Abstract
L3MBTL1, a paralogue of Drosophila tumor suppressor lethal(3)malignant brain tumor (l(3)mbt), binds histones in a methylation state-dependent manner and contributes to higher order chromatin structure and transcriptional repression. It is the founding member of a family of MBT domain-containing proteins that has three members in Drosophila and nine in mice and humans. Knockdown experiments in cell lines suggested that L3MBTL1 has non-redundant roles in the suppression of oncogene expression. We generated a mutant mouse strain that lacks exons 13–20 of L3mbtl1. Markedly reduced levels of a mutant mRNA with an out-of-frame fusion of exons 12 and 21 were expressed, but a mutant protein was undetectable by Western blot analysis. L3MBTL1−/− mice developed and reproduced normally. The highest expression of L3MBTL1 was detected in the brain, but its disruption did not affect brain development, spontaneous movement, and motor coordination. Despite previous implications of L3mbtl1 in the biology of hematopoietic transcriptional regulators, lack of L3MBTL1 did not result in deficiencies in lymphopoiesis or hematopoiesis. In contrast with its demonstrated biochemical activities, embryonic stem (ES) cells lacking L3MBTL1 displayed no abnormalities in H4 lysine 20 (H4K20) mono-, di-, or trimethylation; had normal global chromatin density as assessed by micrococcal nuclease digests; and expressed normal levels of c-myc. Embryonic fibroblasts lacking L3MBTL1 displayed unaltered cell cycle arrest and down-regulation of cyclin E expression after irradiation. In cohorts of mice followed for more than 2 years, lack of L3MBTL1 did not alter normal lifespan or survival with or without sublethal irradiation.
Keywords: Chromatin, Epigenetics, Gene Knockout, Transcription Repressor, Tumor Suppressor, PR-SET7, SFMBT1
Introduction
Proteins containing malignant brain tumor (MBT)3 domains have recently attracted considerable attention because of their emerging role in modulating the organization of cellular DNA in aggregated nucleosomes and chromatin and their important roles in development (1–6). Similar to their better known distant relatives, chromo- and tudor domains, these protein modules selectively bind histone tails depending on the degree of methylation at specific lysine residues (7, 8). MBT domains are highly conserved from Drosophila to mammals, comprise ∼100 amino acid residues, and the structure of multiple family members has been resolved at the atomic level (2, 3, 6, 9–13). Characteristically, MBT domains recognize mono- and dimethylated lysine residues in histone tails but not non- or trimethylated lysines (2–4, 7, 14). Human L3MBTL1, a transcriptional repressor and suspected tumor suppressor gene (15–17), was shown to be capable of compacting nucleosomal arrays by binding mono- and dimethylated histone H4K20 and histone H1bK26 in vitro (4). Thus, MBT domain proteins contribute to the complex organization of chromatin as “readers” and “effectors” of a network of post-translational histone modifications that is critical for the establishment of specific cellular differentiation states (18–20).
MBT domains were first recognized in the cloned gene for the Drosophila mutant lethal(3)malignant brain tumor (l(3)mbt), an embryonic lethal gene associated with transformation of larval brain cells (hence, malignant brain tumor domain) (21). In Drosophila, only two further MBT domain-containing proteins exist, Sex comb on midleg (scm) (6, 22–24) and Sex comb with four MBT domains (sfmbt) (5, 13, 14). Both are members of Polycomb group-related complexes (scm: PRC1 (22); sfmbt: PhoRC (14)) and are required for repression of Hox gene expression during development. Moreover, scm and sfmbt co-occupy Polycomb-responsive elements of target genes in Drosophila and synergize in Polycomb target gene repression (13). Of note, l(3)mbt and sfmbt also appear to synergize in repression of E2F (5). Like l(3)mbt, sfmbt and scm are essential for Drosophila embryonic development, and their disruption results in embryonic lethality (14, 22).
In mice (supplemental Fig. S1) and humans (1), nine genes encoding for proteins with MBT domains are known. Two of them, Scmh1 (25) and Scml2 (26), resemble Drosophila scm in that they contain two MBT domains and a C-terminal sterile α-motif (SAM) domain. Three genes, L3mbtl1 (27), L3mbtl3 (also designated MBT-1 (28)), and L3mbtl4 (supplemental Fig. S1) bear three MBT domains and a C-terminal SAM domain like Drosophila l(3)mbt. Finally, four genes, L3mbtl2 (29), Mbtd1 (9), Sfmbt1, (30), and Sfmbt2 (31), resemble Drosophila sfmbt with four MBT domains. It is uncertain to what extent different mammalian MBT domain-containing proteins have similar functions as might be inferred from their overlapping binding properties to lysine-methylated peptides. So far, only Scmh1 and L3mbtl3 have been targeted in the mouse germ line. Disruption of Scmh1 resulted in defects in spermatogenesis and skeletal development in up to 50% of the homozygous mice, which were otherwise ostensibly normal (32). Disruption of L3mbtl3 resulted in late embryonic or perinatal lethality with anemia and abnormalities in myeloid progenitors but normal development of other organs (28).
Mammalian L3MBTL1 has been shown to function as a transcriptional repressor (15) and physically interact with Tel/Etv6, a key transcriptional regulator in hematopoiesis (33). It was also reported to contribute to the repression of Runx1, another important hematopoietic regulator (34). Echoing its role as a tumor suppressor in Drosophila (5, 21), the location of L3MBTL1 in humans raised suspicions that it might be the critical tumor suppressor responsible for the common loss of chromosome 20q12 in human myeloid hematologic malignancies (15, 16), but this hypothesis was not confirmed in samples from such disorders where L3MBTL1 appeared to be expressed normally (17). Of note, L3MBTL3, L3MBTL2, and SCML2 are mutated in rare cases of medulloblastoma, lending support to the notion that MBT domain proteins may function as tumor suppressors (35). Possibly contributing to tumor suppression, L3MBTL1 was reported to repress c-myc expression in HeLa cells likely by compacting chromatin, an activity that can be demonstrated in vitro with nucleosomal arrays and is dependent on binding to monomethylated H4K20 and mono- or dimethylated H1bK26 (4). L3MBTL1 directly interacts with PR-SET7, which mediates H4K20 monomethylation and enhances repression of transcription by L3MBTL1 (36). Here we report the generation and analysis of mice after disruption of L3mbtl1.
