Abstract
MicroRNAs (miRs) represent a class of endogenous ∼18–25 nucleotide RNAs that regulate gene expression through translational repression by binding to a target mRNA. These miRs regulate several biological functions, such as cell growth, cell differentiation, carcinogenesis, and so on. In a previous report, we have indicated that miR-141 and -200a act as preosteoblast differentiation modulators. In the present study, using microRNA array and in silico analyses, we found that miR-208 is closely involved in preosteoblast differentiation by partially regulating the expression of Ets1 (V-ets erythroblastosis virus E26 oncogene homolog 1), which transactivates osteopontin, runt-related transcription factor 2, parathyroid hormone-related protein, and type I procollagen. Furthermore, the enforced expression of mature miR-208 in murine preosteoblast in MC3T3-E1 cells or primary osteoblast cells remarkably attenuated BMP-2-induced preosteoblast differentiation. In addition, we determined that Ets1 is a target gene of miR-208 by using a sensor luciferase reporter assay. Taken together, these results suggest that the down-regulation of miR-208 in BMP-2-stimulated osteoblast differentiation is an important part of the regulatory machinery involved in early osteogenesis.
Keywords: Bone, Bone Morphogenetic Protein (BMP), Differentiation, Ets Family Transcription Factor, MicroRNA, Ets1, MC3T3-E1, MicroRNA-208, Noncoding RNA, Osteoblast
Introduction
MicroRNAs (miRs)2 represent a class of short (18–25 nucleotide) noncoding single-stranded RNA molecules and have emerged as a new class of regulators in both animal and plant development (1–3). Mature miRs regulate the repression of specific target gene translation and/or promote the degradation of their transcribed mRNAs by binding to the 3′-untranslated regions (UTR) of the target genes. Recently, miRs were found to have implications in diverse biological processes, including cell growth, cell differentiation, apoptosis, carcinogenesis, and diabetes (4–8). In a previous study, we indicated that both miR-141 and -200a regulate BMP-2-stimulated preosteoblast MC3T3-E1 cell differentiation by directly targeting Dlx5 (distal-less homeobox 5) (9). To date, it has already been reported that several miRs, such as miR-26a, miR-29, miR-133, miR-135, miR-206, and miR-2861 regulate osteoblast differentiation (10–14). Thus, it has been strongly suggested that the regulation of osteoblast differentiation by miRs is a notable component of the regulatory machinery.
In this study, we aimed to uncover other miRs critical to osteoblast differentiation. After we had investigated the miR expression profiles in BMP-2-treated MC3T3-E1 cells by miR array analysis, the expression level of miR-208 in BMP-2-treated MC3T3-E1 cells was significantly decreased compared with non-BMP-2-treated cells. To determine the role of miR-208 in osteogenesis, we performed a bioinformatics study and Western blot analysis. In silico analyses showed that Ets1 (V-ets erythroblastosis virus E26 oncogene homolog 1) could be a target gene of miR-208.
The purpose of the present study is to examine the regulation of Ets1 expression by miR-208 in BMP-2-stimulated preosteoblast differentiation in MC3T3-E1 cells. In the present study, we determined that Ets1 is a target gene of miR-208 by using a sensor luciferase reporter assay. Our findings strongly suggest that miRs regulate osteogenic master transcriptional factors in BMP-2-stimulated osteoblast differentiation.
EXPERIMENTAL PROCEDURES
Materials
Bioactive recombinant human BMP-2 was obtained from BioVision Research Products (Mountain View, CA). Eagle's α-minimal essential medium, OPTI-MEM, FBS, and LipofectamimeTM RNAi MAX were obtained from Invitrogen. Alkaline phosphatase (ALP) assay kit LabAssayTM ALP was from WAKO Pure Chemical Industries Ltd. (Osaka, Japan). An ALP staining kit (TRAP and ALP double-stain kit) was obtained from Takara Bio Inc. (Ohtsu, Japan). The antibodies to mouse Ets1, runt-related transcription factor 2 (Runx2), osteopontin (OPN), parathyroid hormone-related protein (PTHrP), and ALP were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Culture
The murine preosteoblast cell line MC3T3-E1 was obtained from RIKEN Cell Bank (Tsukuba, Ibaraki, Japan). Primary murine osteoblast cells, isolated from ICR mouse calvaria, were purchased from Primary Cell Co. Ltd. (Sapporo, Hokkaido, Japan). These cells were cultured in phenol red-free α-minimal essential medium supplemented with 10% heat-inactivated FBS, 100 units/ml of penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. BMP-2 dissolved in PBS was added to the culture medium at a concentration of 300 ng/ml.
MicroRNA Array Hybridization
Total RNA was extracted from the cells by TRIzol containing phenol/guanidium isothiocyanate (Invitrogen) with DNase I treatment. We examined the expression profiles of miRs using Genopal®-MICH chips (Mitsubishi Rayon Co. Ltd., Yokohama, Japan), which is equipped with 180 oligonucleotide DNA probes in hollow plastic fibers for detection of mouse miRs. Hybridization signals were analyzed by using a DNA chip analyzer according to the manufacturer's instructions.
Quantitative RT-PCR
To confirm the reproducibility of miR expression profiles, examined by miR array analysis, we measured their expression levels by using a TaqMan® microRNA reverse transcription kit and a TaqMan® microRNA assay kit (Applied Biosystems, Foster City, CA). Briefly, after the RT of 12.5 ng of total RNA, cDNA was synthesized. The RT products were subjected to qRT-PCR using a Thermal Cycler Dice® real time PCR system TP800 (Takara Bio). The expression levels were normalized to U6, which was used as an internal control and measured by the comparative Ct (ΔΔCt) method. The qRT-PCR consisted of 45 cycles (95 °C for 10 s, 60 °C for 40 s, and 72 °C for 1 s) after an initial denaturation step (95 °C for 10 min).
