Abstract
Cardiac diseases associated with mutations in troponin subunits include hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), and restrictive cardiomyopathy (RCM). Altered calcium handling in these diseases is evidenced by changes in the Ca2+ sensitivity of contraction. Mutations in the Ca2+ sensor, troponin C (TnC), were generated to increase/decrease the Ca2+ sensitivity of cardiac skinned fibers to create the characteristic effects of DCM, HCM, and RCM. We also used a reconstituted assay to determine the mutation effects on ATPase activation and inhibition. One mutant (A23Q) was found with HCM-like properties (increased Ca2+ sensitivity of force and normal levels of ATPase inhibition). Three mutants (S37G, V44Q, and L48Q) were identified with RCM-like properties (a large increase in Ca2+ sensitivity, partial loss of ATPase inhibition, and increased basal force). Two mutations were identified (E40A and I61Q) with DCM properties (decreased Ca2+ sensitivity, maximal force recovery, and activation of the ATPase at high [Ca2+]). Steady-state fluorescence was utilized to assess Ca2+ affinity in isolated cardiac (c)TnCs containing F27W and did not necessarily mirror the fiber Ca2+ sensitivity. Circular dichroism of mutant cTnCs revealed a trend where increased α-helical content correlated with increased Ca2+ sensitivity in skinned fibers and vice versa. The main findings from this study were as follows: 1) cTnC mutants demonstrated distinct functional phenotypes reminiscent of bona fide HCM, RCM, and DCM mutations; 2) a region in cTnC associated with increased Ca2+ sensitivity in skinned fibers was identified; and 3) the F27W reporter mutation affected Ca2+ sensitivity, maximal force, and ATPase activation of some mutants.
Keywords: ATPases, Calcium-binding Proteins, Cardiac Muscle, Cardiac Hypertrophy, Protein-Protein Interactions, Dilated Cardiomyopathy, Restrictive Cardiomyopathy, Hypertrophic Cardiomyopathy, Thin Filament, Troponin C
Introduction
Troponin C is an important Ca2+ buffer of cardiac myofibrils and is sensitive to alterations in the levels of available Ca2+ necessary for contraction. The hypothesis to be tested here is as follows. Can we can make mutations within cTnC that affect the force-pCa relationship, ATPase activation/inhibition, and maximal and basal force consistent with the paradigms associated with the hypertrophic (HCM),2 restrictive (RCM), and dilated (DCM) cardiomyopathies in in vitro functional phenotypes defined below? By doing this, we believe that identifying such mutations will allow us to eventually predict the in vivo phenotypes in knock-in mice.
The Ca2+ sensitivity of contraction can be altered by drugs that increase the Ca2+ affinity of cTnC (1–4) or by HCM mutations located in the thin filament (5–9). RCM mutations in cardiac troponin I (cTnI) and cardiac troponin T (cTnT) also sensitize the myofilament to Ca2+ (10–12). However, to date RCM mutations have not been found in cTnC. These changes heighten the Ca2+ sensitivity of force development and prolong the Ca2+ transient as Ca2+ dissociates more slowly from cTnC (13, 14). This longer duration of the Ca2+ transient acts to maintain the Ca2+ concentrations that are capable of sustaining contraction (15). Mutations linked to RCM in contrast to HCM tend to impair ATPase inhibition in reconstituted systems as well as increase the basal force in skinned fibers (10, 12). HCM mutations generally increase the Ca2+ sensitivity without affecting the ATPase inhibition (16). The functional phenotypes of disease-causing mutations can be correlated with clinical manifestations in patients (17). The HCM and RCM mutations cause diastolic dysfunction (ranging from moderate to severe), and RCM affects an additional parameter because the heart has a “restricted capacity” and is unable to relax fully between subsequent intervals (18). The loss of ATPase inhibitory properties is consistent with this impaired relaxation that is evident in RCM (10–12).