EXPERIMENTAL PROCEDURES
Expression Analysis
For expression analysis by Northern blot and RT-PCR, total RNA was extracted from tissues or cells using TRIzol reagent. Northern blots were probed with full-length cDNAs (IMAGE clone 5708338 for mouse L3mbtl1, IMAGE clone 3962047 for mouse c-myc, and IMAGE clone 6493407 for GAPDH). For RT-PCR, total RNA (1 μg) was generated for first strand DNA synthesis with the SuperScript III kit, and the following primers were used: L3mbtl1-forward, TTC AGC TGG AGC CAG TAC CT; L3mbtl1-reverse, CTC CCA GCA CAC AGA GAG TG (1615 bp); GAPDH-forward, AAC TTT GGC ATT GTG GAA GG; and GAPDH-reverse, TGT GAG GGA GAT GCT CAGT G (599 bp). For detection of the mutant transcript (breakpoint; see Fig. 1h), the following primers were used: L3mbtl1-forwardb, TTC AGC TGG AGC CAG TAC CT; L3mbtl1-reverseb, CTG GGC CTA GCT TGA CAC TC (1261 bp in wild type; 372 bp in KO); L3mbtl4-forward, GAG ACA ACC TAA CCG GAA; L3mbtl4-reverse, TGT CTT CTT GGC TAG AGG CA (1866 bp); L3mbtl2-forward, GCA AGA TCC TGG TAC CTC CA; L3mbtl2-reverse, CGT CAT CAC TGT CAC CAT CC (264 bp); L3mbtl3-forward, GCA GAT GCT CTG GAC ATT CA; L3mbtl3-reverse, CTC GAG CTT CAT CCC AAC TC (204 bp); Scmh1-forward, CTC TGA ACC TCC CAG CAG TC; Scmh1-reverse, AGG TTT TGG GAT GTG CTG AC (277 bp); Scml2-forward, TCC ACT GGG GTA CAC ACT GA; Scml2-reverse, CAA TTT TTC CTG GTG GCT GT (341 bp); Mbtd1-forward, ACT GGT GCC AAA ACA CTT CC; Mbtd1-reverse, GAT TAA GGG GCT GTG CAT GT (225 bp); Sfmbt1-forward, ACC ACA GTT CCC TAC GCA TC; Sfmbt1-reverse, CGA GGG TCT TCT TGT TCT GC (259 bp); Sfmbt2-forward, AGA ACT GTT CCG TCC ACA CC; Sfmbt2-reverse, TTT TCT GTG CTG AGC CCT TT (354 bp); CyclinE1-forward, ATT GGC TAA TGG AGG TGT GC; CyclinE1-reverse, CCA CTT AAG GGC CTT CAT CA (269 bp); c-myc-forward, TGA CCT AAC TCG AGG AGG AGC TGG AAT C; and c-myc-reverse, AAG TTT GAG GCA GTT AAA ATT ATG GCT GAA GC (170 bp). To confirm breakpoint in mutant L3mbtl1, PCR products were cloned using a TOPO TA Cloning® kit, and eight independent clones were subjected to sequencing with identical results (see Fig. 1, i and j).
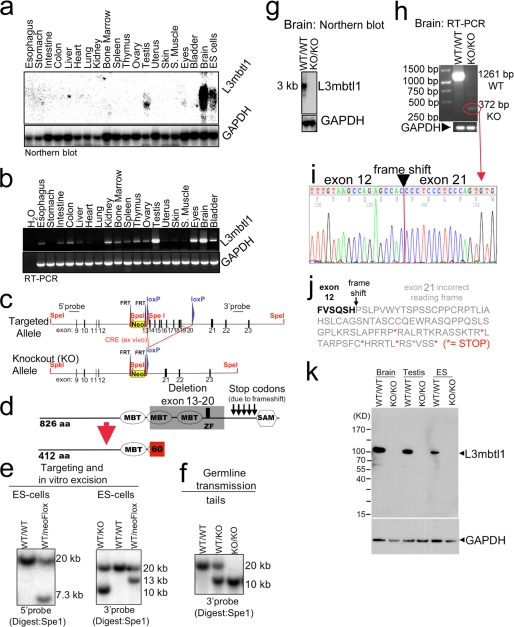
FIGURE 1.
Disruption of L3mbtl1 in mice. a and b, L3mbtl1 expression levels in wild type mice vary among different tissues. S. muscle, skeletal muscle. a, Northern blot analysis demonstrates expression in testes, eyes, and ES cells and most abundantly in the brain. b, expression analysis by RT-PCR reveals widespread, but weaker, expression in other tissues. c and d, strategy for L3mbtl1 disruption (for additional details, see supplemental Fig. S2). d, the knock-out allele lacks the coding region for two MBT domains as well as the zinc finger (amino acids 413 through 708). Additionally, fusion of exon 12 to exon 21 predicts a frameshift that generates several stop codons, so any residual message would not code for the SAM domain but instead express 60 alternative amino acids. e, Southern blot analysis of SpeI-digested DNA from ES cells after targeting and/or Cre-mediated excision of the loxP-flanked regions. As expected, the targeted allele results in a lower band (7.3 kb) compared with the wild type allele (20 kb) using a probe upstream of the targeting vector (5′ probe) (left panel). Correct insertion of the 3′ end was confirmed with a probe downstream of the targeted region (3′ probe; expected bands: wild type, 20 kb; and targeted, 13 kb) (right panel). Excision of the floxed region after transient expression of Cre in vitro results in a 10-kb band (right panel, left lane). f, germ line transmission of the targeted allele documented by Southern blot analysis of tail DNA. g, Northern blot analysis from brains using the entire coding region as a probe demonstrates abundant message in the wild type but no signal in L3mbtl1 knock-out mice. h, RT-PCR analysis for L3mbtl1 using primers upstream and downstream of the deleted region reveals a faint band in the mutant consistent with a shorter message as predicted. i and j, sequence analysis of the PCR product from the mutant in h confirms deletion of exons 13–20 after exon 12 resulting in 60 abnormal residues followed by five stop codons. k, Western blot analysis using an antiserum against a His-tagged fusion protein corresponding to the N-terminal 215 amino acid residues of L3MBTL1 demonstrates absence of the protein in knock-out cells and tissues. Note that a theoretically possible truncated protein (expected size, ∼55 kDa) is not detectable.