To determine the level of Ets1 mRNA, we prepared cDNA from total RNA samples using a PrimeScriptTM reagent kit (Takara Bio). qRT-PCR was performed (Thermal Cycler Dice® TP800) using a SYBER® Premix TaqTM II kit (Takara) and the following primer sets: Ets1 sense, 5′-CTCTCCAGACAGACACCTTGC-3′; Ets1 antisense, 5′-AGCACGGTCACGCACATA-3′; Runx2 sense, 5′-GACGTGCCCAGGCGTATTTC-3′; and Runx2 antisense, 5′-AAGGTGGCTGGGTAGTGCATTC-3′. The cDNA of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The semi-quantitative RT-PCR consisted of 20 cycles (94 °C for 30 s, 57.5 °C for 30 s, and 72 °C for 30 s) after an initial denaturation step (94 °C for 4 min). The PCR products were analyzed by electrophoresis on 2% agarose gels.
Transfection of MC3T3-E1 Cells with miRNAs
MC3T3-E1 cells were seeded in 6-well plates at a concentration of 1 × 105 cells/ml/well on the day before transfection. The mature type of miR-208 (Applied Biosystems; AUAAGACGAGCAAAAAGCUUGU; see Fig. 1A) and its antisense inhibitor (Applied Biosystems)—designed to bind to endogenous specific miRNAs when introduced into cells and inhibit their activities—were transfected using cationic liposomes (RNAiMAX) according to the manufacturer's lipofection protocol. The transfection efficiency was evaluated by the transfection of the cells using Alexa Flour 488 (Molecular Probes); transfection efficiency was greater than 80% (data not shown). Nonspecific control miR (Applied Biosystems) was used as a control for nonspecific effects. The culture medium containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin was changed at 12 h after the transfection. Transfected cells were cultured for 3 days and then reseeded for various experiments at a concentration of 1 × 105 cells/ml.
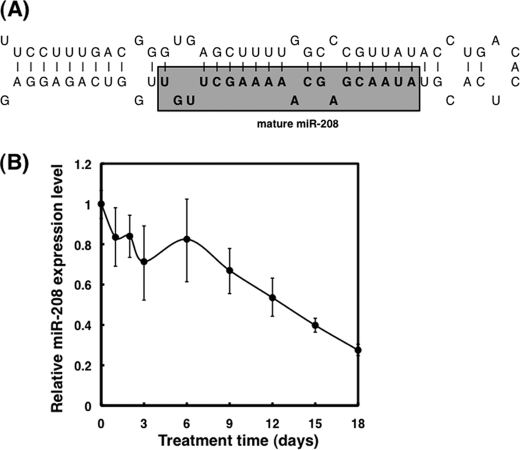
FIGURE 1.
Down-regulation of miR-208 in BMP-2-induced preosteoblast differentiation. A, the sequence of stem-loop structured miR-208. B, the change of miR-208 expression levels in MC3T3-E1 cells until 18 days after BMP-2 stimulation. MC3T3-E1 cells were incubated under BMP-2 stimulation at indicated times after the treatment, and then total RNA was extracted from the cells by TRIzol with DNase I treatment. Expression levels of miR-208 in BMP-2-treated cells were examined by TaqMan® microRNA assay using real time PCR. The relative ratio is shown. The value of the cells before BMP-2 treatment was designated as 1.
ALP Activity and ALP Staining
MC3T3-E1 cells and the transfected MC3T3-E1 cells were inoculated into 24-well plates (1 × 105 cells/ml, 500 μl/well; Nunc, Roskilde, Denmark) and cultured with or without BMP-2, for 3 days. After incubation, the treated cells were washed twice with PBS, and 200 μl of lysis buffer was added to the cell layer and kept on ice for 5 min. The cell lysate was sonicated for 1 min and centrifuged at 1,000 × g, at 4 °C for 10 min. ALP activity was assayed by a spectrophotometric method using a LabAssayTM ALP kit (Wako Pure Chemical Industries Ltd., Osaka, Japan). The absorbance of each well at 405 nm was measured with a microplate reader (Immuno-Mini NJ-2300; Nalge Nunc International K.K., Tokyo, Japan). Then ALP activity was estimated by the using the TRAP and ALP double-staining kit (Takara) according to the manufacturer's protocol.
Alizarin Red Staining
For detection of calcification in differentiated MC3T3-E1 cells, the BMP-2-treated and nontreated cells were washed twice with PBS and fixed with 500 μl of ice-cold 70% ethanol for 10 min. The fixed cells were stained with 500 μl of Alizarin red solution (Sigma).
Western Blotting
For preparation of cell lysate, MC3T3-E1 cells were washed twice with PBS and then harvested. The cell pellet was resuspended in radioimmune precipitation assay buffer containing 25× Complete® and phosphatase inhibitor mixture® (Roche Applied Science). Protein content was measured with a DC protein assay kit (Bio-Rad). Each whole cell lysate was resuspended in SDS-PAGE buffer containing 2% 2-mercaptoethanol and was boiled at 98 °C for 5 min. Twenty micrograms of the protein from each cell lysate were separated by SDS-PAGE by using a 12% polyacrylamide gel and were electroblotted onto a PVDF membrane (GE Healthcare). After blockage of nonspecfic binding sites for 1 h by 5% nonfat milk in TPBS (PBS and 0.1% Tween 20), the membrane was incubated overnight at 4 °C with various primary antibodies. The membrane was then washed three times with TPBS, incubated further with horseradish peroxidase-conjugated secondary antibody at room temperature, and washed again three times with TPBS. Protein bands were detected with enhanced ECL kit (GE Healthcare) and chemiluminescence detector (LAS-4000; Fujifilm).