On the other hand, mutations that cause DCM decrease the Ca2+ sensitivity of force production (19–21) and would shorten the force transient as Ca2+ dissociates more quickly from cTnC. This shortened duration of the force transient would decrease the amount of force-generating cross-bridges that could be recruited during contraction, effectively lowering the amount of force generated (19, 22), which results in systolic dysfunction in patients. The ventricular walls have thinned, and the cardiac output is significantly reduced (23). The lowered force production in skinned fibers correlates with decreased actomyosin activation in the presence of Ca2+ (22).
Targeting the Ca2+ affinity of cTnC directly is more ideal than changing the overall Ca2+ handling and the sarcoplasmic reticulum as a Ca2+ source (24–26). A limitation of many of the drugs available today is that they affect Ca2+ homeostasis but do not directly target cTnC (24). In addition, increased Ca2+ sensitivity has been linked to the susceptibility to arrhythmias that can be a primary cause behind the high incidence of sudden cardiac death in HCM patients (27). Studies conducted with transgenic mice that contain the HCM-cTnT mutations I79N and F110I presented with an increased prevalence of ventricular tachycardia that was paralleled by the degree of myofilament Ca2+ sensitization.
It has been established that mutations in the regulatory proteins (Tm, TnT, TnI, and TnC) have the ability to change the Ca2+ sensitivity of force development. Subsequently, TnC is considered the Ca2+ sensor of contraction. We conclude that TnC would be the ultimate target of these mutations, i.e. the last protein to be affected by this altered function (17). Because Ca2+ sensitization exists as a potential disease mechanism, this suggests that alterations in the Ca2+ regulation of contraction play a distinct role in the disease processes of HCM, RCM, and DCM. Therefore, this study addresses the following question. Can we generate mutations in cTnC not yet discovered in man that alter the functional parameters of contraction and mimic the in vitro phenotypes of HCM, RCM, and DCM previously established in other sarcomeric proteins? If cTnC is directly responsible for the phenotypic properties of these diseases, then it should be possible to recapitulate the cardiomyopathy functional phenotypes in vitro by using mutations in cTnC.
TnC is an EF-hand protein that consists of two globular regions that are connected by a long central helix (28). The two domains contain two EF-hand helix-loop-helix motifs that consist of Ca2+-binding loops and short segments of antiparallel β-sheets between the Ca2+-binding loops of each domain (28, 29). The cTnC N terminus has only one functional low affinity Ca2+-binding site (site II and KCa ∼105 m−1), although the C terminus contains two high affinity Ca2+-binding sites (sites III and IV KCa ∼107 m−1) (30). The N terminus is considered the regulatory domain because binding of Ca2+ to site II initiates contraction. Also, the Ca2+ binding affinity of TnC is influenced by additional members of the troponin (Tn) complex. Cardiac TnC specifically interacts with cTnI and cTnT to make up cardiac (cTn). Located along the thin filament, the cTn complex regulates the contraction of striated muscle, whereby cTnC acts as the Ca2+ sensor, conferring the sensitivity of muscle to Ca2+. Cardiac TnI is the inhibitory subunit and mainly prevents the interaction of actin and myosin in the absence of Ca2+; cTnT binds to Tm and has a role in myosin-actin ATPase activation (31). Within the cTn complex, the cTnC Ca2+-binding sites have a 10-fold increase in Ca2+ affinity than when measured in isolated cTnC (32–34). Studies have shown that TnC is coupled to the activity of the actomyosin ATPase (35, 36). The Ca2+ affinity when translated into Ca2+ sensitivity of force development is dependent on a number of parameters, including cTnC Ca2+ affinity, the members of the Tn complex, the presence of the thin filament (actin and Tm), and myosin cross-bridge formation.
