Abstract
CCN2/connective tissue growth factor is highly expressed in hypertrophic chondrocytes and is required for chondrogenesis. However, the transcriptional mechanisms controlling its expression in cartilage are largely unknown. The activity of the Ccn2 promoter was, therefore, investigated in osteochondro-progenitor cells and hypertrophic chondrocytes to ascertain these mechanisms. Sox9 and T-cell factor (TCF)·lymphoid enhancer factor (LEF) factors contain HMG domains and bind to related consensus sites. TCF·LEF factors are normally repressive but when bound to DNA in a complex with β-catenin become activators of gene expression. In silico analysis of the Ccn2 proximal promoter identified multiple consensus TCF·LEF elements, one of which was also a consensus binding site for Sox9. Using luciferase reporter constructs, the TCF·LEF·Sox9 site was found to be involved in stage-specific expression of Ccn2. Luciferase, electrophoretic mobility shift assay (EMSA), and ChIP analysis revealed that Sox9 represses Ccn2 expression by binding to the consensus TCF·LEF·Sox9 site. On the other hand, the same assays showed that in hypertrophic chondrocytes, TCF·LEF·β-catenin complexes occupy the consensus TCF·LEF·Sox9 site and activate Ccn2 expression. Furthermore, transgenic mice in which lacZ expression is driven under the control of the proximal Ccn2 promoter revealed that the proximal Ccn2 promoter responded to Wnt signaling in cartilage. Hence, we propose that differential occupancy of the TCF·LEF·Sox9 site by Sox9 versus β-catenin restricts high levels of Ccn2 expression to hypertrophic chondrocytes.
Keywords: β-Catenin, Cell Differentiation, Chromatin Immunoprecipitation (ChIP), Connective Tissue, T-cell factor (TCF), Transcription Promoter, Wnt Pathway, Sox9, Cartilage, Chondrogenesis
Introduction
The majority of the mammalian skeleton forms through endochondral ossification, in which osteochondro-progenitor cells aggregate, begin to form an extracellular matrix, and differentiate into chondrocytes. Eventually the cells in the centers of the condensation exit the cell cycle and terminally differentiate. Ultimately a growth plate is formed consisting of slowly dividing (resting) chondrocytes followed by a zone of rapidly proliferating chondrocytes that are aligned into columns and ending with terminally differentiated prehypertrophic and hypertrophic chondrocytes (1). Chondrocyte proliferation and hypertrophy ultimately determine bone length and width. Hypertrophic chondrocytes coordinate multiple aspects of bone formation; they are primarily responsible for establishing the final lengths of bones, recruiting the osteoblasts that will replace the cartilage and stimulating angiogenesis to permit formation of the marrow cavity (1).
Several transcription factors that play essential roles in chondrocyte differentiation have been identified (2). Among the best characterized are the Sox transcription factors, Sox9, L-Sox5, and Sox6, which are required for commitment to the chondrocyte lineage, proliferation, and suppression of premature conversion to hypertrophic chondrocytes (3). Sox proteins contain a DNA binding motif known as a high mobility group (HMG) box. The HMG box binds DNA in the minor groove and bends DNA, conferring on Sox proteins a role in assembly of transcriptional enhancesomes (4). TCF·LEF2 factors also play a role in the decision of the chondrocyte to remain in the cell cycle or to exit and become hypertrophic. As is the case for Sox proteins, TCF·LEF factors contain an HMG box, and they bind the minor groove of DNA. In fact, a subset of TCF·LEF consensus binding sites is also binding sites for Sox proteins. TCF·LEF factors assemble repressor complexes on target genes. However, the binding of the transcription factor β-catenin to the TCF·LEF complex leads to the assembly of an activating complex on the target gene (5). This complex, formed through activation of Wnt signaling pathways, is required for chondrocyte hypertrophy and maturation (6, 7). It has been shown that β-catenin is antagonized and targeted for degradation by Sox9 (6). However, the fact that some TCF·LEF consensus binding sites are also consensus sites for Sox9 suggests that TCF·LEF·β-catenin and Sox9 might differentially occupy these sites, raising the possibility of a novel mechanism by which Sox9 and β-catenin might control differential gene expression.
CCN2/connective tissue growth factor (CCN2) is a matricellular protein with diverse activities. CCN2 is overexpressed in every fibrotic condition described to date, and it promotes the excess extracellular matrix synthesis that is characteristic of all fibrotic diseases (8). The role of CCN2 as a ligand for various integrins, including α5β1, required in chondrocytes, has been well documented (9, 10).
CCN2 is expressed in perichondrium and subjacent chondrocytes at early stages of endochondral bone formation, but once a growth plate is formed, CCN2 is expressed at highest levels in the hypertrophic zone (9–12). Mice lacking Ccn2 die in the immediate perinatal period due to complications of severe skeletal dysplasia; consistent with high levels of CCN2 expression in the hypertrophic zone, Ccn2 mutants exhibit enlarged hypertrophic zones due to premature exit of chondrocytes from the cell cycle and to delayed replacement of hypertrophic chondrocytes by osteoblasts. This delayed ossification is a result in part of decreased transcription of vascular endothelial growth factor (vegf) mRNA (10), thereby leading to delayed invasion by osteoblasts. Moreover, CCN2 can bind to and block VEGF activity (13). Given the importance of CCN2 in chondrocyte hypertrophy specifically and of understanding hypertrophic cartilage-specific gene regulation in general, we investigated the transcriptional mechanisms controlling stage-specific CCN2 expression in the growth plate.
EXPERIMENTAL PROCEDURES
Generation and Analysis of Transgenic Mice
The 4-kb proximal promoter region of the mouse Ccn2 gene was amplified by PCR using the primers 5′ 4-kb (′-CCCACGCGTGAGCAGGCAAATGT-3′) and 3′ 4-kb (5′-CCCCTCGAGTGTGTGAGACTAGG-3′) and subcloned into TA cloning vector (Invitrogen). The 4-kb fragment was further digested with NotI and SpeI restriction enzymes and subcloned into the PstI and BamH1 sites of the transgenic reporter plasmid pNSlacZ, which contains Escherichia coli β-galactosidase gene containing a nuclear localization signal followed by the polyadenylation signal of SV40 large T antigen in pBluescriptSK+. (14). Transgenic mouse lines were generated by pronuclear injection as described previously (15). lacZ staining of paraffin-embedded sections was performed as described (15). Limb organ culture was performed as described previously (16). Control or Wnt-3a-conditioned medium was prepared as described previously (17). Briefly, limbs were maintained in medium conditioned by L-M(TK-) (ATCC CCL-1.3) cells, which stably overexpress Wnt 3A. Control medium was prepared from the parental cell line (ATCC CRL-2648). The CCL-1.3 and CRL-2648 cell lines were obtained from the ATCC.
Cell Culture
The chondrogenic cell line ATDC5 (18) was obtained from the Riken Cell Bank (Tsukuba, Japan). The cells were cultured in maintenance medium consisting of DMEM/Ham's F-12 (1:1) (Invitrogen) supplemented with 5% (v/v) fetal bovine serum (FBS; HyClone Laboratories, Logan, UT), 1% (v/v) penicillin-streptomycin solution (50 units/ml penicillin and 50 μg/ml streptomycin) (Invitrogen), 10 mg/ml human transferrin (Sigma), and 3 × 10−8 m sodium selenite (Sigma). To induce chondrocyte differentiation, cells were cultured in maintenance medium with the addition of 10 μg/ml bovine insulin (Sigma) for 26 days. Cells were grown at 37 °C in a humidified atmosphere of 95% air and 5% CO2, and culture medium was changed every other day.
Preparation of Chicken Primary Chondrocytes
Chicken primary chondrocytes were isolated from the lower (lower third) and upper (upper third) sternum at embryonic days 14 and 18 as described previously (19) and maintained in DMEM (Invitrogen) supplemented with 10% (v/v) FBS (HyClone Laboratories, Logan, UT), 1% (v/v) penicillin-streptomycin solution (50 units/ml penicillin and 50 μg/ml streptomycin). Briefly, the sternums were isolated and digested with 1 mg/ml collagenase (Sigma), 0.0625% trypsin/EDTA solution (Invitrogen) in 1× PBS, 3 times, and the cells were collected from the pools of last two digestions. Cells were passaged not more than 3 times and used within 2 weeks. The differentiation stage of the cells was confirmed by expression of collagen type II and collagen Type X as described under “Results.”
