Abstract
The neonatal Fc receptor, FcRn, is responsible for the long half-life of IgG molecules in vivo and is a potential therapeutic target for the treatment of autoimmune diseases. A family of peptides comprising the consensus motif GHFGGXY, where X is preferably a hydrophobic amino acid, was shown previously to inhibit the human IgG:human FcRn protein-protein interaction (Mezo, A. R., McDonnell, K. A., Tan Hehir, C. A., Low, S. C., Palombella, V. J., Stattel, J. M., Kamphaus, G. D., Fraley, C., Zhang, Y., Dumont, J. A., and Bitonti, A. J. (2008) Proc. Natl. Acad. Sci. U.S.A., 105, 2337–2342). Herein, the x-ray crystal structure of a representative monomeric peptide in complex with human FcRn was solved to 2.6 Å resolution. The structure shows that the peptide binds to human FcRn at the same general binding site as does the Fc domain of IgG. The data correlate well with structure-activity relationship data relating to how the peptide family binds to human FcRn. In addition, the x-ray crystal structure of a representative dimeric peptide in complex with human FcRn shows how the bivalent ligand can bridge two FcRn molecules, which may be relevant to the mechanism by which the dimeric peptides inhibit FcRn and increase IgG catabolism in vivo. Modeling of the peptide:FcRn structure as compared with available structural data on Fc and FcRn suggest that the His-6 and Phe-7 (peptide) partially mimic the interaction of His-310 and Ile-253 (Fc) in binding to FcRn, but using a different backbone topology.
Keywords: Crystal Structure, Drug Design, Immunosupressor, Peptide Interactions, Protein Drug Interactions, FcRn, FcRn Inhibitor, Neonatal Fc Receptor
Introduction
The neonatal Fc receptor, FcRn,2 has been extensively studied for over two decades and is a membrane-bound receptor expressed in a variety of tissues (1, 2, 3). One of the key functions of FcRn is to provide a relatively long plasma half-life to IgG molecules. As an example, the plasma half-life of IgG in normal mice is ∼9 days, but only 1.4 days in FcRn-deficient mice (4). One of the hallmarks of IgG:FcRn binding interaction is its pH dependence, where full binding activity is achieved at pH 6.0, but negligible binding at pH 7.4. This pH selectivity likely contributes to the mechanism of IgG half-life extension by FcRn. Briefly, it is thought that after IgG is taken up nonspecifically into the cell by pinocytosis, IgG is able to bind FcRn in the acidic endosome (pH 6), thereby diverting the IgG molecule from degradation by the lysosome. IgG is then brought back to the cell surface by exocytosis and released into circulation when exposed to extracellular pH 7.4 (1).
Because FcRn function can regulate the half-life of IgG, the receptor represents a possible target for the treatment of humorally mediated autoimmune diseases (5, 6). Selective antagonists of the IgG:FcRn interaction have been studied over the last several years, and examples include monoclonal antibodies (7), mutants of the Fc region of IgG (8), and peptides (9). Our group has focused on studying the effect of anti-FcRn peptides on IgG:FcRn interaction in vitro and their effects on IgG catabolism in vivo. Using peptide phage display screening, we discovered a consensus motif, Gly-His-Phe-Gly-Gly-X-Tyr, where X is preferably a hydrophobic amino acid (10). The motif is enclosed by a cysteine disulfide loop forming an 11-amino acid loop including the cysteines. The peptide was optimized (11, 12) for inhibition of IgG binding to FcRn and stability in plasma, resulting in monomeric peptide SYN1327 (1) and dimeric peptide SYN1436 (2). Remarkably, peptide 2 was able to reduce the IgG concentrations in cynomolgus monkeys by up to 80% with repeated administration, and therefore may be a candidate for the treatment of humorally mediated autoimmune diseases (10).
FcRn is a 52-kDa heterodimeric glycoprotein consisting of a heavy chain and a light chain, β2-microglobulin (β2m). The x-ray crystal structure of the soluble extracellular domain of rat FcRn (herein referred to as srFcRn) was first reported in 1994, and its structure possessed a MHC-class I-like fold where the peptide groove is closed and therefore unable to bind peptides at that site (13). A low resolution x-ray structure of srFcRn in complex with rat Fc (rFc) was also reported in 1994, and the complex displayed an oligomeric ribbon-like structure of srFcRn:rFc molecules with a 2:1 stoichiometry, respectively (14). In 2001, a high resolution structure of the srFcRn:rFc complex was solved using a heterodimeric Fc that precluded the formation of the oligomeric ribbons (15). This structure revealed the details of the specific interaction between the two molecules including the pH-sensitive histidine salt bridges. The extracellular domain of human FcRn (herein referred to as shFcRn), was reported in 2000 and when compared with the structure of rat FcRn, generally resembles the rat structure, but with subtle differences near the Fc binding site such as the packing of certain residues, residue mutations and deletions (16). These subtle changes help rationalize the difference in hIgG binding affinity to human and rat FcRn (17).
Herein we obtained the x-ray co-crystal structure of deglycosylated soluble human FcRn (deg-shFcRn) in complex with peptide 1, a constituent peptide monomer of dimeric peptide 2. In addition, we obtained the x-ray co-crystal structure of deg-shFcRn in complex with dimeric peptide 3, a more soluble analog of peptide dimer 2 (Fig. 1). These data shed light on the peptide mechanism of action, and provide detailed insights into how the peptide is able to bind FcRn with high affinity, and block the IgG:FcRn protein-protein interaction.
FIGURE 1.
Sequence and primary structure of peptides 1–3. Solid lines between Pen and Cys amino acids denote a disulfide bond.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
Peptides 1–3 were synthesized using methodology as described previously (10, 11). Briefly, peptides were synthesized using solid-phase chemistry using standard Fmoc/tBu protocols and purified to >95% purity as determined by reversed-phase HPLC. Identities were confirmed by electrospray mass spectrometry.