We generated 20 strategic mutations in the non-Ca2+-liganding residues and first screened by looking at changes in the Ca2+ sensitivity of force development in cardiac skinned fibers. A few of these mutations were previously reported in the skeletal system (37, 38). In addition, steady-state fluorescence studies have shown that substitution of glutamic acid for nonpolar residues of the nonliganded residues V44Q and L48Q greatly enhances the Ca2+ affinity of cTnC (39). Other Gln substitutions (F20Q, F24Q, V79Q, M80Q, and M81Q) showed various effects on the affinity of the cTnC N terminus for Ca2+. The functional studies were performed with and without the additional mutation F27W. This fluorescent mutant was included so the Ca2+ affinity of the isolated cTnC could be monitored (38). It was generally found that there was a correlation between the Ca2+ affinity of isolated cTnC and the Ca2+ sensitivity in skinned fibers; however, there were a few exceptions to this trend. It is known that additional members of the thin filament can modify the Ca2+ affinity of cTnC; therefore, these mutations may have a more complex mechanism of action (16).
The cTnC mutants E40A and V64Q studied here were previously shown by NMR studies to undergo significant chemical shift changes upon Ca2+ binding to cTnC (40). In addition, the cTnC mutation A23Q located at the interface of cTnI binding was selected for its potential to alter its binding to the regulatory domain through electrostatic interactions. Additional cTnC mutations S37G and T53G were designed to disrupt N-capping, which stabilizes α-helices.
To assess the effects of these mutations, a functional screen was conducted using these cTnC mutants specifically designed to have increased or decreased Ca2+ sensitivity of force production. Also, it was determined if the recovery of maximal and basal force mimicked the HCM, DCM, and RCM functional phenotypes. DCM has the most consistent effect on force recovery, and these mutations generally decrease recovered force in skinned fibers (19). The actomyosin ATPase activities were determined in pCa 4.0 and 8.0 to measure the effects of the mutations on their respective activation and inhibition properties. The increase in the Ca2+ dependence of ATPase activity was measured for seven of the cTnC mutations. The rationale for such experiments stems from the ability of the failing human myocardium to depress ATPase activity; therefore, mutations present in myofilament proteins can influence the Ca2+-activated ATPase rate and its basal activity (41, 42).
In summary, some of the mutations increased or decreased the Ca2+ sensitivity of force development in skinned fibers. Also, it was found that F27W, the mutation used for fluorescence measurements, altered results for several of the mutants. The effects of F27W were primarily on force recovery and ATPase activation. The RCM cTnC candidates that were identified were as follows: S37G, V44Q, and L48Q. The cTnC mutant A23Q displayed effects consistent with the HCM functional phenotype. The ideal cTnC DCM candidates identified in this screen were as follows: E40A and I61Q. In addition, this study identified a specific region in the cTnC regulatory N terminus that directly affects Ca2+ sensitivity. The mutations A23Q, V44Q, M47A, and L48Q are located in this region, and all cause an increase in Ca2+ sensitivity of force development. We show for the first time that the in vitro functional phenotypes and disease states of hypertrophic, dilated, and restrictive cardiomyopathies can be recapitulated by genetic engineering of the Ca2+ sensor of contraction.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Human Cardiac Troponin T, Troponin I, and Troponin C Mutants
The cTnI and cTnT cDNAs were cloned in our laboratory from human cardiac tissue and were used for expression and purification (43, 44). The cDNA for mouse cardiac TnC was previously cloned in our laboratory by RT-PCR using mouse heart total RNA (Stratagene), sequence-specific primers, and the Omniscript RT kit (Qiagen) (45). The mouse cardiac TnC-cDNA was used as a template for PCR using primers designed to produce the specific F27W-containing mutants: F20Q, A23Q, F24Q, S37G, E40A, K43A, V44Q, L48Q, T53G, E55A, L57Q, I61Q, V64Q, F77Q, V79Q, M80Q, M81Q, and R83A. The following mutants were also made with F27W absent: A23Q, S37G, E40A, V44Q, M47A, L48Q, I61Q, V64Q, and V79Q. All subcloned DNA sequences were inserted into the pET3d expression plasmid and sequenced to verify that sequences were correct prior to expression and purification. Standard methods previously used in this laboratory were utilized for the expression and purification of the HcTnCs (12). Briefly, the cTnC-WT and mutants were expressed in Escherichia coli BL21 (DE3) codon plus bacterial cells (Stratagene). The bacterially expressed TnCs were purified over a Q-Sepharose column and eluted with a linear gradient of 0–0.6 m KCl (4 °C) in a buffer containing 50 mm Tris (pH 7.8), 6 m urea, 1 mm EDTA, 1 mm DTT. The purest TnC fractions were then dialyzed into 50 mm Tris-HCl (pH 7.5), 1 mm CaCl2, 1 mm MgCl2, 50 mm NaCl, 1 mm DTT, loaded onto a phenyl-Sepharose column after the addition of 0.5 m AmSO4, and eluted using a Ca2+-free buffer containing 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, and 1 mm DTT at room temperature. Collected fractions were run on SDS-polyacrylamide gels, and the fractions containing the purest most concentrated fractions were pooled yielding >97% pure protein, dialyzed exhaustively against 5 mm NH4HCO3, and then lyophilized. All steps were performed at 4 °C unless otherwise indicated.