Construction of L3mbtl1-targeting Vector
Genomic clones corresponding to the mouse L3mbtl1 locus were isolated from a genomic library prepared from the 129/SvEV mouse strain (Children's Hospital Oakland Research Institute) by screening with a mouse L3mbtl1 probe covering coding exons 17–19. As a source for fragments, a 20-kb L3mbtl1 genomic SpeI fragment that included exons 9–23 was subcloned into the SpeI site of the pBluescript II KS (pBS) vector. To generate the targeting vector, a 4-kb region of genomic DNA (NheI fragment) containing exons 13–20 of the L3mbtl1 gene was blunt end-cloned into the EcoRV site of pBS. Subsequently, the 3′ arm (NheI-HindIII) was introduced into the ClaI site of the same vector by blunt end cloning. To introduce the 3′ loxP site, an adapter (made with the following primers: forward, AGC TTG ATA TCT AAT ATA ACT TCG TAT AAT GTA TGC TAT ACG AAG TTA TTA G; reverse, AGC TCT AAT AAC TTC GTA TAG CAT ACA TTA TAC GAA GTT ATA TTA GAT ATC A) was introduced into the HindIII of pBS just upstream of the 3′ arm. Then, the 5′ arm (XbaI-NheI) was blunt end-cloned upstream into the SacII site of pBS. Finally, the 5′ loxP site and a selectable cassette (PL 451 (37)) containing the genes for neomycin resistance (Neo cassette) flanked by two FRT sites was inserted into the NotI site of pBS just downstream of the 5′ arm. For targeting, the vector was linearized by KpnI.
ES Cell Culture and Generation of Mutant ES Cells and Mice
V6.5 (129SvJaeXC57BL/6; male) ES cells (38) were grown with irradiated mouse embryo fibroblasts (MEFs) on gelatinized tissue culture plates in DMEM supplemented with 15% fetal calf serum, leukemia inhibitory factor (39), non-essential amino acids, l-glutamine, and penicillin/streptomycin. The L3mbtl1-targeting construct was linearized and electroporated into ES cells and subsequently selected in G418 (400 μg/ml). Correctly targeted ES cell clones were identified by Southern blot analysis (see Fig. 1e). Subsequently, the null allele (L3mbtl1+/−) was generated by transient transfection and puromycin selection with a Cre expression plasmid (PAC-Cre) (39). L3mbtl1-targeted clones with confirmed correct karyotypes were injected into C57BL/6 blastocysts to generate chimeras, which were bred with C57/BL6 mice for germ line transmission. Biallelic disruption of L3mbtl1 in ES cells was achieved after selecting correctly targeted heterozygous ES cell clones that tolerated growth in increased neomycin concentrations (2–4 mg/ml) because of crossing over and confirmation of the duplicated targeted allele by Southern blot (see Fig. 4a). After confirmation of the correct karyotype, deletion of the floxed alleles was achieved by transient expression of Cre and was confirmed by Southern blot analysis (see Fig. 4a).
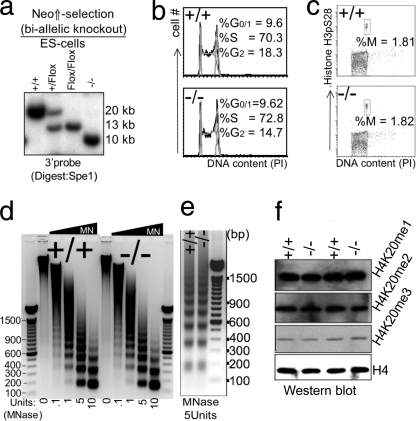
FIGURE 4.
Impact of disruption of L3mbtl1 on ES cells. a, biallelic disruption of L3mbtl1 in ES cells was achieved after selection of clones that had duplicated the targeted allele by crossing over in the presence of increased neomycin concentration (third lane). After confirmation of a normal karyotype (not shown), both alleles of L3mbtl1 were disrupted by transient expression of Cre (right lane). b, no major differences in cell cycle profiles (G1, S, and G2/M) of L3mbtl1−/− ES cells compared with control ES cells. Analysis of cell cycle distribution after staining DNA content with propidium iodide (PI) is shown. c, the G2/M checkpoint is not altered in L3mbtl1−/− ES cells. Simultaneous staining for DNA content and serine 28-phosporylated histone H3 (H3pS28) was used to resolve G2 and M phases of the cell cycle. d and e, analysis of global chromatin structure using limited micrococcal nuclease (MNase) digests of wild type or L3mbtl1−/− ES cells. Micrococcal nuclease cuts DNA that is not occupied by nucleosomes. In dense chromatin or with limited amounts of enzyme, digestion within the internucleosomal spaces is inefficient, resulting in the generation of oligo- and multinucleosomes with distinct sizes (the lowest band represents mononucleosome; higher bands represent oligonucleosome (dimers, trimers, etc.)). In loose (relaxed) chromatin and with high amounts of micrococcal nuclease, fragments representing DNA protected by a single nucleosome are generated preferentially. Note that an equal abundance of the various bands suggests that there is no marked global difference in compaction of chromatin after disruption (e); an equal size of fragments demonstrates that nucleosome spacing is not affected by loss of L3MBTL1. Each panel shows one representative of three independent experiments. DNA markers are shown with sizes of prominent bands labeled. f, global levels of H4K20 mono-, di-, or trimethylation are not affected by the absence of L3MBTL1 in ES cells.
Animal Care and Histopathology
Mice were cared for according to the guidelines and supervision of the Massachusetts General Hospital Subcommittee on Animal Research. All behavioral tests were performed at the University of Massachusetts, Worcester, MA and were approved by the Institutional Animal Care and Use Committee. Blastocyst injection to generate chimeric mice was performed by the Massachusetts General Hospital Gene Targeting Core. Total body γ-irradiation was performed using a 137Cs source (Gammacell 40 Exactor). Kaplan-Meyer survival curves were generated using Prism® version 4 (GraphPad Software); statistical significance was determined with the log rank test. Complete necropsy and histopathological analysis after hematoxylin and eosin staining were performed by the Harvard Medical School Rodent Histopathology Core.
Behavioral Testing
Animals were housed in groups of two to four per cage with food and water ad libitum under a 12-h light/dark cycle (7 a.m. on, 7 p.m. off), and tests were conducted during the light cycle between 7:30 a.m. and 2:30 p.m. For all experiments, adult L3MBTL1-deficient or wild type mice (8–16 weeks old) were used with experimental/control groups matched for age and sex. Motor coordination and learning were assessed using a rotarod apparatus with a five-mouse capacity (UGO Basile North America Inc.). This test consisted of a rotating rod/drum (3-cm diameter) separated by plastic dividers into five compartments (6 cm wide). On day 1, mice were trained to walk on an accelerating rod (4–40 rpm over 300 s) in 10 trials, 300 s each, with 300-s rest time between each trial. On day 2, mice were tested with three trials, 300 s each, with 300-s rest time between each trial. An observer recorded the time and rpm at which a mouse failed the test by falling from the drum or by hanging onto the drum and completing a full rotation. To assess locomotor activity, mice were placed in clean, 19.1 × 29.2 × 12.7-cm shoe box-style cages, and ambulation was measured continuously for 17 h (5 p.m. to 10 a.m.) using a photobeam activity system (San Diego Instruments).