Luciferase Assay
To determine the target region of miR-208 in Ets1, we constructed three kinds of pGL3-Ets1/miR-208 sensor plasmids by inserting a candidate binding site in the 3′-UTR of Ets1 mRNA into the XbaI site of the pGL3 control vector (Promega, Madison, WI). The PCR primer sequences used for construction of the pGL3-Ets1 sensor plasmids are as follows: pGL3-Ets1/miR-208 sensor-A sense, 5′-GCTCTAGAGCCTTGAGTTGTGGATCTACTA-3′; pGL3-Ets1/miR-208 sensor-A antisense, 5′-GCTCTAGAGCCCTTCAATCAAAGTCCATCA-3′; pGL3-Ets1/miR-208 sensor-B sense, 5′-GCTCTAGAGCTTTGATTGAAGGGGTCCA-3′; pGL3-Ets1/miR-208 sensor-B antisense, 5′-GCTCTAGAGCACCCCCTGTCCCCAGATCAGCA-3′; pGL3-Ets1/miR-208 sensor-C sense, 5′-GCTCTAGAGCTACTGAGATCAGAGACCCAA-3′; and pGL3-Ets1/miR-208 sensor-C antisense, 5′-GCTCTAGAGCGATTCCATAGAAGAGCTGCA-3′.
Moreover, we constructed mutant vectors with deleted or mutated miR-208 complementary sites in Ets1 by using a PrimeStarTM mutagenesis basal kit (Takara). The PCR primer sequences used for construction of the mutated pGL3-Ets1/miR-208 sensor-B plasmid are as follows: pGL3-Ets1/miR-208 sensor-B Del sense, 5′-AAAAAATATATGAGGCCGCTCTAAGGTGGTCT-3′; pGL3-Ets1/miR-208 sensor-B Del antisense, 5′-CCTTAGAGCGGCCTCATATATTTTTTTAAGAG-3′; pGL3-Ets1/miR-208 sensor-B Mut sense, 5′-AATATCGTCGGCCGCTCTAAGGTGGTCT-3′; and pGL3-Ets1/miR-208 sensor-B Mut antisense, 5′-CCTTAGAGCGGCCGACGATATTTTTTTA-3′. MC3T3-E1 cells were inoculated into 12-well plates (1 × 105 cells/ml), co-transfected with the sensor vector plasmid or the mutated sensor vector, pRL-SV40 vector, and 40 nm of miR-208 by using LipofectamimeTM RNAiMAX and were then cultured for 2 days. The luciferase activity was measured by using a luciferase assay system (Promega) according to the manufacturer's protocol. The relative luciferase activity was expressed as the ratio of the measured luciferase activity to the control (nonspecific miR).
Statistical Analysis
All of the data were analyzed by one-way analysis of variance and subsequently by Fisher's multiple range test among many groups and by Student's t test between two groups at the significance level of p < 0.05. Statistical analyses were carried out using StatView 5.0 software (SAS Institute, Cary, NC).
RESULTS
Expression Profile of miR-208 in BMP-2-induced Preosteoblast MC3T3-E1 Cell Differentiation
To investigate the expression profiles of miRs in BMP-2-treated MC3T3-E1 cells, we extracted each total RNA from BMP-2-treated and nontreated cells at 24 h after treatment and subjected them to the Genopal®-MICH chips. The expression levels of miR-208 were significantly down-regulated (log ratio, >1.5) between BMP-2-treated and nontreated cells. To confirm the results of miRNA array analysis, we examined the time-dependent expression levels of miR-208 in BMP-2-treated and nontreated cells by TaqMan® assay using a real time PCR. Interestingly, the expression levels of miR-208 in BMP-2-stimulated cells were down-regulated as opposed to those in nontreated cells (Fig. 1B).
Effects of miR-208 on Differentiation in MC3T3-E1 Cells
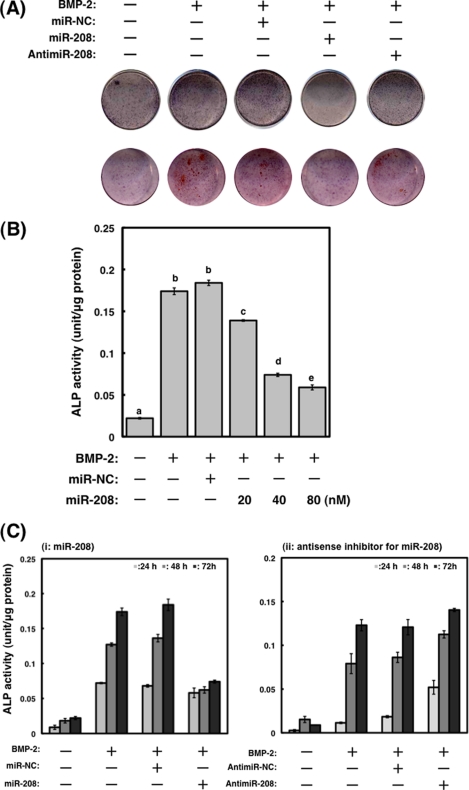
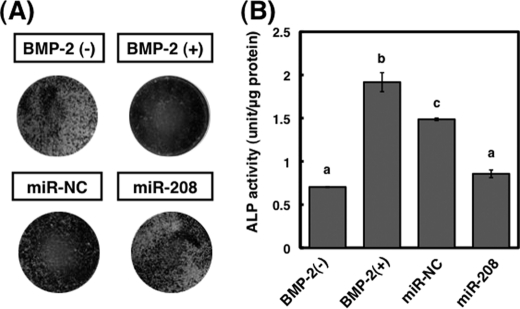
It is well known that ALP activity increases in a time-dependent manner in MC3T3-E1 cells after treatment with BMP-2 (1). To investigate the role of miR-208 during differentiation, we measured ALP activity after the transfection with miR-208 or antisense inhibitor for miR-208. As shown in Fig. 2, ALP activity and mineralization determined by Alizarin red staining in cells transfected with miR-208 were suppressed. On the contrary, compared with BMP-2-treated MC3T3-E1 cells, ALP activity and mineralization in the antisense inhibitor remained unchanged (Fig. 2C, panel ii).