Fiber Preparation and Determination of the Ca2+ Dependence of Force Development
Cardiac tissue was from newly slaughtered pigs obtained at a slaughterhouse. Strips of muscle, 3–5 mm in diameter and ∼5 mm in length, were teased from the papillary muscle of the left ventricle and skinned overnight in a 50% glycerol-relaxing solution that contained low Ca2+ concentrations (10−8 m [Ca2+]free, 1 mm [Mg2+]free, 7 mm EGTA, 2.5 mm MgATP2−, 20 mm MOPS (pH 7.0), 20 mm creatine phosphate, and 15 units/ml creatine phosphokinase, I = 150 mm) and 1% Triton X-100 at −20 °C. Fibers were then transferred to a similar solution without Triton X-100 and kept at −20 °C. A skinned fiber bundle with the diameter of ∼75–100 μm was mounted using stainless steel clips to a force transducer and then immersed in a pCa 8.0 relaxation solution (conditions described above). The contraction solution (pCa 4.0) had the same composition as the pCa 8.0 solution; however, the Ca2+ concentration was 10−4 m, and this was used to measure the initial force. To analyze the Ca2+ dependence of force development, the skinned fiber tension was tested in intermediate Ca2+ solutions ranging from pCa 8.0 to 4.0 and calculated with the pCa calculator program developed in our laboratory (46). The native cTnC was depleted by incubating the fiber in a CDTA (trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid) extracting solution (5 mm CDTA and 25 mm Tris (pH 8.4)) for ∼1.5 h. To evaluate the efficiency of TnC extraction, the residual tension was assessed by activating the fiber in pCa 4.0 solution. Residual tensions of 15% or below from the initial maximal force were considered satisfactory. After that point, fibers were incubated with 28 μm of mutant or WT cTnC diluted in pCa 8.0 for 1 h. To ensure exogenous cTnC reconstitution, the maximal force was verified using pCa 4.0 solution. Data were analyzed using the following equation: % change in force = 100 × ([Ca2+]n/([Ca2+]n + [Ca2+50]n) where “[Ca2+50]” is the free [Ca2+] that produces 50% force and the superscript n stands for the nHill coefficient. All fiber experiments were performed at room temperature.
Basal Force Measurements in Skinned Fibers
The fibers were mounted and skinned with 1% Triton X-100 (in pCa 8), and the initial maximal force was measured. The endogenous porcine cTnC was extracted with 5.5 mm CDTA (pH 8.4). The cTnC-extracted fibers were maximally activated (in pCa 4) to determine the residual force (because of the remaining cTnC). The base lines of the cTnC-extracted fibers were recorded and then incubated with mutant cTnC in pCa 8 solution. After 1 h of cTnC incubation, the maximal force was tested again, and the base line was recorded. The elevation of the base line after cTnC incubation was considered to be an increase in basal force (see supplemental Fig. 1A for further details).
