Abstract
Abnormal activation of calpain is implicated in synaptic dysfunction and participates in neuronal death in Alzheimer disease (AD) and other neurological disorders. Pharmacological inhibition of calpain has been shown to improve memory and synaptic transmission in the mouse model of AD. However, the role and mechanism of calpain in AD progression remain elusive. Here we demonstrate a role of calpain in the neuropathology in amyloid precursor protein (APP) and presenilin 1 (PS1) double-transgenic mice, an established mouse model of AD. We found that overexpression of endogenous calpain inhibitor calpastatin (CAST) under the control of the calcium/calmodulin-dependent protein kinase II promoter in APP/PS1 mice caused a remarkable decrease of amyloid plaque burdens and prevented Tau phosphorylation and the loss of synapses. Furthermore, CAST overexpression prevented the decrease in the phosphorylation of the memory-related molecules CREB and ERK in the brain of APP/PS1 mice and improved spatial learning and memory. Interestingly, treatment of cultured primary neurons with amyloid-β (Aβ) peptides caused an increase in the level of β-site APP-cleaving enzyme 1 (BACE1), the key enzyme responsible for APP processing and Aβ production. This effect was inhibited by CAST overexpression. Consistently, overexpression of calpain in heterologous APP expressing cells up-regulated the level of BACE1 and increased Aβ production. Finally, CAST transgene prevented the increase of BACE1 in APP/PS1 mice. Thus, calpain activation plays an important role in APP processing and plaque formation, probably by regulating the expression of BACE1.
Keywords: Aging, Amyloid, Calpain, Neurodegeneration, Transgenic, Alzheimer Disease, Amyloid Plaque, BACE1, Calpain, Calpastatin
Introduction
Aggregation of amyloid-β (Aβ)3 peptides into compact plaques is a characteristic feature in the pathogenesis of Alzheimer disease (AD) (1, 2). Recently, it is suggested that soluble Aβ oligomers, in the process of aggregation, adversely affect synaptic structure and plasticity (2–9). Aβ peptides are generated in neurons by the sequential proteolytic cleavage of the transmembrane glycoprotein amyloid precursor protein (APP) that is cleaved initially by β-site APP-cleaving enzyme 1 (BACE1, also known as β-secretase) and subsequently by γ-secretase, whose activity is associated with a presenilin (PS)-containing macromolecular complex (10–12), in the transmembrane region of APP (13, 14). Thus, BACE1 has been proposed to be a therapeutic target for AD (15).
Calpains are a family of calcium-activated intracellular cysteine proteases that are involved in many physiological events including long term potentiation (16–18) or neurotoxic insults ranging from ischemia to Alzheimer disease (19–21). Inhibition of calpain by synthetic inhibitors exerts neuroprotection in various models of brain injuries, such as ischemia or excitotoxicity-induced neuronal death (22–24). Interestingly, Aβ aggregation is associated with neuronal and astrocytic calcium dysregulation (25–27). Treatment of cultured cortical neurons with Aβ oligomers caused calcium influx and subsequently calpain activation (21).
A number of proteins have been identified as calpain substrates from various tissues (28). Calpain cleavage of p35, a regulatory partner of cyclin-dependent kinase 5 (Cdk5), generates p25, which causes hyperactivation of Cdk5 (19, 29, 30) and causes phosphorylation of many substrates, including microtubule-associated protein Tau, resulting in the formation of neurofibrillary tangles. Interestingly, induction of p25 resulted in enhanced forebrain Aβ levels before any sign of neuropathology in APP transgenic mice (31), by up-regulating the expression of BACE1 (32). A recent study shows that treatment with synthetic calpain inhibitor improves memory and synaptic transmission in the mouse model of AD (33). Nevertheless, it remains obscure how calpain participates in AD progression.
Here we demonstrate a role for calpain in APP processing and plaque formation. We found that calpain activation increases the level of BACE1, suggesting a mechanism by which calpain promotes APP processing. These results, together with previous findings, indicate that calpain has multiple roles in AD progression, e.g. APP processing, Tau phosphorylation, synaptic dysfunction, and neuronal death.
EXPERIMENTAL PROCEDURES
Mice
The human calpastatin cDNA was subcloned from the plasmid described in our previous study (34) and inserted downstream of the 8.5-kb CaMKII promoter between the NotI and PacI site of a modified pMM279 plasmid (35). The linearized CaMKII-calpastatin fragment between two SalI sites was microinjected into C57BL/6J × FVB/NJ fertilized eggs, which were then reimplanted into pseudopregnant recipient mice. Genotypes were determined by PCR for tail genomic DNA, with primers of the following sequences: 5′-CTCAGAAGCCCCAAGCTCGTCAGTC-3′ (P1 in the CaMKIIα promoter region); 5′-TCCCGATGGTTTATCCGGTTTAGAT-3′ (P2 in the coding region of human calpastatin). hCAST-expressing founders were backcrossed to C57BL/6J mice for more than eight generations. We crossed APPswe/PS1dE9 mice on the B6C3F1/J background (Jackson Laboratory) (36, 37), which express familial AD-causing mutated forms of human APP (APPswe, Swedish familial AD-causing mutation) and presenilin 1 (PS1ΔE9), with CAST Tg mice to generate four genotype offspring: WT, CAST, APP/PS1, and APP/PS1/CAST. Gender-matched littermates were used for comparison whenever possible. All of the experimental protocols followed institutional guidelines for animal care and administration.