Protein Expression and Purification
Soluble human FcRn was expressed using the glutamine synthetase expression system in CHOK1SV cells (Lonza Biologics, Berkshire, UK) and purified as described previously (10,18). The heavy chain of hFcRn was truncated at amino acid 274 to generate the soluble extracellular domain (18). The function of the shFcRn was confirmed by surface plasmon resonance (SPR) analysis of the binding of soluble IgG to immobilized sFcRn. shFcRn bound human IgG at pH 6 with affinities comparable to those reported in the literature and showed very weak or no binding at pH 7.4 (10). The Fc fragment of IgG1 used as a SPR control was expressed recombinantly in CHO cells as described above and purified by protein A affinity chromatography.
Deglycosylation of shFcRn
Soluble human FcRn in PBS was incubated with PNGaseF (NEB Cat. P0705L, 5000 units/mg protein) at 37 °C overnight. SDS-PAGE analysis (4–20%, Tris-Gly) confirmed the loss of several kDa in molecular weight off of the original shFcRn band (26). MES, pH 6 buffer (1 m), was added to the protein to buffer at pH 6. The protein solution was purified by gravity using IgG Sepharose 6 Fast Flow beads (GE Healthcare, Cat. 17-0969-01). The protein was loaded onto the equilibrated column, washed with 50 mm MES, pH 6 buffer, and eluted with 10 mm phosphate pH 7.4 buffer. Elution fractions were analyzed by SDS-PAGE, and fractions containing FcRn were pooled, and dialyzed twice into PBS. The concentration of the protein was determined by measuring the absorbance at 280 nm (Beckman DU-600, 1-cm path length) using the extinction coefficient value of 85,280 m−1 cm−1 and a molecular mass of 42,104 Da. The concentration of deglycosylated FcRn (deg-shFcRn) was 0.82 mg/ml in PBS and frozen at −80 °C.
Surface Plasmon Resonance
SPR measurements were performed using a Biacore 3000 as described previously (10). Briefly, either shFcRn or deg-shFcRn were coated to the dextran surface of a CM5 sensor chip using amine coupling chemistry. The equilibrium response for each peptide dilution at pH 6 was plotted against concentration and analyzed using the steady state affinity model (included in the BiaEval software) to determine Kd values. Equilibrium responses resulting from Fc binding at pH 6 were fit using a model that describes two non-interacting binding sites using nonlinear regression analysis to obtain the two KD values. This model was found previously to best describe the Fc:FcRn binding data with immobilized FcRn (16, 19).
Gel Filtration of deg-shFcRn
Prior to crystallization, deg-shFcRn (0.82 mg/ml) in PBS, pH 7.4, was further purified using a Superdex S200 10/300 eluted with 20 mm Tris-HCl, pH 7.50 and 150 mm NaCl. The purity was over 99% as determined by SDS-PAGE.
Crystallization and Data Collection
The protein sample was concentrated to 12 mg/ml using Amicon Ultra centrifuge filters (10,000 MWCO) and incubated with peptide 1 (1 mm) on ice for 1 h. Co-crystals of deg-shFcRn:peptide 1 were obtained by the sitting-drop vapor-diffusion method using a 96-well Greiner plate: 2 μl of protein:peptide complex were mixed with 2 μl of well solution containing 100 mm sodium phosphate/citric acid (pH 4.2), 20% (w/v) polyethylene glycol 3350 and 8% (v/v) ethanol at 4 °C. Small crystals appeared in 2 days, which were further optimized by multiple rounds of micro- and macro-seeding, resulting in diffraction quality crystals in 7 days. Before data collection, the crystals were transferred into a cryo-protectant solution made up of 10% glycerol in crystallization buffer, after which they were flash-frozen in liquid nitrogen for data collection.
Crystals of the deg-shFcRn:peptide 3 complex were grown using protein and peptide concentrations as described above. The crystallization condition involved 2 μl of protein:peptide complex mixed with 2 μl of crystallization buffer consisting of 100 mm phosphate/citric acid, pH 4.2, 22% (w/v) PEG 1000 and 8% (v/v) ethanol at room temperature. Initial micro crystals were improved by a combination of micro- and macro-seeding, which led to crystals suitable for data collection. The crystals were flash-frozen in liquid nitrogen for data collection. Diffraction image data were collected at the ALS at beam line 4.2.2 (deg-shFcRn with peptide 1) and at the APS at beam line 22-ID (deg-shFcRn with peptide 3).
Structure Determination
Diffraction data for the deg-shFcRn crystal containing peptide 1 were processed using the program d*TREK (20) to a resolution of 2.6 Å. The structure was solved using the molecular replacement program PHASER (21) from the CCP4 program suite (22) using the apo crystal structure (PDB entry: 1EXU) as search model and data to 3.0 Å resolution. Four independent copies of the FcRn protein were placed within the crystal asymmetric unit. When using default program parameters this was a unique solution. There were only two minor packing violations between molecules in the crystal and the final Z-score of 49.2 indicated a correct solution. Strong additional electron densities were observed at equivalent sites in each copy of the protein into which peptide 1 could be modeled. Cycles of minor model refitting were carried out with the MIFit (25) software interspersed with refinement using the REFMAC5 program from the CCP4. A final electron density map of bound peptide 1 is shown in supplemental Fig. S2.