Formation of Troponin Complexes
The purified individual Tn subunits were first dialyzed against 3 m urea, 1 m KCl, 10 mm MOPS, 1 mm DTT, and 0.1 mm phenylmethanesulfonyl fluoride (PMSF) and then twice against the same buffer excluding urea. The protein concentrations of the individual subunits were determined using the Coomassie Plus kit (Pierce) and then mixed in a 1.3:1.3:1 cTnT/cTnI/cTnC molar ratio. After 1 h, the complexes were successively dialyzed against decreasing concentrations of KCl (0.7, 0.5, 0.3, 0.1, 0.05, and 0.025 m). The excess precipitated TnT and TnI that formed during complex formation was removed by centrifugation. Proper stoichiometry was verified by SDS-PAGE before storage of cTn complexes at −80 °C. Ternary cTn complexes were utilized in the actin-Tm-activated myosin-ATPase assays containing the cTnC mutants.
Actin-Tm-activated Myosin-ATPase Assays, Minimum and Maximum ATPase
Porcine cardiac myosin, rabbit skeletal F-actin, and porcine cardiac Tm were prepared as described previously (16). The protein concentrations used for actomyosin ATPase assays were as follows: 0.6 μm porcine cardiac myosin, 3.5 μm rabbit skeletal F-actin, 1 μm porcine cardiac Tm, and 0–2 μm of pre-formed Tn complexes as described above. The buffer conditions for the proteins were as follows: myosin in 10 mm MOPS (pH 7.0), 0.4 m KCl, 1 mm DTT; actin in 10 mm MOPS (pH 7.0), 40 mm KCl; Tm in 10 mm (pH 7.0), 300 mm KCl, 1 mm DTT. The final ionic strength of the reactions was ∼75 mm when considering the ionic contributions from the various protein buffers. The ATPase inhibitory assay was performed in a 0.1-ml reaction mixture (2× values) as follows: 6.76 mm MgCl2, 0.28 μm CaCl2, 3.0 mm EGTA, 2 mm DTT, 23.1 mm MOPS (pH 7.0) at 25 °C. The ATPase activation assay was performed using the same 0.1-ml buffer mixture with (2× values) the following: 6.63 mm MgCl2 and 3.45 mm CaCl2. The ATPase reaction was initiated with the addition of 4 mm ATP and quenched after 20 min using trichloroacetic acid to a final concentration of 35%. The precipitated assay proteins were removed by centrifugation, and the inorganic phosphate concentration in the supernatant, which was released by ATP hydrolysis, was determined according to the method of Fiske and SubbaRow (47).
Ca2+ Sensitivity of the ATPase
pCa curves were performed using 1 μm of pre-formed cTn complex with the same concentrations of porcine β-myosin, rabbit skeletal F-actin, and porcine cardiac Tm as described above. The conditions of the assay varied slightly (2× values), 23 mm MOPS, 4 mm EGTA, 2 mm nitrilotriacetic acid, 7.15–7.48 mm MgCl2 (pH 7.0 at 25 °C), however, in varying pCa solutions, pCa 8.0 (0.113 mm CaCl2); pCa 7.0 (0.899 mm CaCl2); pCa 6.5 (1.915 mm CaCl2); pCa 6.0 (2.982 mm CaCl2); pCa 5.5 (3.639 mm CaCl2); pCa 5.0 (3.971 mm CaCl2); pCa 4.5 (4.278 mm CaCl2); pCa 4.0 (4.913 mm CaCl2), calculated according to methods established in our laboratory (46).
