Abstract
Endogenous pleiotrophin and hepatocyte growth factor (HGF) mediate the neurite outgrowth-promoting activity of chondroitin sulfate (CS)/dermatan sulfate (DS) hybrid chains isolated from embryonic pig brain. CS/DS hybrid chains isolated from shark skin have a different disaccharide composition, but also display these activities. In this study, pleiotrophin- and HGF-binding domains in shark skin CS/DS were investigated. A high affinity CS/DS fraction was isolated using a pleiotrophin-immobilized column. It showed marked neurite outgrowth- promoting activity and strong inhibitory activity against the binding of pleiotrophin to immobilized CS/DS chains from embryonic pig brain. The inhibitory activity was abolished by chondroitinase ABC or B, and partially reduced by chondroitinase AC-I. A pentasulfated hexasaccharide with a novel structure was isolated from the chondroitinase AC-I digest using pleiotrophin affinity and anion exchange chromatographies. It displayed a potent inhibitory effect on the binding of HGF to immobilized shark skin CS/DS chains, suggesting that the pleiotrophin- and HGF-binding domains at least partially overlap in the CS/DS chains involved in the neuritogenic activity. Computational chemistry using molecular modeling and calculations of the electrostatic potential of the hexasaccharide and two pleiotrophin-binding octasaccharides previously isolated from CS/DS hybrid chains of embryonic pig brain identified an electronegative zone potentially involved in the molecular recognition of the oligosaccharides by pleiotrophin. Homology modeling of pleiotrophin based on a related midkine protein structure predicted the binding pocket of pleiotrophin for the oligosaccharides and provided new insights into the molecular mechanism of the interactions between the oligosaccharides and pleiotrophin.
Keywords: Chondroitin Sulfate, Computation, Computer Modeling, Dermatan Sulfate, Glycosaminoglycan, Growth Factors, Heparin Co-factor II, Hepatocyte Growth Factor, Pleiotrophin, Sulfated Oligosaccharides
Introduction
Chondroitin sulfate (CS)5 and dermatan sulfate (DS), synthesized as glycosaminoglycan (GAG) side chains of proteoglycans (1, 2), participate in various biological events (2–7), and their functions are attributed to their structural diversity. CS chains are composed of the repeating disaccharide unit –GlcUA (d-glucuronic acid)-GalNAc-, whereas DS chains are composed of –IdoUA (l-iduronic acid)-GalNAc-, formed from precursor CS chains through the action of glucuronyl C5 epimerase (8). Structural elements of CS and DS are often found in co-polymeric hybrid chains (CS/DS), which tend to exhibit a periodic distribution of GlcUA-containing and IdoUA-containing disaccharide units in a tissue-specific manner (9–11). In biosynthesis, CS, DS, and CS/DS chains are further modified by a number of sulfotransferases at C-2 of GlcUA/IdoUA and/or C-4 and/or C-6 of GalNAc to yield a broad range of related structures including specific patterns of sulfation within oligosaccharide sequences (12). Thus, distinct functional domains of CS/DS chains may be generated in various tissues due to differential expression of modification enzymes. Such structural domains introduced into CS/DS chains in respective tissues may result in different responses to various CS/DS-binding proteins such as heparin-binding growth factors and cytokines (10).
Recent studies showed that CS/DS chains are involved in brain development (10, 11), and CS/DS variants from marine organisms and embryonic pig brain display growth factor-binding and neurite outgrowth-promoting (NOP) activities in vitro (13–18). The presence of oversulfated disaccharides (13–16) and IdoUA are two crucial factors for these functions of CS/DS chains (19, 20), indicating that IdoUA-containing oligosaccharide sequences with specific sulfation patterns may form functional domains for growth factor-binding and NOP activities. However, such functional sequences with a CS/DS hybrid structure are not fully understood.
We have shown that CS/DS chains from embryonic pig brain (E-CS/DS) and their mimicry by those from shark liver (SL-CS/DS) can recruit endogenous pleiotrophin (PTN) and hepatocyte growth factor (HGF) and present them to cultured mouse hippocampal neurons to stimulate the outgrowth of neurites (19, 20). However, the E-CS/DS and SL-CS/DS preparations, although containing similar activity, differ in overall composition, as exemplified by their disaccharide compositions. A series of PTN-binding octasaccharides with unique structures have been isolated from embryonic pig brain (E-CS/DS). These were prepared by digestion with CSase B specific to IdoUA (21), suggesting that the isolated sequences, which lacked IdoUA, were flanked by two IdoUA residues within the CS/DS hybrid chains. Thus, the identification of unique PTN/HGF-binding sequences is likely to explain the presence of similar functional activity in the apparently structurally different E-CS/DS and SL-CS/DS chains.
Recently characterized CS/DS chains from shark skin (SS-CS/DS) have a different composition from the E-CS/DS and SL-CS/DS chains, but strongly interact with various brain-expressed growth factors including PTN, and effectively promote neurite outgrowth (22). Therefore, SS-CS/DS, a potential candidate for a therapeutic agent, is an ideal material with which to identify functional domains. In this study, a novel PTN-binding hexasaccharide was isolated from the SS-CS/DS hybrid chains after digestion with chondroitinase AC-I and its strong binding with HGF was also demonstrated, suggesting that the PTN- and HGF-binding domains in SS-CS/DS overlap partially, if not completely. Intriguingly, the isolated hexasaccharide differed in disaccharide composition and sequence from the PTN-binding octasaccharides isolated from E-CS/DS (21), which prompted us to investigate the three-dimensional structure and electrostatic potential (ESP) distribution of the PTN-binding and non-binding sequences using methods previously reported for comparing multiple CS oligosaccharides recognized by monoclonal antibodies (23, 24). Computational chemistry using molecular modeling, simulation and ESP calculations revealed the three-dimensional structure and distribution of ESP on the molecular surface not only for the oligosaccharides but also for PTN, which allowed us to predict the binding pocket of the PTN molecule and gain new insights into the molecular mechanism by which the oligosaccharides are recognized by PTN.
EXPERIMENTAL PROCEDURES
Materials
The recombinant human (rh)-PTN for analyzing molecular interactions was purchased from RELIA Tech GmbH (Braunschweig, Germany). The rh-PTN for preparing an affinity column was a gift from Dr. Sakuma (Cell Signal Inc., Yokohama, Japan). The rh-hepatocyte growth factor/scatter factor for the interaction analysis was obtained from R&D Systems (Minneapolis, MN). The rh-HGF/scatter factor used for an affinity column was prepared as described previously (25). CSase ABC (EC 4.2.2.4), CSase AC-I (EC 4.2.2.5), standard unsaturated CS disaccharides, and CS-E from squid cartilage were purchased from Seikagaku Corp. (Tokyo, Japan). CSase B was obtained from IBEX Technologies (Montreal, Quebec, Canada). 2-Aminobenzamide (2-AB) was purchased from Nacalai Tesque (Kyoto, Japan). Sodium cyanoborohydride (NaBH3CN) was from Aldrich. EZ-LinkTM biotin-LC-hydrazide was obtained from Pierce. SS-CS/DS was isolated from shark skin as described previously (22).
Affinity Chromatography
SS-CS/DS chains were fractionated by chromatography on a PTN or HGF affinity column. The columns were prepared by coupling rh-PTN (0.5 mg) or rh-HGF (0.6 mg) to a HiTrap N-hydroxysuccinimide-activated column (1 ml) in the presence of excess SS-CS/DS to protect the GAG-binding site on PTN or HGF, as described (19). SS-CS/DS was applied to an affinity column equilibrated with 5 ml of 10 mm Tris-HCl buffer (pH 7.4), and the unbound fraction was reapplied to the column at least 5 times to maximize the immobilization. Subsequently, the column was washed with 3 ml of 10 mm Tris-HCl buffer (pH 7.4) and then eluted with 3 ml of the same buffer containing 0.15 or 0.5 m NaCl. To obtain enough bound materials, multiple affinity fractionations were performed at 4 °C. All fractions were desalted on a PD-10 column (GE Healthcare) and quantified by the carbazole reaction (26).
Analysis of Disaccharide Composition
The disaccharide composition of each SS-CS/DS fraction from affinity chromatography was determined as described previously (19). Briefly, each fraction (1 μg as GAGs) was digested with CSase ABC (27) and labeled with 2-AB (28). Excess 2-AB was removed by extraction with chloroform (29). The 2-AB-labeled digest was analyzed by anion exchange HPLC on an amine-bound PA-03 silica column (YMC-Pack PA, Kyoto, Japan) (30). Identification and quantification of the resulting disaccharides were achieved by comparison with CS-derived authentic unsaturated disaccharides (12).
Molecular Interaction Analysis
Molecular interactions were analyzed based on surface plasmon resonance using a BIAcore J system (BIAcore AB, Uppsala, Sweden). E-CS/DS or SS- CS/DS was immobilized on a sensor chip as described (18). For inhibition experiments, purified SS-CS/DS, its subfractions obtained by PTN-affinity chromatography, and fragments prepared by digestion with CSases, as an inhibitor, were incubated with 100 ng of PTN or 25 ng of HGF in the running buffer (HBS-EP, pH 7.4, BIAcore AB) for 15 min at room temperature prior to injection over the surface of the sensor chip. Other analytical conditions were the same as reported previously (31).
To examine the interaction of HGF with SS-CS/DS, various concentrations of HGF were injected onto the surface of the SS-CS/DS-immobilized sensor chip in the running buffer with a flow rate of 30 μl/min as per the manufacturer's directions. HGF was allowed to interact with SS-CS/DS for 2 min for association and dissociation, respectively, after which the sensor chip was regenerated by injecting 1 m NaCl for 2 min before the next injection. The kinetic parameters were evaluated with BIAevaluation 3.1 software (BIAcore AB) using a 1:1 binding model with mass transfer.