Reagents and Constructs
Thioflavin S was purchased from Sigma. The antibodies used for immunoblotting (IB) or immunostaining (IS) were from: Santa Cruz Biotechnology (calpastatin (H-300, sc-20779, 1:1000 for IB, 1:100 for IS), p35 (sc-820, 1:300), GFP (sc-8334, 1:500), and ERK1 (sc-93, 1:500)), Chemicon (β-actin (monoclonal antibody 1501, 1:10000), spectrin (monoclonal antibody 1622, 1:1000), BACE (antibody 5940, 1:1000), GFAP (monoclonal antibody 360, 1:3000 for IB, 1:1000 for IS), PSD-95 (monoclonal antibody 1596, 1:1000), and synapsin I (antibody 1543, 1:1000 for IB, 1:500 for IS)), Abcam (calpastatin (2G11D6, antibody 3515, 1:1500)), Pierce (AT8, MN1020, 1:2000), Calbiochem (Tau-5, 577801, 1:2000), Upstate (CREB (06-863, 1:2000), phospho-CREB Ser133 (05-807, 1:1000), and Myc (9E10, 05-419, 1:800)), Cell Signaling (phospho-ERK1/2 (9101, 1:1000)), Signet (β-amyloid (4G8, SIG-39220, 1:500 for IS)), or Invitrogen (APP (CT695, 51–2700, 1:1000) and synaptophysin (Z66, 1:4000)). Rabbit anti-m-calpain (1:2000) was introduced in our previous study (34). M-calpain or the mutated form C105S (34) was subcloned into pEGFP-N1 (Clontech) in-frame with enhanced GFP at the HindIII and KpnI sites or into pCAGGS-IRES-GFP (38) at the NotI site. Human calpastatin cDNA (34) was subcloned into CAG-YFP vector, which has dual CAG promoters (39).
Cell Culture, Transfection, and Aβ Treatment
HEK293T cell lines stably expressing APP695myc or APP695 (a kind gift from Dr. Yi-Zheng Wang, Institute of Neuroscience, Chinese Academy of Sciences) were cultured in DMEM, 10% fetal bovine serum (Invitrogen) with G418 (200 μg/ml). Transfection experiments were performed with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Dissociated hippocampal or cortical neurons were prepared as described previously (40). Briefly, hippocampi or cerebral cortex of embryonic day 18 rat embryos were digested with 0.125% trypsin-EDTA for 20 min at 37 °C, followed by trituration with pipettes in the plating medium (DMEM with 10% FBS). Dissociated neurons were transfected by using a nucleofector device (Amaxa, Gaithersburg, MD). Then the neurons were plated onto a 3.5-mm dish coated with poly-d-lysine (0.1 mg/ml). After culturing for 4 h, the media were changed to neuronal culture medium (neurobasal media containing 1% glutamate and 2% B27). Synthetic Aβ1–42 (American Peptide) was dissolved to 200 μm in neurobasal medium (GIBCO) and incubated for 4 days at 37 °C to preaggregate the peptides (41). Neurons at 6 or 7 days in vitro were incubated in the presence of the preaggregated peptide at 5 μm for 24 h.
Brain Extracts and Western Blot
For assessment of calpain cleavage of spectrin or p35 on mice, the brains were dissected immediately and homogenized in a cold lysis buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 1 mm EGTA, 1% Nonidet P-40, a protease inhibitor mixture (0.8 μm aprotinin, 20 μm leupeptin, 10 μm pepstatin A, 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 50 μm bestatin, 15 μm E-64, 1 mm PMSF), and phosphatase inhibitors (10 mm NaF, 0.2 mm sodium orthovanadate, 2 mm sodium pyrophosphate, 2 mm β-glycerophosphate) (19, 33). After clarification by centrifugation (15,700 g at 4 °C for 30 min), supernatant proteins were quantified using Bradford method (Bio-Rad; Cat. #500-0006), and the same amount of proteins (∼50 μg of protein for each sample) were subjected to SDS-PAGE and Western blot analysis with ImageQuant software (GE Healthcare) to measure the intensity of the protein bands.
Immunocytochemistry and Thioflavin S Staining
The mice were fixed by transcardiac perfusion using 4% paraformaldehyde in PBS, pH 7.4. Brain sections (30 μm thickness) were sequentially incubated with individual primary antibodies in PBS containing 1% BSA and 0.3% Triton X-100 overnight at 4 °C. After several washes in PBS, the sections were incubated with biotinylated anti-rabbit or anti-mouse IgG for 3 h at 37 °C and then incubated with Cy3- or Cy2-conjugated streptavidin and counterstained with Hoechst. Fibrillar amyloid deposits were labeled by incubating brain sections for 20 min in a solution of thioflavin S (2 μg/ml in PBS).
Aβ ELISA
Mouse cerebral cortex extracts were treated with 5 m guanidine HCl, 50 mm Tris-HCl, pH 8.0, to solubilize Aβ. Amount of Aβ was analyzed by ELISA according to the manufacturer's instructions (BioSource, Camarillo, CA).
Image Analysis and Immunofluorescence Quantification
Images for the thioflavin S, GFAP, and synapsin staining were acquired with a NIKON E600FN microscope using a 10× objective. The collected images were converted into 8-bit format, and the background was subtracted. An intensity threshold was set and was kept constant for all of the images analyzed with Image-Pro Plus 5.0 software (Media Cybernetic). Plaques within the cerebral cortex, hippocampus, or corpus callosum were measured by standardized fluorescence intensity, and those plaques over 10 μm2 were analyzed for area [(plaque area/total area selected) × 100%] or numbers [(plaque numbers/total area selected (mm2)]. Similarly, the areas of the GFAP-positive region [(GFAP-positive area/total cortical area selected) × 100%] and the synapsin-loss region [(area with synapsin loss/total cortical area selected) × 100%] around plaques were also measured with Image-Pro Plus 5.0 software. The results were obtained from three to six animals in each group (three slices/brain at ∼150-μm spacing) and presented as the means ± S.E. (with p < 0.05 considered as a significant difference, ANOVA supplemented with two-tailed Student's t test).
Morris Water Maze
A hidden platform water maze was used as described previously (42). Gender-matched littermates with various genotypes were used for the behavior tests. A platform (11 cm in diameter) was placed in the center of a specific quadrant of the pool (1.2 m in diameter) and submerged 1 cm underneath the water. The training session consisted of 5 or 7 days, four trials/day. The time the mice spent to locate the hidden platform was recorded to reflect spatial learning ability. After training for 5 days, the platform was removed, and a 60-s probe test was given to trained mice. Time spent in the target quadrant and the number of target platform crossings were recorded to reflect spatial memory.