Plasmids
To create Ccn2 reporter plasmids, PCR was performed using 5′ primers linked with MluI or KpnI (Promega, Madison, WI) restriction enzyme sites and 3′ TS-primer linked with XhoI (Promega) restriction enzyme sites to amplify the fragments: 5′ end 4 kb, CCCACGCGTGAGCAGGCAAATGT; −3.1 kb, CCCACGCGTTTAGAAGTCAAAAC; −2.8 kb, CCCACGCGTCCTTCTGTCCCATC; −2.3 kb, CCCACGCGTCCAACCACTTACCC; −1.8 kb, CCCACGCGTGCTCACACTTCAAG; −1 kb, GGGGTACCGCCGGATTATAACTAGATAC; −0.5 kb, CCCACGCGTCCAAGAGACTACAG; 3′ end-TS, CCCCTCGAGTGTGTGAGACTAGG. To generate the 0.5-kb mutant reporter constructs, a BamHI restriction site was introduced into the TCF·LEF·Sox9 consensus sequence using a 5′-end mutated primer linked with MluI along with the 3′-TS primer to amplify the fragment 0.5 k-TCFmut, GCGGTACCAAGAGACTACAGCCCCGTAAAGAAAAAAAAAAATCCAAggatccGAAAAATATTTTTTTT. The PCR fragments were amplified from the mouse Ccn2 gene (GenBankTM accession number AC099695) and digested with appropriate enzymes and cloned into pGL3basic (Promega). All of the constructs were verified by DNA sequencing and enzyme digestion.
Transfections and Reporter Assays
ATDC5 cells were transfected with the reporter plasmids together with Renilla luciferase plasmid in triplicate using Lipofectamine 2000 (Invitrogen). Luciferase activities were measured by Dual Luciferase Reporter Assay System (Promega) and normalized to Renilla luciferase activity to control for transfection efficacy. All assays were repeated at least three independent times. In some experiments ATCD5 cells were maintained in the presence of Wnt3a- or control-conditioned medium, prepared as described previously (17). In other experiments ATDC5 cells were maintained in the presence of LiCl (8 mm) as described previously (20) or were transfected with a construct (pcDNA-β-catenin S37F) encoding a stabilized form of β-catenin (21).
Chromatin Immunoprecipitation
The entire procedure was performed with the chromatin immunoprecipitation (ChIPs) assay kit (Upstate/Millipore, Billerica, MA) according to the manufacturer's instructions with minor modifications. Briefly, cells were maintained in maintenance medium at low density or treated with insulin for 26 days. Cells were washed with PBS containing proteinase inhibitor mixture (BD Biosciences) and collected and pelleted at 1000 rpm. Cell pellets were resuspended in SDS lysis buffer and incubated on ice for 10 min. Samples were sheared on ice using a Misonix Sonicator 3000 (Misonix, Farmingdale, NY) and resolved on agarose gels to confirm that the average fragment sizes were between 200 and 500 bp. 20 μl of the sonicated samples were saved for total DNA loading controls. DNA concentrations were determined, and equal amounts of chromatin were used for further processing. DNA and proteins were cross-linked with formaldehyde (Sigma) for 10 min at 37 °C; cross-linking was terminated by the addition of glycine (Sigma) at a final concentration 0.125 m. Samples were diluted with ChIP dilution buffer and precleaned with salmon sperm DNA/protein A-agarose, 50% slurry. Precleaned chromatin was incubated with 2 mg of the appropriate antibody at 4 °C for ∼12–16 h. Antibodies used were anti-β-catenin (#9562, Cell Signaling, Danvers, MA), anti-RNA polymerase (#05–623, Millipore, Temecula, CA), anti-Sox9 (#sc-20095), and anti-Tcf4 (#sc-13027). Chromatin-antibody complexes were recovered by sperm DNA/protein A-agarose, 50% slurry, and eluted with 1% SDS and 0.1 m NaHCO3 mixed buffer. Cross-links were reversed with the addition of 0.2 m NaCl at 65 °C for 6 h. The samples were then treated in 0.01 m EDTA, 0.04 m Tris-Cl, pH 6.8, with 30 μg/ml proteinase K at 37 °C for 12–16 h. DNA was recovered by phenol/chloroform extraction and ethanol precipitation. PCR was performed using region-specific primers 1R (5′-GATGAAATATAACTTAGAAA-3′) and 1F (5′-CCGGTACCTGTATTTCAGATAC-3′), 2R (5′-AAGCATTCCTCAAAGAGAAG-3′) and 2F (5′-AGAGCTCACCTCAGAGCCCA-3′), 3R (5′-CAAGAGAACATTCTACCCCA-3′) and 3F (5′-TGATTTCCTCTTTGAGGGCT-3′), 5′ non-specific (5′-AAAGGCTTGGCTGTTC-3′) and 3′ nonspecific (5′-AATGTCCAGATGTACC-3′) for CCN2 analysis and Ihh 1F (5′-GCAACCCACGTCGCAGCCGG-3′) and Ihh 1R (5′-AAGGCTACATTGCCTCCACC-3′) and Ihh 2F (5′-GTACAGAACTAGGGGATGCG-3′) and Ihh 2R (5′-CTTCTGAAGTACCACGAGGA-3′) for Ihh analysis. All PCR products were resolved and visualized on agarose gels. The intensities of images were captured and quantitated by Scion Image camera and software (Scion Image, Frederick, MD).
SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blotting
Western blot analysis was performed with cell extracts from D0 or D26 ATDC5 cells. Whole cell lysates were separated by 10% SDS-PAGE and transferred to nitrocellulose membrane (Bio-Rad). The membrane was incubated with antibody against β-catenin (1:500, #9562), phospho-β-catenin (1:500, #9567), glycogen synthase kinase 3β (1:500, #9315), phospho- glycogen synthase kinase 3β (1:500, #9336) (above are from Cell Signaling, Danvers, MA), Runx2 (1:500, sc-10758), Sox9 (1:500, sc-20095) (Santa Cruz Biotechnology, Santa Cruz, CA), CCN2 (1:500, Abcam, Cambridge, MA), and actin (1:2000, #A2066, Sigma). Antigen-antibody complexes were detected with HRP-conjugated secondary antibodies (#170–6515, Bio-Rad) and visualized with the use of Pierce.
Electrophoretic Mobility Shift Assays
Gel shift assays were performed with the Gel Shift Assay System (Promega) following the manufacturer's protocol. Briefly, nuclear extracts were prepared from D0 and D26 ATDC5 cells following the method of Schreiber and Schaffner (22). Double-stranded nucleotides encompassing the 443-bp consensus TCF·LEF·Sox9 sequence or BamHI-substituted mutation at the TCF·LEF·Sox9 sequence were end-labeled with [γ-32P]ATP and T4 polynucleotide kinase, followed by purification by G25 spin columns. Nuclear extracts were added to labeled oligonucleotide in gel shift binding buffer and incubated for 30 min at room temperature. For competition assays, a 50-fold excess of unlabeled competitor oligonucleotide or nonspecific competitor oligonucleotide was incubated in the binding reaction. For super-shift experiments, 1 μg of the following antibodies was added to the binding reaction for an additional 60 min at room temperature: Tcf4 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), Sox9 (sc-20095; Santa Cruz Biotechnology), and anti-FLAG (1804, Sigma). In some experiments, in vitro translation of Sox9-FLAG (23) was performed using the Retic Lysate IVT Kit (Ambion). 1 or 2 μl of the translation product was used as indicated in the figure legends. DNA-protein complexes were resolved on 4% nondenaturing polyacrylamide gels. The sequences used as probes in the assay were WT TCF·LEF (5′-AAAATCCAAAACAAAGAAAAATATTT-3′) and TCF·LEF mutant (5′-AAAAAAAAAggatccACAAAGAAAAATATTT-3′.
Immunofluorescence
Paraffin-embedded sections were boiled for 15 min in citrate buffer to reverse cross links and unmask epitopes. Sections were blocked with 5% goat serum for 1 h and incubated with anti-CCN2 (CCN2) antibody (Abcam) at a dilution of 1:250 overnight at 4 °C. The following day sections were incubated with secondary antibody (Alexa Fluor 555 or 488 goat anti-rabbit, Invitrogen) for 1 h at room temperature. Sections were counterstained with DAPI (Vectashield, Vector Laboratories, Burlingame, CA).
Semiquantitative Reverse Transcriptase (RT)-PCR
Total RNA was isolated from cultured ATDC5 cells using an RNeasy Mini Kit (Qiagen, Valencia, CA). 0.1 mg of DNase I (Applied Biosystems/Ambion, Austin, TX)-treated total RNA was used for reverse transcription with a SuperScript III First-Strand Synthesis System (Invitrogen) following the manufacturer's protocol. First-strand cDNA synthesis was carried out using 0.1 μg of DNase-treated total RNA in 20 μl of a solution containing first-strand buffer, 50 mg of random hexamers, 0.5 mm dNTPs, 5 mm MgCl2, 10 mm dithiothreitol, 40 units of RNaseOUT and 200 units of reverse transcriptase at 25 °C for 10 min then 50 °C for 50 min. PCR reactions were carried out with primers listed in Table 1.