Complete Blood Counts and Flow Cytometry
For complete blood counts, blood samples (∼50 μl) were obtained by tail vein nicking or retro-orbital sinus puncture, anticoagulated in EDTA, and analyzed using an automated system (Vet Scan HM5). Flow cytometric analysis of bone marrow, spleen, and thymus cells was performed as described previously (33, 40–44).
Western Blot Analysis
For the analysis of H4K20 methylation, histone extraction was performed after three washes with ice-cold PBS, supplemented with 5 mm sodium butyrate, and lysed with a Triton extraction buffer (PBS, 0.5% Triton, 2 mm PMSF, 0.02% NaN3) at a cell density of 107 cells/ml. The lysate was centrifuged at 2000 rpm for 10 min at 4 °C, and the pellet was washed twice with Triton extraction buffer and then resuspended in 0.2 n HCl at a cell density of 4 × 107 cells/ml. Histones were acid-extracted overnight at 4 °C. The next day samples were centrifuged at 2,00 rpm for 10 min at 4 °C, and the supernatant was dialyzed against CH3COOH and H2O. Samples were kept at −80 °C with 20% glycerol. Antibodies for methylated H4K20 were from Upstate. For the analysis of L3MBTL1 protein, nuclear extract preparation was performed as described (45) with minor modifications. Cells (∼107) were trypsinized, washed twice in PBS, resuspended in 5 volumes of hypotonic Roeder A buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, 1 mm PMSF), and incubated for 10 min on ice. Swollen cells were homogenized with 10 strokes in a Dounce homogenizer, and the homogenate was centrifuged for 10 min at 900 × g. The remaining nuclear pellet was washed once in PBS and subsequently homogenized with 15 strokes in Roeder C buffer (20 mm HEPES, pH 7.9, 420 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, 5% glycerine, 0.2 mm EDTA, and 1 mm PMSF). Nuclear extract was cleared at 17,000 × g for 30 min. To preclear cell lysates, normal rabbit IgG and Protein A/G Plus-agarose were added into whole cell lysates. After incubation for 1 h at 4 °C, supernatants were collected by centrifugation at 1000 × g for 5 min at 4 °C. Then, proteins were separated by 4–20% gradient SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were blocked for 60 min in Western blot wash buffer (30 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.25% (v/v) Tween 20) containing 5% (w/v) dry milk and immunostained. The antiserum against the N-terminal 215 amino acids of human L3MBTL1 was from Active Motif (39182, lot 160).
Cell Cycle Analysis
Cell cycle analysis was performed as described (41). The fractions of cells in G1, S, and G2/M phases of the cell cycle were determined with Dean/Jett/Fox analysis (FlowJo software) or by ModFit LT (BD Biosciences). Intracellular staining to separate G2 and M phases was performed using Alexa Fluor 647 anti-histone H3 (Ser(P)-28) from BD Biosciences (558217).
Generation of MEFs
MEFs were prepared from individual embryos bearing L3mbtl1+/+ or L3mbtl1−/− genotypes at embryonic day 13.5. Head and internal organs were removed, and the torso was minced and dispersed in 0.1% trypsin (30 min at 37 °C). Cells were grown for two population doublings and then frozen or used for subsequent experiments. MEFs were maintained in DMEM containing 10% fetal bovine serum and subcultured 1:4 upon reaching confluence.
Irradiation of MEFs
Culture dishes with cells (∼107) were exposed to a single dose of radiation (10 or 20 grays) using a 137Cs source. Analysis was performed after an additional 24 h of culture.
Micrococcal Nuclease Digestion
Nuclei from 1 × 106 ES cells were prepared by pelleting and resuspending cells in buffer containing 10 mm Tris, pH 7.5, 20 mm KCl, 0.5 mm EDTA, 0.5 mm DTT, 0.05 mm spermidine, 0.125 mm spermine, 1 mm PMSF, 0.1% digitonin. Cells were homogenized with 10 strokes in a Bellco all glass Dounce homogenizer (type A pestle), and nuclei were pelleted at 300 × g. Nuclei were then washed once in wash buffer (20 mm HEPES, pH 7.7, 20 mm KCl, 0.5 mm EDTA, 0.5 mm DTT, 1 mm PMSF) followed by a wash in the same buffer additionally containing 300 mm NaCl. Subsequently, nuclei were resuspended in wash buffer supplemented with 300 mm NaCl and 3 mm CaCl2. Chromatin (100 μg of DNA per reaction) was digested for 15 min at room temperature in the presence of 0.05–10 units of micrococcal nuclease (catalog number 10107921001, Roche Applied Science). Following micrococcal nuclease treatment, extracts were supplemented with 5 mm EGTA and 0.05% Nonidet P-40 and centrifuged at 10,000 × g for 15 min at 4 °C. DNA was recovered from each sample by phenol/chloroform extraction and ethanol precipitation and resolved by a 1.2% agarose gel.
RESULTS
L3mbtl1 Is Not Essential for Mouse Development
To begin to explore the role of L3mbtl1 in vivo, we determined its expression pattern. High expression of L3mbtl1, detectable by Northern blot analysis, was only found in the brain, testis, eyes, and ES cells (Fig. 1a). However, RT-PCR analysis revealed that lower levels of mRNA were widely expressed in tissues, including the gastrointestinal system, the hematolymphoid system, kidney, ovary, and bladder (Fig. 1b). To assess the role of L3mbt1 in vivo, we generated knock-out mice (Fig. 1, c–f, and supplemental Fig. S2). The knock-out strategy was designed so that the entire C-terminal part of the protein, including the second and third MBT, zinc finger, and SAM domains, could not be generated even if a mutant mRNA was produced (Fig. 1, c and d). A mutant protein would not be expected to bind methylated histones, which requires the second MBT domain (2–4). The mutant allele was generated in ES cells, transmitted to the germ line after blastocyst injection, and bred to homozygosity (Fig. 1, e and f). L3mbtl1−/− mice were born at expected frequencies, were fertile, and did not display obvious abnormalities. Likewise, complete necropsy and histological analysis of all tissues of three adult mutant mice revealed no abnormalities (data not shown). Northern blot analysis of brain and testis, the organs with the strongest expression, revealed no signal for L3mbtl1 with a probe covering the entire coding region (Fig. 1g). This suggested that a possible truncated mRNA was likely unstable but did not exclude that it was expressed at levels below the detection threshold. Indeed, we were able to detect a very faint band of the expected smaller, mutant message by RT-PCR (Fig. 1h). Cloning and sequencing confirmed the predicted mutation, including frameshift and introduction of stop codons (Fig. 1, i and j). Because an N-terminal protein deficient of all known functional domains of L3mbtl1 might be produced from the mutant allele, we probed for a truncated protein by Western blot analysis using an antiserum against the N-terminal portion of L3mbtl1 (Fig. 1k). Although L3mbtl1 protein could be demonstrated in wild type brain, testis, and ES cells, neither normal L3mbtl1 nor a truncated protein were detectable in the knockout. Thus, we conclude that L3mbtl1 is not required for development in the mouse.