RESULTS
CAST Overexpression Decreases Aβ Plaque Load in APP/PS1 Mice
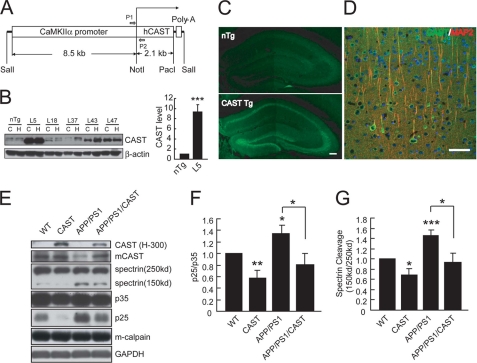
There is a depletion of CAST, the endogenous inhibitor of calpain, in the prefrontal cortex of AD brains (22, 43); this calpastatin decrease may allow calpain activation. We generated transgenic mice overexpressing human CAST under the CaMKIIα promoter (Fig. 1A), which controls gene expression in forebrain regions (35, 44). Brain extracts from hippocampi or cerebral cortex from transgenic lines and littermate controls (nontransgenic, nTg) were analyzed by immunoblot with the H-300 antibody, which recognizes human and mouse calpastatin. Among the five Tg lines, line 5 showed the highest level of CAST expression, with ∼9-fold over nTg mice (Fig. 1B) and thus used for the following experiments. The regional distribution of the overexpressed CAST was analyzed by immunohistochemistry with H-300 antibody. The transgene-derived hCAST was highly expressed in the hippocampus and neocortex (Fig. 1, C and D). Next CAST Tg mice were crossed with APP/PS1 mice. The levels of 150-kDa spectrin and p25, the cleaved fragments of two well established calpain substrates, α-spectrin and p35, respectively, were measured to reflect calpain activity in the brains of littermate mice with different genotypes (Fig. 1, E–G). We found that calpain was activated in the APP/PS1 mice, as reflected by increased levels of 150-kDa spectrin or p25, and overexpression of CAST attenuated spectrin or p35 cleavage in WT or APP/PS1 mice (Fig. 1, E–G), suggesting the inhibition of endogenous calpain by CAST overexpression.
FIGURE 1.
CAST overexpression inhibits calpain in APP/PS1 mice. A, schematic representation of CaMKIIα-hCAST construct used for generating transgenic mice. P1 and P2 are primers used for the genotyping. B, IB for the transgenic human CAST levels with rabbit anti-calpastatin (H-300) antibody in hippocampus (H) and cerebral cortex (C) from 2-month-old nTg or different lines of CAST Tg mice (∼50 μg of protein for each case). Quantification of the blots reveals that the CAST level in the hippocampus of Line 5 (L5) is ∼9-fold of that in nTg. C, hippocampal slices of 2-month-old nTg or CAST Tg (Line 5) were stained with (H-300) antibody. Scale bar, 100 μm. D, double staining with H-300 and MAP2 antibodies in cerebral cortex of hCAST Tg mice (Line 5). Scale bar, 50 μm. E, hippocampal homogenates (∼50 μg of protein) from 12-month-old female littermate mice were subjected to IB with indicated antibodies. mCAST (2G11D6), a monoclonal antibody used to detect endogenous mouse calpastatin. F and G, the ratio of p25/p35 (F) or 150/250-kDa spectrin (G) was quantified to reflect calpain activity, with values from WT mice set as 1.0. Shown are the means ± S.E. (n = 7 animals in each group in F, n = 8 in G). *, p < 0.05; **, p < 0.01; ***, p < 0.001, ANOVA with Student's t test.
Further, we determined the effects of CAST overexpression on the neuropathology of APP/PS1 mice. First, amyloid-β plaques were revealed by staining with anti-amyloid-β antibody 4G8 or thioflavin S, a homogenous mixture of compounds that is used to stain Alzheimer plaques (45) (Fig. 2, A and B). We found that amyloid-β plaque load in the hippocampus, neocortex, and corpus callosum in APP/PS1/CAST mice was markedly reduced compared with gender-matched APP/PS1 littermates (Fig. 2, A–F). The decrease of Aβ plaques was reflected by a reduced area occupied by the Aβ plaques (Fig. 2, C and D) and the number of plaques (Fig. 2, E and F). Moreover, calpastatin overexpression in APP/PS1 mice markedly reduced Aβ levels (Fig. 2G). Thus, inhibition of calpain by overexpressing calpastatin attenuated Aβ production and plaque load in the mouse model of AD.
FIGURE 2.
CAST overexpression decreases amyloid-β plaque in APP/PS1 mice. A and B, brain sections from 10-month-old male APP/PS1 or APP/PS1/CAST littermate mice were co-stained with thioflavin S and 4G8 antibody to label amyloid-β plaques. Shown are representative images of thioflavin S staining (A) and higher magnifications of the boxed areas co-labeled with thioflavin S and 4G8 antibody (B). C–F, quantification of plaque area in the hippocampus, cerebral cortex, or corpus callosum (C and D) and plaque numbers in cerebral cortex (E and F). Shown are the means ± S.E. (n = 6 for APP/PS1, n = 4 for APP/PS1/CAST, in C and E from 8-month-old male mice; n = 4 for APP/PS1, n = 3 for APP/PS1/CAST, in D and F from 10-month-old male mice). Three sections were analyzed for each animal. G, levels of Aβ (sum of Aβ1–40 and Aβ1–42) in cerebral cortex of 6-month-old male mice were measured by ELISA. n = 14 for APP/PS1; n = 13 for APP/PS1/CAST. *, p < 0.05; **, p < 0.01, ANOVA with Student's t test. The scale bars represent 500 μm in A and 100 μm in B.