TABLE 1.
| Species | Target | Forward | Reverse | GenBankTMaccession number |
|---|---|---|---|---|
| Primers used for reverse transcriptase PCR | ||||
| Chicken | CCN2 | CCAGCGTGAAGACGTACAGA | GTCATTGTCTCCAGGGCAGT | NM_204274 |
| Chicken | Col IIa1 | AGAAAGGAATCCAGCCCAAT | ACACCTGCCAGATTGATTCC | NM_204426 |
| Chicken | Col X | AAAATAGTAGACGTTACCTTGACTC | ACATGCATTTACAAATATCGTTAC | |
| Chicken | GAPDH | TATGATGATATCAAGAGGTTAGT | TGTATCCAAACTCATTGTCATAC | NM_204305 |
| Mouse | CCN2 | CAAAGCAGCTGCAAATACCA | GGCCAAATGTGTCTTCCAGT | NM_010217 |
| Mouse | Col II | GGAAAGTCTGGGGAAAGAGG | CAGTCCCTGGGTTACCAGAA | NM_031163 |
| Mouse | Col X | GCCAGGTCTCAATGGTCCTA | AAAAGCAGACACGGGCATAC | NM_009925 |
| Mouse | Runx2 | GAGGGCACAAGTTCTATCTG | CGCTCCGGCCCACAAATCTC | NM_009820 |
| Mouse | GAPDH | CTTGTCATCAACGGGAAGCC | AGACTCCACGACATACTCAGC | XM_983502 |
| Primers used for real-time PCR | ||||
| Mouse | CCN2 | GCAAGGAGTGGGTGTGTG | TGTGTCTTCCAGTCGGTAGG | NM_010217 |
Real Time RT-PCR
RNA and cDNA were prepared as described above. The cDNA mixtures were diluted 5-fold in sterile distilled water, and 1-μl aliquots were subjected to real-time PCR using RT2 qPCR Master Mixes (SuperArray, Frederick, MD). The PCR reactions were performed in 25 μl of a solution containing RT2 qPCR Master Mix and 20 mm specific primers as shown in Table 1. Primers were designed using primer3 software (Whitehead Institute for Biomedical Research, Cambridge, MA). PCR was carried out using a DNA Engine Opticon 2 (MJ Research/Bio-Rad), and the data were analyzed using the MJ Opticon Monitor 3 software. The PCR conditions were 95 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min for 50 cycles, and measurements were taken at the end of the annealing step at 55 °C of each cycle. PCR product specificity was verified by melting curve analysis between 55 and 95 °C. All real-time PCR reactions were performed in triplicate, and the levels of mRNA expression were calculated and normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA at each time point.
Statistical Analysis
All transfection and RT-quantitative PCR experiments were performed with triplicate samples and repeated at least twice. Gel shifts and ChIP analyses were repeated at least twice. Representative results are shown. Data are represented as the mean ± S.E. of the mean, and statistical analysis was performed with Student's t test. A p value of less than 0.05 was considered statistically significant.
RESULTS
CCN2 Expression Is Regulated during Chondrogenic Differentiation of ATDC5 Cells
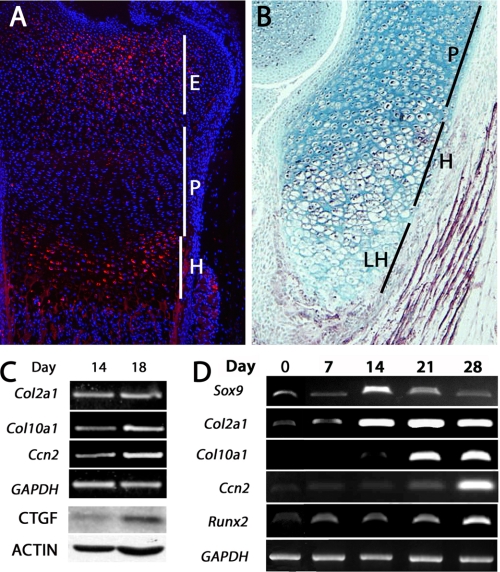
The mechanisms underlying the regulation of Ccn2 mRNA expression in the hypertrophic zone are undefined. Examination of CCN2 protein expression within the growth plate reveals that CCN2 is highly expressed in prehypertrophic and hypertrophic chondrocytes, but mRNA (11) and protein (Fig. 1, A and B) levels decline in the most differentiated cells, the terminal (late) hypertrophic chondrocytes. We confirmed that Ccn2 expression is higher in hypertrophic cells using primary chick sternal chondrocytes. As shown in Fig. 1C, the chondrocyte marker Col2a1 is expressed in cells isolated from day 14 (D14) and day 18 (D18) embryos. However, chondrocytes isolated from D18 embryos highly express the hypertrophic marker Col10a1, whereas chondrocytes isolated from D14 embryos (proliferating stage) express lower levels of Col10a1 (Fig. 1C). Higher levels of Ccn2 mRNA and protein are seen in the D18 hypertrophic chondrocytes (Fig. 1C).
FIGURE 1.
Expression of CCN2 in cartilage. A and B, immunofluorescence and immunohistochemical staining, respectively, of CCN2 expression in E17 (embryonic day 17) (A) and P0 (postnatal day 0) (B) growth plates, show high levels of expression in epiphyseal and hypertrophic chondrocytes, low levels in proliferating chondrocytes, and no expression in terminal hypertrophic cells. C, CCN2 protein and mRNA expression in primary chick chondrocytes is higher in hypertrophic day 18 cells than in proliferating day 14 cells. CTGF, connective tissue growth factor. D, shown is expression of Ccn2 during differentiation of ATDC5 cells. Expression of CCN2 is restricted to hypertrophic stages, which are distinguished from proliferating chondrocytes by expression of Col10a1. E, epiphyseal; H, hypertrophic; LH, late hypertrophic; P, proliferating.
Numerous studies have demonstrated the efficacy of murine chondrocyte ATDC5 cells to study mechanisms underlying chondrocyte differentiation (20, 24–27). ATDC5 cells grown in maintenance medium possess properties of osteochondro-progenitors, as they can be differentiated along either the chondrogenic or osteogenic lineage (20, 24–30). ATDC5 cells grown in the presence of insulin, transferring, and selenium (differentiation medium) over a period of 28 days were utilized as an in vitro model. ATDC5 cells in monolayer in differentiation medium reach confluence on about day 7. Formation of cartilaginous nodules was observed on D14 and well defined cartilaginous nodules were detected by D21; the nodules increased in size through D28. Expression of typical chondrocyte markers verified differentiation and hypertrophy over the 28-day culture period. Expression of Col2a1 mRNA was observed at low levels at D7 and maintained at high levels throughout the remaining culture period (Fig. 1D). Maintenance of Col2a1 expression in late stage cultures has been reported frequently in the literature and most likely reflects the persistence of some cells in a proliferative phase. Expression of Col10a1 transcripts was observed beginning at D21, marking the onset of hypertrophy. Runx2, a marker of terminal hypertrophic chondrocytes, was expressed throughout the culture period but at the highest levels at D28. Thus, based on the expression of these markers, cultures at D0 represent osteochondro-progenitors, up to D14 represent the proliferative phase of chondrocyte differentiation, D21 the onset of hypertrophic differentiation, and D28 cultures represent more mature hypertrophic cells. Thus, Ccn2 expression correlates with the onset of hypertrophy in ATDC5 cells and recapitulates the time course of expression in cartilage in vivo.
A 0.5-kb Proximal Ccn2 Promoter Region Contains a Stage-specific Responsive Element
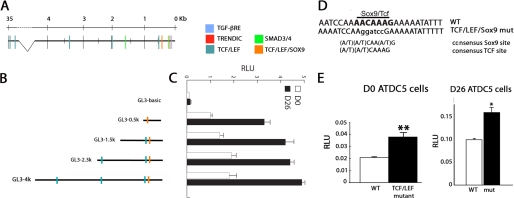
We used the ECR Browser (31) to find evolutionary conserved regions of at least 100 bp of greater than 50% identity between the human, mouse, and chicken Ccn2 genomic regions spanning 40 kb on either side of the protein coding region. We found an evolutionary conserved region ∼35 kb upstream of the transcription start site of Ccn2 and a second within the 4-kb proximal promoter region; a third evolutionary conserved region was found immediately 3′ to the transcription termination site (data not shown). Comparison of the mouse and human genomes revealed that the majority of the 4-kb proximal Ccn2 promoter region was conserved (data not shown). Therefore, we performed a TRANSFAC analysis of this region (32) that revealed multiple conserved consensus binding sites (data not shown). Among these, binding sites for TCF·LEF, Smad2/3, and Sox5/6/9 were found, all of which are known to regulate chondrocyte proliferation and differentiation (33) (Fig. 2A).