Diffraction data for the deg-shFcRn crystal containing peptide 3 was processed using the HKL software (23) to a resolution of 3.1 Å. This structure was solved by molecular replacement with the PHASER program using the protein components of the structure containing peptide 1, finding four copies of the protein in the crystal asymmetric unit. This was the only acceptable solution produced by the program. There were no crystal packing violations and the final Z-score of 28.7 was very strongly indicative of a correct solution. Although not included in the search model, density at two adjacent peptide binding sites in the better ordered pair of proteins was clearly visible and somewhat scattered densities were observed at the corresponding sites in the two more poorly ordered copies of the protein. Despite the relatively low resolution of this structure, the shape of the stronger pair of densities was well-fit by two copies of peptide 1 in the conformation previously modeled, with a disordered linker region. This structure model for the two peptide units was docked into the density and the resulting protein-peptide complex was lightly refined with REFMAC5. Based on a data completeness threshold value of 75% the nominal resolution for this structure determination is ∼3.3 Å (Table 2). A final electron density map of bound peptide 3 is shown in supplemental Fig. S2.
TABLE 2.
Data collection and refinement statistics
Statistical values given in parenthesis refer to the highest resolution bin.
| Data collection | deg-shFcRn:Peptide 1 | deg-shFcRn:Peptide 3 |
|---|---|---|
| Space group | C2221 | P21 |
| Unit cell (Å) | 114.3, 118.3, 248.3 | 68.1, 158.4, 82.5, ß = 90.1° |
| Resolution (Å) | 41.1-2.6 | 50.0-3.1 |
| Wavelength (Å) | 1.0 | 1.0 |
| Observations | 363,112 | 83,929 |
| Unique reflections | 51,434 | 29,051 |
| Completeness (%) | 98.7 (97.8) | 90.4 (58.0) |
| I/σ(I) | 8.4 (2.9) | 7.7 (1.1) |
| Rmerge | 0.115 (0.435) | 0.167 (0.635) |
| Refinement | ||
| Resolution (Å) | 40.5-2.6 | 36.6-3.3 |
| Reflections | 51,365 | 23,973 |
| Rwork | 0.261 | 0.314 |
| Rfree | 0.325 | 0.395 |
| No. of protein copies | 4 | 4 |
| No. of protein and peptide atoms | 11398 | 11241 |
| No. of waters | 183 | 0 |
| Wilson B-factor (Å2) | 37.7 | 88.8 |
| Average B-factors (Å2) | ||
| Protein | 32.9 | 111.7 |
| Peptide | 32.5 | 105.5 |
| Water | 25.5 | - |
| RMS deviations from ideality | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles (°) | 0.961 | 1.184 |
| Ramachandran plot outliers (%) | 0.3 | 2.3 |
All processed and reduced reflection data within the specified resolution limits were used in the structure refinement. Assignment of Ramachandran plot outliers (Table 2) is based on phi-psi distributions determined from the Richardson laboratory (24) using a threshold for outlier detection incorporating 99.95% of their reference data.
The structures of peptides 1 and 3 in complex with deg-shFcRn and diffraction data have been deposited with the RCSB Protein Data Bank (entry IDs: 3M17 and 3M1B, respectively).
Structure Analysis
Accessible surface area calculations (25) were carried out using the version of the program AREAIMOL from the CCP4 program suite using a probe radius of 1.4 Å. Shape complementarity was analyzed using the program SC (28) with a probe radius of 1.7 Å. Structure superposition calculations and molecular graphics images were created using MIFit (26).
The superposition of the protein structure in complex with peptide 1 with the apo structure (PDB entry: 1EXU) was carried out using all 354 common Cα atoms from chain A (heavy chain) and B (β2-microglobin domain).
The docking model for the human FcRn:Fc complex from the rat form (15) (PDB entry: 1I1A) was developed by superimposing equivalent Cα atoms in the FcRn:Fc interface region from our FcRn model containing peptide 1 onto the FcRn component of the complex and also superimposing a human Fc structure (PDB entry: 1DN2) onto the Fc component of the complex. The interface residues used in these calculations were defined to be those identified by Martin et al. (see Table 3 in Ref. 15) as the 20 interacting amino acids in the rat FcRn:Fc complex.
RESULTS
Deglycosylated Human FcRn
The soluble extracellular domain of human FcRn (shFcR) was expressed in CHOK1SV cells and purified as described previously (10). Initial attempts to crystallize shFcRn in the presence of peptides 1 and 2 were unsuccessful. Although the carbohydrate in srFcRn was found to contribute to Fc binding via Asn-128, this glycosylation site is not present in human FcRn (15). We hypothesized that the glycosylation pattern of shFcRn may be hindering the formation of the co-crystals, and that removal of all shFcRn glycosylation would enhance crystal formation. Based on the available structures of shFcRn and srFcRn:rFc, the glycosylation site at Asn-102 is distant from the Fc:FcRn binding interface (15), and also the srFcRn:srFcRn dimerization contact points (33). Therefore, this modification was not expected to compromise the FcRn-peptide, FcRn-Fc interactions. Treatment of shFcRn with PNGase generated the Asn-102-deglycosylated analog of shFcRn (deg-shFcRn, supplemental Fig. S1). The binding affinity of peptide 1 and Fc to both shFcRn and deg-shFcRn were determined by surface plasmon resonance (SPR), and were unchanged in comparison to the glycosylated form of FcRn. (Table 1) (see supplemental materials). This confirmed that the N-linked carbohydrates of human FcRn do not contribute to peptide or Fc binding.
TABLE 1.