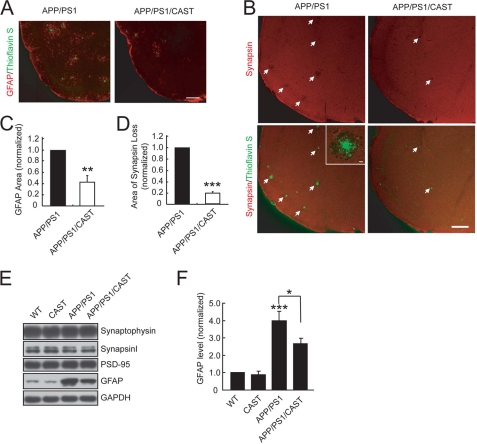
In the brains of AD patients and the AD mouse model, there are remarkable activated astrocytes around the area of Aβ plaques (46, 47). In line with this notion, a number of GFAP-positive areas were observed around the plaques (Fig. 3A). Protein levels of GFAP in the cerebral cortex were also found to be markedly increased in APP/PS1 mice (Fig. 3, E and F). Interestingly, the area covered by GFAP-positive regions and the elevation of GFAP levels was reduced in the cerebral cortex of APP/PS1/CAST mice (Fig. 3, A, C, E, and F). The reduction in GFAP areas and levels may be due to the decrease in Aβ production and area/number of plaques. Aβ plaques have also been shown to cause synaptic disruption (3, 4, 6). As shown in the inset of Fig. 3B, the intensity of synapsin, a presynaptic vesicle protein, was decreased at regions around Aβ plaques in the cerebral cortex, suggesting a loss of synapse. This reduction of synapsin was in part prevented by CAST overexpression (Fig. 3, B and D). However, unlike that of GFAP, the total levels of synaptic proteins, such as synaptophysin, synapsin, and PSD95, did not exhibit significant changes in APP/PS1 mice, compared with WT control mice, without or with CAST expression (Fig. 3E). This may be due to local synaptic abnormalities rather than global synaptic changes, as demonstrated in a previous study (48). Nevertheless, calpain inhibition by overexpressing CAST prevented Aβ deposition, astrocyte activation, and synapse loss.
FIGURE 3.
CAST overexpression prevents synapse loss and gliosis in APP/PS1 mice. Brain sections from 10-month-old APP/PS1 or APP/PS1/CAST littermate mice were co-stained with thioflavin S and anti-GFAP antibody to mark reactive astrocytes (A) or with anti-synapsin to reveal synapse loss (white arrows) (B) around plaques. Inset in B shows the decrease of synapsin signal around the Aβ plaque, indicating synaptic disruption. C and D, quantification for the area of GFAP-positive regions (C) or synapsin loss around plaques (D), with the values from APP/PS1 mice set as 1.0. Shown are the means ± S.E. (n = 4 animals of each group in C, n = 3 in D). Three sections were analyzed for each animal. E, homogenates of cerebral cortex (∼50 μg of protein) from 12-month-old mice were subjected to IB with indicated antibodies. F, GFAP level in E was quantified with values from WT mice set as 1.0. Shown are the means ± S.E. from seven mice/genotype. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ANOVA with Student's t test. The scale bars show 200 μm (A and B) and 10 μm (inset of B).
CAST Overexpression Blocks Tau Phosphorylation and Improves Spatial Learning and Memory of APP/PS1 Mice
AD is also characterized by intracellular neurofibrillary tangles containing hyperphosphorylated Tau (14, 49). Tau is phosphorylated at over 38 serine/threonine residues by several kinases, including ERK and Cdk5 (50). Hyperphosphorylated Tau is observed in the APP/PS1 mice brain after the onset of Aβ plaques (51). Calpain is known to activate Cdk5 by the cleavage of Cdk5 activator p35 to p25, which has a stronger activity toward Cdk5 (21). Among the many Tau phosphoepitopes, phosphorylation at AT8 epitopes mediated by ERK1/2 and Cdk5 (19) is often found in the AD brain and used as a pathogenic marker for AD. We found that the level of AT8-positive phosphorylated Tau was markedly increased in the brain of APP/PS1 mice, compared with WT control mice (Fig. 4A, lanes 1 and 3). Interestingly, this increase was prevented by CAST overexpression (Fig. 4, A and B). Thus, calpain inhibition impeded Tau phosphorylation in APP/PS1 mice.
FIGURE 4.
CAST overexpression blocks Tau phosphorylation and promotes spatial learning of APP/PS1 mice. A–F, hippocampal homogenates (∼50 μg of protein) from 12-month-old female littermate mice were subjected to IB with indicated antibodies. Shown are representative blots against phosphorylated Tau (AT8) or total Tau (Tau-5) (A), phosphorylated CREB (p-CREB) or total CREB (C), and phosphorylated ERK (p-ERK) or total ERK (E), respectively. The values for AT8/Tau-5 (B), p-CREB/CREB (D), or p-ERK/ERK (F) were quantified with values from WT mice set as 1.0. Shown are the means ± S.E. (n = 6 animals of each group in B, n = 5 in D, and n = 6 in F). G–L, overexpression of hCAST in APP/PS1 mice prevents learning and memory deficits in the Morris water maze. Mice at different ages were analyzed (G–I, ∼7-month-old female mice; J–L, ∼15-month-old male mice) for escape latency (G and J) after training for different days. After training for 5 days, a probe test was given to determine the percentage of time spent in the target quadrant (H and K) or number of target crossings (I and L) within 60 s. The number of animals in each group is indicated in the histograms (G and J). *, p < 0.05; **, p < 0.01; ***, p < 0.001, ANOVA with Student's t test.
Previous studies have shown that phosphorylation of two memory-associated proteins, ERK and the CREB protein, is decreased in the transgenic mouse model of AD (33, 52, 53). Consistently, we found that ERK1/2 or CREB phosphorylation was decreased in APP/PS1 mice compared with control mice (Fig. 4, C–F). However, these decreases were prevented by CAST overexpression (Fig. 4, C–F). The reverse relationship between the level of AT8 and p-ERK suggests that the changes of AT8 may not be mediated by ERK; rather, these changes are probably mediated by Cdk5.