FIGURE 2.
Effects of transfection with mature and antisense inhibitor for miR-208 on differentiation in MC3T3-E1 cells. A, differentiation and mineralization in MC3T3-E1 cells transfected with miR-208 or antisense inhibitor for miRNA-208 were observed by ALP and Alizarin red staining after treatment with BMP-2. Negative control miR-NC (miR-NC) was designed to have no significant sequence similarity to mouse, rat, or human transcription products. B, the increased expression of miR-208 in the cells by the transfection significantly suppressed the ALP activity. The cells were treated with BMP-2 (300 ng/ml) for 3 days. The values of each group were expressed as the means ± S.E. of three separate experiments. Mean values with different letters are significantly different (p < 0.01, one-way analysis of variance followed by Fisher's multiple-range test). C, effects of miR-208 or antisense inhibitor for miRNA-208 on BMP-2-stimulated ALP activities in MC3T3-E1 cells. The cells transfected with miR-208 significantly suppressed the ALP activity following BMP-2 stimulation (panel i), but antisense inhibitor for miRNA-208 was unchanged compared with those of BMP-2-treated MC3T3-E1 cells (panel ii). The values of each group were expressed as the means ± S.E. of three separate experiments.
Expression of Osteogenic Master Transcriptional Factor Ets1 Is Regulated by MicroRNA-208
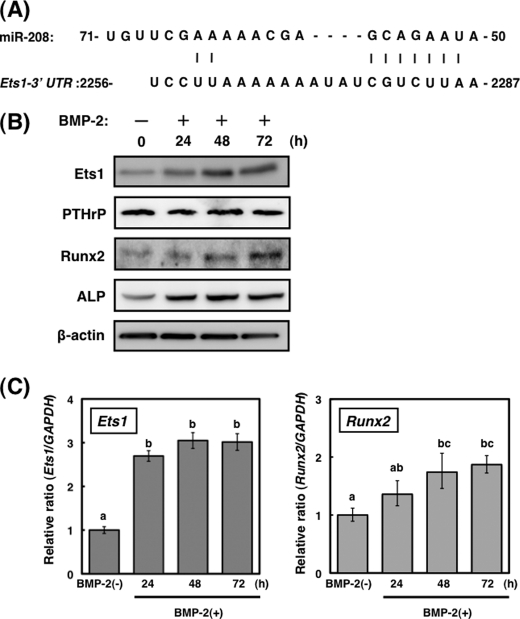
To identify the target genes of miR-208 in osteogenesis, we searched for candidate genes using the miRBase Target version 5 and miRanda databases. miR-208 targets 823 genes, and osteogenic master transcriptional factor Ets1 is included as a target gene (Fig. 3A). BMP-2 stimulation caused a considerable increase in the protein expression levels of Ets1 up to 48 h (Fig. 3B), whereas the levels of Ets1 mRNA examined by real time PCR remained nearly unchanged from 24–48 h (Fig. 3C). Furthermore, the levels of Runx2 and ALP proteins that are well known as osteoblast differentiation markers were significantly increased by BMP-2 stimulation (Fig. 3B). However, the mRNA level of PTHrP, which is also modulated by Ets1, was not changed by BMP-2 stimulation.
FIGURE 3.
Expression of Ets1 in BMP-2-stimulated osteoblast differentiation. A, nucleotide sequences of miR-208 and their complementation to Ets1. B, time course of Ets1, PTHrP, Runx2, and ALP expressions in BMP-2-treated MC3T3-E1 cells (20 μg of protein/lane). A representative blot from three independent experiments is shown. C, time course of Ets1 and Runx2 mRNA expressions in BMP-2-treated MC3T3-E1 cells. The values of each group were expressed as the means ± S.E. of three separate experiments. Mean values with different letters are significantly different (p < 0.01, one-way analysis of variance followed by Fisher's multiple-range test).