Surface plasmon resonance data comparing the binding affinities of shFcRn and deg-shFcRn to peptide 1 and human Fc at pH 6
| Analyte | shFcRn | deg-shFcRn | |
|---|---|---|---|
| nm | nm | ||
| Peptide 1 | 39 | 40 | |
| Human Fca | KD1 | 415 ± 62 (50 ± 6) | 466 ± 132 (53 ± 6) |
| (n = 4) | (n = 5) | ||
| KD2 | 7.5 ± 0.8 (50 ± 6) | 8.8 ± 2.6 (47 ± 6) | |
| (n = 4) | (n = 5) | ||
Structure of Peptide 1:deg-shFcRn
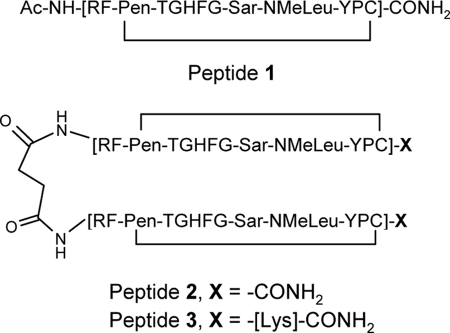
The x-ray crystal structure of peptide 1 in complex with deg-shFcRn was determined to 2.6 Å resolution and contained four proteins and four peptides in the asymmetric unit (Table 2), with each copy possessing similar structure. Peptide 1 is bound to FcRn at the site implicated in Fc binding (Fig. 2A). Each peptide covers an area of ∼360 Å2 on the surface of the FcRn molecule, and ∼51% of this area is comprised of non-polar atom types. The shape complementary (28) index for the protein-peptide interaction is ∼0.83. Since there is no published structure of human FcRn (hFcRn) in complex with human Fc (hFc), we modeled the hFcRn:hFc complex by overlaying a crystal structure of the hFcRn protein (this work) onto the homologous acids in the structure of a rat FcRn:rat Fc complex (PDB entry: 1I1A) (Fig. 2B). The modeling results indicate that peptide 1 does bind at the Fc binding site (vide infra).
FIGURE 2.
Comparison of the deg-shFcRn:peptide 1 structure and a model of human FcRn:Fc showing that the peptide and Fc occupy the same binding site. A, crystal structure of peptide 1 in complex with deg-shFcRn at 2.6 Å resolution depicting the binding location of peptide 1 on FcRn. B, model of the human FcRn:Fc complex obtained by overlaying a crystal structure of the human FcRn protein (this work) and the rat FcRn:Fc complex (PDB entry: 1I1A). For both figures, FcRn is depicted in cartoon form, and the peptide or Fc is represented by a purple and blue tube, respectively.
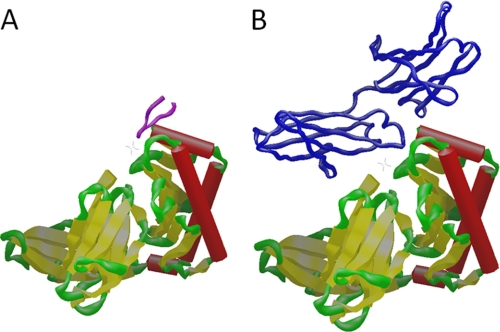
The structure of deg-shFcRn bound to peptide 1 was nearly identical to that of the apo structure of shFcRn (PDB: 1EXU) (16) with a rms difference of 0.96 Å (Fig. 3). These data are consistent with the rFc:srFcRn structure where no major conformational changes in srFcRn were observed upon rFc binding (15). In the peptide 1:deg-shFcRn structure, there was only a small shift in the loop around amino acid Lys-85 which appears to be both pulled toward the peptide by ∼3 Å and somewhat reordered so that Tyr-11 from the peptide approaches Gly-83 in FcRn and Phe-7 (amide NH) from the peptide makes a water-mediated interaction with Gly-86 (CO) from FcRn (Fig. 3). In addition, amino acids 52–59 in deg-shFcRn, which are distant from the peptide 1 binding site, are now visible and were previously disordered in structure 1EXU (16).
FIGURE 3.
A, overlay of the main chain atoms of the crystal structure of peptide 1 (purple) in complex with deg-shFcRn (green) with the apo structure (PDB entry: 1EXU, blue). The loop region from amino acid 52–59 (red) was not seen in the apo crystal structure (1EXU) but is resolved in the crystal form containing peptide 1. B, close-up of the main chain atoms of the conformation of the loop region around Gly-84 from deg-shFcRn that is modified in response to peptide binding relative to the conformation seen in the apo structure. The peptide 1 side-chains of His-6 and Phe-7 are also shown. An ordered water molecule is bound between the amide proton from Gly-86 on deg-shFcRn and the amide carboxyl from Phe-7 on the peptide as shown by the dashed line.
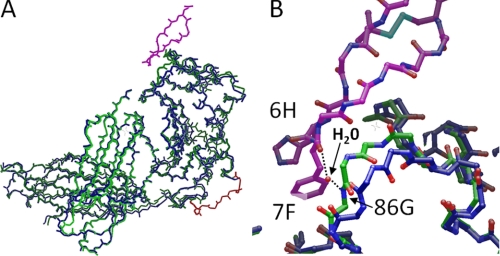
The three-dimensional structure of peptide 1 is stabilized by four internal hydrogen bonds, Pen3(NH)-Tyr11(CO), Gly5(NH)-Sar9(O), and Tyr11(NH)-Pen3(CO), Cys13(NH)-Arg1(CO) as well as a disulfide bridge between Pen3 and Cys-13. This suggests that the peptide may be well-structured prior to binding to FcRn. Three peptide residues are responsible for the majority of the contact with deg-shFcRn: His-6, Phe-7, and Tyr-11 (Fig. 4). The side-chain of His-6 forms both a stacked interaction with the side-chain of Trp-131, and a hydrogen bond/ionic interaction with Glu-115. The amide protons of both His-6 and Phe-7 on peptide 1 form putative hydrogen bonds with the side-chain of Glu-133. In addition, the side-chain of Phe-7 makes hydrophobic contact with the side chains of Leu-112 and Trp-131 from FcRn, as well as an edge-face interaction with His-6. Tyr-11 is also at the contact surface, and its phenolic hydroxyl makes an apparent end-on-end hydrogen bond with Tyr-88 of FcRn. Amino acids from the β2m domain of FcRn are too far from the peptide to form direct interactions, with Ile1 making the closest approach at a distance ∼4.5 Å.
FIGURE 4.
Key amino acids responsible for the binding of peptide 1 (purple) to deg-shFcRn (yellow).