Neurite Outgrowth Promotion Assays
These assays were carried out as described previously (15, 32). The immunostained cells on each coverslip were scanned and digitized with a ×20 objective lens on an optical microscope (BH-2, Olympus, Tokyo, Japan) equipped with a digital camera (HC-300Z/OL, Olympus). One hundred clearly isolated cells with at least one neurite longer than the cell body were chosen at random to determine the length of the longest neurite using morphological analysis software (Mac SCOPE; Mitani Corp., Tokyo, Japan). At least three independent experiments per parameter or condition were carried out.
Effects of CSase Digestion on the PTN Binding and NOP Activities of the SS-CS/DS Fraction with High Affinity for PTN
The SS-CS/DS fraction eluted with 10 mm Tris-HCl buffer (pH 7.4) containing 0.5 m NaCl, which is designated a high affinity fraction (SS-CS/DS-H) with the highest level of NOP activity, was digested with CSase ABC, AC-I, or B, and the PTN-binding and NOP activities of these digests were evaluated based on their inhibitory activity against the binding of PTN to E-CS/DS and the NOP activity, as described under “Surface Plasmon Resonance Analysis” and “Neurite Outgrowth Promotion Assays,” respectively. Upon enzymatic treatment, an aliquot (20 μg) of SS-CS/DS-H was digested with 20 mIU of each CSase in a total volume of 100 μl of the appropriate buffer at 37 °C for 60 min as described previously (33), and then the reaction mixture was boiled at 100 °C for 1 min to denature the enzyme.
Preparation of Oligosaccharides from SS-CS/DS-H
SS-CS/DS-H (1 mg as GAG) was dissolved into 500 μl of 50 mm Tris-HCl buffer (pH 7.3), and exhaustively digested with 100 mIU of CSase AC-I at 37 °C for 24 h. The digest was fractionalized on a SuperdexTM Peptide HR column (GE Healthcare), which was eluted with 0.2 m NH4HCO3 at a flow rate of 0.4 ml/min, being monitored with a UV-visible detector at 232 nm. Oligosaccharide peaks were individually collected and desalted by repeated lyophilization.
Preparation of the Minimal PTN-binding Hexasaccharide Fraction
The CSase AC-I digest of SS-CS/DS-H (100 μg) was subjected to chromatography on a PTN affinity column as described above. Three fractions, eluted stepwise with 10 mm Tris buffer (pH 7.4) containing 0, 0.15, and 0.5 m NaCl, were individually analyzed using gel filtration under the same conditions as used under “Preparation of Oligosaccharides from SS-CS/DS-H.” According to the profiles of their gel filtrations, hexasaccharides represented the minimum size of SS-CS/DS oligosaccharides with PTN-binding capacity (see text). To prepare PTN-binding hexasaccharides, the size-defined hexasaccharide fraction, which was prepared as described under “Preparation of Oligosaccharides from SS-CS/DS-H,” was subfractionated on a PTN affinity column under the same conditions used above.
Subfractionation of the PTN-binding Hexasaccharides by Anion Exchange HPLC
The PTN-binding hexasaccharide fraction was subfractionated by anion exchange HPLC on an amine-bound silica PA-03 column. The hexasaccharide fraction was loaded on a column equilibrated with 311 mm NaH2PO4, washed with 311 mm NaH2PO4 for 5 min, and then eluted with a linear gradient from 311 to 803 mm NaH2PO4 over 90 min followed by another linear gradient from 803 to 1,000 mm NaH2PO4 over 10 min at a flow rate of 1.0 ml/min at room temperature. The eluates were monitored by measuring absorbance at 232 nm, and a major peak (M) eluted at 69.8 min was collected and desalted with a SuperdexTM Peptide column as described above.
Sequencing of the Major Minimal PTN-binding Hexasaccharide
The major fraction (M) of the minimal PTN-binding hexasaccharides was sequenced at a low picomole level by CSase digestion in conjunction with HPLC (28). Briefly, to identify the disaccharide composition, an aliquot (3 pmol) of fraction M was digested with CSase ABC or B, and labeled with 2-AB. The 2-AB was then extracted with chloroform. For sequencing, fraction M (30 pmol) was labeled with 2-AB and purified by paper chromatography, and then an aliquot (3 pmol) of 2-AB-labeled M was treated with CSase ABC, AC-II, or B. The CSase ABC digest was labeled with 2-AB again to tag the disaccharide released from the non-reducing end. All these preparations were individually analyzed by anion exchange HPLC on a PA-03 column eluted with a linear gradient from 16 to 1,000 mm NaH2PO4 over a period of 90 min monitored by fluorescent detection.
Computation of ESP for the Isolated Hexa- and Authentic Octasaccharides by the Density Functional Theory (DFT)
The initial conformations of the isolated hexa- and authentic octasaccharides were constructed using the Accelrys Materials Studio module. The dihedral angles (ϕ, ψ) were initially set to reproduce the most probable structures of oligosaccharides as reported recently for experimental and theoretical studies of multiple oligosaccharide sequences recognized by monoclonal antibodies (23, 24). As for molecular charges, all the sulfate groups in the oligosaccharides were treated as –SO3−, whereas all the carboxyl groups were treated as -COO− to reflect the structures under physiological pH. The initial geometries of the oligosaccharides were optimized using quantum mechanical approaches based on DFT to obtain structures stable in water. The DMol3 module (35, 36) in the Accelrys Materials Studio was used for these full geometry optimization calculations. The exchange correlation function chosen for the DFT calculations was Perdew-Burke-Ernzerhof (37) based on generalized gradient approximation. Perdew-Burke-Ernzerhof is frequently utilized in DFT calculations based on its strong physical background and reliable numerical performance. The basis set used was Double Numerical plus d-functions, which comprises one numerical function for each core atomic orbital, two numerical functions for each valence atomic orbital, and a polarization d-function for all the non-hydrogen atoms. The core treatment employed all electrons to depict the electronic structures for the oligosaccharides despite the high cost. The effect of water was taken into account by adopting the COnductor-like Solvent MOdel (38, 39), and permittivity was set to 78.4, which corresponds to the permittivity of water at 25 °C (40). The tolerance value of the Self-consistent Field was 1.0 × 10−5 in electronic density, and the convergence threshold for full geometry optimization was 2.0 × 10−5 hartree in energy. The ESP charges and zones for the optimized geometries of the oligosaccharides were calculated.
Homology Modeling and ESP Calculations for the PTN Protein Molecule
The electrostatic properties of the PTN protein were also computed to investigate the intermolecular interactions between the oligosaccharides and PTN. First, a partial PTN sequence (Cys99–Cys141) corresponding to the three-dimensional structure of midkine (Cys62–Cys104), which is the same gene family member as PTN and has a heparin-binding site, extracted after a pairwise sequence alignment (supplemental Fig. S1) was carried out between the sequences of entry p63089 in the pdb_seqres data base and the 1MKC entry in the Protein Data Bank. The midkine structure of the 1MKC entry, which has 54.5% sequence identity and 77.3% similarity to the extracted PTN sequence and has already been determined by NMR spectroscopy (41), was used as a mold for PTN. Homology modeling for the extracted PTN sequence was performed to obtain the unreported three-dimensional structure of PTN using the Discovery MODELER module (42–45) of the Accelrys Discovery Studio. The most stable three-dimensional structure of PTN with the best Discrete Optimized Protein Energy score (47) was chosen after 100 stable candidates were generated using MODELER. Second, the energy minimization for the three-dimensional structure of PTN modeled by MODELER was calculated using the Discovery Studio CHARMm module (48–50), which is versatile and widely used for molecular mechanics and dynamics incorporating original force fields and widely applicable to broad classes of biomolecular systems. In the energy minimization by the force field CHARMm, the root mean square gradient, steps, and implicit solvent dielectric constants used were 0.001, 5000, and 80, respectively, and an implicit solvent model of water was incorporated for Generalized Born with Molecular Volume (51). Third, the ESP zones of PTN, which were mapped to the solvent accessible surface area, were calculated using the DMol3 module under the same conditions as those for the oligosaccharides, except that the basis set for PTN was double numerical. This lacks the d-functions from the double numerical plus d-functions used for oligosaccharides, and is computationally less demanding and better suited to PTN, which is much larger than the oligosaccharides.
RESULTS
Affinity Fractionation of SS-CS/DS
SS-CS/DS chains interact with PTN with high affinity (KD = 7.6 ± 5.5 nm) and promote the outgrowth of neurites of hippocampal neurons in vitro (22). PTN also mediates the NOP activity of E-CS/DS (18). To investigate the correlation between the PTN-binding and NOP activities of the SS-CS/DS chains, the purified SS-CS/DS preparation (22) was subjected to affinity chromatography using a PTN-immobilized column to isolate the high affinity fraction containing the functional PTN-binding domain in the SS-CS/DS chains. It yielded an unbound (SS-CS/DS-U), a low affinity (SS-CS/DS-L) and a high affinity (SS-CS/DS-H) fraction, which were eluted stepwise with 10 mm Tris-HCl buffer (pH 7.4) alone, and the buffer containing 0.15 and 0.5 m NaCl, respectively. The proportion of these fractions, determined by the carbazole reaction for uronic acid, was 23.3, 51.7, and 25.0%, respectively (Table 1).
TABLE 1.
Disaccharide composition of SS-CS/DS subfractions obtained by affinity chromatography using a PTN-immobilized column
The purified SS-CS/DS preparation was separated into three subfractions using a PTN-immobilized column, as described under “Experimental Procedures.” The proportion of these subfractions was determined by the carbazole reaction (26), and the disaccharide composition of each subfraction was analyzed by digestion with CSase ABC followed by anion-exchange HPLC after 2-AB labeling, as described under “Experimental Procedures.”
| Unsaturated disaccharide | Subfractions |
||
|---|---|---|---|
| SS-CS/DS-U | SS-CS/DS-L | SS-CS/DS-H | |
| mol % | |||
| ΔO: ΔHexUA-GalNAc | 27.9 | 7.8 | 6.2 |
| ΔC: ΔHexUA-GalNAc(6S) | 36.5 | 34.7 | 15.9 |
| ΔA: ΔHexUA-GalNAc(4S) | 30.7 | 44.1 | 56.8 |
| ΔD: ΔHexUA(2S)-GalNAc(6S) | 1.1 | 3.6 | 3.2 |
| ΔB: ΔHexUA(2S)-GalNAc(4S) | 0.2 | 1.3 | 9.4 |
| ΔE: ΔHexUA-GalNAc(6S,4S) | 3.6 | 8.5 | 8.5 |
| Sulfation degreea | 0.77 | 1.06 | 1.15 |
| Proportion of subfractions (%) | 23.3 | 51.7 | 25.0 |
a Sulfation degree was calculated as the average number of sulfate groups per disaccharide unit.