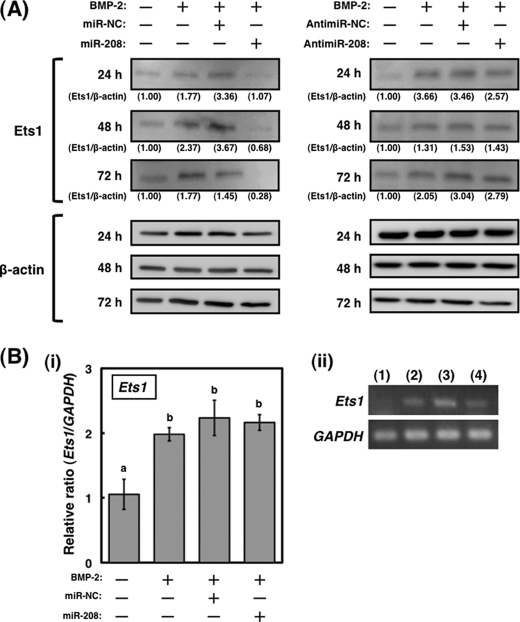
The transfection of the cells with miR-208 induced the down-regulation of Ets1 protein expression at the indicated times (Fig. 4A, left panels), whereas the antisense inhibitor for miR-208 did not affect Ets1 protein expression (Fig. 4A, right panels). As judged by using a qRT-PCR, the levels of Ets1 mRNA in the cells transfected with miR-208 remained unchanged compared with BMP-2-stimulated cells (Fig. 4B, panel (i)). Thus, the increase in Ets1 expression may be due to the drop in the levels of miR-208 after the BMP-2 treatment (Fig. 1B). These findings suggest that miR-208 regulates the expression of Ets1 at a translational level.
FIGURE 4.
Expression of Ets1 is translationally regulated by miR-208 in osteoblast differentiation. A, expression profile of Ets1 protein in MC3T3-E1 cells transfected with miR-208 or antisense inhibitor miR-208 after treatment (treatment time was 24, 48, and 72 h, 20 μg of protein/lane). A representative Western blot from three independent experiments is shown. B, panel (i), the changes of Ets1 mRNA expression in the transfected cells with miR-208 were examined by qRT-PCR. GAPDH was used as an internal control. The values of each group were expressed as the means ± S.E. of three separate experiments. Mean values with different letters are significantly different (p < 0.01, one-way analysis of variance followed by Fisher's multiple-range test). Panel (ii), the relative level of Ets1 mRNA in the transfected cells was normalized to GAPDH. Lane 1, BMP-2 (−); lane 2, BMP-2(+); lane 3, BMP-2(+) (300 ng/ml) and miR-negative control; lane 4, BMP-2(+) and miR-208 (40 nm).
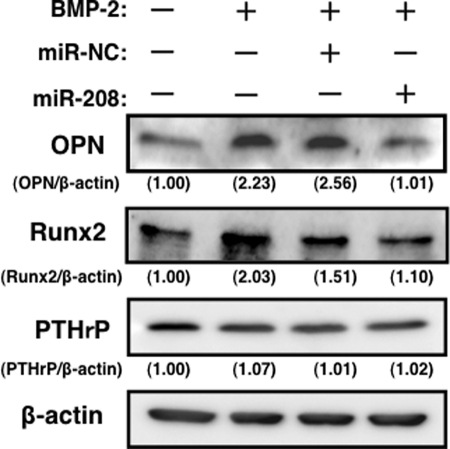
The protein expression levels of OPN, PTHrP, and Runx2 following BMP-2 stimulation were further examined. As shown in Fig. 5, the expression level of OPN at day 10 was significantly decreased in cells transfected with miR-208 compared with both control miR-NC and BMP-2-stimulated cells. Additionally, the expression level of Runx2 at day 3 was slightly decreased in cells transfected with miR-208 compared with both control miR-NC and BMP-2-stimulated cells. On the other hand, the expression level of PTHrP, for which mRNA levels are also regulated by Ets1, remained unchanged in the cells transfected with miR-208 (Fig. 5).
FIGURE 5.
Expression of OPN, Runx2, and PTHrP in the cells transfected with miR-208 osteoblast differentiation. OPN, Runx2, and PTHrP expression in BMP-2-treated MC3T3-E1 cells (20 μg of protein/lane). A representative Western blot from three independent experiments is shown.
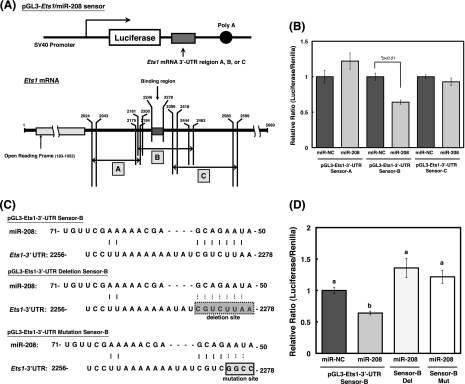
Ets1 Is a Target of miR-208
To determine the target region of miR-208 for Ets1 mRNA, we constructed three kinds of luciferase reporter plasmids (A, B, and C) with or without the possible binding region (numbers in Mus musculus E26 avian leukemia oncogene 1, 5′ domain (Ets1), transcript variant a, mRNA: 2246–2278) in 3′-UTR of Ets1 mRNA (Fig. 6A). The levels of luciferase activity in pGL3-Ets1/miR-208 sensor-B, which contains the 2246–2278 region of the 3-UTR of Ets1, were lower than those in the nonspecific control sensor vector (Fig. 6B). However, the levels of luciferase activity in pGL3-Ets1/miR-208 sensor-A/C, which lacks the 2246–2278 region of the 3′-UTR of Ets1, were not decreased compared with those in the nonspecific control sensor vector (p < 0.01). We attempted to confirm these results by using two different mutants of the miR-208-binding site in pGL3-Ets1/miR-208 sensor-B (Fig. 6C). The levels of luciferase activity in cells transfected with the pGL3-Ets1/miR-208 sensor-B (wild type) were significantly reduced, whereas the activity level of sensor-B mutants were not reduced (Fig. 6D). From these results, it is possible that the target region of Ets1 (which we proposed) is a binding site for miR-208.
FIGURE 6.