The rescue of ERK and CREB phosphorylation shown above and the prevention of synapse loss (Fig. 3, B and D) by CAST overexpression prompted us to investigate the learning and memory performance of these mice. We compared the spatial learning performance of wild type and transgenic mice in the Morris water maze assay (42, 54). In normal acquisition of a visible platform, two groups of mice showed no difference in the speed and latency period to locate the platform (data not shown), suggesting that CAST overexpression had no effects on sensory motor or motivational performance. Next, we determined the effects of APP/PS1 transgene, CAST transgene, and number of training days on the spatial learning behavior, as reflected from escape latency toward a hidden platform, by using three-way repeated measure analysis of variance (ANOVA). We found that increasing days of training significantly improved learning performance in all groups (p < 0.001 in Fig. 4, G and J), whereas APP/PS1 transgene exhibited a marked impairment effect (p < 0.001 in Fig. 4, G and J) and CAST transgene exhibited a marked improvement effect (p < 0.01 in Fig. 4G and p < 0.05 in Fig. 4J) on learning behavior. Statistically, there was a significant interaction between APP/PS1 and CAST transgene (p < 0.001 in Fig. 4G and p < 0.05 in Fig. 4J). Specifically, after 4 or 5 days of training, the APP/PS1/CAST mice spent less time in locating the submerged escape platform compared with APP/PS1 mice at ∼7 months of age (Fig. 4G) or 15 months of age (Fig. 4J), as reflected by significantly reduced escape latency across trials. Thus, impaired spatial learning ability in APP/PS1 mice can be ameliorated by CAST overexpression. Next, a probe test was performed to determine the spatial memory of these mice. As expected, APP/PS1 mice showed defects in spatial memory, as reflected by reduced time spent in the target quadrant (Fig. 4, H and K) and a decreased number of crossings across the target (Fig. 4, I and L). Interestingly, these memory defects were prevented by the overexpression of CAST, in either young animals (Fig. 4, H and I) or relatively old animals (Fig. 4, K and L). Single CAST transgenic mice exhibited no difference in the ability of locating the hidden platform (Fig. 4, G and J) or spatial memory in the probe test (Fig. 4, H, I, K, and L). Thus, CAST transgene is able to alleviate the learning and memory deterioration in APP/PS1 mice.
CAST Overexpression Decreases APP Processing in APP/PS1 Mice
Having demonstrated the effects of CAST overexpression on plaque formation and other pathological or behavior defects of APP/PS1 mice, we next investigated how these effects were achieved. First, we determined the APP processing in APP/PS1 mice without or with CAST overexpression. As shown in Fig. 5A, cleaved C-terminal fragments of APP and β- and α-CTF were observed in the brain lysates of APP/PS1 mice. The increase in β-CTF reflects the activation or up-regulation of BACE1, consistent with the findings that BACE1 protein and activity are elevated in the AD brain (55–57). In agreement with these findings, BACE1 levels were also found to be increased in the brain lysates of APP/PS1 mice (Fig. 5A). However, overexpression of CAST caused a reduction in the levels of β-CTF (Fig. 5, A and B) as well as BACE1 (Fig. 5, A and C), although it had no effect on APP levels (Fig. 5A). Thus, calpain activation may contribute to BACE1 expression.
FIGURE 5.
Calpain activation up-regulates BACE1 levels. A–C, hippocampal homogenates (∼50 μg of protein) of 12-month-old littermate female mice with different genotypes were probed in IB with antibody against BACE1 or C-terminal APP, which recognizes β- or α-CTF and full-length APP. The levels of β-CTF (B) or BACE1 (C) were quantified with values from APP/PS1 mice or WT mice set as 1.0, respectively. Shown are the means ± S.E. from six mice/genotype. *, p < 0.05; **, p < 0.01, ANOVA with Student's t test. D, cultured cortical neurons at day in vitro 6 were treated with 5 μm preaggregated Aβ1–42 peptides for indicated times. BACE1 levels were determined by IB with β-actin as loading control. E, primary neurons transfected with CAST or vehicle vector (control) were treated with 5 μm preaggregated Aβ1–42 peptide for 24 h. The BACE1 levels were determined in IB and quantitatively analyzed (n = 4, **, p < 0.01, ANOVA with Student's t test). F, HEK293 stable cell lines expressing APP-Myc were transfected with GFP-tagged m-calpain or a mutated form of m-calpain, C105S, which loses proteolytic activity. Cell lysates (∼50 μg of protein) were subjected to IB with antibodies against Myc (for the detection of APP, CTFs, and AICD), or GFP (for the detection of transfected calpain or C105S), BACE1, or β-actin. AICD, APP intracellular domain. G, APP-expressing HEK293 cells were transfected with pCAGGS-IRES-GFP plasmids that encode m-calpain or C105S. After 36 h, the media were changed to fresh serum-free DMEM, and after another 12 h, the media were collected and measured for the concentration of Aβ1–42 or Aβ1–40 by ELISA. Shown are the means ± S.E. from three experiments. *, p < 0.05; ***, p < 0.001, ANOVA with Student's t test. H, the model illustrating the role of calpain in up-regulating BACE1 and promoting APP processing. Note that calpain mediates the feedback loop between Aβ production and BACE1 expression.
Calpain Activation Up-regulates BACE1 Levels
A number of neurotoxic factors, including Aβ (21), can activate calpain, raising the possibility that there is feedback between BACE1 expression and Aβ production. To test this hypothesis, we treated cultured cortical neurons with Aβ peptides and found that this treatment caused up-regulation of BACE1 (Fig. 5D). Interestingly, Aβ-induced BACE1 increase was attenuated by overexpression of CAST (Fig. 5E). Thus, Aβ-induced BACE1 up-regulation depends on calpain. To directly determine the role of calpain in BACE1 expression, we overexpressed m-calpain in heterologous cells. We found that the levels of BACE1 and APP CTFs were concomitantly increased with overexpression of m-calpain, compared with the catalytic inactive form of m-calpain C105S, in APP695myc-expressing HEK293 cells (Fig. 5F). In agreement with this result, the amount of Aβ observed in the culture media of APP695-expressing HEK293 cells was higher in calpain-transfected cells than in C105S-transfected cells (Fig. 5G). Together, all of these results support the conclusion that calpain activation leads to BACE1 up-regulation, which in turn promotes APP processing and Aβ production.