FIGURE 2.
Identification of putative regulatory sites in the Ccn2 promoter region. A, schematic diagram of the mouse Ccn2 promoter region, showing consensus binding sites for TCF·LEF and Smad3/4 factors, which are known to regulate Ccn2 expression in other cell types. The TGF-β response element (TGF-βRE), shown to function as a basal element (56), the TRENDIC site, proposed to be involved in regulation of Ccn2 expression in cartilage (46), and an overlapping TCF·LEF·Sox9 binding site located 449–440 bp upstream of the transcription start site are illustrated. B, shown are schematic diagrams of the Ccn2-luciferase reporter constructs used throughout the paper. C, shown are relative expression levels of Ccn2-luciferase reporter constructs in ATDC5 D0 osteochondro-progenitors versus hypertrophic D26 chondrocytes. RLU, relative luciferase units. D, shown is the sequence of the Sox9·TCF·LEF site at −443 bp in the CCN2 promoter. E, shown are relative luciferase activity levels of 0.5-kb WT and 0.5-kb (TCF·Sox mut) constructs in D0 and D26 ATDC5 chondrocytes. *, p < 0.05; **, p < 0.005.
We generated serial deletions to search for regulatory elements within the 4-kb proximal Ccn2 fragment that confer higher expression in hypertrophic than in proliferating chondrocytes (Fig. 2B). Promoter activity was observed in both osteochondro-progenitors (D0) and hypertrophic (D26) cells (Fig. 2C). Promoter activity was seen for all Ccn2 constructs in D0 cells, but all of the promoter fragments were more active in D26 than in D0 cells; the maximum -fold induction (3.3-fold) from D0 to D26 was seen for the 0.5-kb Ccn2 promoter fragment. These findings imply that there are stage-specific regulatory elements residing in the 4-kb proximal Ccn2 promoter region, and elements residing in 0.5-kb promoter appear to be primarily responsible for this stage-specific induction.
A TCF·LEF Consensus Site in the Ccn2 Proximal 0.5-kb Promoter Region Is Involved in Stage-specific Gene Expression
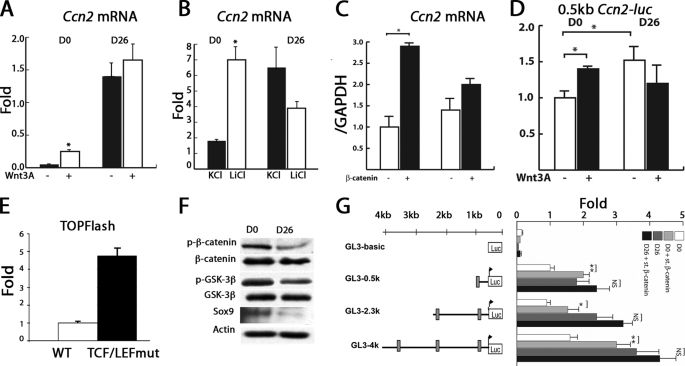
As discussed above, three TCF·LEF consensus sequences were found within the 4-kb proximal promoter region of Ccn2, residing 443, 542, 1963, and 3491 bp upstream of the transcriptional start site (Fig. 2A). CCN2 is induced by canonical Wnt/β-catenin signaling in several cell types (34, 35). Moreover, canonical Wnt/β-catenin signaling pathways are required for chondrocyte hypertrophy (6, 7). The TCF·LEF·Sox9 site at −443 bp (Fig. 2D), thus, attracted our attention because it resides in the 0.5-kb region that retains stage-specific activity. Introduction of a mutation that destroys the TCF·LEF·Sox9 site at −443 bp (Fig. 2E) into the 0.5-kb Ccn2 proximal promoter led to elevated luciferase activity levels in D0 osteochondro-progenitors (Fig. 2E). The TCF·LEF mutant also exhibited higher activity than the WT construct in D26 cells (Fig. 2E). Involvement of β-catenin in the regulation of Ccn2 induction in chondrocytes was supported by the effects of Wnt3A-conditioned medium; Ccn2 mRNA levels increased ∼4-fold after Wnt3A treatment in D0 cells (Fig. 3A). A similar result was seen with LiCl, indicating that the canonical Wnt pathway is responsible for this induction (Fig. 3B). Neither Wnt3A nor LiCl increased Ccn2 mRNA expression in hypertrophic D26 cells. In accordance, expression of stabilized β-catenin also led to induction of Ccn2 mRNA expression at D0 but not at D26 (Fig. 3C). Similarly, expression of the 0.5kb Ccn2 proximal promoter was increased by Wnt3a in D0 cells but not in D26 cells (Fig. 3D). Consistent with this observation, the activity of the TCF·LEF reporter plasmid TOPflash was higher in D26 cells (Fig. 3E), indicating that levels of endogenous β-catenin are already elevated in hypertrophic chondrocytes. We performed Western blotting experiments to test this hypothesis. In accordance with previous studies (36), the amount of total β-catenin was indistinguishable at both stages, but there was more of the phosphorylated (targeted for degradation) form in D0 osteochondro-progenitors than in hypertrophic D26 cells; this was correlated with the presence in D0 cells of relatively more activated (unphosphorylated) glycogen synthase kinase 3β (GSK-3β), which phosphorylates and targets β-catenin for destruction compared with total glycogen synthase kinase 3β (Fig. 3F).
FIGURE 3.
Ccn2 is a Wnt-responsive gene in chondrocytes. A, Ccn2 mRNA levels in ATDC5 (D0 (osteochondro-progenitor) and D26 (hypertrophic) chondrocytes treated with Wnt3A or control-conditioned medium are shown. Data are expressed as -fold induction, determined by real time RT-PCR. B, relative levels of Ccn2 mRNA expression induced by KCl versus LiCl in D0 versus D26 ATDC5 chondrocytes are shown. C, relative levels of Ccn2 mRNA expression induced by overexpression of stabilized β-catenin are shown. D, relative levels of activity of the 0.5 kb Ccn2 proximal promoter construct in the presence or absence of Wnt3A in D0 versus D26 ATDC5 cells are shown. E, relative levels of TOPflash luciferase activity in D0 versus D26 ATDC5 chondrocytes are shown. F, levels of expression of total and p-β-catenin (targeted for degradation), p-glycogen synthase kinase (p-GSK; activated) and total glycogen synthase kinase, and Sox9 in lysates from D0 and D26 ATDC5 cells were analyzed by Western blot. G, shown are relative luciferase activity levels of Ccn2 promoter activity in the presence or absence of stabilized β-catenin in D0 and D26 ATDC5 chondrocytes. *, p < 0.05.
In D0 ATDC5 cells, luciferase activity driven by the 0.5-kb Ccn2 fragment that contains the TCF·LEF site at −443 bp was enhanced 2-fold by overexpressed stabilized β-catenin (Fig. 3G). The longer constructs, which include the remaining TCF·LEF consensus sequences, did not exhibit any significant differences in the -fold induction compared with the 0.5-kb fragment (Fig. 3G). In hypertrophic D26 cells, there was a trend toward enhanced activity of the Ccn2 promoters in response to overexpressed β-catenin, but the level of activation was not significant in most experiments (Fig. 3G). Thus, the −443 bp TCF·LEF site may be a component of a stage-specific enhancer controlling Ccn2 expression in hypertrophic chondrocytes.
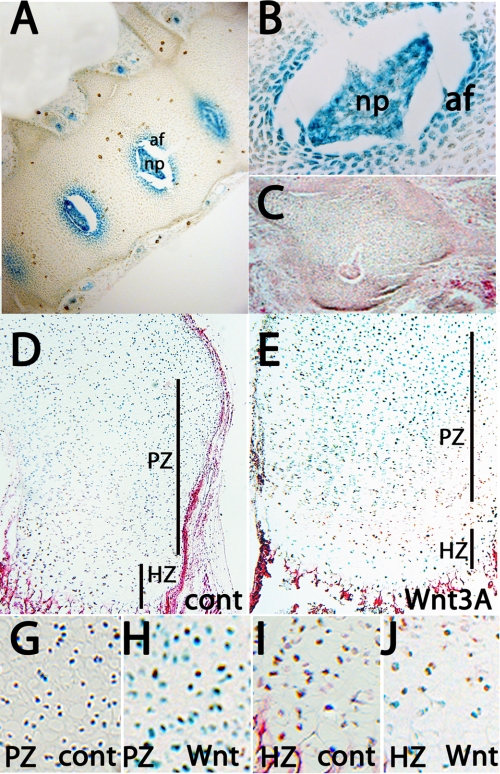
We tested whether the 4-kb proximal promoter region is involved in Ccn2 expression in cartilage in vivo by generating transgenic mice in which expression of nuclear-localized lacZ is driven under the control of the 4-kb proximal Ccn2 promoter. Very similar patterns of expression were detected in all three independent stable lines (data not shown). The strongest expression in cartilage was detected in the inner annulus of the developing intervertebral discs (Fig. 4, A–C). Strong lacZ expression was also seen in the nucleus pulposus. This pattern of transgene expression is in excellent agreement with endogenous Ccn2 expression in intervertebral discs (37). lacZ expression was not detected in growth plate cartilage in midgestation, newborn, or adult mice (data not shown).