Identification of the target gene of miRNA-208 by luciferase reporter assay. A, schematic representation of the sensor vectors used in the luciferase assay for identification of target region in Ets1 for miR-208. Construction of three kinds (squares A, B, and C) of luciferase reporter plasmids including the 3′-UTR of Ets1 mRNA. B, each sensor vector, pRL-SV 40, and miR-NC or -208 were co-transfected into MC3T3-E1 cells. After 24 h, firefly and Renilla luciferase activities were measured by the using the luciferase assay system (Promega) according to the manufacturer's protocol. The activities were calculated as a ratio of firefly to Renilla luciferase activity. The values of each group were expressed as the means ± S.E. of three separate experiments. *, significant difference at p < 0.01 by Student's t test. p < 0.05 is significant. C, the deleted or mutated region corresponding to the seed sequence of miR-208 sensor vector were made from pGL3-miR-208 sensor-B. D, MC3T3-E1 cells were co-transfected with the pGL3-Ets1/miR-208 sensor-B, deleted pGL3-Ets1/miR-208 sensor-B (Del), or mutated pGL-Ets1/miR-208 sensor-B (Mut) and miR-NC or -208. The activities were calculated as a ratio of firefly to Renilla luciferase activity. The values of each group were expressed as the means ± S.E. of three separate experiments. Mean values with different letters are significantly different (p < 0.01, one-way analysis of variance followed by Fisher's multiple-range test).
Effects of miR-208 on Differentiation in Primary Murine Osteoblast Cells
We examined the role of miR-208 during the BMP-2-induced osteoblast differentiation in primary murine osteoblast cells. As shown in Fig. 7, the ALP activities in cells transfected with miR-208 were significantly suppressed. These results suggest that the regulation of miR-208 in BMP-2-stimulated osteoblast differentiation is physiologically relevant.
FIGURE 7.
Effects of transfection with mature and antisense inhibitor for miR-208 on differentiation in primary murine osteoblast cells. A, the differentiation in primary murine osteoblast cells transfected with miR-208 was observed by ALP staining after treatment with BMP-2. Negative control miR-NC (miR-NC) was designed to have no significant sequence similarity to mouse, rat, or human transcription products. B, the increased expression of miR-208 by the transfection significantly suppressed the ALP activity. The cells were treated with BMP-2 (300 ng/ml) for 3 days. The values of each group were expressed as the means ± S.E. of three separate experiments. Mean values with different letters are significantly different (p < 0.01, one-way analysis of variance followed by Fisher's multiple-range test).
Effects of miR-208 on the Activation of ERK1/2
To gain further insight into the mechanisms underlying regulation of Ets1 expression by miR-208, we examined the activation of ERK1/2 in MC3T3-E1 cells and miR-208-transfected MC3T3-E1 cells. It is well known that ERK1/2 regulates Ets1 activation (15). The levels of ERK1/2 activation by BMP-2 stimulation in miR-208-transfected MC3T3-E1 cells at the indicated times were almost similar to those of MC3T3-E1 cells (Fig. 8, A and B). Additionally, the levels of ERK1/2 activation by osteoblast differentiation medium stimulation in MC3T3-E1 cells also remained unchanged compared with those of MC3T3-E1 or miR-208-transfected MC3T3-E1 cells (Fig. 8C).
FIGURE 8.
Changes in the activation of ERK1/2 by miR-208 during BMP-2 stimulation in MC3T3-E1 cells by Western blot analysis. A, the levels of activated ERK by BMP-2 stimulation in MC3T3-E1 cells (5 μg protein/lane). B, the levels of activated ERK1/2 by BMP-2 stimulation in MC3T3-E1 cells transfected with miR-208. C, the levels of activated ERK by osteoblast differentiation medium (phenol red-free α-minimal essential medium-including 100 μg/ml of l-ascorbic acid, 10 mm of β-glycerophosphate, and 10 mm of HEPES buffer) stimulation in MC3T3-E1 cells. A representative blot from three independent experiments is shown.
DISCUSSION
In a previous study, we indicated that both miR-141 and -200a regulate BMP-2-stimulated preosteoblast MC3T3-E1 cell differentiation by directly targeting Dlx5 (9). In this study, we demonstrated for the first time that miR-208 regulates BMP-2-stimulated mouse preosteoblast differentiation by targeting Ets1. The Ets1 proto-oncogene, a member of the Ets family, was originally found in a study of the avian E26 retrovirus genome (15–17). The Ets transcription factor family is closely involved in regulation of cell proliferation (18), differentiation (19), metastasis (20), apoptosis (21), and angiogenesis (22). The Ets transcriptional factors also play critical roles in osteogenesis (23). Vary et al. (24) indicated that Ets1 is highly expressed during the proliferation stages in BMP-2-stimulated MC3T3-E1 cells. Ets1 expression is also modulated by PKC, p38 MAPK, retinoic acid, Ets1/AP-1 interaction, and the activation of the Ras/Raf/MEK/ERK1/2 signaling pathway. Consequently, this leads to the transactivation of target genes, such as OPN, PTHrP, Runx2, and tenascin-C (a polymorphic high molecular mass extracellular matrix glycoprotein secreted from osteoblasts in osteogenesis), and type I procollagen (15, 20, 25–31). The OPN and Runx2 expressions in miR-208-transfected MC3T3-E1 cells following BMP-2 stimulation were suppressed compared with that of control cells, whereas the expression of PTHrP remained unchanged. As shown in Fig. 3B, PTHrP was constantly expressed in MC3T3-E1 cells and was not changed by BMP-2 stimulation. Thus, we consider that the expressional regulations of OPN and Runx2 by miR-208 were preferentially carried out over PTHrP expression. This is likely because the excess PTHrP expression is not a necessary response in BMP-2-stimulated MC3T3-E1 cells. Therefore, the level of PTHrP in miR-208-transfected MC3T3-E1 cells might not have changed, either. The enforced expression of miR-208 in MC3T3-E1 or primary murine osteoblast cells significantly attenuated BMP-2-stimulated osteoblast differentiation, suggesting that the down-regulation of miR-208 is an important common phenomenon for osteoblast differentiation.