DISCUSSION
Aberrant calpain activation has been observed in the brain of AD patients (58). Interestingly, there is a depletion of calpastatin, the endogenous inhibitor of calpain, in the prefrontal cortex of AD brains (22, 43); this calpastatin decrease may allow calpain activation. In this study, we demonstrate a role for calpain in BACE1 expression in the transgenic mouse model of AD, cultured primary neurons, and APP-expressing heterologous cells. We found that inhibition of calpain reduced the levels of BACE1 and attenuated Aβ deposition. In contrast, activation of calpain caused opposite effects. Thus, calpain has multiple roles in AD progression, e.g. BACE1 expression, Tau phosphorylation, synaptic dysfunction, and neuronal death.
Previous studies have shown that the levels or activity of BACE1 are increased upon a variety of stress stimulation, such as oxidative stress (59), hypoxia (60), ischemia (61), and traumatic brain injury (62). In the AD brain, the levels and activity of BACE1 are elevated by ∼2-fold (55–57), and up-regulation or activation of BACE1 may initiate or accelerate AD pathogenesis. Interestingly, p25 induction has been shown to induce production and accumulation of Aβ in vivo (31), probably by transcriptional regulation of BACE1 expression (32), but how p25 is produced is unclear. Given the aberrant calpain activation in the brain of AD patients (58) and the known cleavage of p35 to p25 by neurotoxicity-activated calpain (21), we have investigated the role of calpain in AD progression. Synthetic calpain inhibitors have been shown to improve memory and synaptic transmission but have no effects on Aβ deposition in APP/PS1 mice (33). Given that synthetic calpain inhibitors may modulate other proteases that may influence neurodegeneration processes, we have investigated the role of calpain by overexpressing CAST in APP/PS1 mice. We found that APP/PS1/CAST mice exhibited reduced amyloid plaque burdens and less extent for the loss of synapse and spatial learning deficits. Interestingly, the levels of BACE1 and the corresponding β-CTF of APP were also decreased in APP/PS1/CAST mice, suggesting that calpain activation promotes BACE1 expression and APP processing. Consistently, overexpressing m-calpain increased BACE1 levels in APP-expressing HEK293 cells. Two previous studies showed that calpain activation increased α-CTF production from APP (63, 64). In line with this notion, we found that levels of α-CTF were decreased in APP/PS1/CAST mice (Fig. 5A) and calpain up-regulation caused an increase in α-CTF, as well as β-CTF (Fig. 5F). Our demonstration that calpain activation by Aβ promotes BACE1 expression, together with previous reports of p25 regulation of BACE1 (31, 32), led to a model shown in Fig. 5H. We propose that calpain activation triggered by various stress stimulations or Aβ itself may increase the level of BACE1, which in turn promotes processing of APP and generation of Aβ.
A number of neurological insults activate calpain, which causes synaptic dysfunction and neuronal degeneration (28). Enhanced calpain activity triggered by calcium influx or the loss of calpastatin causes cleavage of functional proteins in the brain (60, 65–67). Inhibition of calpain has been suggested to prevent neuronal degeneration induced by multiple neurotoxtic factors (22). Thus, calpain acts to promote AD progression in multiple steps. In addition to the role in later neuronal dysfunction, calpain plays an important role in APP processing, prior to the observable neurodegeneration, and could be a promising target for AD prevention and treatment.
Acknowledgments
We are grateful to Y. Huang and Y. Q. Ding for assistance with CAST transgenic mice. We thank Dr. Y. Z. Wang for providing the pCAGGS-IRES-GFP plasmid and the APP-expressing HEK293 stable cell line. The ION Imaging Facility provided technical assistance for microscope analysis.
This work was supported by National Natural Science Foundation of China Grants 30721004 and 30825013, National Basic Research Program Grant 2006CB806600 and 2011CB809002, Key State Research Program of China Grant 2006CB943900, Chinese Academy of Sciences Grant KSCX2-YW-R-102, and Program of Shanghai Subject Chief Scientist Grant 08XD14050.
- Aβ
- amyloid-β
- BACE1
- β-site APP-cleaving enzyme 1
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- PS1
- presenilin 1
- CAST
- calpastatin
- CaMKII
- calcium/calmodulin-dependent protein kinase II
- CREB
- cAMP-response element-binding protein
- Cdk5
- Cyclin-dependent kinase 5
- CTF
- C-terminal fragment
- IB
- immunoblotting
- IS
- immunostaining
- ANOVA
- analysis of variance
- Tg
- transgenic
- nTG
- nontransgenic
- GFAP
- glial fibrillary acidic protein.