Peptide 1 is found at a crystal contact and forms a short β-sheet, primarily through interactions with the peptide backbone of Phe2 on both peptides (CO-NH × 2). This peptide-peptide interaction appears to be a crystallization artifact rather than a potential model for the dimeric peptide 3. Here the two copies of peptide 1 are arranged in anti-parallel fashion with a small overlapping region such that the two N termini are ∼13 Å apart, which is greater than the possible span of the linker region in peptide 3.
Comparison of Peptide 1 with Human Fc
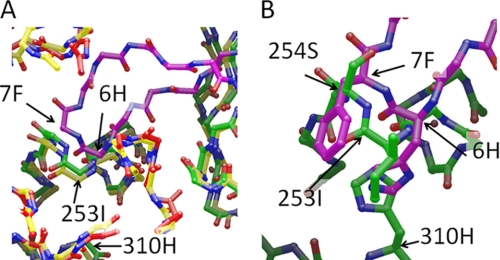
Peptide 1 binds to FcRn at the same general binding site as Fc, despite having no sequence homology to Fc. Because there is no available structure of the human FcRn in complex with human IgG, a comparison of the peptide 1 binding conformation and interactions with those forming the human FcRn:human Fc complex can be approached by using the atomic resolution structure of the rat FcRn:rat Fc complex (15) (PDB entry: 1I1A). A docking model for the human FcRn:Fc complex was created by superimposing both our human deg-shFcRn structure and an atomic resolution model of human Fc (for example, PDB ID: 1DN2) onto the structure of the rat FcRn:rat Fc complex by using the positions of the Cα atoms of key residues at the interface. These models indicate that the position of the His-6 side-chain from peptide 1 occupies the same approximate location as the side chain His-310 from hFc. However, the peptide backbone is positioned in a completely different location from protein backbone atoms in hFc and does not appear to map onto the hFc structure (Fig. 5). Furthermore, it appears that both His-6 and Phe-7 from the peptide have taken the place of critical hFc residue Ile-253 in packing against Trp-131, albeit with different backbone topology given that His-310 and Ile-253 are from discontiguous loops in hFc. From the rat Fc:FcRn structure (15), it was observed that the amide NH of Ile253 (hFc) forms a putative hydrogen bond with Glu-133 in rat FcRn. Interestingly, the amide protons of both His-6 and Phe-7 also appear to form this putative hydrogen bonding interaction with Glu-133.
FIGURE 5.
Overlay of peptide 1 with the docking model for the human FcRn:Fc complex. A, main chain bonds for the template structure (rat FcRn:Fc complex) (15) are colored yellow, the human FcRn protein with peptide 1 bound (this work) is shown with main chain bonds colored red, and the human Fc structure is shown with main chain bonds colored green. The main chain atoms from the peptide are shown in purple. The main chain traces for the human Fc and FcRn components align well with the rat FcRn:Fc complex in this region. B, close-up of all atom detail of the peptide in the region of His-6/Phe-7 with the aligned human Fc structure. Amino acids His-310 and Ile-253 are positioned so as to be approximately comparable to His-6 and Phe-7 but the main chain trace is different.
Structure of Peptide 3:deg-shFcRn
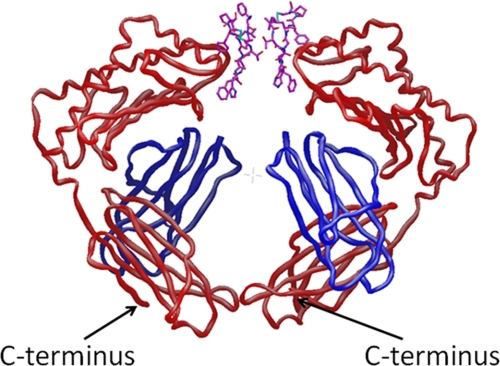
Dimeric anti-FcRn peptide 2 was active in a transgenic mouse model of IgG catabolism, whereas monomeric peptide 1 was inactive at similar doses (10). Therefore, it was of great interest to obtain the crystal structure of dimeric peptide 2 in complex with FcRn to help understand its mechanism of action. Because of solubility limitations with peptide 2, the equipotent (26) analog peptide 3, with one additional C-terminal Lys residue on each C termini, was used in crystallization studies. Peptide 3 was crystallized in complex with deg-shFcRn and the structure was determined to 3.3 Å resolution with molecular replacement using the existing peptide 1:deg-shFcRn structure as a model. The asymmetric unit contains 4 copies of deg-shFcRn, and one copy of peptide 3 bridging two molecules of deg-shFcRn (Fig. 6). Although crystals only diffracted to limited resolution, the structure data clearly demonstrate that one peptide 3 molecule can bind to two deg-shFcRn proteins at the same time. This is consistent with previous SPR data (10) with peptide 2, which suggested such a 2:1 binding interaction and helps explain the dramatic boost in in vitro activity for the peptide dimers over that of the peptide monomers (12). The linker consisting of Arg-succinyl-Arg-Phe is not visible in this structure due to inherent flexibility in this region and/or the limited resolution of the data. The Lys residues, added to the C terminus of peptide 2 to enable crystallization, were not near the binding interaction, consistent with previous structure-activity data for the C terminus of the peptide (11). Note that the two C termini of deg-shFcRn dimer are arranged side-by-side at the opposite ends of peptide binding. In full-length FcRn, the C terminus leads to a putative transmembrane domain that was removed for the purposes of crystallization. Hence, this packing arrangement could be functionally significant and is consistent with the possibility that peptide 3 can bridge two FcRn molecules on intracellular membranes.
FIGURE 6.
Crystal structure of peptide 3 in complex with deg-shFcRn to 3.3 Å resolution. The traces of the FcRn heavy chain is shown in red, the chain traces of the β2-microglobin domains is shown in blue, and the atomic model for peptide 3 is shown with bonds drawn in purple. The linker connecting the two peptide monomers, consisting of Arg-succinyl-Arg-Phe, is not visible in this structure due to inherent flexibility in this region and/or the limited resolution of the data.