Normal CNS Development and Function in L3mbtl1−/− Mice
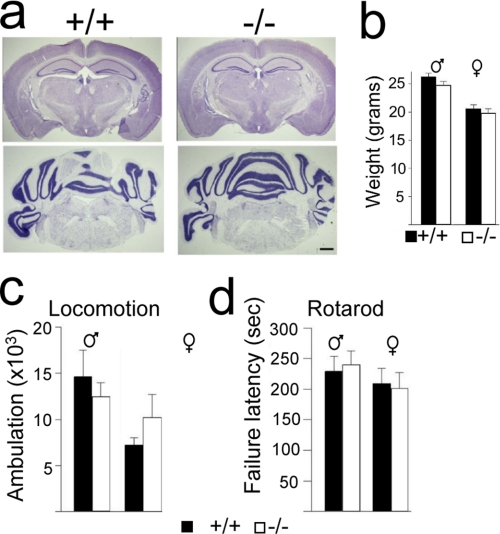
In light of the high expression level of L3MBTL1 in the brain, we investigated the brain and neurological function after its loss. However, detailed analysis of brain morphology and cytoarchitecture revealed no abnormalities (Fig. 2a). Moreover, mice lacking L3MBTL1 showed no obvious behavioral abnormalities in reproduction or feeding (data not shown) and maintained normal weights (Fig. 2b). Likewise, formal assessment of spontaneous movement (Fig. 2c) and motor coordination (Fig. 2d) suggested that CNS function was well preserved in the absence of L3MBTL1. Thus, L3MBTL1 is not essential for brain development and function.
FIGURE 2.
Disruption of L3mbtl1 is compatible with ostensibly normal CNS development and function. a, normal brain morphology and cytoarchitecture in L3mbtl1−/− mice. Representative 15–20-μm-thick Nissl-stained coronal sections display normal morphology and cytoarchitecture in forebrain (upper panels) and hindbrain (lower panels) of 6–8-week-old mutants (right) and littermate controls (left). Scale bar, 0.5 mm. b, unaltered body weight after disruption of L3mbtl1 (mean ± standard error (S.E.), n = 8–15 animals/group; black bars, control; white bars, mutants). c and d, behavioral testing. c, locomotor activity is not perturbed in the absence of L3MBTL1. Shown is the total ambulation over 17 h tracked with a photobeam (5 p.m. to 10 a.m.; summed ambulation, means ± S.E., n = 4–6 animals/group). d, rotarod motor coordination performance is not perturbed in L3mbtl1−/− mice. The bar graph shows latency to failure in seconds (see “Experimental Procedures”; day 2, three-trial mean ± S.E., n = 5).
Disruption of L3mbtl1 Does Not Result in Abnormal Hematopoiesis or Lymphopoiesis
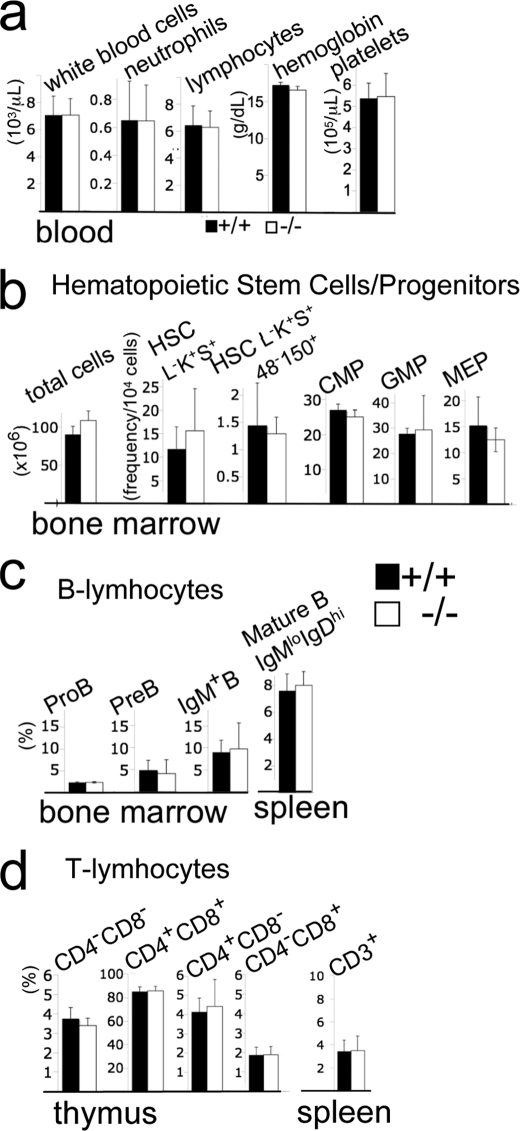
L3MBTL1 has been linked to two major regulators of hematopoiesis as it was reported to interact with Tel/Etv6 (15) and was implicated in repression of Runx1 (34). mRNA expression for L3mbtl1 in bone marrow, spleen, and thymus was low but detectable by RT-PCR (Fig. 1b). Nevertheless, blood counts (Fig. 3a), the frequencies of major bone marrow progenitor and stem cell populations (Fig. 3b), and lymphopoiesis in bone marrow, thymus, and spleen (Fig. 3, c and d) were entirely normal. Thus, L3MBTL1 is dispensable for steady state lympho- or hematopoiesis.
FIGURE 3.
Disruption of L3mbtl1 does not perturb hematopoiesis or lymphopoiesis. a, normal blood cell counts in the absence of L3MBTL1 (n = 5, mean and standard deviation (S.D.)). b, total bone marrow cell counts (number per two femurs, tibiae, and fibulae) and immuno-flow cytometric analysis of major stem cell and myeloid progenitor population demonstrate normal hematopoiesis in L3mbtl1−/− mice. Hematopoietic stem cells (HSC) were identified as lineage marker− (L−), cKit+ (K+), Sca1+ (S+) cells with or without additional SLAM markers CD48 (48−) and CD150 (150+). Frequencies of common myeloid progenitor (CMP; L−K+S−CD34+FCγR−), granulocyte-monocyte progenitors (GMP; L−K+S−CD34+FCγR+), and megakaryocyte-erythrocyte progenitors (MEP; L−K+S−CD34−FCγR−) are shown. b and c, no major abnormalities in lymphopoiesis in the absence of L3MBTL1. c, frequencies (%) of B cells and their progenitors. Pro-B cells (IgM−, B220+, CD43+), pre-B cells (IgM−, B220+,CD43−), and IgM+ bone marrow cells as well as mature spleen B cells (IgMlow, IgDhi) were determined as shown previously. d, frequencies of T cell precursors in the thymus and mature T cells in the spleen were determined as described. Bar graphs represent averages and S.D. Black bars, wild type littermates (n = 3); white bars, L3mbtl1−/− mice (n = 3). No differences were significant (<0.05) by two-tailed Student t test.