REFERENCES
- 1.Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 2.Haass C., Selkoe D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 3.Stern E. A., Bacskai B. J., Hickey G. A., Attenello F. J., Lombardo J. A., Hyman B. T. (2004) J. Neurosci. 24, 4535–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knowles R. B., Wyart C., Buldyrev S. V., Cruz L., Urbanc B., Hasselmo M. E., Stanley H. E., Hyman B. T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5274–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Amore J. D., Kajdasz S. T., McLellan M. E., Bacskai B. J., Stern E. A., Hyman B. T. (2003) J. Neuropathol. Exp. Neurol. 62, 137–145 [DOI] [PubMed] [Google Scholar]
- 6.Lombardo J. A., Stern E. A., McLellan M. E., Kajdasz S. T., Hickey G. A., Bacskai B. J., Hyman B. T. (2003) J. Neurosci. 23, 10879–10883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brendza R. P., Bacskai B. J., Cirrito J. R., Simmons K. A., Skoch J. M., Klunk W. E., Mathis C. A., Bales K. R., Paul S. M., Hyman B. T., Holtzman D. M. (2005) J. Clin. Invest. 115, 428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Alloza M., Robbins E. M., Zhang-Nunes S. X., Purcell S. M., Betensky R. A., Raju S., Prada C., Greenberg S. M., Bacskai B. J., Frosch M. P. (2006) Neurobiol. Dis. 24, 516–524 [DOI] [PubMed] [Google Scholar]
- 9.Koffie R. M., Meyer-Luehmann M., Hashimoto T., Adams K. W., Mielke M. L., Garcia-Alloza M., Micheva K. D., Smith S. J., Kim M. L., Lee V. M., Hyman B. T., Spires-Jones T. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y. M., Xu M., Lai M. T., Huang Q., Castro J. L., DiMuzio-Mower J., Harrison T., Lellis C., Nadin A., Neduvelil J. G., Register R. B., Sardana M. K., Shearman M. S., Smith A. L., Shi X. P., Yin K. C., Shafer J. A., Gardell S. J. (2000) Nature 405, 689–694 [DOI] [PubMed] [Google Scholar]
- 11.Li Y. M., Lai M. T., Xu M., Huang Q., DiMuzio-Mower J., Sardana M. K., Shi X. P., Yin K. C., Shafer J. A., Gardell S. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6138–6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esler W. P., Kimberly W. T., Ostaszewski B. L., Diehl T. S., Moore C. L., Tsai J. Y., Rahmati T., Xia W., Selkoe D. J., Wolfe M. S. (2000) Nat. Cell Biol. 2, 428–434 [DOI] [PubMed] [Google Scholar]
- 13.Selkoe D. J. (2000) Ann. N.Y. Acad. Sci. 924, 17–25 [DOI] [PubMed] [Google Scholar]
- 14.Selkoe D. J. (2001) Physiol. Rev. 81, 741–766 [DOI] [PubMed] [Google Scholar]
- 15.Vassar R., Kovacs D. M., Yan R., Wong P. C. (2009) J. Neurosci. 29, 12787–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staubli U., Larson J., Thibault O., Baudry M., Lynch G. (1988) Brain Res. 444, 153–158 [DOI] [PubMed] [Google Scholar]
- 17.Oliver M. W., Baudry M., Lynch G. (1989) Brain Res. 505, 233–238 [DOI] [PubMed] [Google Scholar]
- 18.Carafoli E., Molinari M. (1998) Biochem. Biophys. Res. Commun. 247, 193–203 [DOI] [PubMed] [Google Scholar]
- 19.Patrick G. N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L. H. (1999) Nature 402, 615–622 [DOI] [PubMed] [Google Scholar]
- 20.Nixon R. A. (2000) Ann. N.Y. Acad. Sci. 924, 117–131 [PubMed] [Google Scholar]
- 21.Lee M. S., Kwon Y. T., Li M., Peng J., Friedlander R. M., Tsai L. H. (2000) Nature 405, 360–364 [DOI] [PubMed] [Google Scholar]
- 22.Rao M. V., Mohan P. S., Peterhoff C. M., Yang D. S., Schmidt S. D., Stavrides P. H., Campbell J., Chen Y., Jiang Y., Paskevich P. A., Cataldo A. M., Haroutunian V., Nixon R. A. (2008) J. Neurosci. 28, 12241–12254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartus R. T., Elliott P. J., Hayward N. J., Dean R. L., Harbeson S., Straub J. A., Li Z., Powers J. C. (1995) Neurol. Res. 17, 249–258 [DOI] [PubMed] [Google Scholar]
- 24.Cao G., Xing J., Xiao X., Liou A. K., Gao Y., Yin X. M., Clark R. S., Graham S. H., Chen J. (2007) J. Neurosci. 27, 9278–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busche M. A., Eichhoff G., Adelsberger H., Abramowski D., Wiederhold K. H., Haass C., Staufenbiel M., Konnerth A., Garaschuk O. (2008) Science 321, 1686–1689 [DOI] [PubMed] [Google Scholar]
- 26.Kuchibhotla K. V., Goldman S. T., Lattarulo C. R., Wu H. Y., Hyman B. T., Bacskai B. J. (2008) Neuron 59, 214–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchibhotla K. V., Lattarulo C. R., Hyman B. T., Bacskai B. J. (2009) Science 323, 1211–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 29.Ahlijanian M. K., Barrezueta N. X., Williams R. D., Jakowski A., Kowsz K. P., McCarthy S., Coskran T., Carlo A., Seymour P. A., Burkhardt J. E., Nelson R. B., McNeish J. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2910–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patzke H., Maddineni U., Ayala R., Morabito M., Volker J., Dikkes P., Ahlijanian M. K., Tsai L. H. (2003) J. Neurosci. 23, 2769–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruz J. C., Kim D., Moy L. Y., Dobbin M. M., Sun X., Bronson R. T., Tsai L. H. (2006) J. Neurosci. 26, 10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen Y., Yu W. H., Maloney B., Bailey J., Ma J., Marié I., Maurin T., Wang L., Figueroa H., Herman M., Krishnamurthy P., Liu L., Planel E., Lau L. F., Lahiri D. K., Duff K. (2008) Neuron 57, 680–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trinchese F., Fa' M., Liu S., Zhang H., Hidalgo A., Schmidt S. D., Yamaguchi H., Yoshii N., Mathews P. M., Nixon R. A., Arancio O. (2008) J. Clin. Invest. 118, 2796–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F., Qian L., Yang Z. H., Huang Y., Ngo S. T., Ruan N. J., Wang J., Schneider C., Noakes P. G., Ding Y. Q., Mei L., Luo Z. G. (2007) Neuron 55, 247–260 [DOI] [PubMed] [Google Scholar]
