repair of AP sites, to prevent mutagenesis and replication fork collapse and to ensure the availability of a robust replication template.
Here we show that the HSV-1 UDG, UL2, associates with the viral replisome via an interaction with UL30. This interaction supports the notion of a viral repair complex that may be involved in replication-coupled BER to ensure genome maintenance during lytic replication and in the emergence of the virus from neuronal latency. These scenarios will be discussed.
EXPERIMENTAL PROCEDURES
Enzymes and Reagents
HSV-1 UL30 was expressed in Spodoptera frugiperda cells and purified as described (44). The experiments described here used UL30 fraction V. HSV-1 UL2 was purified from Escherichia coli as a thioredoxin and V5-tagged fusion protein (UL2-Trx-V5) as described (43). E. coli pol I, bacteriophage T4 polynucleotide kinase, and bovine serum albumin (BSA) were obtained from New England Biolabs. Sequencing grade porcine trypsin was obtained from Promega. Unlabeled deoxyribonucleoside 5′-triphosphates (disodium salts) were purchased from GE Healthcare. [γ-32P]ATP (3,000 Ci/mmol) and [α-32P]dCTP (3,000 Ci/mmol) were from PerkinElmer Life Sciences. Deoxy-[5-3H]uridine triphosphate (18.5 Ci/mmol) was from Amersham Biosciences. The anti-UL30 (SUR60) and anti-UL42 (SUR33)-specific rabbit sera were as described previously (44). Anti-V5, Alexa Fluor 488 (green) goat anti-mouse, and Alexa Fluor 568 (red) goat anti-rabbit IgG were obtained from Invitrogen. Goat anti-rabbit and goat anti-mouse IgG conjugated to HRP were purchased from ICN and Sigma, respectively. Phosphonoacetic acid (PAA) and EZview red protein A affinity gel were purchased from Sigma. Fifty μg of purified UL30 and 110 μg of aldolase (Sigma) were covalently coupled to agarose beads using the MicroLink (AminoLink plus micro immobilization) protein coupling kit from Pierce as recommended by the manufacturer.
Nucleic Acids
Oligonucleotides PBAZ3 (31-mer), PBAZ4 (16-mer,) and PBAZ7 (31-mer) were as described previously (42, 43). PBAZ10 (31-mer) was synthesized by Operon Biotechnologies and is identical to PBAZ3 except that the T at position 9 was replaced with a U. Where indicated, oligonucleotides were 5′-32P-labeled with T4 polynucleotide kinase and [γ-32P]ATP followed by removal of unincorporated nucleotides using Microspin G-25 columns (GE Healthcare). Primer templates (PBAZ4:PBAZ3 and PBAZ4:PBAZ10) were constructed by annealing 5′-32P-labeled PBAZ4 with a 1.25-fold molar excess of unlabeled oligonucleotide in 20 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 0.1 m NaCl by heating to 95 °C for 5 min followed by gradual cooling to <30 °C. Activated calf thymus DNA was purchased from Sigma. [3H]Uracil-containing DNA was prepared as described in the supplemental material.
Plasmids encoding the seven essential HSV-1 replication genes under the control of the CMV immediate early promoter were those described by Heilbronn and zur Hausen (45) and kindly provided by Dr. Gary Hayward (Johns Hopkins University). The HSV-1 oriS-containing plasmid pGEM822 was as described previously (46). pcDNA3.1D/V5-His-/lacZ was provided by Invitrogen.
pcDNA-UL2 in which UL2 is cloned downstream of the CMV immediate early promoter as a V5-tagged protein was constructed by inserting the UL2 open reading frame (encoding a 36.3-kDa 334 amino acid protein starting at the first methionine of the UL2 gene) into pcDNA3.1D/V5-His-TOPO (Invitrogen). The insert was amplified by PCR from HSV-1 strain KOS genomic DNA using the GC-RICH PCR system from Roche Applied Science and purified for cloning as described previously for pET102-UL2 (43). Progeny plasmid was screened for the presence of the correct insert and confirmed by DNA sequencing of both strands using the T7 promoter and T7 transcription termination/reverse primers.
Purification of HSV-1 pol
The HSV-1 pol was purified from virus-infected cells as described previously (11). After the gel filtration step on Superose 12, pol fractions were pooled, diluted to 50 mm NaCl, and injected onto a MonoQ HR 5/5 column (GE Healthcare) that had been equilibrated with 20 mm HEPES-NaOH, pH 7.5, 10% glycerol, 0.2 mm EDTA, 1 mm dithiothreitol (DTT), and protease inhibitors (100 μm phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 0.7 μg/ml pepstatin). The column was developed at 0.5 ml/min with a 10-ml linear gradient of 50 mm–0.5 m NaCl, and 0.2-ml fractions were collected. The peak of pol activity, eluting at ∼0.45 m NaCl, was pooled, diluted to 50 mm NaCl, and injected onto a 1-ml Resource Q column (GE Healthcare) that had been equilibrated with 20 mm HEPES-NaOH, pH 7.5, 10% glycerol, 0.2 mm EDTA, 1 mm DTT, and protease inhibitors as above. The column was developed at 0.5 ml/min with a 15-ml linear gradient of 50 mm–0.6 m NaCl, and 0.2-ml fractions were collected. The peak of protein elution was analyzed by SDS-PAGE and for UDG and pol activity (see Fig. 1). The concentration of pol was calculated using an extinction coefficient of 139,070 m−1 cm−1 at 280 nm calculated from the amino acid sequences of UL30 and UL42 (47). It should be noted that the specific activity of nucleotide incorporation of 100 nm pol was equivalent to that of 5 nm purified recombinant UL30.