FIGURE 4.
Expression of 4 kb Ccn2-lacZ in skeletal tissues. A–C, lacZ expression in annulus fibrosis and nucleus pulposus of the intervertebral discs at E16.5 is shown. B is a higher magnification image, and C is an intervertebral disc from a non-transgenic control. D and E, 4-kb Ccn2-lacZ expression in P2 metatarsals maintained in the presence of control (D) or Wnt3A-conditioned (E) medium for 48 h is shown. G–J, higher magnification images of proliferating (G and H) and hypertrophic (I and J) zones from metatarsals cultured for 48 h in control (G and I) or Wnt3A-conditioned (H and J) medium are shown. Af, annulus fibrosis; np, nucleus pulposus; HZ, hypertrophic zone; PZ, proliferative zone. Cont, control.
However, incubation of isolated P0 limbs for 2 days in culture medium containing 10% serum induced lacZ staining in hypertrophic chondrocytes (Fig. 4, D and I), suggesting that a factor present in serum has a permissive role in expression of the 4-kb Ccn2 promoter in the growth plate. Moreover, lacZ expression was considerably stronger in growth plates cultured in the presence of Wnt3a (Fig. 4E). Wnt3a did not alter levels of lacZ expression in the hypertrophic zone but up-regulated lacZ expression in proliferating chondrocytes (Fig. 4, G and H). The lower sensitivity of hypertrophic chondrocytes to exogenous Wnt3a most likely reflects the fact that levels of endogenous Wnt signaling are already high in hypertrophic chondrocytes (6). These results are consistent with the in vitro studies (Fig. 3) showing that the proximal Ccn2 promoter is active in D26 hypertrophic chondrocytes and can be activated in D0 osteoprogenitor cells by Wnt signaling. Taken together, these results suggest that the 4-kb proximal Ccn2 promoter contains sequences that drive Wnt-inducible gene expression in vivo, but additional regions in the Ccn2 promoter must also be required.
Sox9 Represses CCN2 Promoter Activity in Proliferating Cells
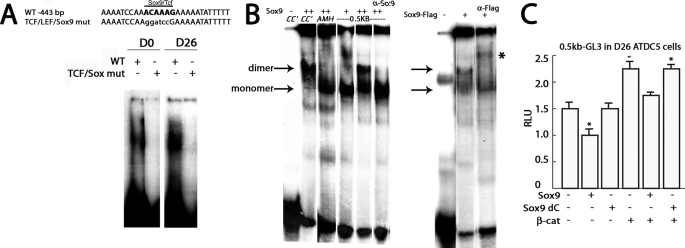
Sox9 is highly expressed in proliferating chondrocytes and at lower levels in hypertrophic cells (38) (Figs. 1D and 3D). Because the TCF·LEF site at −443 bp in the Ccn2 promoter is also a consensus binding site for Sox9 (Fig. 2D), we tested whether this site was occupied in chondrocytes using EMSA. A shifted band was also observed in the presence of nuclear extract from osteochondro-progenitor D0 and hypertrophic D26 cells for an oligonucleotide containing the −443 bp TCF·LEF·Sox9 site from the Ccn2 proximal promoter, and mutation of this site prevented complex formation, which could indicate binding by either Sox9 and/or TCF·LEF factors (Fig. 5A). Sox9 binds as a dimer to palindromic consensus sites in promoters that it activates in chondrocytes (39) but binds predominantly as a monomer to the anti-Müllerian hormone promoter, which is repressed by Sox9 (40). The consensus binding site at −443 bp in the Ccn2 promoter is a monomeric site. Therefore, we tested whether Sox9 binds to the TCF·LEF·Sox9 site at −443 bp in the Ccn2 proximal promoter and whether it binds as a dimer or monomer. As described previously, overexpressed Sox9 bound as a dimer to an oligonucleotide containing the consensus Sox9 binding site from the type II collagen (Col2a1) promoter CC′ (39) (Fig. 5B). In accordance with previous studies, Sox9 bound as a monomer to the anti-Müllerian hormone promoter (AMH, Fig. 5B). Incubation of an oligonucleotide containing the TCF·LEF·Sox9 consensus site at −443 bp in the Ccn2 proximal promoter with Sox9 led to a shifted band consistent with Sox9 binding as a monomer; incubation with supraphysiological levels of Sox9 causes a further shift consistent with dimer formation, but the majority of the shifted oligonucleotide migrated at a position consistent with monomeric binding of Sox9 (Fig. 5B). The slower mobility band was prevented from forming by incubation with an antibody against Sox9 (Fig. 5B). It is conceivable that the anti-Sox9 antibody recognizes an epitope that is accessible in the dimeric complex but not the monomeric complex. In contrast, a supershift was obtained using a FLAG-tagged version of Sox9 and α-FLAG antibody (Fig. 5B). In this case the FLAG epitope is presumably accessible to the antibody when Sox9-FLAG binds as a monomer. In summary, these findings indicate that Sox9 binds to the TCF·LEF·Sox9 consensus site at −443 bp in the Ccn2 promoter, primarily as a monomer.
FIGURE 5.
Analysis of Sox9 binding and activity. A, shown the sequence of an oligonucleotide spanning the −443-bp TCF·LEF·Sox9 site from the 0.5-kb CCN2 promoter and the corresponding mutant. EMSA analysis of nuclear extracts of ATCD5 cells with oligonucleotides in which the TCF·LEF·Sox9 (TCF·Sox mut) consensus site has been mutated shows that the mutation prevents formation of a shifted band. B, shown is EMSA of the oligonucleotide shown in A spanning the TCF·LEF·Sox9 binding site at −443 bp in the Ccn2 proximal promoter in the presence of in vitro translated Sox9. CC′ and anti-Müllerian hormone are control oligos containing a palindromic Sox9 binding site from the Col2a1 gene and a monomeric binding site from the anti-Müllerian hormone promoter, respectively (39, 40). +, 1 μl of in vitro translated product; ++, 2 μl of in vitro translated product. Sox9 binds to the 443-bp site at a mobility consistent with monomer binding at the lower concentration (+) of in vitro translated product and with a mobility consistent with both monomer and dimer formation at the higher concentration (++) of in vitro translated product. Incubation with Sox9 antibody prevented formation of the slower mobility band. Shifted bands are indicated by arrows. C, shown are relative luciferase activities (RLU) of 0.5-kb Ccn2 proximal promoter in the presence or absence of WT Sox9 or Sox9 dC. β-cat, β-catenin.
It has been suggested that Sox9 acts as a repressor when bound to the Anti-Müllerian Hormone promoter as a monomer (40). To determine whether binding of Sox9 to the TCF·LEF·Sox9 site at −443 bp mediated Ccn2 repression, we tested whether a Sox9 DNA binding dominant negative mutant (C-terminal-truncated form of Sox9 that can bind to DNA but has no transactivation activity; Sox9 dC) (23) is able to repress activity of the 0.5-kb proximal Ccn2 promoter. As shown in Fig. 5C, WT Sox9 represses promoter activity, but Sox9 dC does not. Moreover, WT Sox9 can counteract the stimulatory effects of overexpressed β-catenin, but Sox9 dC does not.