Tryptophan Fluorescence of cTnC
The cTnCs containing F27W were dialyzed into fluorescence buffer containing 2 mm EGTA, 5 mm NTA, 120 mm MOPS, 90 mm KCl. Before each titration, 1.25 mm MgCl2 and 1 mm fresh DTT solution were added. Steady-state fluorescence measurements were performed in a Jasco 6500 spectrofluorimeter, and tryptophan fluorescence was excited at 285 nm emission and detected at 338 nm. The values were normalized so that 100% fluorescence corresponds to the highest fluorescence intensity in the presence of Ca2+. Data were fitted to the Hill equation that accounted for fluorescence changes due to the Ca2+ binding to the N-terminal sites.
Circular Dichroism Measurements
Far-UV CD spectra were collected using a 1-mm path quartz cell in a Jasco J-720 spectropolarimeter. Spectra were recorded at 195–250 nm with a bandwidth of 1 nm at a speed of 50 nm/min and a resolution of 0.5 nm at room temperature (∼20 °C). Ten scans were averaged, and no numerical smoothing was applied. The optical activity of the buffer was subtracted from relevant protein spectra. Mean residue ellipticity ([θ]MRE, in degree·cm2/dmol) for the spectra was calculated utilizing the same Jasco system software and using the following equation: [θ]MRE = [θ]/(10 × Cr × l), where [θ] is the measured ellipticity in millidegrees; Cr is the mean residue molar concentration, and l is the path length in centimeters (16). Protein concentrations were determined by the Biuret reaction using bovine serum albumin as a standard. The CD experiments were performed using three different conditions: apo (1 mm EGTA, 20 mm MOPS, 100 mm KCl (pH 7.0)); Mg2+ (1 mm EGTA, 20 mm MOPS, 100 mm KCl, 2.075 mm MgCl2 (pH 7.0), yielding a free [Mg2+] of 2 mm), and Ca2+ (1 mm EGTA, 20 mm MOPS, 100 mm KCl, 2.075 mm MgCl2, 1.096 mm CaCl2 (pH 7.0), yielding a free [Ca2+] of 0.1 mm and 2 mm free [Mg2+]). The experimental protein concentration for the WT and each mutation was 0.2 mg/ml.
Three-dimensional Modeling
The cTnCs mutations were modeled in Protein Data Bank (PDB) files as depicted using PyMol software. PyMol is an open source molecular visualization program that allows the manipulation of PDB files that contain molecular coordinates from x-ray crystallography- or NMR-solved structures. The program allows mutagenesis of selected residues, portrays potential side chain interactions, and hydrogen bonding that may occur due to changes in the nature and proximity of the side chains.
Statistical Analysis
The experimental results were reported as × ± S.E., and analyzed for significance using Student's t test at p < 0.05.
RESULTS
Effects of the cTnC Mutations on the Ca2+ Sensitivity and Maximal Tension in Skinned Muscle Fibers
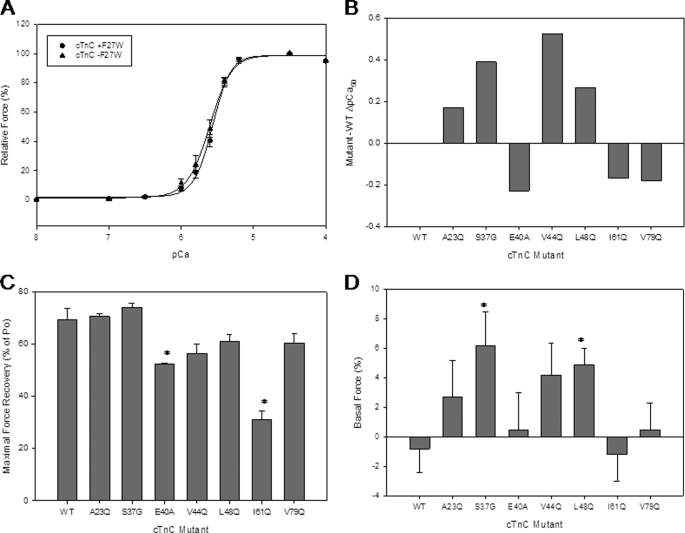