- 35.Tsien J. Z., Chen D. F., Gerber D., Tom C., Mercer E. H., Anderson D. J., Mayford M., Kandel E. R., Tonegawa S. (1996) Cell 87, 1317–1326 [DOI] [PubMed] [Google Scholar]
- 36.Jankowsky J. L., Slunt H. H., Gonzales V., Jenkins N. A., Copeland N. G., Borchelt D. R. (2004) Neurobiol. Aging 25, 885–892 [DOI] [PubMed] [Google Scholar]
- 37.Jankowsky J. L., Slunt H. H., Ratovitski T., Jenkins N. A., Copeland N. G., Borchelt D. R. (2001) Biomol. Eng. 17, 157–165 [DOI] [PubMed] [Google Scholar]
- 38.Jia Y., Zhou J., Tai Y., Wang Y. (2007) Nat. Neurosci. 10, 559–567 [DOI] [PubMed] [Google Scholar]
- 39.Saito T., Nakatsuji N. (2001) Dev. Biol. 240, 237–246 [DOI] [PubMed] [Google Scholar]
- 40.Zhang X., Zhu J., Yang G. Y., Wang Q. J., Qian L., Chen Y. M., Chen F., Tao Y., Hu H. S., Wang T., Luo Z. G. (2007) Nat. Cell Biol. 9, 743–754 [DOI] [PubMed] [Google Scholar]
- 41.Nicholson A. M., Ferreira A. (2009) J. Neurosci. 29, 4640–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vorhees C. V., Williams M. T. (2006) Nat. Prot. 1, 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nixon R. A. (2003) Ageing Res. Rev. 2, 407–418 [DOI] [PubMed] [Google Scholar]
- 44.Leissring M. A., Farris W., Chang A. Y., Walsh D. M., Wu X., Sun X., Frosch M. P., Selkoe D. J. (2003) Neuron 40, 1087–1093 [DOI] [PubMed] [Google Scholar]
- 45.LeVine H., 3rd. (1999) Methods Enzymol. 309, 274–284 [DOI] [PubMed] [Google Scholar]
- 46.Itagaki S., McGeer P. L., Akiyama H., Zhu S., Selkoe D. (1989) J. Neuroimmunol. 24, 173–182 [DOI] [PubMed] [Google Scholar]
- 47.Frautschy S. A., Horn D. L., Sigel J. J., Harris-White M. E., Mendoza J. J., Yang F., Saido T. C., Cole G. M. (1998) J. Neurosci. 18, 8311–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai J., Grutzendler J., Duff K., Gan W. B. (2004) Nat. Neurosci. 7, 1181–1183 [DOI] [PubMed] [Google Scholar]
- 49.Imahori K., Uchida T. (1997) J. Biochem. 121, 179–188 [PubMed] [Google Scholar]
- 50.Shelton S. B., Johnson G. V. (2004) J. Neurochem. 88, 1313–1326 [DOI] [PubMed] [Google Scholar]
- 51.Kurt M. A., Davies D. C., Kidd M., Duff K., Howlett D. R. (2003) Neurobiol. Dis. 14, 89–97 [DOI] [PubMed] [Google Scholar]
- 52.Dineley K. T., Westerman M., Bui D., Bell K., Ashe K. H., Sweatt J. D. (2001) J. Neurosci. 21, 4125–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Q. L., Harris-White M. E., Ubeda O. J., Simmons M., Beech W., Lim G. P., Teter B., Frautschy S. A., Cole G. M. (2007) J. Neurochem. 103, 1594–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morris R. (1984) J. Neurosci. Methods 11, 47–60 [DOI] [PubMed] [Google Scholar]
- 55.Fukumoto H., Cheung B. S., Hyman B. T., Irizarry M. C. (2002) Arch. Neurol. 59, 1381–1389 [DOI] [PubMed] [Google Scholar]
- 56.Yang L. B., Lindholm K., Yan R., Citron M., Xia W., Yang X. L., Beach T., Sue L., Wong P., Price D., Li R., Shen Y. (2003) Nat. Med. 9, 3–4 [DOI] [PubMed] [Google Scholar]
- 57.Li R., Lindholm K., Yang L. B., Yue X., Citron M., Yan R., Beach T., Sue L., Sabbagh M., Cai H., Wong P., Price D., Shen Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3632–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saito K., Elce J. S., Hamos J. E., Nixon R. A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2628–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamagno E., Bardini P., Obbili A., Vitali A., Borghi R., Zaccheo D., Pronzato M. A., Danni O., Smith M. A., Perry G., Tabaton M. (2002) Neurobiol. Dis. 10, 279–288 [DOI] [PubMed] [Google Scholar]
- 60.Zhang X., Zhou K., Wang R., Cui J., Lipton S. A., Liao F. F., Xu H., Zhang Y. W. (2007) J. Biol. Chem. 282, 10873–10880 [DOI] [PubMed] [Google Scholar]
- 61.Wen Y., Onyewuchi O., Yang S., Liu R., Simpkins J. W. (2004) Brain Res 1009, 1–8 [DOI] [PubMed] [Google Scholar]
- 62.Blasko I., Beer R., Bigl M., Apelt J., Franz G., Rudzki D., Ransmayr G., Kampfl A., Schliebs R. (2004) J. Neural. Transm. 111, 523–536 [DOI] [PubMed] [Google Scholar]
- 63.Chen M., Durr J., Fernandez H. L. (2000) Biochem. Biophys. Res. Commun. 273, 170–175 [DOI] [PubMed] [Google Scholar]
- 64.Chen M., Fernandez H. L. (2004) Biochem. Biophys. Res. Commun. 316, 332–340 [DOI] [PubMed] [Google Scholar]
- 65.Wang K. K. (2000) Trends Neurosci. 23, 20–26 [DOI] [PubMed] [Google Scholar]
- 66.Abe K., Takeichi M. (2007) Neuron 53, 387–397 [DOI] [PubMed] [Google Scholar]
- 67.Jang Y. N., Jung Y. S., Lee S. H., Moon C. H., Kim C. H., Baik E. J. (2009) J. Neurosci. 29, 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]