Sox9 and TCF·LEF·β-Catenin Complexes Have Opposing Effects on Ccn2 Expression via Binding to the −443-bp TCF·LEF·Sox9 Site
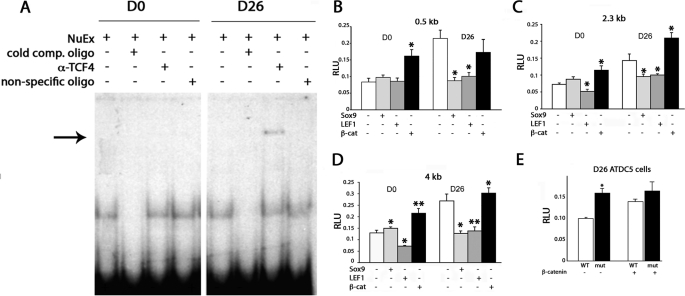
The above results suggested that Sox9 represses Ccn2 proximal promoter activity in D0 osteochondro-progenitors but that TCF·LEF·β-catenin complexes activate Ccn2 promoter activity in hypertrophic D26 chondrocytes. TCF4 is expressed in chondrocytes and is required for β-catenin binding (41, 42). Therefore, we performed a supershift assay to test whether TCF4 could be detected in complexes binding to an oligo containing the −443-bp TCF·LEF·Sox9 site in the Ccn2 proximal promoter. A band supershifted by an anti-TCF4 antibody was detected in extracts from hypertrophic D26 cells but not in D0 osteochondro-progenitors (Fig. 6A). Because the 443-bp TCF·LEF and Sox9 binding sites overlap, it was not possible to mutate them individually. However, as previously discussed, complex formation was not observed with a probe in which the TCF·LEF·Sox9 site was destroyed by site-directed mutagenesis (Fig. 5A).
FIGURE 6.
Regulation of Ccn2 expression by Sox9 and β-catenin. A, EMSAs using nuclear extracts from D0 osteochondro-progenitor and hypertrophic D26 ATDC5 cells show a shifted band that is supershifted (arrow) by an anti-TCF4 antibody in D26 cells. B–D, expression of the 0.5-kb (B), 2.3-kb (C), and 4-kb (D) Ccn2-luciferase constructs in D0 versus D26 ATDC5 chondrocytes in the presence of overexpressed Sox9, LEF1, or stabilized β-catenin. E, shown are relative luciferase activity levels of 0.5-kb WT and 0.5-kb (TCF·Sox mut) constructs in D26 ATDC5 chondrocytes in the presence (+) or absence (−) of overexpressed stabilized β-catenin. *, p < 0.05; **, p < 0.005. RLU, relative luciferase units.
Having demonstrated that Sox9 binds the Ccn2 proximal promoter in D0 ATDC5 cells and TCF4 binds this promoter in D26 ATDC5 cells, co-transfection assays were performed to test how Sox9 and TCF·LEF·β-catenin complexes affect Ccn2 promoter activity (Fig. 6, B–D). Sox9 overexpression had little effect in D0 osteochondro-progenitors, where endogenous levels of Sox9 are already high, but had a repressive effect in hypertrophic D26 cells, where endogenous Sox9 levels are low. In contrast, overexpression of β-catenin activated all three constructs in D0 cells (Fig. 6, B–D).
β-Catenin activates gene expression at TCF·LEF sites through formation of complexes containing TCF·LEF cofactors. In the absence of β-catenin, TCF·LEF factors act as repressors (5, 43). In accordance, overexpression of LEF1 in D0 osteochondro-progenitors, where endogenous levels of β-catenin are low, led to repression of activity of the 2.3- and 4-kb constructs (Fig. 6, B and C); there was no repression of the 0.5-kb construct (Fig. 6A). Taken together, these results are consistent with the hypothesis that the 0.5-kb Ccn2 construct is not repressed by overexpressed TCF·LEF in D0 osteochondro-progenitors because high levels of repressive Sox9 already occupied the −443-bp TCF·LEF·Sox9 site. Similarly, the fact that the −443-bp TCF·LEF·Sox9 site is not responsive to overexpressed β-catenin in D26 hypertrophic cells is consistent with the hypothesis that this site is already occupied by endogenous TCF·LEF·β-catenin complexes in hypertrophic chondrocytes. This notion is consistent with the finding that the 443-bp site is the only TCF·LEF site in the 4-kb Ccn2 proximal promoter region that overlaps with a consensus Sox9 binding site (Fig. 2A and data not shown). As expected by the fact that the additional TCF·LEF sites in the 2.3- and 4-kb Ccn2 promoters are not consensus sites for Sox9 and, therefore, may not already be occupied in D0 cells, the 2.3- and 4-kb Ccn2 promoters are repressed by overexpressed LEF1 in D0 cells.
The TCF·LEF mutant exhibited higher activity than the WT construct in D26 cells (Fig. 6E). As expected, when TCF·LEF factors are unable to bind, the mutated construct was no longer responsive to overexpressed stabilized β-catenin (Figs. 2E and 6E). Taken together, these findings suggest that a major function of the −443-bp TCF·LEF·Sox9 site is to repress Ccn2 expression in osteochondro-progenitors, and this repression involves Sox9.
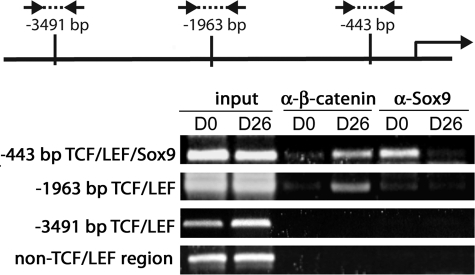
Because Sox9 levels are higher at D0 than D26 (Fig. 3D), we speculated that Sox9 occupies the −443-bp TCF·LEF·Sox9 site in the proximal Ccn2 promoter in D0 osteochondro-progenitors but not in hypertrophic D26 chondrocytes. Similarly, we suspected that in hypertrophic D26 cells, when levels of Sox9 are low but β-catenin is stabilized, β-catenin forms a complex with TCF·LEF factors at the −443-bp site to permit Ccn2 promoter activity. In agreement, ChIP experiments showed that endogenous Sox9 strongly binds to the −443-bp TCF·LEF·Sox9 site in D0 osteochondro-progenitors but not in D26 hypertrophic cells (Fig. 7). Furthermore, Sox9 binding to the TCF·LEF site at −1963 bp is minimal, and Sox9 binding to the −3491 bp site is undetectable, as predicted based on the lack of overlapping consensus Sox9 binding sites in these regions. Endogenous β-catenin binds strongly to the −443-bp and −1963-bp TCF·LEF sites in hypertrophic D26 cells but only weakly in D0 cells (Fig. 7). These results show that repressive Sox9 at D0 and activating β-catenin·TCF·LEF at D26 differentially occupy the −443-bp TCF·LEF·Sox9 site in the Ccn2 promoter.
FIGURE 7.
Occupancy of consensus TCF·LEF sites within the Ccn2 promoter analyzed by ChIP analysis in D0 osteochondro-progenitor and hypertrophic D26 ATDC5 chondrocytes. ChIP analysis of β-catenin and Sox9 binding to the regions encompassing the −443-bp consensus TCF·LEF·Sox9 site and the TCF·LEF consensus sites at −1963 and −3491 bp is shown. The diagram shows relative positions of the primers spanning the consensus TCF·LEF sites in the proximal Ccn2 promoter. The arrow marks the transcription start site. See “Experimental Procedures” for primer sequences. *, p < 0.05; **, p < 0.005.
The ability of Sox9 to bind to the −443-bp TCF·LEF·Sox9 site but not the −1963-bp and −3491-bp TCF·LEF elements may underlie the ability of overexpressed LEF1 to repress activity of the 2.3- and 4-kb Ccn2 promoters but not the 0.5-kb promoter at D0 (Fig. 6, B–D); prior occupancy of the −443-bp site by Sox9 may abrogate the effects of overexpressed LEF1. These results suggest that Sox9 represses Ccn2 expression in osteochondro-progenitors by binding to the −443-bp TCF·LEF·Sox site. When β-catenin levels rise and Sox9 levels fall in hypertrophic chondrocytes, β-catenin binds to the −443-bp TCF·LEF·Sox9 site to permit Ccn2 gene expression.