In an attempt to elucidate the regulatory machinery underlying the modulation of BMP-2-stimulated preosteoblast differentiation by miR-208, we investigated its effect on ERK activation in BMP-2-stimulated MC3T3-E1 cells. Ets1 contains two phosphorylation sites: threonine 38 and an array of serines within the exon VII domain. Phosphorylation of threonine 38 by ERK1/2 activates Ets1, whereas phosphorylation of the exon VII domain by calcium/calmodulin-dependent protein kinase and myosin light chain kinase inhibits Ets1 DNA binding activity (7). In a recent report, Foulds et al. (30) also indicated that Ras/mitogen-activated protein kinase (MAPK) signaling activates Ets1 and Ets2 by cAMP-responsive element-binding protein-binding protein/p300 recruitment. The data presented in Fig. 8 show that ERK1/2 was activated by BMP-2 stimulation in either MC3T3-E1 cells or the miR-208-transfected cells. Additionally, the behavior of the activation of ERK1/2 in normal MC3T3-E1 cells was similar to that of the miR-208-transfected cells. Li et al. (12) reported that miR-208 expression is down-regulated by osteogenesis-inducing medium stimulation in MC3T3 cells. We also observed the activation of ERK1/2 in MC3T3-E1 cells, as well as BMP-2 stimulation (Fig. 8C) by this particular osteogenesis-inducing medium stimulation. Thus, the activation of ERK1/2 in the osteoblast differentiation stage was irrelevant to the intracellular levels of miR-208. From these observations, it is highly probable that the down-regulation of OPN and Runx2 expression, which are transcriptionally regulated in part by Ets1, were mainly due to the direct suppression of Ets1 expression by miR-208 at the translational level.
Ets1 also regulates ring finger protein 11 (RNF11) expression in osteogenesis in MC3T3-E1 cells (33–35). RNF11 enhances the Smad4-dependent TGFβ signaling pathway (26). Moreover, Azmi et al. (36) reported that RNF11 interacts with the Hect family of E3 ubiquitin ligases, and Smad-ubiquitin regulatory factors (Smurf) 1 and 2, which are negative regulators of both BMP and TGFβ signaling pathways. It is well known that Smurf1 regulates osteoblast differentiation by enhancing proteasomal degradation of R-Smads and Runx2 (37). On the other hand, Smurf2 preferentially regulates Smad1 and 2 (32). Therefore, it was presumed that Ets1/RNF11/Smurf signaling acts as one of the key mechanisms of osteogenesis (Fig. 9). As a consequence, it was considered that the down-regulation of miR-208 was a critical process in BMP signaling during osteoblast differentiation. However, further study is required to clarify the relationship between miRs and the BMP signaling pathway.
FIGURE 9.
A scheme showing the regulation of miR-208 on BMP-2-stimulated osteoblast differentiation. The enforced expression of miR-208 significantly blocked BMP-2-stimulated preosteoblast differentiation by suppression of Ets1 expression at the translational level. Gao et al. (34) reported that RNF11 expression is regulated by Ets1. RNF11 regulates Smurf-dependent R-Smad degradation and TGFβ signaling by Smad4 activation.
In conclusion, we have found that miR-208 regulates BMP-2-stimulated mouse preosteoblast differentiation by targeting Ets1. Thus, miR-208 should be considered an important candidate as an osteoblast differentiation molecular target for the development of preventive or therapeutic agents against osteogenic disorders.
This work was supported by Grant-in-Aid for Young Scientists (B) 21700715 from the Ministry of Education, Culture, Sports, Science and Technology.
- miR
- microRNA
- ALP
- alkaline phosphatase
- BMP
- bone morphogenetic protein
- OPN
- osteopontin
- PTHrP
- parathyroid hormone-related protein
- RNF11
- ring finger protein 11
- Runx2
- runt-related transcription factor 2
- UTR
- untranslated region(s)
- qRT
- quantitative RT.