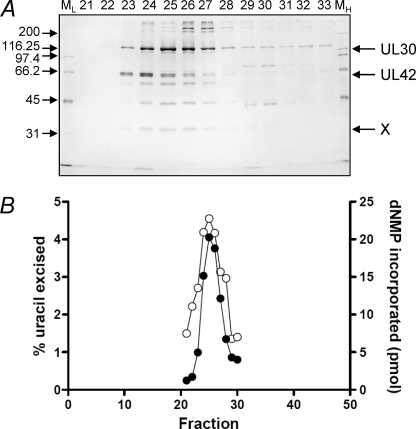
FIGURE 1.
Co-purification of HSV-1 DNA polymerase and uracil DNA glycosylase. A, fractions eluting from the Resource Q column were resolved by 10% SDS-PAGE followed by silver staining. The positions of markers (ML and MH, Bio-Rad low and high range standards, respectively), UL30 and UL42 are as indicated. X indicates the position of an ∼36-kDa polypeptide. B, the specified fractions were assayed for pol (●) and UDG (○) activities as described under “Experimental Procedures.”
Mass Spectrometry
The species labeled X in Fig. 1A was excised from a Coomassie Blue G-250-stained gel, processed as described in the supplemental material, and analyzed by liquid chromatography electrospray ionization quadrupole time of flight mass spectrometry (LC-ESI-QTOF-MS) by the Mass Spectrometry Laboratory of the Department of Chemistry and Biochemistry at Florida Atlantic University. The results were analyzed using the Mascot search engine, which revealed, with high probability-based Mowse scores, peptides derived from UL30, UL42, and trypsin, as well as two HSV-1 UL2 peptides: ANVPPPPSLR and VVIIGQDPYHHPGQAHGLAFSVR.
Cells, Transfection, Infection, Extracts, Protein Binding, and Indirect Immunofluorescence Microscopy
Vero and 293 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal calf serum (Lonza), 1% penicillin-streptomycin, and 1% l-glutamine at 37 °C and 5% CO2. Transfections were performed using Lipofectamine 2000 (Invitrogen). Infections with HSV-1 were with strain KOS. Details of the transfections, extract preparation, and protein binding experiments are described in the supplemental material.
Co-localization of UL2 and UL30 was examined by indirect immunofluorescence microscopy as described in the supplemental material. Briefly, Vero cells grown on coverslips were transfected with the indicated combinations of plasmids, treated with PAA, fixed, and probed with anti-V5 or anti-UL30 followed by detection with Alexa Fluor-conjugated secondary antibodies. The cells were observed using a Leica DM IRB inverted microscope at the University of Miami Analytical Imaging Core Facility.
Enzyme Assays
pol activity eluting from the Resource Q column was assayed in 10-μl reactions containing 20 mm HEPES-NaOH, pH 7.5, 0.1 m NaCl, 1 mm DTT, 5% glycerol, 2.5 mm MgCl2, 0.1 mg/ml BSA, 1 μg of activated calf thymus DNA, 50 μm dATP, dGTP, and dTTP, and 20 μm [α-32P]dCTP (∼10 Ci/mmol). After 30 min at 37 °C, incorporation was measured using the DE81 paper binding method (44).
UDG activity eluting from the Resource Q column was assayed in 10-μl reactions containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 1 mm EDTA, and 1 μg of [3H]uracil-containing DNA. After 60 min at 37 °C, the DNA was precipitated with TCA, and acid-soluble radioactivity was determined by scintillation counting.
UDG activity was also determined by measuring alkaline cleavage of oligonucleotide PBAZ7, which contains a U at position 17. Standard reactions (10 μl) contained 25 mm HEPES-NaOH, pH 7.5, 1 mm DTT, 2.5 mm MgCl2, 50 μg/ml BSA, 10 nm 5′-32P-labeled PBAZ7, and the indicated concentrations of protein. After 10 min at 37 °C, NaOH was added to 0.2 m followed by 15 min at 75 °C. Reactions were combined with an equal volume of stop buffer (95% formamide, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol) and heated for 3 min at 75 °C prior to electrophoresis through 16% polyacrylamide-8 m urea gels in glycerol tolerant gel buffer (89 mm Tris, pH 9.0, 28.5 mm taurine and 0.5 mm EDTA). Reaction products were visualized and quantified by storage phosphor analysis with a GE Healthcare Storm 820 using ImageQuant version 5.2. Cleavage is expressed as a percentage of total radioactivity.
Standard primer extension reactions (10 μl) contained 25 mm HEPES-NaOH, pH 7.5, 25 mm NaCl, 1 mm DTT, 2.5 mm MgCl2, 50 μg/ml BSA, 100 μm dNTP, and 10 nm 5′-32P-labeled PBAZ4 annealed to PBAZ3 (T-containing template) or PBAZ10 (U-containing template) as indicated. Reactions were initiated by the addition of protein, incubated for 5 min at 37 °C, and terminated by the addition of 5 μl of stop buffer and then analyzed as described for the UDG assay described above. Primer extension products are expressed as a percentage of total radioactivity.
RESULTS
The HSV-1 Uracil DNA Glycosylase Co-purifies with the Viral DNA Polymerase
In a search for factors that associate with the HSV-1 pol, we examined proteins that co-purified with the viral pol when purified from HSV-1-infected cells. The purification of the HSV-1 pol has been described previously and entails fractionation of nuclear proteins from HSV-1-infected cells (see “Experimental Procedures”). In this study, the final step in the purification was anion exchange chromatography on Resource Q. Fig. 1A shows a silver-stained gel of the polypeptides eluting around the protein peak. An ∼36-kDa species labeled X exhibited a similar elution profile to the UL30 and UL42 subunits and was therefore identified as a candidate co-purifying protein. The identity of this protein was investigated by LC-ESI-QTOF-MS of trypsin-digested protein. This analysis revealed several peptides that are part of the ∼36-kDa HSV-1 UDG, UL2 (see “Experimental Procedures”). Subsequent biochemical analysis showed that UDG and pol activity eluting from the Resource Q column are coincident (Fig. 1B). Co-purification of HSV-1 UDG and pol suggests that UL2 interacts with the viral replicase and prompted further investigations.