Sox9 and β-Catenin May Differentially Regulate Expression of Multiple Hypertrophic Cartilage-specific Genes
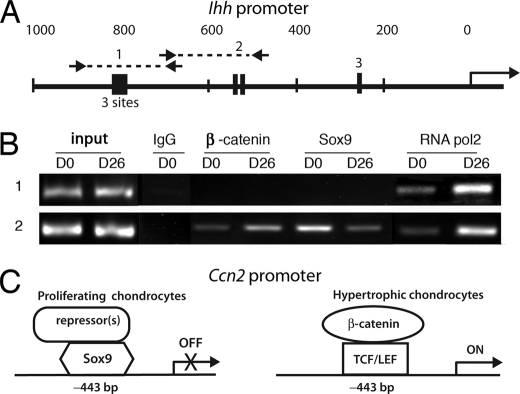
Indian hedgehog (Ihh) is expressed in prehypertrophic chondrocytes and is required for chondrocyte maturation in vivo (44). Little is known about the transcriptional regulation of Ihh expression in the growth plate, but canonical Wnt signaling in prehypertrophic chondrocytes activates Ihh transcription, and ChIP analysis revealed TCF·LEF consensus sites capable of binding to TCF·LEF·β-catenin complexes (45). However, whether or not these or other consensus TCF·LEF sites might also serve as binding sites for Sox9 was not determined. The promoter sequence of the mouse Ihh gene was, therefore, examined for consensus TCF·LEF motifs to explore the possibility that Sox9 and β-catenin might differentially bind to any of these sites in osteochondro-progenitors versus hypertrophic chondrocytes in a manner similar to that observed for the −443-bp site in the Ccn2 promoter. Several potential TCF·LEF sites were found in the Ihh proximal promoter region in addition to those identified previously (45) (Fig. 8A; supplemental Fig. 1). ChIP analyses of two Ihh promoter regions harboring these sites were performed to investigate whether Sox9 and β-catenin bind differentially in chondro-progenitor versus hypertrophic chondrocytes (Fig. 8B). Sox9 was found to be associated with one of these regions (region 2) of the Ihh promoter in D0 cells (Fig. 8B and data not shown). Higher levels of Sox9 binding were seen in D0 than in D26 cells in region 2; reciprocally, β-catenin binding was higher at D26 using the region 2 primers. Although consensus TCF·LEF sites were also seen in region 1, no binding of either β-catenin or Sox9 was seen in this region. Additional studies will be required to determine the relevance of these findings to the expression of Ihh. Nevertheless, the results suggest that Sox9 may act as a repressor of both Ihh and Ccn2 expression in chondro-progenitors, whereas β-catenin activates expression of both genes in hypertrophic chondrocytes.
FIGURE 8.
Differential occupancy of the Ihh promoter by β-catenin and Sox9 in osteochondro-progenitor versus hypertrophic chondrocytes. A, shown is a schematic diagram of the mouse Ihh proximal promoter region. Arrows mark the primers used to amplify regions 1 and 2. The primer sequences are indicated under “Experimental Procedures.” TCF·LEF consensus sites are represented as black boxes. Three TCF·LEF consensus sequences separated by 1–5 bp are present in region 1. A, the TCF·LEF site identified previously by Später et al. (45) is shown as region 3. B, ChIP analysis of Ihh promoter occupancy in D0 versus D26 ATDC5 chondrocytes is shown. ChIP for RNApol2 confirms activity of the Ihh promoter is higher in D26 cells than in D0 cells. C, model depicting proposed interactions of Sox9, TCF·LEF, and β-catenin at the −443 bp site in the Ccn2 proximal promoter is shown. In osteochondro-progenitors, a repressive Sox9 complex occupies the −443-bp site. In hypertrophic chondrocytes, stabilized β-catenin levels rise and Sox9 levels decrease, permitting formation of an activating β-catenin·TCF·LEF complex.
DISCUSSION
CCN2 is essential for chondrocyte proliferation and maturation (10). Ccn2 is expressed in proliferating chondrocytes at low levels and at much higher levels in hypertrophic chondrocytes. Ccn2 knock-out mice exhibit chondrodysplasia encompassing reduced proliferation and extracellular matrix production (9, 10). Ccn2 mutants also exhibit expanded hypertrophic zones as a result of defective clearance of terminal chondrocytes (10).
Studies of the regulation of Ccn2 expression in chondrocytes are limited. A cis-regulatory element within the proximal 110-bp Ccn2 promoter region has been reported to control cell type-specific transcriptional activity (46). The discovered sequence, TRENDIC, enhanced Ccn2 promoter activity in chondrocytes compared with fibroblasts. Matrix metalloproteinase 3 (MMP3) was shown to bind to TRENDIC to activate Ccn2 expression (47). However, Mmp3-deficient mice do not exhibit apparent cartilage phenotypes (48). The transcription factor c-Maf is expressed in hypertrophic cells and is required for chondrocyte differentiation (49). Omoteyama et al. (50) demonstrated that c-Maf induces Ccn2 expression in fibroblasts, but binding sites for c-Maf were not identified, and the study did not determine whether c-Maf regulates Ccn2 expression in chondrocytes. Retinoic acid (RA) up-regulates CCN2 expression in proliferating and hypertrophic chondrocytes by activating mitogen-activated protein kinase activities (51), but whether or not Ccn2 is a direct target of RA receptors is as yet unknown.
We demonstrated here that the relative levels of Sox9 and β-catenin regulate stage-specific Ccn2 expression in chondrocytes. Sequences within the 4-kb proximal Ccn2 promoter fragment drove Wnt-inducible Ccn2 expression in chondrocytes in vitro and in vivo. TCF·LEF consensus sequences were found in the 4-kb Ccn2 promoter fragment, and ChIP analysis revealed that the −443 bp site, which is a consensus binding site for both TCF·LEF and Sox9, was associated with Sox9 in D0 osteochondro-progenitors and with β-catenin in hypertrophic D26 chondrocytes. In agreement, reporter constructs containing these sites were repressed by Sox9 and activated by β-catenin. A model representing the proposed actions of these transcription factors at the consensus TCF·LEF·Sox9 site at −443 bp on the Ccn2 promoter is shown in Fig. 8C.
The ability of Sox9 to function as a transcriptional activator has been well characterized. Its function as a potential repressor is far less well understood. Previous studies have shown that β-catenin and Sox9 have opposing activities in chondrogenesis (6, 52). This antagonism involved cytoplasmic interaction between β-catenin and Sox9, promoting ubiquitin-mediated degradation of the β-catenin/Sox complex. In other systems, Sox9 antagonizes Wnt pathways by inducing the expression of transcriptional repressors of Wnt signaling (53). We cannot rule out either of these possibilities with respect to Ccn2 expression. However, the observation that Sox9 binds to the Ccn2 promoter raises the possibility of a more direct mechanism. Sox proteins are related to TCF·LEFs and bind to similar consensus sequences (54). The possibility that Sox9 and β-catenin·TCF·LEF complexes might antagonize each other by competing for occupancy of promoter binding sites has been suggested previously but not demonstrated (55). Our data support this possibility for Ccn2 regulation; ChIP analysis revealed that binding of β-catenin and Sox9 are mutually exclusive at the −443-bp site. Moreover, a similar relationship was seen for Sox9 and β-catenin binding to a site on the Ihh promoter, suggesting that Sox9 may exert a direct repressive function on the expression of multiple hypertrophic genes whose expression is activated by TCF·LEF·β-catenin complexes. The repressive function of Sox9 is most likely mediated by its ability to recruit repressors, possibly histone deacetylases, to the Ccn2 promoter in osteochondro-progenitor cells. Thus, repression by Sox9 and activation by β-catenin via binding to a common TCF·LEF·Sox9 consensus site may represent a general mechanism for regulating the expression of genes expressed in hypertrophic cartilage.
Supplementary Material
Acknowledgments
We thank Dr. Buer Song, Dr. Marcos Pacheco, and Gavin Gabreski for technical assistance. We are grateful to Drs. Paul Benya and Sotirios Tetradis for advice during the execution of the experiments and for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AR052686.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- TCF
- T-cell factor
- LEF
- lymphoid enhancer factor
- CCN2
- Cyr61-CTGF-Nov 2
- CTGF
- connective tissue growth factor
- lacZ
- β-galactosidase
- D-
- day.