Similar to the peptide 1 structure, no significant interactions were observed between the paired deg-shFcRn proteins. In addition, only minor interactions occurred between the two cyclized peptide domains of peptide 3, including possible hydrogen bonds between Thr4A(βOH)-Phe2B(CO), Thr4A(CO)-Thr4B(βOH) and a hydrophobic interaction between the side chains of Pen3A and 10MeLeuB.
Correlation of X-ray Structural Data with SAR Data
Structure-activity relationships (SAR) for a series of monomeric peptides using in vitro assays has been reported and suggested the residues and molecular interactions important for peptide binding to shFcRn (11). The x-ray crystallographic data are consistent with those data, and aid our understanding and interpretation of the SAR data. For example, substitutions at His-6 of peptide 1 favored amino acids with side-chains that are both aromatic and positively charged, such as para-guanylphenylalanine. This preference correlates with the aromatic stacked interaction of the His-6 side-chain with Trp-131 and the hydrogen bonding/charged interaction with Glu-115. Tyr-11 was also found to contribute significantly to the peptide affinity for FcRn as removal of the phenolic moiety resulted in a loss in binding affinity of >40-fold. Indeed, the phenolic hydroxyl is involved in a hydrogen bonding interaction with the phenolic hydroxyl of Tyr-88 on FcRn. Also, when backbone amide protons in peptide 1 were substituted with methyl groups at Gly-5, His-6, Phe-7, and Tyr-11, a decrease in binding to FcRn was observed. From the current structure, these data correlate with the putative hydrogen bonds Gly5(NH)-Sar9(O), His6(NH)-Glu133(CϵOO), Phe7(NH)-Glu133(CϵOO), Tyr11(NH)-Pen3(CO), respectively.
The substitution of Pen3 for Cys-3 in the optimization of peptide 1 for binding to shFcRn led to an increase in binding affinity of ∼20-fold, and was a particularly useful substitution in improving the binding affinity of the peptide motif, although the reason for the boost in affinity was not understood at the time. From the structures, Pen3 does not appear to have direct contacts with shFcRn, and therefore likely contributes to the binding affinity by generating more favorable and stabilizing torsional angles for the rest of the peptide including nearby hydrogen bond Tyr11(NH)-Pen3(CO).
It was determined previously that truncation of the amino acids on the N-terminal side of the disulfide loop resulted in a reduction in in vitro activity, whereas truncation of residues on the C-terminal side of the disulfide loop resulted in no change in in vitro activity (11). The observation of the Cys13(NH)-Arg1(CO) internal hydrogen correlate well with the fact that removal of Arg-1 further reduces binding activity, by removing this structure-stabilizing hydrogen bond.
One residue in peptide 1, which could not be substituted with l-Ala or d-Ala without >50-fold loss in activity was Gly-5. This suggested that steric limitations may prevent further functionalization at this site. The x-ray structure confirms this hypothesis, as both α protons are directed into the FcRn surface with minimal space available for a possible side-chain.
The SAR data also indicated a preference for hydrophobic amino acids at the Phe-2 and Leu-10 positions. The side-chain of Leu-10 is ∼5 Å away from the side-chain of Phe-2, and therefore such a hydrophobic interaction could enhance the stability of the overall peptide structure, and hence its activity.
Lastly, peptide dimers were linked by connecting the N termini of their constituent monomers (peptide 3) but also in a N-to-C-terminal and C-to-C-terminal fashion (12). The SAR data suggested that both termini are in close proximity since there was little difference between the in vitro data of the differently oriented analogs. Indeed, the peptide:deg-shFcRn structures indicate that both the N and C termini are in close proximity.
DISCUSSION
The anti-FcRn peptides were originally discovered by peptide phage display, and chemically optimized for FcRn binding using extensive structure-activity relationships prior to the availability of the structures reported herein. Chemically optimized peptide dimer 2 was able to inhibit FcRn in vivo and can reduce endogenous levels of IgG in cynomolgus monkeys by up to 80%, and therefore may be of use in the treatment of IgG-mediated autoimmune diseases. The x-ray structures of both optimized peptides 1 and 3 in complex with human FcRn reveal the atomic details of the peptide:FcRn interactions, and how the peptides inhibit the binding of the natural ligand IgG (Fc). Prior in vitro data suggested that the peptides were competitive inhibitors of the hIgG:shFcRn interaction, and that the peptides may bind to the same site on FcRn as does Fc. Indeed, the structures of peptide 1 and 3 demonstrate that this is the case.
One of the most remarkable aspects of the structures of peptides 1 and 3 in complex with deg-shFcRn is that only three amino acid residues (His-6, Phe-7, Tyr-11) make up the vast majority of the binding interaction with FcRn. The smaller buried surface area of 360 Å2 is able to effectively compete with the Fc:FcRn protein-protein interface of 1870 Å2 (rat) (15), and has near-equivalent (peptide 1) or higher affinity (peptide 2) for shFcRn than does Fc (10). The majority of the amino acids comprising peptides 1 and 3 are present to direct and constrain those three amino acids into a precise conformation for binding to FcRn. The calculation of shape complementarity (28) shows that peptide 1 makes a better match (Sc, 0.83) to the surface topography of the human FcRn molecule than does Fc bound to FcRn (Sc, 0.71) in proteins from rat. These data demonstrate that only key “hotspots” (29) need to be targeted on the FcRn:IgG protein-protein interface for effective binding/inhibition, and is reminiscent of other peptide-protein interactions where a large protein binding partner can be replaced by smaller peptides by targeting such key hot-spots (30, 31). These structures further support the possibility of discovering small molecule inhibitors to the interaction (32).