The Interaction between UL2 and the HSV-1 pol Is Mediated by the UL30 Subunit
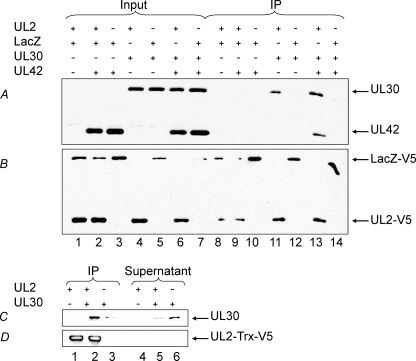
To ascertain whether UL2 indeed interacts with the HSV-1 pol, we investigated co-immunoprecipitation of UL2 and pol in cells that were transfected with various combinations of V5-tagged UL2 or LacZ and UL30 and UL42. Fig. 2, A and B, show that anti-V5 antibody was able to co-precipitate V5-tagged UL2 and UL30 (lane 11). UL30 was not precipitated when UL2 was substituted with V5-tagged LacZ (lane 12). Moreover, the UL42 subunit was not co-precipitated with UL2. However, given the tight association between the UL30 and UL42 subunits, UL42 was co-precipitated by UL2 when UL30 was present (lane 13). These results indicate that the HSV-1 UDG does indeed associate with the pol and that this interaction is mediated via the UL30 subunit.
FIGURE 2.
Co-immunoprecipitation of UL30 with UL2. Transfection, immunoprecipitation with anti-V5, and immunoblotting were performed as described under “Experimental Procedures.” A and B, co-immunoprecipitation of UL30 with UL2 in transfected cells. Aliquots of the indicated input fractions (Input; lanes 1-7) and immunoprecipitates (IP; lanes 8-14) were probed with combined anti-UL30/UL42 rabbit antisera (A) or anti-V5 (B). C and D, co-immunoprecipitation of purified UL30 with purified UL2. Aliquots of the indicated immunoprecipitates (IP; lanes 1-3) and supernatants (Supernatant; lanes 4-6) were probed with anti-UL30 rabbit serum (C) or anti-V5 (D). The combinations of proteins are indicated, as are the positions of UL30, UL42, UL2-V5, UL2-Trx-V5, and LacZ-V5. LacZ-V5 was included as a nonspecific control and used to balance the total amount of DNA in the transfections.
The interaction between UL2 and UL30 was investigated further by co-precipitation of the purified proteins. Fig. 2, C and D, show that anti-V5 antibody was able to precipitate UL30 only in the presence of purified V5-tagged UL2 (lane 2), with a concomitant decrease in the amount of soluble UL30 (lane 5).
Next, we examined whether UL2 could bind to immobilized UL30 when compared with a control protein, in this case aldolase. Fig. 3 shows that UL2 was retained by UL30 but not aldolase beads, eluting at 1 m NaCl (compare lanes 2–4 and lanes 7–9). The physical interactions between UL2 and UL30 demonstrated in these experiments support and confirm the results of the co-immunoprecipitation experiment with transfected cell extracts.
FIGURE 3.
Purified UL2 binds to immobilized UL30. Purified UL2-Trx-V5 was applied to UL30 (lanes 1-4) or aldolase beads (lanes 6-9) as described under “Experimental Procedures.” Flow-through (FT), and consecutive 1 m NaCl eluates (E1, E2, and E3) were analyzed by immunoblotting using anti-V5. The position of UL2-Trx-V5 is as indicated. M (lane 5), purified UL2-Trx-V5 control.
The Interaction between UL2 and pol Occurs in HSV-1-infected Cells
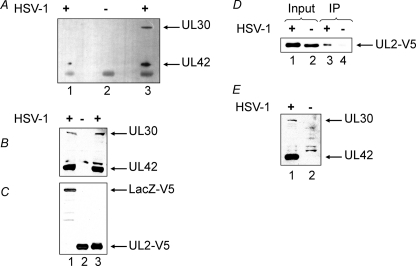
In the first instance, to demonstrate that UL2 interacts with the viral pol in virus-infected cells, we investigated whether UL2 could co-precipitate the subunits of the viral pol in HSV-1-infected cells. Because UL2-specific antibodies were unavailable to us, cells were transfected with either V5-tagged UL2 or V5-tagged LacZ and superinfected with HSV-1. The data in Fig. 4, A–C, show that anti-V5 antibody co-precipitated UL30 and UL42 only in cells that were transfected with V5-tagged UL2 and superinfected with HSV-1 (Fig. 4A, lane 3). No UL30 or UL42 was co-precipitated when UL2 was substituted with V5-tagged LacZ (Fig. 4A, lane 1) or in the presence of V5-tagged UL2 but without HSV-1 infection (Fig. 4A, lane 2).
FIGURE 4.