After digestion with CSase ABC, the three subfractions showed Δ4,5HexUAα1–3GalNAc (ΔO unit), Δ4,5HexUAα1-3GalNAc(6S) (ΔC unit), Δ4,5HexUAα1–3GalNAc(4S) (ΔA unit), Δ4,5HexUA(2S)α1–3GalNAc(6S) (ΔD unit), Δ4,5HexUA(2S)α1- 3GalNAc(4S) (ΔB unit), and Δ4,5HexUAα1–3GalNAc(4S,6S) (ΔE unit) with different proportions of the disaccharide components among the fractions (Table 1) (for abbreviations of disaccharide units, also see Ref. 11), where 2S, 4S, 6S, and Δ4,5HexUA represent 2-O-sulfate, 4-O-sulfate, 6-O-sulfate, and 4,5-unsaturated hexuronic acid, respectively. The proportions of ΔO and ΔC units generally decreased with an increase in the affinity for PTN, whereas the proportions of ΔA and ΔB units markedly increased, which may indicate that the PTN-binding domain of the SS-CS/DS chains contains parental disaccharides (A/iA and B/iB) of ΔA and ΔB units, where A, iA, B, and iB represent GlcUAβ1–3GalNAc(4S) (A unit), IdoUAα1–3GalNAc(4S) (iA unit), GlcUA(2S)β1–3GalNAc(4S) (B unit), and IdoUA(2S)α1–3GalNAc(4S) (iB unit), respectively (11).
PTN-binding and NOP Activities of the SS-CS/DS Subfractions
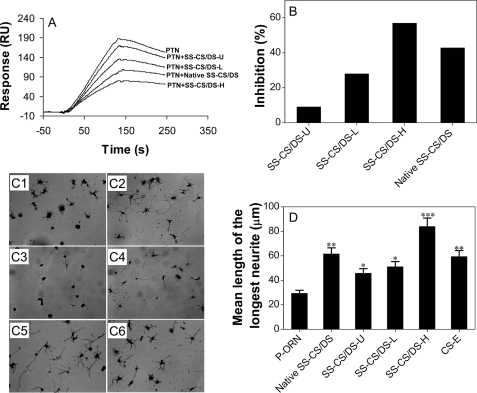
The PTN-binding activity of the SS-CS/DS subfractions was compared with that of the E-CS/DS chains, using a BIAcore system, where E-CS/DS was immobilized on a sensor chip and the SS-CS/DS subfractions were used as inhibitors for the binding of PTN to E-CS/DS as detailed under “Experimental Procedures.” SS-CS/DS-H and SS-CS/DS-L showed stronger and weaker inhibitory activity, respectively, than E-CS/DS, and SS-CS/DS-U showed negligible activity (Fig. 1, A and B). These results suggest that SS-CS/DS-H contains a PTN-binding domain, which may closely resemble the PTN-binding domains of E-CS/DS, and may form a similar structural basis for the NOP activity of SS-CS/DS.
FIGURE 1.
Comparison of the PTN-binding activity and NOP activity of SS-CS/DS subfractions. A, inhibition of the binding of PTN to CS/DS isolated from embryonic pig brain (E-CS/DS) was evaluated using SS-CS/DS subfractions as inhibitors. A fixed concentration of PTN (100 ng) was mixed with native SS-CS/DS, SS-CS/DS-U, SS-CS/DS-L, or SS-CS/DS-H at a final concentration of 0.5 μg/ml and incubated for 15 min. Binding of PTN to immobilized E-CS/DS was evaluated using a BIAcore system as described under “Experimental Procedures,” and the sensorgrams are shown. B, the results shown in panel A are expressed as relative percentages of inhibition compared with the binding of PTN to E-CS/DS in the absence of inhibitors. C, NOP activities. E16 mouse hippocampal neuronal cells were grown for 24 h on various substrates prepared by pre-coating poly-dl-ornithine (C1) and subsequently CS-E (a positive control) (C6), native SS-CS/DS (C2), SS-CS/DS-U (C3), SS-CS/DS-L (C4), or SS-CS/DS-H (C5), fixed, and immunostained for microtubule-associated protein 2 (52) and neurofilaments (53). One hundred randomly selected individual neurons were used to measure the mean length of the longest neurite under each set of conditions. D, the values obtained from two separate experiments are expressed as the mean ± S.E. Mann-Whitney's U test was used to evaluate the significance of differences between the means (*, p < 0.5; **, p < 0.01; and ***, p < 0.001).
We recently demonstrated a positive correlation between the PTN-binding and NOP activities of the E-CS/DS chains (19). To investigate the NOP activity of the SS-CS/DS subfractions, neurite outgrowth promotion assays were carried out using hippocampal neuronal cells isolated from E16 mouse embryos. SS-CS/DS-U and SS-CS/DS-L exhibited significant yet weaker activity than the unfractionated SS-CS/DS and CS-E (a positive control) (14), whereas SS-CS/DS-H exhibited markedly stronger activity (Fig. 1, C5 and D). These results are consistent with the finding that the PTN-binding activity of E-CS/DS is involved in its NOP activity (19).
Effects of Digestion with CSase on the PTN-binding and NOP Activities of SS-CS/DS-H
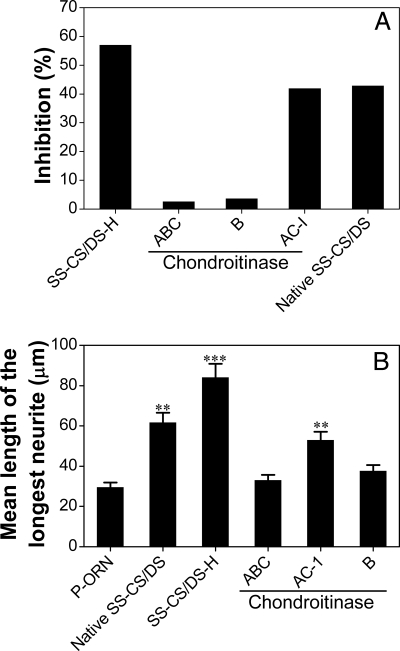
To investigate whether the CS or DS domains of the SS-CS/DS-H chains contribute to their biological activities, effects of digestion of SS-CS/DS-H with various CSases on the growth factor-binding and NOP activities were examined. The activity of SS-CS/DS-H to inhibit the binding of PTN to the E-CS/DS chains was almost completely eliminated by digestion with CSase ABC or B, but it was only partially affected by digestion with CSase AC-I (Fig. 2A). Neurite outgrowth promotion assays of the digests gave similar results, namely almost complete inhibition with a CSase ABC or B digest and less inhibition with a CSase AC-I digest (Fig. 2B). These results together suggest that DS domains, which are specifically degraded by CSase B (54), play a major role in the PTN-binding and NOP activities of the SS-CS/DS-H chains. In contrast, the CS domains, which are selectively digested by CSase AC-I (55), play a significant yet minor role in these activities. Therefore, CSase AC-I was used to digest the SS-CS/DS-H chains to isolate oligosaccharide fragments containing the putative PTN-binding sequences.
FIGURE 2.
Effects of digestion with CSases on the activities of SS-CS/DS-H. SS-CS/DS-H was digested with CSase ABC, AC-I, or B, and each digest was evaluated for inhibition of the binding of PTN to E-CS/DS (A) and NOP activity (B) under the same conditions as described in the legend to Fig. 1, A–B and C–D, respectively.
Preparation of Size-defined Oligosaccharides
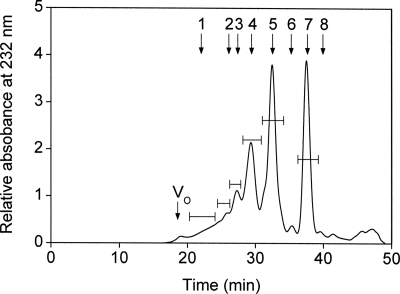
SS-CS/DS-H was exhaustively digested with CSase AC-I and fractionated by gel filtration on a SuperdexTM Peptide column, which revealed that the SS-CS/DS-H chains were extensively digested into oligosaccharides (Fig. 3) despite the fact that CSase AC-I had little effect on the PTN-binding activity of SS-CS/DS-H (Fig. 2A). The oligosaccharides were eluted as a series of well resolved peaks corresponding to di- to octasaccharides as main products with some larger, poorly resolved minor fragments (Fig. 3). The chromatographic pattern reflects a highly mixed distribution of the GlcUA and IdoUA residues along the SS-CS/DS-H chains, which display complex CS/DS hybrid structures as previously reported for the purified SS-CS/DS fraction (22). These results together imply that the high PTN-binding activity of SS-CS/DS-H is attributable to the CS/DS hybrid domain rather than discrete CS or DS regions. The individual oligosaccharide fractions were pooled as indicated by bars, and desalted by repeated lyophilization.
FIGURE 3.
Preparation of oligosaccharides from SS-CS/DS-H. SS-CS/DS-H (1 mg) was exhaustively digested with CSase AC-I and then separated into its constituent oligosaccharides by gel filtration chromatography on a SuperdexTM Peptide column, as described under “Experimental Procedures.” Oligosaccharide peaks corresponding to di- to octasaccharides, and combined oligosaccharides larger than octasaccharides were individually pooled. Vo and Vt represent the void volume and the total volume (24 ml), respectively. The elution positions of the following standard oligosaccharides (56) are indicated by arrows: 1, oligosaccharides ≥12-mer (12-mer and larger); 2, CS-decasaccharides; 3, CS-octasaccharides; 4, CS-hexasaccharides; 5, CS-tetrasaccharides; 6, disulfated CS-disaccharides; 7, monosulfated CS-disaccharides; 8, unsulfated CS-disaccharides.