The first level at which the cTnC mutants were screened was in cardiac skinned fibers. This was considered the most physiologically relevant assay available to evaluate the consequences of the cTnC mutant proteins. The mutant cTnCs were incorporated into the cTnC-depleted porcine cardiac fibers, and their effects on Ca2+ sensitivity and force recovery were determined. Fig. 1A compares the Ca2+ sensitivity of force development obtained in depleted cardiac skinned fibers reconstituted with cTnC plus and minus the F27W mutation. First, it was determined which mutations cause an increase or decrease in Ca2+ sensitivity. Of the 18 mutants evaluated, only the following 4 mutants were found to significantly increase the Ca2+ sensitivity of force development: A23Q, S37G, V44Q, and L48Q. (see Fig. 1B and Table 1, not all data shown). Ten mutations decreased the Ca2+ sensitivity of force development and were reported as negative pCa50 values (see Table 1 and data not shown). Four mutations did not significantly change the Ca2+ sensitivity of tension development as follows: F24Q, K43A, E55A, and R83A (data not shown). Experiments were first performed with mutant cTnCs that contained the F27W mutation that allowed for the measurements of Ca2+ affinity by monitoring the Ca2+-dependent changes in fluorescence from the lone tryptophan (Trp). It became evident that the F27W probe altered the functional profiles of the mutants that were studied; therefore, the data discussed in this paper will be from experiments performed with eight of the cTnC mutants that lack F27W. See Fig. 1, B and C, for skinned fiber results that contain the mutants in the (−F27W) background. The functional data results and Ca2+ affinity values for mutants containing F27W are reported in Table 1.
FIGURE 1.
Ca2+ dependence of force development in porcine skinned fibers reconstituted with the cTnC mutants. A, comparison of skinned fiber results obtained by reconstitution of skinned fibers with cTnC (−F27W) versus cTnC (+F27W). The fibers were depleted of endogenous cTnC by CDTA treatment and reconstituted with mutant mouse cTnCs. B, skinned fiber ΔpCa50 (Mutant pCa50−WT pCa50) from cTnC mutants (−F27W). Data graphed where endogenous pCa50 is utilized as a control from which the experimental pCa50 is subtracted. Data are shown where WT = 0. C, maximal force recovery for cTnC mutants (−F27W) reconstituted in skinned fibers. The % force recovered values are a comparison of the amount of force produced in endogenous pCa curves, and the value was obtained by the experimental pCa curves. D, basal force data are shown for cTnC mutants (−F27W) and plotted as % of increased base line after cTnC reconstitution. The basal force was calculated by dividing the increased base line by the maximal force recovery (see supplemental Fig. 1A). Each point represents an average number of experiments performed and is expressed as mean ± S.E. *, p < 0.05 compared with wild type.
TABLE 1.
Summary of the functional data obtained for the cTnC mutants in the +F27W and −F27W backgrounds
Numbers in parentheses indicate the number of experiments. − indicates −F27W, and + indicates +F27W.