Co-immunoprecipitation of pol with UL2 in HSV-1-infected cells. Transfection, infection, immunoprecipitation, and immunoblotting were performed as described under “Experimental Procedures.” A–C, immunoprecipitation with anti-V5. A, immunoprecipitates probed with a combined anti-UL30/UL42 rabbit serum. B, input fractions probed with combined anti-UL30/UL42 rabbit sera. C, input fractions probed with anti-V5. For each panel, lane 1 represents aliquots of immunoprecipitates (A) or inputs (B and C) from cells transfected with LacZ-V5 and superinfected with HSV-1. Lane 2 represents aliquots of immunoprecipitates (A) or inputs (B and C) from cells transfected with UL2-V5 and mock-infected with HSV-1. Lane 3 represents aliquots of immunoprecipitates (A) or inputs (B and C) from cells transfected with UL2-V5 and superinfected with HSV-1. D and E, immunoprecipitation with anti-UL42 serum. D, aliquots of the indicated input fractions (Input; lanes 1 and 2) and immunoprecipitates (IP; lanes 3 and 4) were probed with anti-V5. E, aliquots of HSV-1-infected (lane 1) and mock-infected (lane 2) input fractions were probed with combined anti-UL30/UL42 rabbit sera. Infection with HSV-1 is indicated with a +. The positions of UL30, UL42, LacZ-V5, and UL2-V5 are as indicated.
To further demonstrate this interaction in virus-infected cells, we examined the ability of anti-UL42 serum to precipitate UL2 in cells that were transfected with V5-tagged UL2 and superinfected with HSV-1. Fig. 4, D and E, show that anti-UL42 serum only co-precipitated UL2 in HSV-1-infected cells (Fig. 4D, lane 3) in which UL42 and UL30 were expressed (Fig. 4E, lane 1). Similar experiments using anti-UL30 serum were unsuccessful due to the inability of these antibodies to immunoprecipitate UL30.
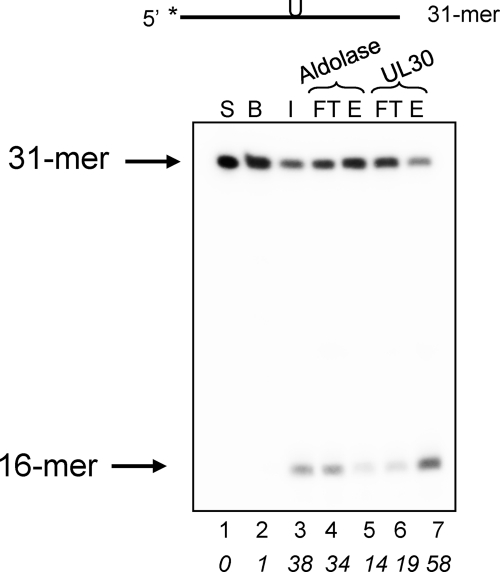
Next, we investigated whether UL2 produced in HSV-1-infected cells could bind to immobilized UL30. In this experiment, extract was prepared from HSV-1-infected cells and applied to UL30 or aldolase beads (Fig. 5). Because UL2-specific antibodies were unavailable to us, UDG activity was determined biochemically by measuring alkaline cleavage of abasic DNA generated through UDG action. The data show that the extract exhibited UDG activity (Fig. 5, lane 3). Importantly, four times more UDG activity was eluted from the UL30 beads than from the aldolase control beads (compare lanes 5 and 7), indicating that UDG activity, and by inference UL2, is specifically retained by UL30. This conclusion is supported by the finding that uninfected cell extracts exhibited a much lower specific activity for UDG that was not significantly bound to UL30 beads (data not shown). Collectively, the findings in this section indicate that UL2 and UL30 interact in the context of an HSV-1 infection.
FIGURE 5.
UDG activity in HSV-1-infected cell extract binds to immobilized UL30. Two hundred-μl aliquots of extract from HSV-1-infected cells were applied to aldolase (lanes 4 and 5) or UL30 (lanes 6 and 7) beads, and the input (I), flow-through (FT), and 1 m NaCl eluates (E) were assayed for UDG activity using 4 nm 5′-32P labeled PBAZ7 as described under “Experimental Procedures.” S, substrate; B, substrate with buffer. The positions of 31-mer substrate and 16-mer product are as indicated. The numbers in italics below each lane represent the percentage of cleavage of substrate. The asterisk denotes the position of the 5′ [32P] label.
UL2 and UL30 Co-localize to Viral Prereplicative Sites
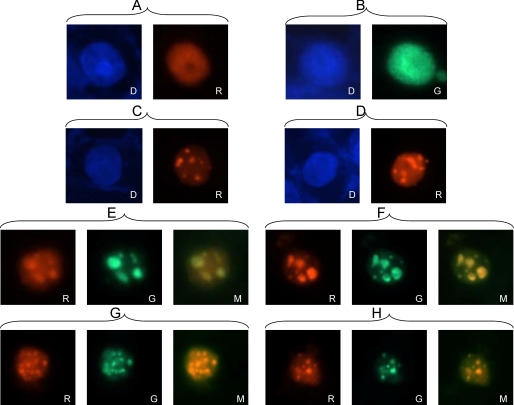
It is well documented that the essential viral replication proteins organize into discrete nuclear foci, termed prereplicative sites, that mature into viral DNA replication compartments (25–29). Although the HSV-1 pol is not a critical initiator of this assembly process, it is recruited to these sites prior to the onset of DNA replication (27, 48). We took advantage of prereplicative site formation to investigate whether UL2 also localizes to these foci. Therefore, Vero cells were transfected with the seven essential viral replication genes, a plasmid containing oriS and PAA, to arrest DNA replication at the stage of prereplicative sites. Fig. 6 shows that both UL30 and UL2, when expressed individually, localized throughout the nuclei of transfected cells (Fig. 6, A and B). Fig. 6, C and D, recapitulate previous findings of the seven essential replication proteins assembling at prereplicative sites in the presence of an oriS-containing plasmid and treatment with PAA. In this case, prereplicative sites were visualized by immunofluorescence using anti-UL30 serum and an Alexa Fluor 568-labeled secondary antibody (red fluorescence). We next examined whether UL2 co-localizes to the prereplicative sites by including V5-tagged UL2 with the essential replication proteins and visualizing its localization with an Alexa Fluor 488-labeled secondary antibody (green fluorescence). Fig. 6, E–H, are four examples that show that at this resolution, the red (UL30) and green (UL2) fluorescent foci are coincident, as indicated in the merged images. Therefore, UL2 appears to associate with the viral replication apparatus at prereplicative sites, presumably via its interaction with UL30.