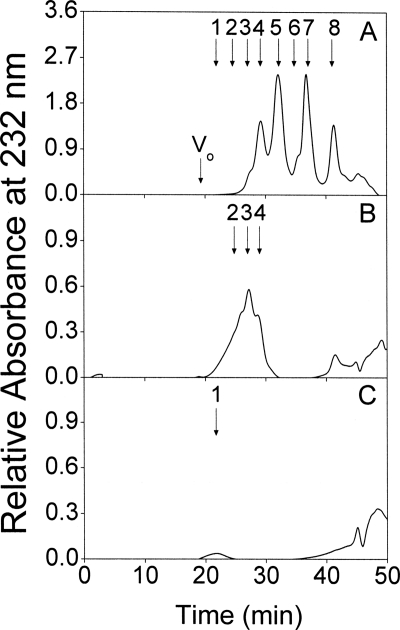
Determination of the Minimal Size of the PTN-binding Oligosaccharides
The minimum size of the SS-CS/DS-H oligosaccharides, which retain the ability to bind PTN, was estimated by affinity chromatography of a CSase AC-I digest on a PTN-immobilized column. The CSase AC-I digest of SS-CS/DS-H was eluted with 10 mm Tris-HCl buffer (pH 7.4) containing zero, 0.15, and 0.5 m NaCl. Gel filtration chromatography of the unbound fraction showed that it contained most of the di- and tetrasaccharides as well as a majority of hexasaccharides (Fig. 4A). In contrast, a minority of the hexasaccharides and most of the octa- and decasaccharides were detected in the 0.15 m fraction (Fig. 4B). In the 0.5 m fraction, only trace amounts of dodecasaccharides and larger oligosaccharides were recovered (Fig. 4C). Taken together, these results showed that the minimal oligosaccharide able to bind to PTN was a hexasaccharide.
FIGURE 4.
Determination of the minimal size of CS/DS oligosaccharides that bind to PTN. The CSase AC-I digest of SS-CS/DS-H (100 μg) was applied to a PTN-immobilized column equilibrated with 10 mm Tris-HCl buffer (pH 7.4) and eluted stepwise using the same buffer containing zero, 0.15, and 0.5 m NaCl. The zero m (A), 0.15 m (B), and 0.5 m (C) fractions were analyzed individually by gel filtration chromatography on a SuperdexTM Peptide column, being monitored by measuring absorbance at 232 nm as described under “Experimental Procedures.” For the elution positions of standard oligosaccharides indicated by arrows, see the legend to Fig. 3.
Preparation of the PTN-binding Hexasaccharide Fraction
Because the minimal PTN binding size was a hexasaccharide, the bulk hexasaccharide fraction obtained in Fig. 3 was subfractionated using a PTN column into unbound, and 0.15 and 0.5 m NaCl eluted fractions as described above. The majority (95%) of hexasaccharides were recovered in the unbound fraction (supplemental Fig. S2A), and although no hexasaccharide was detected in the 0.5 m fraction (supplemental Fig. S2C), a small yet significant amount (5%) was obtained in the 0.15 m fraction (supplemental Fig. S2B). The main peak with a shoulder, as marked by a bar in supplemental Fig. S2B, was pooled and used for subsequent purification.
Isolation and Characterization of the Major Minimal PTN-binding Sequence
To isolate the major PTN-binding sequence, the PTN-binding hexasaccharide shown in supplemental Fig. S2B was further fractionated by anion exchange HPLC (supplemental Fig. S3). High sensitivity was required for detection due to the limited amount (∼10 μg) of the key hexasaccharide fraction, and although it resulted in an unstable baseline, a sharp major oligosaccharide peak (fraction M) was clearly detected together with several minor peaks (supplemental Fig. S3). Fraction M was used for the subsequent characterization.
To determine the sugar composition and the number of sulfate groups, fraction M was first analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry in a positive ion mode. A molecule-related ion signal of the protonated complex of the major component in fraction M with a basic peptide (Arg-Gly)15 was detected at m/z 4749.49 (supplemental Fig. S4). The difference (1535.74 Da) between the m/z 4749.49 of the complex and the m/z 3213.75 of the (Arg-Gly)15 is the observed mass of the major compound in fraction M, which corresponded to ΔHexUA1HexUA2HexNAc3(OSO3)5, where HexUA and HexNAc represent hexuronic acid (GlcUA or IdoUA) and N-acetyl-d-hexosamine, respectively. The molecular mass of 1538.26 Da calculated for the deduced composition is consistent with the unsaturated pentasulfated hexasaccharide.
Disaccharide composition was analyzed next. Fraction M was digested with CSase ABC, labeled with 2-AB, and analyzed by anion exchange HPLC, which showed ΔA and ΔB units in a molar ratio of 1.0:2.0 (Fig. 5B), suggesting that fraction M was composed of one GlcUAβ1–3GalNAc(4S) or IdoUAα1–3GalNAc(4S) unit and two GlcUA(2S)β1–3GalNAc(4S) or IdoUA(2S)α1–3GalNAc(4S) units, from which the ΔA and ΔB units were derived. CSase B was used to test for IdoUA in the CSase AC-I-resistant fraction. Interestingly, the digest obtained with CSase B specific to GalNAc-IdoUA linkages gave the same chromatographic profile (Fig. 5C) as the CSase ABC digest. In contrast, CSase AC-II specific to GalNAc-GlcUA linkages did not act on fraction M at all (Fig. 5D). These results together suggest that the two internal uronic acid residues are IdoUA rather than GlcUA.
FIGURE 5.
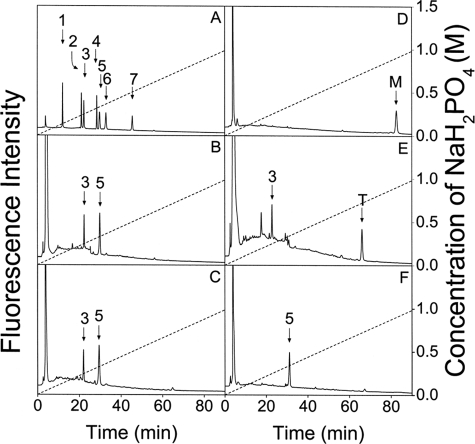
Sequencing analysis of the major minimal PTN-binding SS-CS/DS hexasaccharide fraction. The major fraction (M) of the minimal PTN-binding hexasaccharides, which was eluted at 69.8 min under supplemental Fig. S3, was analyzed. The disaccharide composition of fraction M was determined by comparing the elution positions of the 2-AB-labeled unsaturated disaccharides produced by CSases ABC (B) and B (C) with those of authentic 2-AB-derivatized unsaturated CS disaccharides indicated by arrows (A). 1, ΔO; 2, ΔC; 3, ΔA; 4, ΔD; 5, ΔB; 6, ΔE; and 7, ΔT (Δ4,5HexUA(2S)α1–3GalNAc(4S,6S)). For the abbreviations, ΔO, ΔA, ΔB, ΔC, ΔD, and ΔE, see Table 1 and Ref. 11. The sequencing analysis was achieved by digestion with various CSases, followed by anion exchange HPLC on an amine-bound silica PA-03 column using a NaH2PO4 gradient. Fraction M was labeled with 2-AB, and digested with CSase AC-II (D), ABC (E), or B (F), and each digest was analyzed by anion exchange HPLC. The CSase ABC digest was labeled with 2-AB again to determine the disaccharide located at the non-reducing end of the hexasaccharide.
An aliquot of 2-AB-labeled M was digested with CSase ABC and labeled with 2-AB again, which showed ΔA and unsaturated oligosaccharide T in a molar ratio of 1:1 (Fig. 5E). It has been shown that 2-AB derivatization renders tetrasaccharides at the reducing end of oligosaccharides resistant to the action of CSase ABC (28). Hence, ΔA should have been released from the non-reducing end of the major compound in fraction M, whereas oligosaccharide T (Fig. 5E) should be an unsaturated tetrasaccharide composed of two ΔB units derived from the reducing end. The digestion of 2-AB-labeled M with CSase B indeed released 2-AB-labeled ΔB, confirming that a CSase B-sensitive iB unit was located at the reducing end of the major compound in fraction M (Fig. 5F). Based on these results, it was concluded that the major compound in fraction M has an unsaturated hexasaccharide sequence, ΔA-iB-iB (Δ4,5HexUAα1–3GalNAc(4S)β1–4IdoUA(2S)α1- 3GalNAc(4S)β1–4IdoUA(2S)α1–3GalNAc(4S)).
Demonstration of Interactions of SS-CS/DS with HGF
PTN and HGF can mediate the NOP activity of CS/DS chains (19, 20), indicating a possible overlap of the PTN- and HGF-binding sequences in the CS/DS chains. Preliminary studies showed that HGF strongly interacted with the SS-CS/DS chains immobilized on a sensor chip in a BIAcore system (data not shown) as in the case of PTN (22). To investigate the disaccharide composition required for the binding of HGF, SS-CS/DS was fractionated by affinity chromatography using an HGF-immobilized column, which was eluted stepwise with 10 mm Tris-HCl buffer (pH 7.4) containing 0, 0.15, and 0.5 m NaCl. These fractions displayed a similar profile to the fractions eluted from the PTN column under the same chromatographic conditions, namely, proportions of ΔA and ΔB units were significantly greater in the high affinity fraction (0.5 m) than the unbound or low affinity fraction (0.15 m) (Table 2). The results suggest that precursors A/iA and B/iB of the two disaccharide units ΔA and ΔB also play key roles in the HGF-binding domains as shown above for the PTN-binding domain of the SS-CS/DS chains. The much higher binding of SS-CS/DS to HGF than to PTN (Tables 1 and 2) is assumed to reflect less demanding structural requirements such as lower sulfation degrees and shorter sizes for the binding to HGF than to PTN (see also “Discussion”).
TABLE 2.