Prior co-crystallization attempts of the FcRn:Fc complex (rat, mouse, and human FcRn, wild-type Fc) led to low resolutions structures of FcRn:Fc molecules that revealed general packing interactions and a stoichiometry of 2:1, but precluded high-resolution structural information of the Fc:FcRn binding interface (14,15). Two different forms of FcRn:Fc dimers were evident in these structures. The first form, named the “lying down” complex, is a structure where FcRn dimerizes via its α3/β2m domains and the carbohydrate attached to residue Asn-225, and is in complex with one side of bivalent Fc (CH2/CH3 domain). The other form, named the “standing up” complex, uses bivalent Fc to bridge two FcRn molecules at each Fc binding site. It is not known if one form of the complex is more relevant than the other, or if they co-exist as observed in the x-ray oligomeric ribbon structures (33). However, it was suggested (14) that the lying-down was the more relevant complex for FcRn binding to Fc, primarily due to 1) site-directed mutagenesis data that suggested that FcRn dimers (e.g. without a bridging ligand) bind preferentially to IgG and 2) modeling data that suggested that the Fab arms of IgG would be unable to bind to FcRn at the cell surface in the standing-up complex because of steric crowding at the membrane surface, but easily accommodated in the lying-down complex. The arrangement of FcRn dimers observed in the structure of peptide 3:deg-shFcRn is consistent with the “standing up” complex, and is likely relevant to the mechanism by which peptide 3 and similar anti-FcRn peptide dimers inhibit FcRn function in vivo. Therefore, it may be possible that the anti-FcRn dimers and Fc use different orientations of FcRn to form their 2:1 complexes. Note that peptide monomer 1 was not active in a mouse model of IgG catabolism, even at elevated doses (10), which suggests that such a 2:1 complex may be necessary for inhibition of the IgG-FcRn recycling pathway. However, because the dimeric peptides were >100-fold more potent than their constituent monomers in vitro (12), it is possible that more potent monomeric peptides would also be active in the mouse model for IgG catabolism. Lastly, the ability of the monomeric and dimeric peptides to enter the endosome is unknown, and may also contribute to differences in monomeric and dimeric peptide activities in vivo. However, cellular uptake by nonspecific pinocystosis, thought to be the primary route of entry into the cell by IgG, should be similar for monomeric and dimeric peptides. The possibility exists that there are differences in receptor-mediated uptake from the transient expression of FcRn at the cell surface at the time of IgG exocytosis by FcRn (pH 7.4). Because peptide dimer 2 binds >200-fold more tightly to FcRn at pH 7.4 than peptide monomer 1 (10), such differences in uptake by this mechanism may be expected.
One of the hallmarks of the Fc:FcRn binding interaction is its pH dependence; Fc has full binding activity toward FcRn at pH 6, but minimal binding at pH 7.4. It was demonstrated previously that there were no significant differences in the structure of FcRn when crystallized at pH 6.5 versus pH 8 (34). These data suggested that the pH dependence of the interaction can be attributed to the chemical properties of the proteins themselves, and not to conformational or allosteric effects at different pH. Structural data (15,16) and other site-directed mutational studies (27) implicated Glu-115 and Asp-130 on human FcRn (Glu-117 and Glu-132 in rat FcRn) as being responsible for the pH-dependent binding to residues His-310 and His-435 on Fc. A proton on the histidine imidazole side-chain can be titrated between pH 6 and 7.4 and results in an ionic molecular switch for binding to FcRn. Interestingly, peptide 1 also binds to one of these residues (Glu-115) with His-6 and also possesses a pH-dependent binding interaction with FcRn, albeit with only ∼5-fold reduction in affinity at pH 7.4 versus pH 6 (10).
The structures of peptide 1 and 3 in complex with deg-shFcRn correlate well with the structure-activity relationships that were studied extensively (11,12). As mentioned previously, methylation of certain backbone amide bonds led to losses in in vitro activity, and these data correlate with the structural data indicating that those amide protons are involved in binding interactions either with FcRn or within the peptide itself. In contrast, introduction of N-methyl moieties at positions Sar9 and NMeLeu10 each improved the peptide binding affinity for FcRn by ∼2–3-fold (11). The structure of peptide 1 indicates that both additional N-methyl groups are solvent exposed, and are not involved in any significant interaction with either peptide 1 itself, or with FcRn. The source of the affinity improvement may be due to preclusion of alternate lower affinity peptide folds that may have involved amide protons from amino acids 9 (Sar) and 10 (NMeLeu). The elimination of amide protons at positions 9 and 10 removes competition for some of the available hydrogen bonding interactions, likely allowing more favorable hydrogen bonding interactions to occur. For example, by removing the possibility of the NMeLeu10 amide proton from forming a hydrogen bonding interaction, it favors the adjacent Sar9 amide carbonyl to form the internal hydrogen bond with Gly-5 (Gly5(NH)-Sar9(O)).
Lastly, one of the most specific interactions observed by SAR was the phenolic hydroxyl of Tyr-11, where removal of this hydroxyl group resulted in >40-fold loss in in vitro activity (11). Indeed, the observation of the Tyr11-Tyr88 (OH-OH) interaction validates this previous data. The analogous residue in rat FcRn is Phe-88, and this lack of phenolic moiety in rat FcRn likely contributes to the limited binding affinity of the anti-FcRn peptide family for rat FcRn (10).
In conclusion, the structures of peptides 1 and 3 in complex with deg-shFcRn were determined using x-ray crystallography. It was found that monomeric peptide 1 binds to deg-shFcRn in the same general location as Fc, confirming that the anti-FcRn peptides are competitive inhibitors of the human IgG:human FcRn interaction. The structure of dimeric peptide 3 demonstrates that the peptide can bind simultaneously to two deg-shFcRn protein molecules and may be relevant to its mechanism of FcRn inhibition in vivo and to the design of future FcRn inhibitors.