FIGURE 6.
Co-localization of UL30 and UL2 to viral prereplicative sites. Indirect immunofluorescence microscopy of transfected cells was performed as described under “Experimental Procedures.” D, DAPI staining; R, red fluorescence (UL30); G, green fluorescence (UL2); M, merged red/green fluorescence. A, cells transfected with UL30. B, cells transfected with UL2. C and D, cells transfected with UL30, the remaining six essential viral replication proteins, oriS-containing plasmid, and PAA treatment. E–H, cells transfected with UL2, UL30, the remaining six essential viral replication proteins, oriS-containing plasmid, and PAA treatment.
The Association of UL2 with the HSV-1 pol Causes DNA Synthesis to Stall at Resulting Abasic Sites
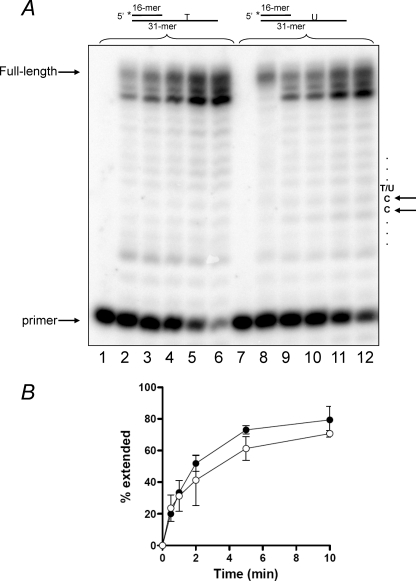
To investigate the functional consequences of the interaction between UL2 and UL30, we examined the ability of the HSV-1 pol to utilize templates that contained either a thymine or a uracil residue in in vitro primer extension reactions. In the first instance, we investigated whether a template uracil impacted primer extension by UL30 alone. Fig. 7 shows that there was no significant difference in the ability of UL30 to extend a primer up to and beyond a site-specific uracil when compared with the identical substrate containing a thymine at that position. Importantly, no pausing or stalling at the template uracil or upstream of it was observed. It should be noted that this particular template 31-mer was not efficiently extended to the end, instead generating a 29-mer as the primary extension product. Identical products were formed when the template was extended with Klenow or exonuclease-deficient Klenow pol (data not shown). Therefore, this appears to be a characteristic of this particular sequence and is inconsequential to our analysis.
FIGURE 7.
Template uracil does not affect DNA synthesis by UL30. Primer extension reactions were performed with 4 nm UL30 for the times indicated on either the T-containing (lanes 1–6) or the U-containing templates (lanes 7–12) as described under “Experimental Procedures.” A, reaction products. Lanes 1-6 and 7-12, 0, 0.5, 1, 2, 5, and 10 min of incubation. The positions of primer and full-length product and the relevant sequence of the template are as indicated. The asterisk denotes the position of the 5′ [32P] label. B, quantitation of the data from three independent experiments. ●, T-containing template; ○, U-containing template. Error bars indicate S.E.
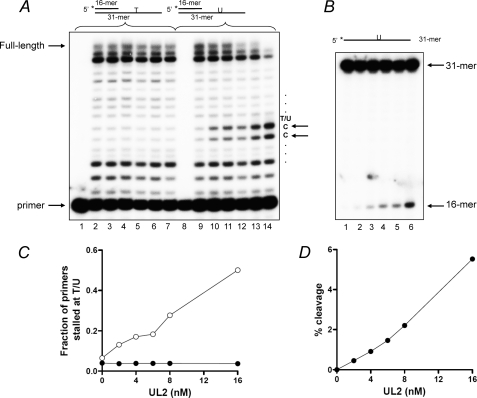
Importantly, when primer extension reactions were performed in the presence of increasing concentrations of UL2, there was notable stalling at the two positions preceding the template uracil (positions −1 and −2) but not with the template thymine (Fig. 8A, compare lanes 7 and 14). This observation is presumably due to the conversion of uracil to abasic sites, which were previously shown to be non-instructional for UL30 (39), resulting in futile cycles of incorporation and excision around that site, manifested by the two prominent extension products at −1 and −2. A similar observation was also made with UL30 attempting to bypass bulky cisplatin-induced d(GpG) and N-2-acetylaminofluorene adducts, resulting in the accumulation of extension products at positions −1 to −3, preceding the lesion (49, 50). It should also be noted that the level of stalling correlates well with the amount of UDG activity observed with the concentrations of UL2 used in this experiment (Fig. 8, B and D). Moreover, quantitation of the data shows that at the highest concentration of UL2 (16 nm), of the primers extended (∼30% of total), 50% stalled at positions −1 and −2 with the uracil-containing template, whereas there was no stalling with the thymine-containing template (Fig. 8C).
FIGURE 8.