Disaccharide composition of SS-CS/DS subfractions obtained by affinity chromatography using an HGF-immobilized column
The purified SS-CS/DS preparation was loaded on an HGF-immobilized column and separated into three subfractions (unbound fraction, 0.15 m fraction, and 0.5 m fraction) by stepwise elution with 10 mm Tris-HCl buffer (pH 7.4) containing 0, 0.15, or 0.5 m NaCl, as described in the text. The proportion of these subfractions and the disaccharide composition of each subfraction were analyzed as described in the legend to Table I.
| Unsaturated disaccharide | Subfractions |
||
|---|---|---|---|
| Unbound | 0.15 m | 0.5 m | |
| mol % | |||
| ΔO: ΔHexUA-GalNAc | 35.6 | 9.9 | 5.8 |
| ΔC: ΔHexUA-GalNAc(6S) | 34.6 | 36.4 | 22.7 |
| ΔA: ΔHexUA-GalNAc(4S) | 26.4 | 42.4 | 52.6 |
| ΔD: ΔHexUA(2S)-GalNAc(6S) | 1.0 | 3.1 | 3.8 |
| ΔB: ΔHexUA(2S)-GalNAc(4S) | 0.2 | 2.0 | 9.7 |
| ΔE: ΔHexUA-GalNAc(6S,4S) | 2.2 | 6.2 | 8.4 |
| Sulfation degreea | 0.68 | 1.01 | 1.14 |
| Proportion of subfractions (%) | 7.9 | 50.5 | 41.6 |
a Sulfation degree was calculated as the average number of sulfate groups per disaccharide unit.
Demonstration of the HGF-binding Capacity of the PTN-binding Hexasaccharide
Based on the evidence above and recent findings that CS/DS isolated from embryonic pig brain and shark liver recruited endogenous PTN and HGF to stimulate neurite outgrowth in cultured mouse hippocampal neurons (19, 20), we speculated that the major compound in fraction M may bind HGF. To explore this possibility, the minimum size required for binding HGF was investigated by evaluating the inhibitory activities of the CSase AC-I-produced SS-CS/DS oligosaccharide fractions (Fig. 3) against the binding of HGF to the SS-CS/DS chains immobilized on the surface of a sensor chip in the BIAcore J system. As shown in Fig. 6A, the hexasaccharide was the smallest size able to inhibit the binding of HGF to SS-CS/DS: there was no inhibition by the di- and tetrasaccharide fractions and stronger inhibition by the octasaccharide and larger fractions. Notably, fraction M, which was purified after PTN affinity chromatography, displayed much stronger (more than 50-fold) inhibition of HGF binding to SS-CS/DS at a low dose of 10 pmol than the inhibition of a SS-CS/DS hexasaccharide mixture (Fig. 3) at a much higher dose (500 pmol) (Fig. 6B). These results suggest that the major PTN-binding hexasaccharide ΔA-iB-iB in fraction M has the capacity to bind HGF as well. Hence, it was concluded that the PTN and HGF binding sequences in the SS-CS/DS chains contain some common determinants and may overlap.
FIGURE 6.
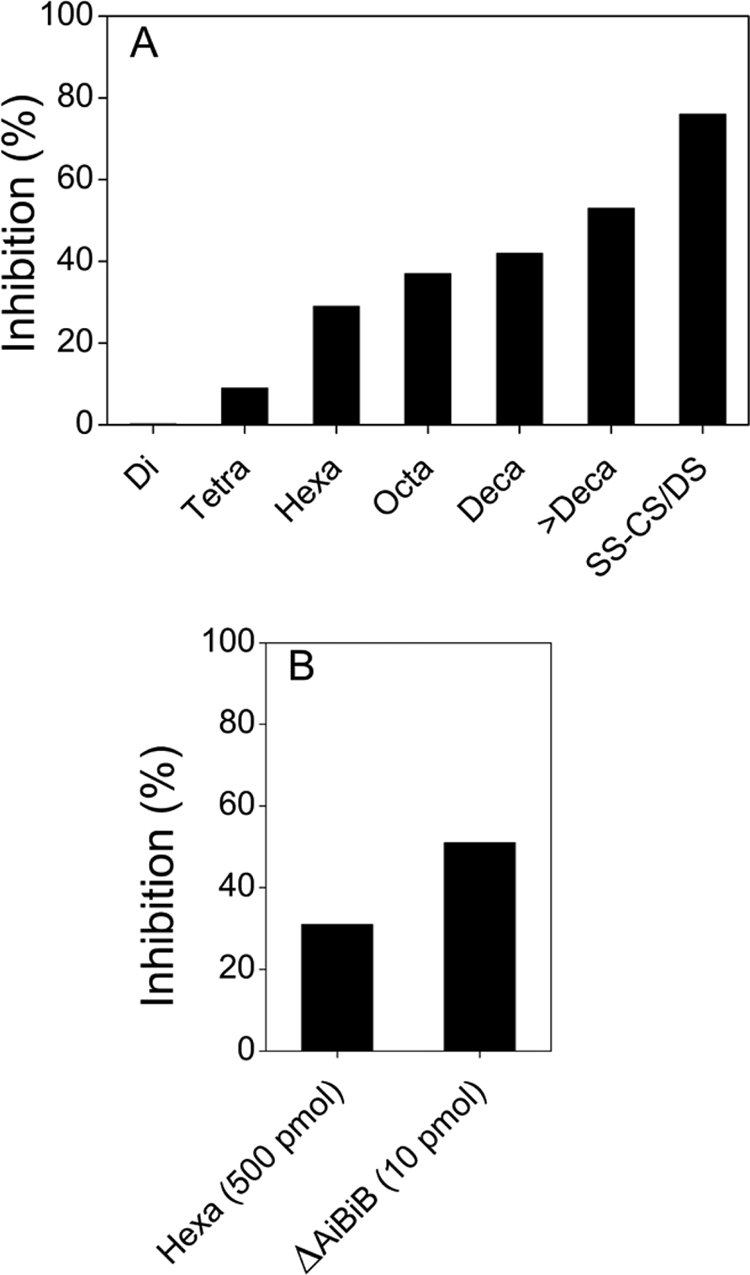
Inhibitory effect of SS-CS/DS oligosaccharides and fraction M on the binding of PTN to immobilized SS-CS/DS. A fixed concentration of HGF (50 ng) was mixed with each size-defined SS-CS/DS oligosaccharide fraction produced by CSase AC-I, the isolated hexasaccharide ΔA-iB-iB, or the parental SS-CS/DS polysaccharide fraction, and incubated for 15 min. Interactions of the HGF in each sample with immobilized E-CS/DS were analyzed using a BIAcore system as described under “Experimental Procedures.” A, the inhibitory activity of a series of SS-CS/DS oligosaccharide fractions (di-, tetra-, hexa-, octa-, deca-, and >decasaccharide (larger than deca-)) and native SS-CS/DS (10 μg/ml each) was compared. B, the inhibitory activity of fraction M (10 pmol) was compared with that of the parental SS-CS/DS hexasaccharide mixture (500 pmol) obtained in Fig. 3.
Comparison of the Conformations, ESP Charges and Maps for the Isolated PTN-binding Hexa- and Authentic PTN-binding and Non-binding Octasaccharides
In previous studies we demonstrated the effectiveness of molecular modeling and simulation, especially the calculation of ESP, for investigating molecular recognition (23, 24). In this study, more advanced computational methods were applied not only to the oligosaccharides but also to PTN to obtain detailed information on the molecular interactions despite the complexity and cost of calculations. DFT, which gives greater accuracy than the semiempirical molecular orbital method used previously (24), was employed as described under “Experimental Procedures,” because it can derive more complete electronic structures and more accurate conformational properties for both oligosaccharides and PTN.
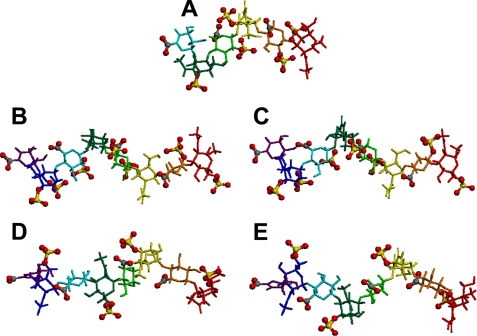
The five CS/DS oligosaccharides were fully geometry optimized as described under “Experimental Procedures,” and the predicted energy-minimal conformation of each oligosaccharide and the dihedral angles (ϕ, ψ) between the adjacent constituent sugar residues are shown in Fig. 7 and supplemental Table S1, respectively. The differences of the ϕ and ψ values between the adjacent residues of the two PTN-non-binding octasaccharides (ΔC-C-A-C and ΔC-A-A-A) were in a range of 2.4–13.3 and 0.5–18.2, whereas those of the two PTN-binding octasaccharides (ΔE-D-A-D and ΔE-D-iA-D) were 3.6–30.7 and 4.9–44.8, respectively. Hence, the conformations of the non-binding octasaccharides are very similar to each other, and the differences of the values of the dihedral angles between the adjacent residues of the binding octasaccharides were not as large (less than 30) either, except for the ϕ of glycosidic linkage 8-7 and the ψ linkages 3-2 and 6-5 (supplemental Table S1). It is likely that the concatenation of the ESP zone of the binding oligosaccharides (Fig. 8, A–C) more easily occurs in cooperation with carboxyl groups having similar negative charges to sulfate groups (supplemental Tables S2 and S3). In fact, the sulfate groups of the binding oligosaccharides with more sulfate groups than the non-binding ones are facing toward similar directions (Fig. 7). It seems more difficult for the non-binding octasaccharides having a smaller number of sulfate groups to concatenate the ESP zone (Fig. 8, D and E) compared with the binding octasaccharides.
FIGURE 7.