Cell Cycle Progression, Global Chromatin Density, and H4K20 Methylation in ES Cells Are Not Altered in Absence of L3mbtl1
ES cells display comparatively high levels of L3mbtl1 mRNA (Fig. 1a). We generated ES cells with biallelic loss of L3mbtl1 (Fig. 4a). These were morphologically normal, showed unaltered colony formation in culture, and displayed normal cell cycle profiles (data not shown and Fig. 4, b and c). In light of the unique potential of L3mbt1 to compact nucleosomal arrays in vitro (4), we assessed chromatin density in ES cells using micrococcal nuclease (Fig. 4, d and e). With this approach, global differences in chromatin density have been demonstrated with ES cells that lack three of the six isoforms of the linker histone H1 (including H1e, the homologue of H1.4, which binds L3MBTL1) (4, 46). However, neither chromatin density nor nucleosome spacing was altered in the absence of L3MBTL1 (Fig. 4, d and e). As mono- and dimethylated forms of histone H4K20 bind L3MBTL1 (4) and have the potential to influence the progressive methylation of this site (36), we investigated whether absence of L3mbtl1 perturbed the levels of mono-, di-, or trimethylation of H4K20 by Western blot analysis in ES cells. However, L3MBTL1 did not appear to influence the global methylation state of H4K20 (Fig. 4f). Thus, ES cells do not require L3mbtl1 to maintain normal global chromatin density or H4K20 methylation. This does not rule out that L3MBTL1 influences these parameters at specific genomic sites.
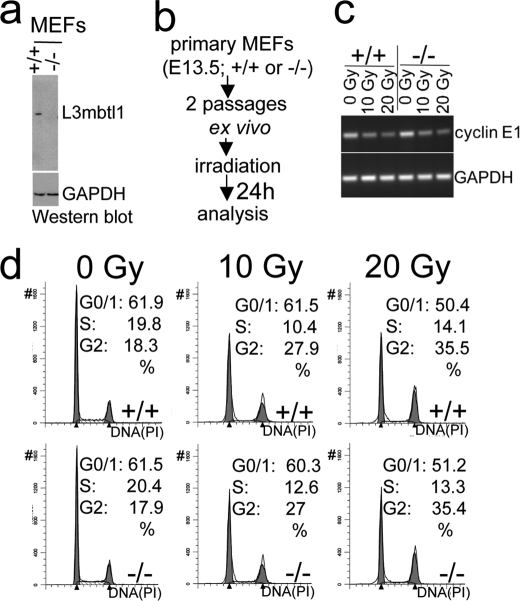
Lack of L3MBTL1 in MEFs Is Not Associated with Altered Cyclin E Expression or Cell Cycle Arrest in Response to Irradiation
L3MBTL1 has been reported previously to bind to the promoter of cyclin E, and it was inferred that it repressed expression (4, 36). Cyclin E is a target gene of E2F that promotes cell cycle progression at the G1/S checkpoint and is down-regulated in the context of RB-mediated cell cycle arrest after irradiation in MEFs (47). Therefore, we generated MEFs from L3mbtl1+/+ and L3mbtl1−/− embryos and exposed them to escalating doses of γ-irradiation (10 and 20 grays) (Fig. 5b). Surprisingly, L3MBTL1 expression did not appear to influence cyclin E expression at base line or affect the dose-dependent down-regulation of cyclin E after irradiation (Fig. 5c). Consistently, both L3mbtl1+/+ and L3mbtl1−/− MEFs showed the expected cell cycle block following irradiation, i.e. decreased G1/S progression and accumulation in G2 (Fig. 5d). Thus, loss of L3mbtl1 does not affect the G1/S checkpoint in response to irradiation.
FIGURE 5.
MEFs lacking L3MBTL1 display unaltered cell cycle arrest in response to irradiation. a, MEFs from L3mbtl−/− embryos lack L3MBTL1 protein. b, scheme of experiment. c, RT-PCR before and after irradiation demonstrates dose-dependent, diminished expression of cyclin E after irradiation in the presence or absence of L3MBTL1. d, cell cycle analysis reveals dose-dependent arrest in the cell cycle irrespective of L3mbtl1 gene status. Gy, grays; PI, propidium iodide.
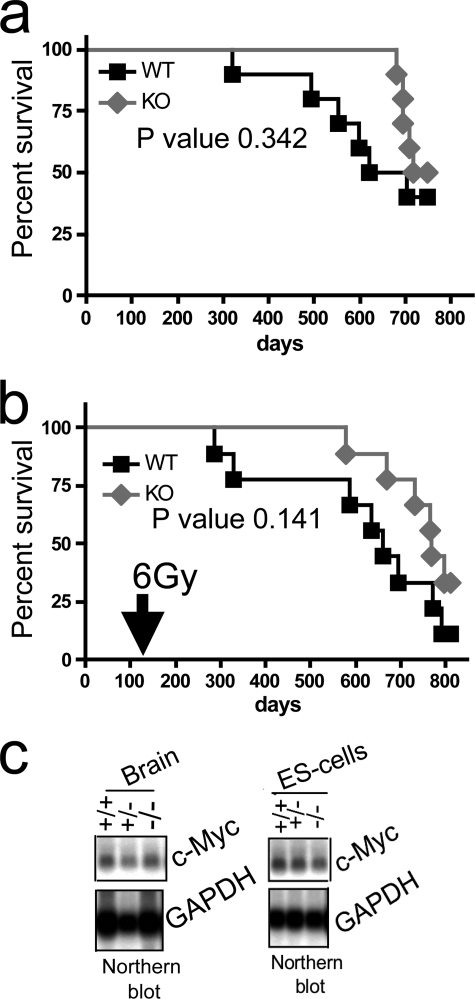
Lack of L3MBTL1 Does Not Alter Normal Lifespan or Survival after Sublethal Irradiation
l(3)mbt is considered a tumor suppressor in Drosophila (5), and MBT domain family proteins are subject to rare mutations in medulloblastoma (35). Therefore, we assessed the impact of absence of L3MBTL1 on lifespan and survival after low dose irradiation in cohorts of mice (Fig. 6, a and b). Untreated mice did not exhibit altered mortality after more than 2 years of observation. Similarly, there was no difference in survival after sublethal irradiation (6 grays), which is in contrast to, for example, findings in p53 heterozygous mice, which succumb to tumor formation within less than a year after exposure to a similar dose (48). In agreement with this, we did not find differences in c-myc expression after L3MBTL1 disruption in any mouse tissue or in ES cells (Fig. 6c and data not shown), suggesting that in the mouse L3MBTL1 is not essential for the suppression of c-myc. This is in contrast with findings in HeLa cells where shRNA-mediated knockdown of L3MBTL1 leads to increased c-myc expression (4). Thus, we conclude that L3mbtl1 does not function as a general tumor suppressor in mice. However, we cannot exclude that its loss may enhance tumor growth in the context of specific oncogenes or in the molecular pathogenesis of selected tumors.