UL2 causes UL30 to stall upstream of a template uracil. Primer extension reactions were performed with 4 nm UL30 and the indicated concentrations of UL2 on either the T-containing (lanes 1–7) or the U-containing templates (lanes 8–14) as described under “Experimental Procedures.” A, primer extension reactions. Lanes 1 and 8, substrate only; lanes 2-7 and 9-14, reactions with 0, 2, 4, 6, 8, and 16 nm UL2. B, UDG activity of UL2 was assayed using the 5′-32P labeled 31-mer as described under “Experimental Procedures.” Lanes 1-6, 0, 2, 4, 6, 8, and 16 nm UL2. The positions of primer, full-length product, 31-mer substrate, and 16-mer product are as indicated. The asterisk denotes the position of the 5′ [32P] label. C, quantitation of the fraction of primers stalled upstream (positions −1 and −2) of template T (●) or U (○) from the data in A. D, quantitation of the data shown in B.
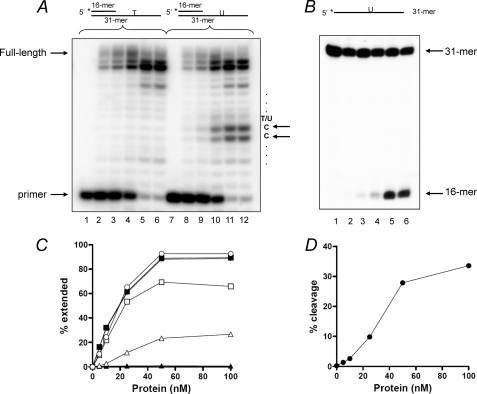
The consequence of UL2 associating with the viral replisome on the replication of uracil-containing templates was further examined using the viral pol purified from HSV-1-infected cells, which contains UL30 and UL42 in complex with UL2. Fig. 9 shows that with the thymine-containing template, increasing concentrations of the UL30-UL42-UL2 complex led to efficient extension of the primer, generating the 29-mer primary product without stalling at the template thymine (lanes 1-6). However, with the uracil-containing template, stalling at positions −1 and −2 was evident with increasing concentrations of the complex (lanes 7-12). The degree of stalling correlated well with the UDG activity of the complex (Fig. 9, B and D). Quantitation of the data shows that although the total amount of primer extended is the same for both the thymine-containing and the uracil-containing templates, only ∼66% of the primers are extended beyond the template uracil with ∼33% stalling at positions −1 and −2, whereas 100% of the extended primers reach beyond the template thymine (Fig. 9C). Overall, the experiments described in this section demonstrate that the UDG activity of UL2, either via the addition of purified UL2 or contained within the pol purified from HSV-1-infected cells, converts template uracil residues into abasic sites that are non-instructional for the pol, causing it to stall and leading to futile cycles of incorporation and excision at positions −1 and −2 with respect to the adduct.
FIGURE 9.
DNA polymerase from HSV-1-infected cells stalls upstream of template uracil. Primer extension reactions were performed with the indicated concentration of HSV-1 DNA polymerase-UL2 complex purified from HSV-1-infected cells on either the T-containing (lanes 1–6) or the U-containing templates (lanes 7–12) as described under “Experimental Procedures.” A, primer extension reactions. Lanes 1-6 and 7-12, reactions with 0, 5, 10, 25, 50, and 100 nm protein. B, UDG activity of UL2 was assayed using the 5′-32P labeled 31-mer as described under “Experimental Procedures.” Lanes 1-6, 0, 5, 10, 25, 50, and 100 nm protein. The positions of primer, full-length product, 31-mer substrate, and 16-mer product are as indicated. The asterisk denotes the position of the 5′ [32P] label. C, quantitation of the data in A. Filled symbols, T-containing templates; open symbols, U-containing templates. ●, ○, total extension; ■, □, extension beyond template T or U; ▴, ▵, stalling upstream (positions −1 and −2) of template T or U. D, quantitation of the data shown in B.
DISCUSSION
The findings reported here demonstrate that HSV-1 UDG (UL2) associates with the viral replisome via an interaction with the catalytic subunit of the pol (UL30) both in vitro and in vivo in the context of a viral infection and by its localization to viral nuclear prereplicative sites. The consequences of this interaction are that DNA synthesis by the pol stalls at template uracils due to their conversion to non-instructional abasic sites.
Initially, our studies were motivated by a search for factors that associate with the pol to promote coupled leading and lagging strand synthesis at the viral DNA replication fork. UL2 was therefore a candidate factor with a potential effect on pol activity. Hence, we examined the effect of UL2 on both the processivity and the rate of UL30 in the presence and absence of UL42. However, our experiments did not show any effect of UL2 on DNA synthesis. Moreover, we were unable to show that UL30 had any specific effect on the UDG activity of UL2. This is in contrast to the situation in vaccinia virus in which an interaction between the pol and UDG has also been demonstrated (51, 52). In this virus, the UDG is essential independently of its function as a UDG (53). In fact, in support of our initial hypothesis, it was shown that the UDG conferred increased processivity on the pol, suggesting that it performs a critical replication function rather than as an essential BER component (51, 52, 54).
The interaction between UDG and pol has also been described for two other herpesviruses, human cytomegalovirus and Varicella-Zoster virus (55–57). However, in both reports, the UDG associates not with the catalytic subunit of the pol, as shown here for HSV-1, but rather with the pol accessory subunit (human cytomegalovirus UL44 and Varicella-Zoster virus Orf 16). Consistent with our findings, Strang and Coen (58) recently described that the interaction between the human cytomegalovirus UDG (UL114) and pol is also mediated via the pol catalytic subunit (UL57) in vitro, although in vivo the interaction obviously entails the pol holoenzyme. In a manner somewhat analogous to vaccinia virus, in human cytomegalovirus, mutants in the UDG (UL114) exhibit a replication defect, with a reduction in early phase DNA synthesis and an impairment in the transition to late phase DNA replication (55, 59, 60). In fact, Mocarski and colleagues (60) proposed a model in which the UDG introduces random cleavage sites in the viral genome that act as pol-priming sites to allow the transition from early stage θ to late stage σ or rolling circle replication.