REFERENCES
- 1.Ambros V. (2003) Cell 113, 673–676 [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 3.Shukla L. I., Chinnusamy V., Sunkar R. (2008) Biochim. Biophys. Acta. 1779, 743–748 [DOI] [PubMed] [Google Scholar]
- 4.Greene S. B., Gunaratne P. H., Hammond S. M., Rosen J. M. (2010) J. Cell Sci. 123, 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao C., Sun G., Li S., Lang M. F., Yang S., Li W., Shi Y.Proc. Natl. Acad. Sci. 107, 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Incoronato M., Garofalo M., Urso L., Romano G., Quintavalle C., Zanca C., Iaboni M., Nuovo G., Croce C. M., Condorelli G. (2010) Cancer Res. 70, 3638–3646 [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto Y., Akiyama Y., Otsubo T., Shimada S., Yuasa Y. (2010) Carcinogenesis 31, 777–784 [DOI] [PubMed] [Google Scholar]
- 8.Herrera B. M., Lockstone H. E., Taylor J. M., Ria M., Barrett A., Collins S., Kaisaki P., Argoud K., Fernandez C., Travers M. E., Grew J. P., Randall J. C., Gloyn A. L., Gauguier D., McCarthy M. I., Lindgren C. M. (2010) Diabetologia 53, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh T., Nozawa Y., Akao Y. (2009) J. Biol. Chem. 284, 19272–19279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luzi E., Marini F., Sala S. C., Tognarini I., Galli G., Brandi M. L. (2008) J. Bone Miner. Res. 23, 287–295 [DOI] [PubMed] [Google Scholar]
- 11.Li Z., Hassan M. Q., Volinia S., van Wijnen A. J., Stein J. L., Croce C. M., Lian J. B., Stein G. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13906–13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., Hassan M. Q., Jafferji M., Aqeilan R. I., Garzon R., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2009) J. Biol. Chem. 284, 15676–15684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inose H., Ochi H., Kimura A., Fujita K., Xu R., Sato S., Iwasaki M., Sunamura S., Takeuchi Y., Fukumoto S., Saito K., Nakamura T., Siomi H., Ito H., Arai Y., Shinomiya K. I., Takeda S. (2010) Proc. Natl. Acad. Sci. U.S.A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Xie H., Liu W., Hu R., Huang B., Tan Y. F., Xu K., Sheng Z. F., Zhou H. D., Wu X. P., Luo X. H. (2009) J. Clin. Invest. 119, 3666–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittmer J. (2003) Mol. Cancer 2, 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trojanowska M. (2000) Oncogene 19, 6464–6471 [DOI] [PubMed] [Google Scholar]
- 17.Yordy J. S., Muise-Helmericks R. C. (2000) Oncogene 19, 6503–6513 [DOI] [PubMed] [Google Scholar]
- 18.Qi M. C., Hu J., Zou S. J., Chen H. Q., Zhou H. X., Han L. C. (2008) Int. J. Oral Maxillofac. Surg. 37, 453–458 [DOI] [PubMed] [Google Scholar]
- 19.John S. A., Clements J. L., Russell L. M., Garrett-Sinha L. A. (2008) J. Biol. Chem. 283, 951–962 [DOI] [PubMed] [Google Scholar]
- 20.Wai P. Y., Mi Z., Gao C., Guo H., Marroquin C., Kuo P. C. (2006) J. Biol. Chem. 281, 18973–18982 [DOI] [PubMed] [Google Scholar]
- 21.Pei H., Li C., Adereth Y., Hsu T., Watson D. K., Li R. (2005) Cancer Res. 65, 7205–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei G., Srinivasan R., Cantemir-Stone C. Z., Sharma S. M., Santhanam R., Weinstein M., Muthusamy N., Man A. K., Oshima R. G., Leone G., Ostrowski M. C. (2009) Blood 114, 1123–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raouf A., Seth A. (2000) Oncogene. 19, 6455–6463 [DOI] [PubMed] [Google Scholar]
- 24.Vary C. P., Li V., Raouf A., Kitching R., Kola I., Franceschi C., Venanzoni M., Seth A. (2000) Exp. Cell Res. 257, 213–222 [DOI] [PubMed] [Google Scholar]
- 25.Newberry E. P., Willis D., Latifi T., Boudreaux J. M., Towler D. A. (1997) Mol. Endocrinol. 11, 1129–1144 [DOI] [PubMed] [Google Scholar]
- 26.Sato M., Morii E., Komori T., Kawahata H., Sugimoto M., Terai K., Shimizu H., Yasui T., Ogihara H., Yasui N., Ochi T., Kitamura Y., Ito Y., Nomura S. (1998) Oncogene 17, 1517–1525 [DOI] [PubMed] [Google Scholar]
- 27.Raouf A., Li V., Kola I., Watson D. K., Seth A. (2000) Oncogene 19, 1969–1974 [DOI] [PubMed] [Google Scholar]
- 28.Cataisson C., Gordon J., Roussière M., Abdalkhani A., Lindemann R., Dittmer J., Foley J., Bouizar Z. (2003) Mol. Cell. Endocrinol. 204, 155–168 [DOI] [PubMed] [Google Scholar]
- 29.Jinnin M., Ihn H., Asano Y., Yamane K., Trojanowska M., Tamaki K. (2004) Oncogene 23, 1656–1667 [DOI] [PubMed] [Google Scholar]
- 30.Foulds C. E., Nelson M. L., Blaszczak A. G., Graves B. J. (2004) Mol. Cell. Biol. 24, 10954–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Hassan M. Q., Xie R. L., Hawse J. R., Spelsberg T. C., Montecino M., Stein J. L., Lian J. B., van Wijnen A. J., Stein G. S. (2009) J. Biol. Chem. 284, 3125–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connor M. K., Azmi P. B., Subramaniam V., Li H., Seth A. (2005) Mol. Cancer Res. 3, 453–461 [DOI] [PubMed] [Google Scholar]
- 34.Gao Y., Ganss B. W., Wang H., Kitching R. E., Seth A. (2005) Exp. Cell Res. 304, 127–135 [DOI] [PubMed] [Google Scholar]
- 35.Azmi P. B., Seth A. K. (2009) Anticancer Res. 29, 2253–2263 [PubMed] [Google Scholar]
- 36.Azmi P., Seth A. (2005) Eur. J. Cancer 41, 2549–22560 [DOI] [PubMed] [Google Scholar]
- 37.Kaneki H., Guo R., Chen D., Yao Z., Schwarz E. M., Zhang Y. E., Boyce B. F., Xing L. (2006) J. Biol. Chem. 281, 4326–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]