Full geometry optimized structures of the five oligosaccharide. Each conformation of the isolated oligosaccharides is shown. A, ΔA-iB-iB; B, ΔE-D-A-D; C, ΔE-D-iA-D; D, ΔC-C-A-C; and E, ΔC-A-A-A. Different colored stick models for each sugar and ball and stick model for sulfate and carboxyl groups were used to make it easy understand the relative positions between sugar rings and sulfate and carboxyl groups. Red and yellow balls represent oxygen and sulfur atoms, respectively, of the sulfate and carboxyl groups. Oligosaccharides are displayed with the nonreducing on the left and the reducing end on the right.
FIGURE 8.
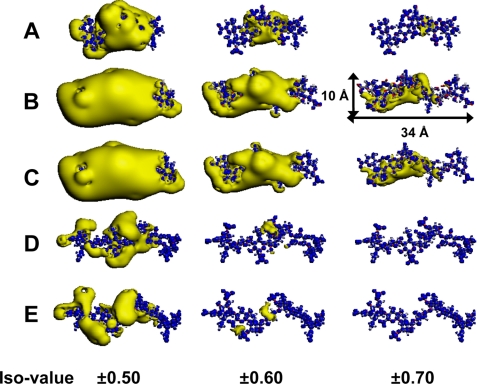
Calculated ESP distribution of the five isolated oligosaccharides. The distribution of ESP was calculated for each isolated oligosaccharide, and electronegative (yellow) and electropositive (blue) zones are shown. A, ΔA-iB-iB; B, ΔE-D-A-D; C, ΔE-D-iA-D; D, ΔC-C-A-C; and E, ΔC-A-A-A. The putative PTN-binding oligosaccharides, ΔE-D-A-D and ΔE-D-iA-D, have spreading electronegative zones at iso values of −0.5 to −0.7 and even ΔA-iB-iB had a small electronegative zone at −0.7. In contrast, the non-binding octasaccharides, ΔC-C-A-C and ΔC-A-A-A, had no electronegative zones at −0.7. The spread of the electronegative zones for the PTN-binding oligosaccharides was larger than that for the non-binding oligosaccharides at the same iso values. The iso values are 0.5, 0.6, and 0.7 eV for the iso surface of the electropositive zones and −0.5, −0.6, and −0.7 eV for that of the electronegative zones.
ESP calculations were first applied to the isolated hexasaccharide ΔA-iB-iB and two putative PTN-binding octasaccharides previously isolated from E-CS/DS (ΔE-D-A-D, ΔE-D-iA-D), where the D unit stands for GlcUA(2S)β1–3GalNAc(6S) (21). In addition, two PTN-non-binding octasaccharides (ΔC-C-A-C and ΔC-A-A-A) (21) were also analyzed as negative controls, where the C unit stands for GlcUAβ1–3GalNAc(6S). These PTN-binding and non-binding octasaccharides were isolated after digestion of E-CS/DS with CSase B. However, it was not clear whether either one, or both ΔE-D-A-D and ΔE-D-iA-D could bind PTN at that time, because they could not be separated from each other by chromatography. It was more likely, however, that the purified fraction contained ΔE-D-A-D rather than ΔE-D-iA-D, which would have been degraded by CSase B. The ESP atomic point charges of sulfate groups and carboxyl groups in the five oligosaccharides, which make large contributions in the ESP maps, are summarized in supplemental Tables S2 and S3. The negative net charges of both the sulfate and carboxyl groups had similar values ranging from −0.77 to −0.86 and −0.72 to −0.85, respectively. Interestingly, the results revealed only a slight difference in the negative net charges between the sulfate and carboxyl groups. The sulfate group of the A unit of ΔE-D-A-D had a slightly larger negative net charge (−0.82) than that (−0.86) of the iA unit of ΔE-D-iA-D, whereas the negative net charges of the carboxyl groups in the A and iA units were opposite (−0.80 versus −0.75). The electronegative zones of the ESP maps at iso values of ±0.5, ±0.6, and ±0.7, which interacted with PTN, were calculated based on DFT for the five oligosaccharides, and are displayed in Fig. 8.
The ESP distribution showed spreading of the electronegative zones around both the sulfate and carboxyl groups having negative clouds of oxygen atoms, whereas the electropositive zones were not spread in the five oligosaccharides. Interestingly, the ESP maps revealed that not only the optimized three-dimensional structure but also the electronegative zones of the putative PTN-binding octasaccharides (ΔE-D-A-D and ΔE-D-iA-D) had very similar shapes (Fig. 8, B and C), suggesting that the predicted criteria for molecular recognition showed little difference between the two octasaccharides containing the A unit with GlcUA and iA unit with IdoUA, and that both octasaccharides most likely can bind PTN. The electronegative zones of the non-binding octasaccharides (ΔC-C-A-C and ΔC-A-A-A) were also very similar to each other and different from those of the binding octasaccharides (Fig. 8, D and E). The hexasaccharide ΔA-iB-iB also had an electronegative zone, although with a narrower range than that of the binding octasaccharides (Fig. 8A), which may reflect its relatively weak affinity for PTN. The putative PTN-binding oligosaccharides, ΔE-D-A-D and ΔE-D-iA-D, have spreading electronegative zones at iso values of −0.5 to −0.7 and even ΔA-iB-iB had a small electronegative zone at −0.7. In contrast, the non-binding octasaccharides, ΔC-C-A-C and ΔC-A-A-A, had no electronegative zones at −0.7. The spread of the electronegative zones was greater for the PTN-binding oligosaccharides than for the non-binding oligosaccharides at the same iso values. Snapshots of the calculated ESP distribution of the five isolated oligosaccharides every 90 degrees at iso values of ±0.5, ±0.6, and ±0.7 are shown under supplemental Fig. S5, 1–3, respectively. The overall results (Fig. 8 and supplemental Fig. S5, 1–3) showed that the spreading of the electronegative zone of ESP distribution was large for ΔE-D-A-D and ΔE-D-iA-D, medium for ΔA-iB-iB, and small for ΔC-C-A-C and ΔC-A-A-A at each negative iso value. Thus, the differences predicted from the ESP maps showed similarity in the shapes and spreading patters shared by the PTN-binding oligosaccharides, and differences from the non-binding oligosaccharides.
Homology Modeling and ESP Maps for PTN Protein
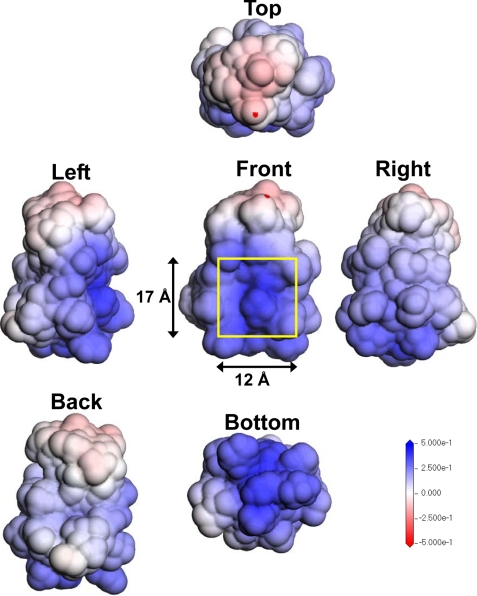
The homology modeling by MODELER was also preprocessed for the PTN sequence based on the related protein midkine, which is the same gene family member and has a sequence 54.5% identical with and 77.3% similar to that of PTN. The three-dimensional model of PTN with the best score (supplemental Fig. S6) was fully optimized by energy minimization using the Discovery Studio CHARMm module as described under “Experimental Procedures.” The distribution of ESP was calculated for the optimized PTN structure to predict the binding pocket of PTN for the sulfated oligosaccharides. The height and width of PTN were ∼30 and 24 Å, respectively (supplemental Fig. S6). On the other hand, the four octasaccharides were ∼34 Å long and 10 Å wide (Fig. 8B). The ESP zones of PTN mapped on the solvent accessible surface area using the DMol3 module are displayed from five different directions in Fig. 9. The sugar-binding site predicted by the ESP calculations based on DFT is shown in Fig. 9 (front), and had a positive surface in the ESP-mapped solvent accessible surface area and potentially complemented the spreading negative zones on the ESP maps of the oligosaccharides. The candidate sugar-binding site of PTN is ∼17 Å in length and 12 Å in width. Thus, analysis of the ESP zone mapped on the solvent accessible surface area by DFT also identified the binding pocket without difficulty, which formed a concave, electrostatic positive surface with a good structural match and reasonable size to bind the preferred oligosaccharides.
FIGURE 9.
ESP zone mapped to the solvent accessible surface area of PTN. The ESP zones of PTN, which were mapped to the solvent accessible surface area, were determined using ab initio DFT under the same conditions as those for the oligosaccharides except for the basis set. The basis set used for PTN was double numerical because PTN is much larger in size than the oligosaccharides. The solvent accessible surface area is 1.4 Å (the radius of a water molecule) from the van der Waals surface. The six visualized images indicate individual ESP distributions corresponding to the top, bottom, front, back, left, and right sides of the PTN molecule, respectively. The range of the values for contour maps is 0.5 eV (blue) for the electropositive surface and −0.5 eV (red) for the electronegative surface. The yellow square frame and the scales show the size of the cavity of the PTN molecule.
DISCUSSION
The NOP activity of CS/DS chains is mediated by PTN and HGF (19, 20), and depends on the presence of disulfated disaccharide units and IdoUA (13–20). Previously, we isolated several PTN-binding octasaccharide sequences such as ΔC-C-D-C, ΔA-C-D-C and/or ΔC-A-D-C, ΔD-C-D-C, ΔC-D-D-C and/or ΔC-D-iD-C, as well as ΔE-D-A-D and/or ΔE-D-iA-D (iD unit stands for IdoUA(2S)α1–3GalNAc(6S)) have been isolated from E-CS/DS by us (21). In this study, we isolated another novel minimal PTN-binding oligosaccharide, ΔA-iB-iB, from SS-CS/DS chains, which possessed HGF-binding activity as well, providing evidence for the hypothesis that PTN and HGF have overlapping binding domains to mediate the NOP activity of CS/DS chains.