In slight contrast to UDG performing a direct DNA replication function, in human cells, the nuclear UDG (UNG2) and other BER components have been shown to associate with the replication fork apparatus including RP-A, proliferating cell nuclear antigen, MCM7, pol α, pol δ, pol ϵ, and DNA ligase 1 (61). Succinctly, in this case, the authors propose that BER and replication complexes interact to coordinate these two processes. Similarly, we propose that the association of UL2 with the viral replisome reported here is indicative of coupling of BER with the replication process. This notion is supported by our recent reports in which we demonstrated that UL30 possesses AP and 5′-deoxyribose phosphate lyase activities (42) and demonstrated the capacity of certain HSV-1 enzymes to promote BER in vitro (43).
It is easy to envision a scenario in which BER is necessary to maintain the viral genome both during lytic replication and as the virus emerges from latency in postmitotic neurons during which the quiescent genome is susceptible to spontaneous base loss and deamination. Hence, BER would be required to repair AP sites, which occur at a frequency of 2.8–5.9 per viral genome (39). BER would also be required to prevent uracil accumulation, which has been shown to impact the recognition of the viral origins by the origin-binding protein (62) and to prevent mutation fixation. In the context of reactivation from latency, BER may play a critical role to provide a robust replication template following potentially long periods of quiescence. In this regard, UL2 mutants exhibit reduced neurovirulence and a decreased reactivation frequency (63). Thus, BER action in HSV-1 may be important for viral reactivation following quiescence in neuronal cells that exhibit lower levels of cellular genome surveillance (64).
In summary, our data show that the HSV-1 UDG interacts with the viral replisome. Although it is not unexpected that UL2 and UL30 interact as part of a BER complex given that UL30 possesses both lyase and pol activities, this association is also indicative of a scenario in which a DNA replication-BER complex is engaged in concurrent DNA replication and genome surveillance/repair. This scenario has been proposed for archaea in which pol show the ability to recognize template uracil as a means of genome surveillance (65). Excision of unusual bases, in this case uracil, must be tightly coupled to the repair of resulting AP sites to prevent fork stalling because this would otherwise necessitate repair via homologous recombination. Indeed, the abundance of recombination intermediates seen during HSV-1 replication (31) and the capacity of the virus to perform recombination-mediated replication (35) may well derive from this. The findings reported here in combination with our recent work on viral BER (43) set the stage for future work to investigate whether additional repair factors associate with the replisome and to establish a functional relationship between DNA replication and repair in HSV-1.
This work was supported, in whole or in part, by National Institutes of Health Grant GM62643 (to P. E. B.)
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental procedures.
- pol
- polymerase
- AP
- apurinic/apyrimidinic
- BER
- base excision repair
- PAA
- phosphonoacetic acid
- UDG
- uracil DNA glycosylase.
REFERENCES
- 1.Boehmer P. E., Nimonkar A. V. (2003) IUBMB Life 55, 13–22 [DOI] [PubMed] [Google Scholar]
- 2.Boehmer P. E., Villani G. (2003) Prog. Nucleic Acid Res. Mol. Biol. 75, 139–171 [DOI] [PubMed] [Google Scholar]
- 3.Knipe D. M., Cliffe A. (2008) Nat. Rev. Microbiol. 6, 211–221 [DOI] [PubMed] [Google Scholar]
- 4.McGeoch D. J., Dalrymple M. A., Dolan A., McNab D., Perry L. J., Taylor P., Challberg M. D. (1988) J. Virol. 62, 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C. A., Nelson N. J., McGeoch D. J., Challberg M. D. (1988) J. Virol. 62, 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehmer P. E., Lehman I. R. (1997) Annu. Rev. Biochem. 66, 347–384 [DOI] [PubMed] [Google Scholar]
- 7.Lehman I. R., Boehmer P. E. (1999) J. Biol. Chem. 274, 28059–28062 [DOI] [PubMed] [Google Scholar]
- 8.Purifoy D. J., Lewis R. B., Powell K. L. (1977) Nature 269, 621–623 [DOI] [PubMed] [Google Scholar]
- 9.Quinn J. P., McGeoch D. J. (1985) Nucleic Acids Res. 13, 8143–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knopf K. W. (1979) Eur. J. Biochem. 98, 231–244 [DOI] [PubMed] [Google Scholar]
- 11.Crute J. J., Lehman I. R. (1989) J. Biol. Chem. 264, 19266–19270 [PubMed] [Google Scholar]
- 12.Vaughan P. J., Purifoy D. J., Powell K. L. (1985) J. Virol. 53, 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo M. L., Jackwood D. H., Murphy M., Marsden H. S., Parris D. S. (1988) J. Virol. 62, 2874–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez T. R., Lehman I. R. (1990) J. Biol. Chem. 265, 11227–11232 [PubMed] [Google Scholar]
- 15.Gallo M. L., Dorsky D. I., Crumpacker C. S., Parris D. S. (1989) J. Virol. 63, 5023–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottlieb J., Marcy A. I., Coen D. M., Challberg M. D. (1990) J. Virol. 64, 5976–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komazin-Meredith G., Mirchev R., Golan D. E., van Oijen A. M., Coen D. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10721–10726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crute J. J., Tsurumi T., Zhu L. A., Weller S. K., Olivo P. D., Challberg M. D., Mocarski E. S., Lehman I. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 2186–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruckner R. C., Crute J. J., Dodson M. S., Lehman I. R. (1991) J. Biol. Chem. 266, 2669–2674 [PubMed] [Google Scholar]