FIGURE 6.
Disruption of L3mbtl1 does not affect lifespan with or without sublethal dose irradiation. a, Kaplan-Meyer survival curves of L3mbtl1−/− (n = 10) and littermate L3mbtl1+/+ control mice (n = 10) reveal no significant difference in lifespan. b, single dose (6 grays (Gy)), non-lethal total body irradiation of cohorts of L3mbtl1−/− (n = 10) and littermate L3mbtl1+/+ control mice (n = 9) at 3 months of age results in no significant difference in survival; p values were calculated by log rank test. c, no alteration of global c-myc expression levels in L3mbtl1−/− brains or ES cells as analyzed by Northern blot.
DISCUSSION
In light of the documented roles of mammalian L3MBTL1 in ex vivo systems (4, 15, 34, 36), it is surprising that loss of L3mbtl1 does not provoke phenotypes even after long observation. Our strategy for inactivating L3mbtl1 ensures the disruption of all known functional domains even if a truncated mutant protein expressed at levels below the detection level of our Western blot was produced. Specifically, a possible mutant protein would lack the second MBT domain of L3MBTL1 that binds methylated histones (4), the hinge region that binds PR-SET7 together with the MBT domains (36), and the SAM domain that is critical for protein interactions and repression (15). Moreover, we demonstrated that the mRNA of mutant L3MBTL1 is expressed only at very low levels that are exclusively detectable by PCR as a faint band, and a mutant protein could not be detected by Western blot analysis (Fig. 1). It is possible that L3MBTL1-mediated chromatin compaction and gene repression are required in cryptic contexts or in responses to challenges that we have not investigated. More likely, the expansion of the family of MBT domain-containing proteins in evolution (see supplemental Fig. S1) has resulted in overlap of function and redundancy. This may be advantageous in higher organisms because this family of proteins is so important in development and tumor suppression that loss of each individual member in Drosophila is catastrophic (14, 21, 24). Consistent with this interpretation, disruption of Scmh1 resulted only in mild phenotypes with partial penetrance (32), and disruption of L3mbtl3 ostensibly only affected the hematopoietic system despite wide expression (28). It is uncertain which other MBT family protein(s) may be functionally replacing L3MBTL1 in the mutant mice. Obvious candidates are L3mbtl3 and L3mbtl4 because of their structural resemblance to L3MBTL1 (see supplemental Fig. S1) and because both are widely expressed (Ref. 28 and supplemental Figs. S3 and S4). It is also possible that the less homologous members of the MBT domain family might functionally replace L3MBTL1 in specific contexts as binding to methylated histone peptides is largely overlapping within the entire family. In agreement with this, all MBT domain family members are expressed in the tissues with the most pronounced L3MBTL1 expression (i.e. brain, testis, and ES cells; supplemental Fig. S4). Moreover, in Drosophila, l(3)mbt and sfmbt synergize in repression of E2F target genes (5), and scm and sfmbt synergize in Polycomb target gene repression (13). Of note, we have also targeted Sfmbt1 in mice, and its disruption does not cause immediate phenotypes even in combination with the L3mbtl1 mutant presented here.4 Resolution of the biology of the MBT domain family of proteins in mammals is likely to require the generation and combination of different mouse mutants.
Supplementary Material
Acknowledgments
We thank Lee Zou, Johnathan Whetstine, and Nick Dyson for helpful advice and discussion. We are grateful to Laura Prickett and Kat Folz-Donahue from the Harvard Stem Cell Institute Flowcore at Massachusetts General Hospital and Frederick A. Schroeder for expert assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HD 048489 from the NICHD (to S. A.). This work was also supported by a V Foundation Scholar Award and a Kimmel Foundation Scholar Award (to R. M.), a contribution from the Ellison Foundation to Massachusetts General Hospital (MGH) startup funds (to H. H.), and the Harvard Stem Cell Institute (to H. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
J. Qin and H. Hock, unpublished data.
- MBT
- malignant brain tumor
- l(3)mbt
- lethal(3)malignant brain tumor
- scm
- Sex comb on midleg
- sfmbt
- Sex comb with four MBT domains
- SAM
- sterile α-motif
- pBS
- pBluescript II KS
- MEF
- mouse embryo fibroblast.