Catlow et al. (57) recently studied the recognition of different GAGs, HS, and DS, by HGF using a wide array of natural and modified GAGs, and suggested that no specific isomer in either GAG is vital for interaction and activity, and that both IdoUA and sulfate density are required. Deakin et al. (58) further revealed that a minimum binding sequence is a disulfated trisaccharide comprised of a flexible internal IdoUA flanked by monosulfated hexosamine residues. The structural feature of the hexasaccharide isolated in this study is in complete agreement with the notion suggested by Deakin et al. (58). In the light of their findings, however, it should be noted that our hexasaccharide does not represent the minimum size for the binding of HGF and possibly of PTN, and that more sulfated oligosaccharides are more likely to bind more strongly to the proteins. Presumably, the flexibility of the sugar chains afforded by the two IdoUA residues in our hexasaccharide are crucial for the binding to two different growth factors. Notably, however, the octasaccharide, ΔE-D-A-D, which do not contain IdoA, was revealed to have a very similar conformation and ESP map to those of its isomer ΔE-D-iA-D, and most likely can bind PTN, suggesting that IdoA is not always an essential factor for the binding to functional proteins. As discussed below, the molecular shape with the ESP surface is the structural basis or the entity for the “overall organization of functional GAG domains,” which has been discussed by Lindahl and colleagues (59).
An IdoUA(2S) residue in the heparan sulfate of N-syndecan is essential for the binding of PTN and induction of neurite outgrowth (60). iB units containing IdoUA(2S) are abundant in CS/DS chains from the developing mouse cerebellum (61) and play a key role in the PTN-binding and NOP activities of CS/DS chains from embryonic pig brain (19) and shark liver (20). These findings suggest that CS/DS chains containing iB units may exert neuritogenic activity by recruiting PTN and mimicking the activity of heparan sulfate of N-syndecan. The identification of the PTN-binding hexasaccharide ΔA-iB-iB in this study directly demonstrated the critical role of the iB unit in the interaction with PTN. HGF binds to and activates the tyrosine kinase receptor cMet, and is also a versatile signal for developing neurons (62). GAGs are essential co-receptors for the activation of cMet (63). Our study using primary mouse hippocampal cells showed that SL-CS/DS hybrid chains recruited endogenous HGF to stimulate the outgrowth of neurites of hippocampal neurons probably through cMet (20). It is worth noting that heparin and heparin oligosaccharides stimulate the production of HGF at the post-translational level (64, 65), suggesting an additional possibility, that SS-CS/DS and ΔA-iB-iB also possess the HGF-inducing activity.
Because the requirement of IdoUA(2S) differed between previous (57) and the present findings and the structure of ΔA-iB-iB also looked different from that of a series of previously isolated D-unit-containing octasaccharides (21), it was hypothesized that the PTN-binding sequences must have a similar three-dimensional structure and electronic properties. Thus, in this study, the oligosaccharide-binding site in PTN was predicted by deriving the three-dimensional structure and the electrostatic features of the ESP distributions. This was carried out not only for the five oligosaccharides (ΔA-iB-iB, ΔE-D-A-D, ΔE-D-iA-D, ΔC-C-A-C, and ΔC-A-A-A), but also for PTN using advanced computational methods including homology modeling with amino acid sequence alignment, energy minimization with a hydration effect by molecular mechanics, and full geometry optimization with a solvent effect by DFT. This provided additional information on the structure and electronic properties of the octasaccharides as reported previously (24). As shown in Fig. 8 and supplemental Fig. S5, the ESP distributions of the PTN-binding oligosaccharides were clearly different particularly at the non-reducing ends from those of the non-binding oligosaccharides. It seems that PTN recognizes the features identified by the ESP shapes of the oligosaccharides having disulfated disaccharide units such as iB, D, and ΔE (also see the first paragraph of the “Discussion”). In addition, the ESP analysis demonstrated that the negative iso-surface is spread in restricted areas surrounding the sulfate groups of a disulfated disaccharide(s). The ESP distributions in the five isolated oligosaccharides showed features likely to enable electrostatic interactions with proteins or monoclonal antibodies, focusing on the spatial distribution of the electronegative zones and as exemplified by the recognition of the octasaccharides by the anti-CS monoclonal antibodies (23, 24, 66).
Previously, we showed that two analogous octasaccharide sequences ΔD-C-C-C and ΔC-C-A-D, but not another one ΔC-A-D-C, were recognized by the monoclonal antibody WF6 (24). It seems, however, in the present case, the number and localization of sulfate groups as well as the flexible IdoUA appear to be more crucial factors for the electrostatic surface. Yet, this study still provided additional important information on the electronic profiles such as shapes and spreading pattern of the electronegative zone, which are caused by the positions of sulfate and carboxyl groups in the conformation of oligosaccharides.
PTN, also known as heparin-binding growth-associated molecule, binds to heparin-type glycans as well to regulate the neurite extension and plasticity of hippocampal neurons (68). The minimum size of the PTN-binding site of heparin has been reported to be a 12–16-mer (69). In this study, the PTN affinity chromatography of SS-CS/DS-H-derived oligosaccharides showed that the 6–10-mer fractions retained the capacity to bind PTN but their affinity was weaker than that of the undigested polysaccharide chains, and 12-mer was the minimum oligosaccharide with affinity comparable with that of the parental polysaccharides. This may imply that in addition to a PTN-binding core oligosaccharide such as ΔΑ-iB-iB, extended sugar residues are required for maximal interaction with PTN as in the case of heparan sulfate (68). It remains to be determined whether heparan sulfate-type and CS/DS-type oligosaccharides bind to the same pocket of PTN, and if there is significant similarity in the three-dimensional structure and electronic properties between the two types of oligosaccharides.
This study also showed that SS-CS/DS is a potential candidate for a therapeutic agent as the expression of PTN, the related protein midkine, and HGF is increased in various human tumors, making them promising as targets for tumor therapy (34, 46). Importantly, the structural identification of the common sugar-binding domain for PTN and HGF as well as the sugar counterpart by the methods used here has provided a fundamental structural platform for the drug design of sugar or non-sugar small molecules, which may avoid any side effects of the parental polysaccharides. The obvious next step of this computational study is a docking simulation experiment of the protein and the corresponding sugar sequence(s).
Supplementary Material
Acknowledgments
We thank Accelrys K. K. for use of the Accelrys Material Studio and Discovery Studio and Prof. Timothy Hardingham (Manchester University) for critical reading of the manuscript.
This work was supported in part by Grants-in-aid for Exploratory Research 17659020 (to K. S.), Scientific Research B 16390026, 18390030, and 20390019 (to K. S.), Scientific Research on Priority Areas 14082207 (to K. S.), the Future Drug Discovery and Medical Care Innovation Program (to K. S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), and the Human Frontier Science Program RGP18/2005 (to K. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S3.
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- GAG
- glycosaminoglycan
- IdoUA
- l-iduronic acid
- GlcUA
- d-glucuronic acid
- HexUA
- hexuronic acid
- HexNAc
- N-acetyl-d-hexosamine
- Δ4,5HexUA
- 4,5-unsaturated hexuronic acid
- 2S, 4S, and 6S
- 2-O-sulfate, 4-O-sulfate, 6-O-sulfate, respectively
- A, B, C, D, and E units
- GlcUAβ1–3GalNAc(4S), GlcUA(2S)β1–3GalNAc(4S), GlcUAβ1–3GalNAc(6S), and GlcUA(2S)β1–3GalNAc(6S), and GlcUAβ1–3GalNAc(4S,6S), respectively
- iA, iB, iC, iD, and iE units
- IdoUAα1–3GalNAc(4S), IdoUA(2S)α1–3GalNAc(4S), IdoUAα1–3GalNAc(6S), IdoUA(2S)α1–3GalNAc(6S), and IdoUAα1–3GalNAc(4S,6S), respectively
- ΔA, ΔB, ΔC, ΔD, and ΔE units
- Δ4,5HexUAα1–3GalNAc(4S), Δ4,5HexUA(2S)α1–3GalNAc(4S), Δ4,5HexUAα1–3GalNAc(6S), Δ4,5HexUA(2S)α1–3GalNAc(6S), and Δ4,5HexUAα1–3GalNAc(4S,6S), respectively
- PTN
- pleiotrophin
- rh-PTN
- recombinant PTN
- HGF
- hepatocyte growth factor
- CSase
- chondroitinase
- 2-AB
- 2-aminobenzamide
- NOP
- neurite outgrowth-promoting
- E-CS/DS
- CS/DS from embryonic pig brain
- SL-CS/DS
- CS/DS from shark liver
- SS-CS/DS
- CS/DS from shark skin
- SS-CS/DS-U, SS-CS/DS-L, and SS-CS/DS-H
- SS-CS/DS fractions with no, low, or high affinity to PTN, respectively
- DFT
- density functional theory
- ESP
- electrostatic potential.