- 20.Fierer D. S., Challberg M. D. (1992) J. Virol. 66, 3986–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makhov A. M., Boehmer P. E., Lehman I. R., Griffith J. D. (1996) EMBO J. 15, 1742–1750 [PMC free article] [PubMed] [Google Scholar]
- 22.He X., Lehman I. R. (2000) J. Virol. 74, 5726–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aslani A., Olsson M., Elias P. (2002) J. Biol. Chem. 277, 41204–41212 [DOI] [PubMed] [Google Scholar]
- 24.Makhov A. M., Lee S. S., Lehman I. R., Griffith J. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 898–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan M. P., Chen L. B., Knipe D. M. (1984) Cell 36, 857–868 [DOI] [PubMed] [Google Scholar]
- 26.Lukonis C. J., Weller S. K. (1996) J. Virol. 70, 1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liptak L. M., Uprichard S. L., Knipe D. M. (1996) J. Virol. 70, 1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkham J., Coen D. M., Weller S. K. (1998) J. Virol. 72, 10100–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor T. J., McNamee E. E., Day C., Knipe D. M. (2003) Virology 309, 232–247 [DOI] [PubMed] [Google Scholar]
- 30.Budzowska M., Kanaar R. (2009) Cell Biochem. Biophys. 53, 17–31 [DOI] [PubMed] [Google Scholar]
- 31.Severini A., Scraba D. G., Tyrrell D. L. (1996) J. Virol. 70, 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nimonkar A. V., Boehmer P. E. (2002) J. Biol. Chem. 277, 15182–15189 [DOI] [PubMed] [Google Scholar]
- 33.Nimonkar A. V., Boehmer P. E. (2003) J. Biol. Chem. 278, 9678–9682 [DOI] [PubMed] [Google Scholar]
- 34.Nimonkar A. V., Boehmer P. E. (2003) Nucleic Acids Res. 31, 5275–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nimonkar A. V., Boehmer P. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuven N. B., Willcox S., Griffith J. D., Weller S. K. (2004) J. Mol. Biol. 342, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muylaert I., Elias P. (2010) J. Biol. Chem. 285, 13761–13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes D. E., Lindahl T. (2004) Annu. Rev. Genet. 38, 445–476 [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y., Song L., Stroud J., Parris D. S. (2008) DNA Repair 7, 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullaney J., Moss H. W., McGeoch D. J. (1989) J. Gen. Virol. 70, 449–454 [DOI] [PubMed] [Google Scholar]
- 41.Williams M. V. (1984) J. Biol. Chem. 259, 10080–10084 [PubMed] [Google Scholar]
- 42.Bogani F., Boehmer P. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11709–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogani F., Chua C. N., Boehmer P. E. (2009) J. Biol. Chem. 284, 16784–16790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boehmer P. E. (1996) Methods Enzymol. 275, 16–35 [DOI] [PubMed] [Google Scholar]
- 45.Heilbronn R., zur Hausen H. (1989) J. Virol. 63, 3683–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sampson D. A., Arana M. E., Boehmer P. E. (2000) J. Biol. Chem. 275, 2931–2937 [DOI] [PubMed] [Google Scholar]
- 47.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 48.Goodrich L. D., Schaffer P. A., Dorsky D. I., Crumpacker C. S., Parris D. S. (1990) J. Virol. 64, 5738–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villani G., Tanguy Le Gac N., Wasungu L., Burnouf D., Fuchs R. P., Boehmer P. E. (2002) Nucleic Acids Res. 30, 3323–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arana M. E., Song L., Tanguy Le Gac N., Parris D. S., Villani G., Boehmer P. E. (2004) DNA Repair 3, 659–669 [DOI] [PubMed] [Google Scholar]
- 51.Stanitsa E. S., Arps L., Traktman P. (2006) J. Biol. Chem. 281, 3439–3451 [DOI] [PubMed] [Google Scholar]
- 52.Ishii K., Moss B. (2002) Virology 303, 232–239 [DOI] [PubMed] [Google Scholar]
- 53.De Silva F. S., Moss B. (2003) J. Virol. 77, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Silva F. S., Moss B. (2008) Virol. J. 5, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prichard M. N., Lawlor H., Duke G. M., Mo C., Wang Z., Dixon M., Kemble G., Kern E. R. (2005) Virol. J. 2, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., Fossum E., Haas J. (2006) Science 311, 239–242 [DOI] [PubMed] [Google Scholar]
- 57.Ranneberg-Nilsen T., Dale H. A., Luna L., Slettebakk R., Sundheim O., Rollag H., Bjørås M. (2008) J. Mol. Biol. 381, 276–288 [DOI] [PubMed] [Google Scholar]
- 58.Strang B. L., Coen D. M. (2010) J. Gen. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prichard M. N., Duke G. M., Mocarski E. S. (1996) J. Virol. 70, 3018–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Courcelle C. T., Courcelle J., Prichard M. N., Mocarski E. S. (2001) J. Virol. 75, 7592–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parlanti E., Locatelli G., Maga G., Dogliotti E. (2007) Nucleic Acids Res. 35, 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Focher F., Verri A., Verzeletti S., Mazzarello P., Spadari S. (1992) Chromosoma 102, S67–S71 [DOI] [PubMed] [Google Scholar]
- 63.Pyles R. B., Thompson R. L. (1994) J. Virol. 68, 4963–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson D. M., 3rd, McNeill D. R. (2007) Neuroscience 145, 1187–1200 [DOI] [PubMed] [Google Scholar]
- 65.Greagg M. A., Fogg M. J., Panayotou G., Evans S. J., Connolly B. A., Pearl L. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9045–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]