REFERENCES
- 1.Kronenberg H. M. (2003) Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi T., Kronenberg H. (2005) Endocrinology 146, 1012–1017 [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre V., Smits P. (2005) Birth Defects Res. C Embryo Today 75, 200–212 [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre V., Dumitriu B., Penzo-Méndez A., Han Y., Pallavi B. (2007) Int. J. Biochem. Cell Biol. 39, 2195–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoppler S., Kavanagh C. L. (2007) J. Cell Sci. 120, 385–393 [DOI] [PubMed] [Google Scholar]
- 6.Akiyama H., Lyons J. P., Mori-Akiyama Y., Yang X., Zhang R., Zhang Z., Deng J. M., Taketo M. M., Nakamura T., Behringer R. R., McCrea P. D., de Crombrugghe B. (2004) Genes Dev. 18, 1072–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day T. F., Guo X., Garrett-Beal L., Yang Y. (2005) Dev. Cell 8, 739–750 [DOI] [PubMed] [Google Scholar]
- 8.Shi-Wen X., Leask A., Abraham D. (2008) Cytokine Growth Factor Rev. 19, 133–144 [DOI] [PubMed] [Google Scholar]
- 9.Nishida T., Kawaki H., Baxter R. M., Deyoung R. A., Takigawa M., Lyons K. M. (2007) J. Cell Commun. Signal. 1, 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivkovic S., Yoon B. S., Popoff S. N., Safadi F. F., Libuda D. E., Stephenson R. C., Daluiski A., Lyons K. M. (2003) Development 130, 2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oka M., Kubota S., Kondo S., Eguchi T., Kuroda C., Kawata K., Minagi S., Takigawa M. (2007) J. Histochem. Cytochem. 55, 1245–1255 [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi T., Nishida T., Shimo T., Kobayashi K., Kubo T., Tamatani T., Tezuka K., Takigawa M. (2000) Endocrinology 141, 264–273 [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto G., Inoki I., Fujii Y., Aoki T., Ikeda E., Okada Y. (2002) J. Biol. Chem. 277, 36288–36295 [DOI] [PubMed] [Google Scholar]
- 14.Kothary R., Clapoff S., Darling S., Perry M. D., Moran L. A., Rossant J. (1989) Development 105, 707–714 [DOI] [PubMed] [Google Scholar]
- 15.Brugger S. M., Merrill A. E., Torres-Vazquez J., Wu N., Ting M. C., Cho J. Y., Dobias S. L., Yi S. E., Lyons K., Bell J. R., Arora K., Warrior R., Maxson R. (2004) Development 131, 5153–5165 [DOI] [PubMed] [Google Scholar]
- 16.Retting K. N., Song B., Yoon B. S., Lyons K. M. (2009) Development 136, 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R., 3rd, Nusse R. (2003) Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 18.Atsumi T., Miwa Y., Kimata K., Ikawa Y. (1990) Cell Differ. Dev. 30, 109–116 [DOI] [PubMed] [Google Scholar]
- 19.Pateder D. B., Ferguson C. M., Ionescu A. M., Schwarz E. M., Rosier R. N., Puzas J. E., O'Keefe R. J. (2001) J. Cell. Physiol. 188, 343–351 [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki Y., Kugimiya F., Chikuda H., Kamekura S., Ikeda T., Kawamura N., Saito T., Shinoda Y., Higashikawa A., Yano F., Ogasawara T., Ogata N., Hoshi K., Hofmann F., Woodgett J. R., Nakamura K., Chung U. I., Kawaguchi H. (2008) J. Clin. Invest. 118, 2506–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubinfeld B., Robbins P., El-Gamil M., Albert I., Porfiri E., Polakis P. (1997) Science 275, 1790–1792 [DOI] [PubMed] [Google Scholar]
- 22.Schreiber E., Schaffner W. (1989) Somat. Cell Mol. Genet. 15, 591–603 [DOI] [PubMed] [Google Scholar]
- 23.Lefebvre V., Huang W., Harley V. R., Goodfellow P. N., de Crombrugghe B. (1997) Mol. Cell. Biol. 17, 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiyama H., Hiraki Y., Shigeno C., Kohno H., Shukunami C., Tsuboyama T., Kasai R., Suzuki F., Konishi J., Nakamura T. (1996) J. Bone Miner. Res. 11, 22–28 [DOI] [PubMed] [Google Scholar]
- 25.Enomoto H., Enomoto-Iwamoto M., Iwamoto M., Nomura S., Himeno M., Kitamura Y., Kishimoto T., Komori T. (2000) J. Biol. Chem. 275, 8695–8702 [DOI] [PubMed] [Google Scholar]
- 26.Usui M., Xing L., Drissi H., Zuscik M., O'Keefe R., Chen D., Boyce B. F. (2008) J. Bone Miner. Res. 23, 314–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hata K., Nishimura R., Muramatsu S., Matsuda A., Matsubara T., Amano K., Ikeda F., Harley V. R., Yoneda T. (2008) J. Clin. Invest. 118, 3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman B., Gigout L. I., Sudre L., Grant M. E., Wallis G. A. (2001) J. Cell Biol. 154, 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akiyama H., Kanno T., Ito H., Terry A., Neil J., Ito Y., Nakamura T. (1999) J. Cell. Physiol. 181, 169–178 [DOI] [PubMed] [Google Scholar]
- 30.Brown A. J., Alicknavitch M., D'Souza S. S., Daikoku T., Kirn-Safran C. B., Marchetti D., Carson D. D., Farach-Carson M. C. (2008) Bone 43, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovcharenko I., Nobrega M. A., Loots G. G., Stubbs L. (2004) Nucleic Acids Res. 32, W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) Nucleic Acids Res. 34, D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldring M. B., Tsuchimochi K., Ijiri K. (2006) J. Cell. Biochem. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 34.Si W., Kang Q., Luu H. H., Park J. K., Luo Q., Song W. X., Jiang W., Luo X., Li X., Yin H., Montag A. G., Haydon R. C., He T. C. (2006) Mol. Cell. Biol. 26, 2955–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labbé E., Lock L., Letamendia A., Gorska A. E., Gryfe R., Gallinger S., Moses H. L., Attisano L. (2007) Cancer Res. 67, 75–84 [DOI] [PubMed] [Google Scholar]
- 36.Enomoto-Iwamoto M., Kitagaki J., Koyama E., Tamamura Y., Wu C., Kanatani N., Koike T., Okada H., Komori T., Yoneda T., Church V., Francis-West P. H., Kurisu K., Nohno T., Pacifici M., Iwamoto M. (2002) Dev. Biol. 251, 142–156 [DOI] [PubMed] [Google Scholar]
- 37.Erwin W. M., Ashman K., O'Donnel P., Inman R. D. (2006) Arthritis Rheum. 54, 3859–3867 [DOI] [PubMed] [Google Scholar]
- 38.Ng L. J., Wheatley S., Muscat G. E., Conway-Campbell J., Bowles J., Wright E., Bell D. M., Tam P. P., Cheah K. S., Koopman P. (1997) Dev. Biol. 183, 108–121 [DOI] [PubMed] [Google Scholar]
- 39.Sock E., Pagon R. A., Keymolen K., Lissens W., Wegner M., Scherer G. (2003) Hum. Mol. Genet. 12, 1439–1447 [DOI] [PubMed] [Google Scholar]
- 40.De Santa Barbara P., Bonneaud N., Boizet B., Desclozeaux M., Moniot B., Sudbeck P., Scherer G., Poulat F., Berta P. (1998) Mol. Cell. Biol. 18, 6653–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhold M. I., Naski M. C. (2007) J. Biol. Chem. 282, 3653–3663 [DOI] [PubMed] [Google Scholar]
- 42.Dong Y. F., Soung do Y., Chang Y., Enomoto-Iwamoto M., Paris M., O'Keefe R. J., Schwarz E. M., Drissi H. (2007) Mol. Endocrinol. 21, 2805–2820 [DOI] [PubMed] [Google Scholar]
- 43.Brantjes H., Barker N., van Es J., Clevers H. (2002) Biol. Chem. 383, 255–261 [DOI] [PubMed] [Google Scholar]
- 44.St.-Jacques B., Hammerschmidt M., McMahon A. P. (1999) Genes Dev. 13, 2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Später D., Hill T. P., O'sullivan R. J., Gruber M., Conner D. A., Hartmann C. (2006) Development 133, 3039–3049 [DOI] [PubMed] [Google Scholar]
- 46.Eguchi T., Kubota S., Kondo S., Kuboki T., Yatani H., Takigawa M. (2002) Biochem. Biophys. Res. Commun. 295, 445–451 [DOI] [PubMed] [Google Scholar]
- 47.Eguchi T., Kubota S., Kawata K., Mukudai Y., Uehara J., Ohgawara T., Ibaragi S., Sasaki A., Kuboki T., Takigawa M. (2008) Mol. Cell. Biol. 28, 2391–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Page-McCaw A., Ewald A. J., Werb Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacLean H. E., Kim J. I., Glimcher M. J., Wang J., Kronenberg H. M., Glimcher L. H. (2003) Dev. Biol. 262, 51–63 [DOI] [PubMed] [Google Scholar]
- 50.Omoteyama K., Ikeda H., Imaki J., Sakai M. (2006) Biochem. Biophys. Res. Commun. 339, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 51.Shimo T., Koyama E., Sugito H., Wu C., Shimo S., Pacifici M. (2005) J. Bone Miner. Res. 20, 867–877 [DOI] [PubMed] [Google Scholar]
- 52.Topol L., Chen W., Song H., Day T. F., Yang Y. (2009) J. Biol. Chem. 284, 3323–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bastide P., Darido C., Pannequin J., Kist R., Robine S., Marty-Double C., Bibeau F., Scherer G., Joubert D., Hollande F., Blache P., Jay P. (2007) J. Cell Biol. 178, 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wegner M. (1999) Nucleic Acids Res. 27, 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang C., Basta T., Jensen E. D., Klymkowsky M. W. (2003) Development 130, 5609–5624 [DOI] [PubMed] [Google Scholar]
- 56.Holmes A., Abraham D. J., Chen Y., Denton C., Shi-wen X., Black C. M., Leask A. (2003) J. Biol. Chem. 278, 41728–41733 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.