REFERENCES
- 1.Rodén L. (1980) in The Biochemistry of Glycoproteins and Proteoglycans (Lennarz W. J. ed) pp. 491–517, Plenum Publishing Corp., New York [Google Scholar]
- 2.Poole A. R. (1986) Biochem. J. 236, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hascall V. C. (1977) J. Supramol. Struct. 7, 101–120 [DOI] [PubMed] [Google Scholar]
- 4.Ruoslahti E. (1988) Annu. Rev. Cell. Biol. 4, 229–255 [DOI] [PubMed] [Google Scholar]
- 5.Maimone M. M., Tollefsen D. M. (1990) J. Biol. Chem. 265, 18263–18271; Correction (1991) J. Biol. Chem. 266, 14830 [PubMed] [Google Scholar]
- 6.Trowbridge J. M., Gallo R. L. (2002) Glycobiology 12, 117R–25R [DOI] [PubMed] [Google Scholar]
- 7.Li F., Ten Dam G. B., Murugan S., Yamada S., Hashiguchi T., Mizumoto S., Oguri K., Okayama M., van Kuppevelt T. H., Sugahara K. (2008) J. Biol. Chem. 283, 34294–34304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silbert J. E., Sugumaran G. (2002) IUBMB Life 54, 177–186 [DOI] [PubMed] [Google Scholar]
- 9.Cheng F., Heinegård D., Malmström A., Schmidtchen A., Yoshida K., Fransson L. A. (1994) Glycobiology 4, 685–696 [DOI] [PubMed] [Google Scholar]
- 10.Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., Kitagawa H. (2003) Curr. Opin. Struct. Biol. 13, 612–620 [DOI] [PubMed] [Google Scholar]
- 11.Sugahara K., Mikami T. (2007) Curr. Opin. Struct. Biol. 17, 536–545 [DOI] [PubMed] [Google Scholar]
- 12.Sugahara K., Yamada S. (2000) Trends Glycosci. Glycotech. 12, 321–349 [Google Scholar]
- 13.Nadanaka S., Clement A., Masayama K., Faissner A., Sugahara K. (1998) J. Biol. Chem. 273, 3296–3307 [DOI] [PubMed] [Google Scholar]
- 14.Clement A. M., Sugahara K., Faissner A. (1999) Neurosci. Lett. 269, 125–128 [DOI] [PubMed] [Google Scholar]
- 15.Hikino M., Mikami T., Faissner A., Vilela-Silva A. C., Pavão M. S., Sugahara K. (2003) J. Biol. Chem. 278, 43744–43754 [DOI] [PubMed] [Google Scholar]
- 16.Purushothaman A., Fukuda J., Mizumoto S., ten Dam G. B., van Kuppevelt T. H., Kitagawa H., Mikami T., Sugahara K. (2007) J. Biol. Chem. 282, 19442–19452 [DOI] [PubMed] [Google Scholar]
- 17.Nandini C. D., Mikami T., Ohta M., Itoh N., Akiyama-Nambu F., Sugahara K. (2004) J. Biol. Chem. 279, 50799–50809 [DOI] [PubMed] [Google Scholar]
- 18.Bao X., Nishimura S., Mikami T., Yamada S., Itoh N., Sugahara K. (2004) J. Biol. Chem. 279, 9765–9776 [DOI] [PubMed] [Google Scholar]
- 19.Bao X., Mikami T., Yamada S., Faissner A., Muramatsu T., Sugahara K. (2005) J. Biol. Chem. 280, 9180–9191 [DOI] [PubMed] [Google Scholar]
- 20.Li F., Shetty A. K., Sugahara K. (2007) J. Biol. Chem. 282, 2956–2966 [DOI] [PubMed] [Google Scholar]
- 21.Bao X., Muramatsu T., Sugahara K. (2005) J. Biol. Chem. 280, 35318–35328 [DOI] [PubMed] [Google Scholar]
- 22.Nandini C. D., Itoh N., Sugahara K. (2005) J. Biol. Chem. 280, 4058–4069 [DOI] [PubMed] [Google Scholar]
- 23.Blanchard V., Chevalier F., Imberty A., Leeflang B. R., Basappa, Sugahara K., Kamerling J. P. (2007) Biochemistry 46, 1167–1175 [DOI] [PubMed] [Google Scholar]
- 24.Pothacharoen P., Kalayanamitra K., Deepa S. S., Fukui S., Hattori T., Fukushima N., Hardingham T., Kongtawelert P., Sugahara K. (2007) J. Biol. Chem. 282, 35232–35246 [DOI] [PubMed] [Google Scholar]
- 25.Nakamura T., Nishizawa T., Hagiya M., Seki T., Shimonishi M., Sugimura A., Tashiro K., Shimizu S. (1989) Nature 342, 440–443 [DOI] [PubMed] [Google Scholar]
- 26.Bitter T., Muir H. M. (1962) Anal. Biochem. 4, 330–334 [DOI] [PubMed] [Google Scholar]
- 27.Saito H., Yamagata T., Suzuki S. (1968) J. Biol. Chem. 243, 1536–1542 [PubMed] [Google Scholar]
- 28.Kinoshita A., Sugahara K. (1999) Anal. Biochem. 269, 367–378 [DOI] [PubMed] [Google Scholar]
- 29.Kawashima H., Atarashi K., Hirose M., Hirose J., Yamada S., Sugahara K., Miyasaka M. (2002) J. Biol. Chem. 277, 12921–12930 [DOI] [PubMed] [Google Scholar]
- 30.Sugahara K., Okumura Y., Yamashina I. (1989) Biochem. Biophys. Res. Commun. 162, 189–197 [DOI] [PubMed] [Google Scholar]
- 31.Deepa S. S., Yamada S., Zako M., Goldberger O., Sugahara K. (2004) J. Biol. Chem. 279, 37368–37376 [DOI] [PubMed] [Google Scholar]
- 32.Faissner A., Clement A., Lochter A., Streit A., Mandl C., Schachner M. (1994) J. Cell Biol. 126, 783–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugahara K., Ohkita Y., Shibata Y., Yoshida K., Ikegami A. (1995) J. Biol. Chem. 270, 7204–7212 [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto K., Nakamura T., Sakai K., Nakamura T. (2008) Proteomics 8, 3360–3370 [DOI] [PubMed] [Google Scholar]
- 35.Delley B. (1990) J. Chem. Phys. 92, 508–517 [Google Scholar]
- 36.Delley B. (2000) J. Chem. Phys. 113, 7756–7764 [Google Scholar]
- 37.Perdew J. P., Burke K., Ernzerhof M. (1996) Phys. Rev. Lett. 77, 3865–3868 [DOI] [PubMed] [Google Scholar]
- 38.Klamt A., Schüürmann G. (1993) J. Chem. Soc. Perkin Trans. 25, 799–805 [Google Scholar]
- 39.Delley B. (2006) Mol. Simul. 32, 117–123 [Google Scholar]
- 40.Fernández D. P., Mulev Y., Goodwin A. R., Sengers J. M. H. (1995) J. Phys. Chem. Ref. Data 24, 33–70 [Google Scholar]
- 41.Iwasaki W., Nagata K., Hatanaka H., Inui T., Kimura T., Muramatsu T., Yoshida K., Tasumi M., Inagaki F. (1997) EMBO J. 16, 6936–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) in Current Protocols in Bioinformatics (Baxevanis A. D., Stein L. D., Stormo G. D., Yates J. R., III eds) Suppl. 15, pp. 5.6.1–5.6.30, John Wiley & Sons, Inc., New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 44.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 45.Fiser A., Do R. K., Sali A. (2000) Protein Sci. 9, 1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muramatsu T. (2002) J. Biochem. 132, 359–371 [DOI] [PubMed] [Google Scholar]
- 47.Shen M. Y., Sali A. (2006) Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comp. Chem. 4, 187–217 [Google Scholar]
- 49.MacKerell A. D., Jr., Brooks B., Brooks C. L., 3rd, Nilsson L., Roux B., Won Y., Karplus M. (1998) in The Encyclopedia of Computational Chemistry (Schleyer P. v. R. ed) Vol. 1, pp. 271–277, John Wiley & Sons, Chichester UK [Google Scholar]
- 50.Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C. S., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) J. Comp. Chem. 30, 1545–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee M., Salsbury F., Brooks C. (2002) J. Chem. Phys. 116, 10606–10614 [Google Scholar]
- 52.Cáceres A., Banker G. A., Binder L. (1986) J. Neurosci. 6, 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debus E., Weber K., Osborn M. (1983) Differentiation 25, 193–203 [DOI] [PubMed] [Google Scholar]
- 54.Michelacci Y. M., Dietrich C. P. (1975) Biochem. J. 151, 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamagata T., Saito H., Habuchi O., Suzuki S. (1968) J. Biol. Chem. 243, 1523–1535 [PubMed] [Google Scholar]
- 56.Maeda N., He J., Yajima Y., Mikami T., Sugahara K., Yabe T. (2003) J. Biol. Chem. 278, 35805–35811 [DOI] [PubMed] [Google Scholar]
- 57.Catlow K. R., Deakin J. A., Wei Z., Delehedde M., Fernig D. G., Gherardi E., Gallagher J. T., Pavão M. S., Lyon M. (2008) J. Biol. Chem. 283, 5235–5248 [DOI] [PubMed] [Google Scholar]
- 58.Deakin J. A., Blaum B. S., Gallagher J. T., Uhrín D., Lyon M. (2009) J. Biol. Chem. 284, 6311–6321 [DOI] [PubMed] [Google Scholar]
- 59.Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell. Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kinnunen T., Raulo E., Nolo R., Maccarana M., Lindahl U., Rauvala H. (1996) J. Biol. Chem. 271, 2243–2248 [DOI] [PubMed] [Google Scholar]
- 61.Mitsunaga C., Mikami T., Mizumoto S., Fukuda J., Sugahara K. (2006) J. Biol. Chem. 281, 18942–18952 [DOI] [PubMed] [Google Scholar]
- 62.Maina F., Klein R. (1999) Nat. Neurosci. 2, 213–217 [DOI] [PubMed] [Google Scholar]
- 63.Deakin J. A., Lyon M. (1999) J. Cell. Sci. 112, 1999–2009 [DOI] [PubMed] [Google Scholar]
- 64.Matsumoto K., Tajima H., Okazaki H., Nakamura T. (1993) J. Biochem. 114, 820–826 [DOI] [PubMed] [Google Scholar]
- 65.Sakiyama R., Fukuta K., Matsumoto K., Furukawa M., Takahashi Y., Nakamura T. (2007) J. Biochem. 141, 653–660 [DOI] [PubMed] [Google Scholar]
- 66.Deepa S. S., Yamada S., Fukui S., Sugahara K. (2007) Glycobiology 17, 631–645 [DOI] [PubMed] [Google Scholar]
- 67.Deleted in proof
- 68.Raulo E., Tumova S., Pavlov I., Pekkanen M., Hienola A., Klankki E., Kalkkinen N., Taira T., Kilpelaïnen I., Rauvala H. (2005) J. Biol. Chem. 280, 41576–41583 [DOI] [PubMed] [Google Scholar]
- 69.Fath M., VanderNoot V., Kilpeläinen I., Kinnunen T., Rauvala H., Linhardt R. J. (1999) FEBS Lett. 454, 105–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.