REFERENCES
- 1.Bonasio R., Lecona E., Reinberg D. (2010) Semin. Cell Dev. Biol. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H., Fischle W., Wang W., Duncan E. M., Liang L., Murakami-Ishibe S., Allis C. D., Patel D. J. (2007) Mol. Cell 28, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min J., Allali-Hassani A., Nady N., Qi C., Ouyang H., Liu Y., MacKenzie F., Vedadi M., Arrowsmith C. H. (2007) Nat. Struct. Mol. Biol. 14, 1229–1230 [DOI] [PubMed] [Google Scholar]
- 4.Trojer P., Li G., Sims R. J., 3rd, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S. D., Wang Y. H., Reinberg D. (2007) Cell 129, 915–928 [DOI] [PubMed] [Google Scholar]
- 5.Lu J., Ruhf M. L., Perrimon N., Leder P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9381–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm C., de Ayala Alonso A. G., Rybin V., Steuerwald U., Ly-Hartig N., Fischle W., Müller J., Müller C. W. (2007) EMBO Rep. 8, 1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurer-Stroh S., Dickens N. J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C. P. (2003) Trends Biochem. Sci. 28, 69–74 [DOI] [PubMed] [Google Scholar]
- 9.Eryilmaz J., Pan P., Amaya M. F., Allali-Hassani A., Dong A., Adams-Cioaba M. A., Mackenzie F., Vedadi M., Min J. (2009) PLoS One 4, e7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y., Nady N., Qi C., Allali-Hassani A., Zhu H., Pan P., Adams-Cioaba M. A., Amaya M. F., Dong A., Vedadi M., Schapira M., Read R. J., Arrowsmith C. H., Min J. (2009) Nucleic Acids Res. 37, 2204–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathyamurthy A., Allen M. D., Murzin A. G., Bycroft M. (2003) J. Biol. Chem. 278, 46968–46973 [DOI] [PubMed] [Google Scholar]
- 12.Wang W. K., Tereshko V., Boccuni P., MacGrogan D., Nimer S. D., Patel D. J. (2003) Structure 11, 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm C., Matos R., Ly-Hartig N., Steuerwald U., Lindner D., Rybin V., Müller J., Müller C. W. (2009) EMBO J. 28, 1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klymenko T., Papp B., Fischle W., Köcher T., Schelder M., Fritsch C., Wild B., Wilm M., Müller J. (2006) Genes Dev. 20, 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boccuni P., MacGrogan D., Scandura J. M., Nimer S. D. (2003) J. Biol. Chem. 278, 15412–15420 [DOI] [PubMed] [Google Scholar]
- 16.Li J., Bench A. J., Vassiliou G. S., Fourouclas N., Ferguson-Smith A. C., Green A. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7341–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacGrogan D., Kalakonda N., Alvarez S., Scandura J. M., Boccuni P., Johansson B., Nimer S. D. (2004) Genes Chromosomes Cancer 41, 203–213 [DOI] [PubMed] [Google Scholar]
- 18.Campos E. I., Reinberg D. (2009) Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 19.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wismar J., Löffler T., Habtemichael N., Vef O., Geissen M., Zirwes R., Altmeyer W., Sass H., Gateff E. (1995) Mech. Dev. 53, 141–154 [DOI] [PubMed] [Google Scholar]
- 22.Bornemann D., Miller E., Simon J. (1996) Development 122, 1621–1630 [DOI] [PubMed] [Google Scholar]
- 23.Peterson A. J., Mallin D. R., Francis N. J., Ketel C. S., Stamm J., Voeller R. K., Kingston R. E., Simon J. A. (2004) Genetics 167, 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon J., Chiang A., Bender W. (1992) Development 114, 493–505 [DOI] [PubMed] [Google Scholar]
- 25.Berger J., Kurahashi H., Takihara Y., Shimada K., Brock H. W., Randazzo F. (1999) Gene 237, 185–191 [DOI] [PubMed] [Google Scholar]
- 26.Montini E., Buchner G., Spalluto C., Andolfi G., Caruso A., den Dunnen J. T., Trump D., Rocchi M., Ballabio A., Franco B. (1999) Genomics 58, 65–72 [DOI] [PubMed] [Google Scholar]
- 27.Koga H., Matsui S., Hirota T., Takebayashi S., Okumura K., Saya H. (1999) Oncogene 18, 3799–3809 [DOI] [PubMed] [Google Scholar]
- 28.Arai S., Miyazaki T. (2005) EMBO J. 24, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wismar J. (2001) FEBS Lett. 507, 119–121 [DOI] [PubMed] [Google Scholar]
- 30.Usui H., Ichikawa T., Kobayashi K., Kumanishi T. (2000) Gene 248, 127–135 [DOI] [PubMed] [Google Scholar]
- 31.Frankenberg S., Smith L., Greenfield A., Zernicka-Goetz M. (2007) BMC Dev. Biol. 7, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takada Y., Isono K., Shinga J., Turner J. M., Kitamura H., Ohara O., Watanabe G., Singh P. B., Kamijo T., Jenuwein T., Burgoyne P. S., Koseki H. (2007) Development 134, 579–590 [DOI] [PubMed] [Google Scholar]
- 33.Hock H., Meade E., Medeiros S., Schindler J. W., Valk P. J., Fujiwara Y., Orkin S. H. (2004) Genes Dev. 18, 2336–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sims J. K., Houston S. I., Magazinnik T., Rice J. C. (2006) J. Biol. Chem. 281, 12760–12766 [DOI] [PubMed] [Google Scholar]
- 35.Northcott P. A., Nakahara Y., Wu X., Feuk L., Ellison D. W., Croul S., Mack S., Kongkham P. N., Peacock J., Dubuc A., Ra Y. S., Zilberberg K., McLeod J., Scherer S. W., Sunil Rao J., Eberhart C. G., Grajkowska W., Gillespie Y., Lach B., Grundy R., Pollack I. F., Hamilton R. L., Van Meter T., Carlotti C. G., Boop F., Bigner D., Gilbertson R. J., Rutka J. T., Taylor M. D. (2009) Nat. Genet. 41, 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalakonda N., Fischle W., Boccuni P., Gurvich N., Hoya-Arias R., Zhao X., Miyata Y., Macgrogan D., Zhang J., Sims J. K., Rice J. C., Nimer S. D. (2008) Oncogene 27, 4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P., Jenkins N. A., Copeland N. G. (2003) Genome Res. 13, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eggan K., Akutsu H., Loring J., Jackson-Grusby L., Klemm M., Rideout W. M., 3rd, Yanagimachi R., Jaenisch R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rideout W. M., 3rd, Hochedlinger K., Kyba M., Daley G. Q., Jaenisch R. (2002) Cell 109, 17–27 [DOI] [PubMed] [Google Scholar]
- 40.Foudi A., Hochedlinger K., Van Buren D., Schindler J. W., Jaenisch R., Carey V., Hock H. (2009) Nat. Biotechnol. 27, 84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hock H., Hamblen M. J., Rooke H. M., Schindler J. W., Saleque S., Fujiwara Y., Orkin S. H. (2004) Nature 431, 1002–1007 [DOI] [PubMed] [Google Scholar]
- 42.Hock H., Hamblen M. J., Rooke H. M., Traver D., Bronson R. T., Cameron S., Orkin S. H. (2003) Immunity 18, 109–120 [DOI] [PubMed] [Google Scholar]
- 43.Eminli S., Foudi A., Stadtfeld M., Maherali N., Ahfeldt T., Mostoslavsky G., Hock H., Hochedlinger K. (2009) Nat. Genet. 41, 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindler J. W., Van Buren D., Foudi A., Krejci O., Qin J., Orkin S. H., Hock H. (2009) Cell Stem Cell 5, 43–53 [DOI] [PubMed] [Google Scholar]
- 45.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan Y., Nikitina T., Zhao J., Fleury T. J., Bhattacharyya R., Bouhassira E. E., Stein A., Woodcock C. L., Skoultchi A. I. (2005) Cell 123, 1199–1212 [DOI] [PubMed] [Google Scholar]
- 47.Brugarolas J., Moberg K., Boyd S. D., Taya Y., Jacks T., Lees J. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao J. H., Wu D., DelRosario R., Castellanos A., Balmain A., Perez-Losada J. (2008) Oncogene 27, 6596–6600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.