Supplementary Material
Acknowledgments
We thank the protein production and peptide synthesis groups at Biogen Idec/Syntonix for technical assistance and Dr. Alan Bitonti for helpful discussions. Crystal data collection assistance was provided by Jay Nix (ALS) and Rick Walter (Shamrock Structures).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (codes 3M17 and 3M1B) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FcRn
- neonatal Fc receptor
- deg-shFcRn
- deglycosylated soluble human FcRn
- SPR
- surface plasmon resonance
- Pen
- L-penicillamine amino acid
- Sar
- sarcosine
- NMeLeu
- N-methylleucine
- SAR
- structure-activity relationships
- PDB
- Protein Data Bank.
REFERENCES
- 1.Roopenian D. C., Akilesh S. (2007) Nat. Rev. Immunol. 7, 715–725 [DOI] [PubMed] [Google Scholar]
- 2.Ghetie V., Ward E. S. (2000) Annu. Rev. Immunol. 18, 739–766 [DOI] [PubMed] [Google Scholar]
- 3.Andersen J. T., Sandlie I. (2009) Drug Metab. Pharmacokinet. 24, 318–332 [DOI] [PubMed] [Google Scholar]
- 4.Roopenian D. C., Christianson G. J., Sproule T. J., Brown A. C., Akilesh S., Jung N., Petkova S., Avanessian L., Choi E. Y., Shaffer D. J., Eden P. A., Anderson C. L. (2003) J. Immunol. 170, 3528–3533 [DOI] [PubMed] [Google Scholar]
- 5.Liu Z., Roopenian D. C., Zhou X., Christianson G. J., Diaz L. A., Sedmak D. D., Anderson C. L. (1997) J. Exp. Med. 186, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Z., Lennon V. A. (1999) N. Engl. J. Med. 340, 227–228 [DOI] [PubMed] [Google Scholar]
- 7.Liu L., Garcia A. M., Santoro H., Zhang Y., McDonnell K., Dumont J., Bitonti A. J. (2007) J. Immunol. 178, 5390–5398 [DOI] [PubMed] [Google Scholar]
- 8.Vaccaro C., Zhou J., Ober R. J., Ward E. S. (2005) Nat. Biotechnol. 23, 1283–1288 [DOI] [PubMed] [Google Scholar]
- 9.Low S. C., Mezo A. R. (2009) AAPS J. 11, 432–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mezo A. R., McDonnell K. A., Hehir C. A., Low S. C., Palombella V. J., Stattel J. M., Kamphaus G. D., Fraley C., Zhang Y., Dumont J. A., Bitonti A. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2337–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mezo A. R., McDonnell K. A., Castro A., Fraley C. (2008) Bioorg. Med. Chem. 16, 6394–6405 [DOI] [PubMed] [Google Scholar]
- 12.McDonnell K. A., Low S. C., Hoehn T., Donnelly R., Palmieri H., Fraley C., Sakorafas P., Mezo A. R. (2010) J. Med. Chem. 53, 1587–1596 [DOI] [PubMed] [Google Scholar]
- 13.Burmeister W. P., Gastinel L. N., Simister N. E., Blum M. L., Bjorkman P. J. (1994) Nature 372, 336–343 [DOI] [PubMed] [Google Scholar]
- 14.Burmeister W. P., Huber A. H., Bjorkman P. J. (1994) Nature 372, 379–383 [DOI] [PubMed] [Google Scholar]
- 15.Martin W. L., West A. P., Jr., Gan L., Bjorkman P. J. (2001) Mol. Cell 7, 867–877 [DOI] [PubMed] [Google Scholar]
- 16.West A. P., Jr., Bjorkman P. J. (2000) Biochemistry 39, 9698–9708 [DOI] [PubMed] [Google Scholar]
- 17.Ober R. J., Radu C. G., Ghetie V., Ward E. S. (2001) Int. Immunol. 13, 1551–1559 [DOI] [PubMed] [Google Scholar]
- 18.Gastinel L. N., Bjorkman P. J. #5, 623,053 U.S. Patent.
- 19.Vaughn D. E., Bjorkman P. J. (1997) Biochemistry 36, 9374–9380 [DOI] [PubMed] [Google Scholar]
- 20.Pflugrath J. W. (1999) Acta Cryst. D55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 21.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project, Number 4 (1994) Acta Cryst. D50, 760–763 [Google Scholar]
- 23.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24.Lovell S. C., Davis I. W., Arendal W. B., III, de Bakker P. I. W., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins Struct. Funct. Genet. 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 25.Lee B., Richards F. M. (1971) J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 26.MIFit 8 Manual (2008) Rigaku Americas Corporation, The Woodlands, TX [Google Scholar]
- 27.Vaughn D. E., Milburn C. M., Penny D. M., Martin W. L., Johnson J. L., Bjorkman P. J. (1997) J. Mol. Biol. 274, 597–607 [DOI] [PubMed] [Google Scholar]
- 28.Lawrence M. C., Colman P. M. (1993) J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 29.Clackson T., Wells J. A. (1995) Science 267, 383–386 [DOI] [PubMed] [Google Scholar]
- 30.DeLano W. L., Ultsch M. H., de Vos A. M., Wells J. A. (2000) Science 287, 1279–1283 [DOI] [PubMed] [Google Scholar]
- 31.Livnah O., Stura E. A., Johnson D. L., Middleton S. A., Mulcahy L. S., Wrighton N. C., Dower W. J., Jolliffe L. K., Wilson I. A. (1996) Science 273, 464–471 [DOI] [PubMed] [Google Scholar]
- 32.Wells J. A., McClendon C. L. (2007) Nature 450, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 33.Raghavan M., Bjorkman P. J. (1996) Annu. Rev. Cell Dev. Biol. 12, 181–220 [DOI] [PubMed] [Google Scholar]
- 34.Vaughn D. E., Bjorkman P. J. (1998) Structure 6, 63–73 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.