| Mutant | Skinned fiber |
Reconstituted ATPase |
Isolated TnC (F27W) |
|||
|---|---|---|---|---|---|---|
| pCa50 | nHill | pCa50 | nHill | pCa50 | nHill | |
| WT− | 5.66 ± 0.01 | 3.54 ± 0.12 (7) | 5.54 ± 0.05 | 2.30 ± 0.94 (8) | ||
| WT+ | 5.64 ± 0.01 | 2.23 ± 0.02 (9) | 5.57 ± 0.06 | 2.34 ± 0.79 (8) | 5.48 ± 0.02 | 1.08 ± 0.04 (5) |
| HCM | ||||||
| A23Q− | 5.85 ± 0.02a | 1.43 ± 0.24 (5)a | 6.01 ± 0.02a | 1.70 ± 0.65 (4) | ||
| A23Q+ | 5.76 ± 0.02a | 1.66 ± 0.06 (8)a | 5.79 ± 0.09a | 1.41 ± 0.17 (8)a | 5.98 ± 0.08a | 0.72 ± 0.10 (4)a |
| RCM | ||||||
| S37G− | 6.05 ± 0.01a | 2.43 ± 0.15 (6)a | 6.70 ± 0.02a | 1.83 ± 0.71 (6) | ||
| S37G+ | 5.81 ± 0.03a | 1.78 ± 0.13 (8)a | 6.69 ± 0.05a | 1.50 ± 0.22 (8) | 5.47 ± 0.02 | 0.93 ± 0.04 (4) |
| V44Q− | 6.19 ± 0.03a | 1.73 ± 0.16 (6)a | 6.73 ± 0.13a | 1.72 ± 0.81 (5) | ||
| V44Q+ | 6.04 ± 0.02a | 1.39 ± 0.04 (8)a | 6.36 ± 0.05a | 2.22 ± 0.50 (8) | 6.29 ± 0.07a | 0.36 ± 0.03 (4)a |
| L48Q− | 5.93 ± 0.02a | 2.89 ± 0.33 (6) | 6.19 ± 0.06a | 1.40 ± 0.27 (8) | ||
| L48Q+ | 6.02 ± 0.01a | 1.58 ± 0.08 (9)a | 6.12 ± 0.05a | 1.48 ± 0.20 (8) | 6.13 ± 0.05a | 0.56 ± 0.04 (4)a |
| DCM | ||||||
| E40A− | 5.43 ± 0.01a | 2.55 ± 0.06 (6)a | 5.22 ± 0.11a | 1.79 ± 0.74 (6) | ||
| E40A+ | 5.43 ± 0.01a | 1.66 ± 0.85 (7) | 5.43 ± 0.04a | 2.00 ± 0.38 (8) | 5.16 ± 0.08a | 1.49 ± 0.37 (4) |
| 61Q− | 5.52 ± 0.01a | 3.11 ± 0.24 (5) | 5.39 ± 0.06a | 2.04 ± 0.55 (4) | ||
| I61Q+ | 5.42 ± 0.02a | 1.42 ± 0.10 (10)a | 5.51 ± 0.05 | 1.65 ± 0.32 (8) | 5.42 ± 0.07 | 0.95 ± 0.05 (4) |
| V79Q− | 5.47 ± 0.01a | 2.77 ± 0.10 (6)a | 5.34 ± 0.08a | 3.15 ± 1.27 (6) | ||
| V79Q+ | 5.44 ± 0.02a | 1.99 ± 0.17 (5) | 5.47 ± 0.03a | 1.27 ± 0.15 (8)a | 5.30 ± 0.03a | 0.74 ± 0.05 (4)a |
a p < 0.05 compared with the WT at same condition.
The force recovery in skinned fibers was significantly decreased for the cTnC mutants E40A and I61Q as seen in Fig. 1C. The cTnC mutants A23Q and S37G showed a slight tendency toward increased force recovery, although the effect did not reach significance compared with WT. The skinned fiber force recovery values for a couple of the mutants A23Q and S37G were significantly higher in the absence of F27W (data not shown).
The Hill coefficient nH measures the steepness of the force-pCa relationship and reports how activation occurs upon Ca2+ binding and the degree of communication between adjacent regulatory units. Muscle activation is a highly cooperative process, and the slope of the force- pCa relationship gives an indication of how the mutations affect the contractile apparatus. Several mutations A23Q, S37G, V44Q and L48Q decreased the nH values compared with the WT (see Table 1). These mutations were identified in a specific region of cTnC that also was associated with Ca2+ sensitization and exists within a cluster of residues.
Effects of the cTnC Mutations on Basal Force in Skinned Muscle Fibers
The basal force measurements examined the effect of the cTnC mutants on the ability of skinned fibers to undergo relaxation at pCa 8.0. The base line was increased for several mutants, and this can be considered an increase in the basal force that occurred upon reconstitution of the mutant cTnC into skinned fibers. For a schematic of the procedure and how the measurements were performed, see supplemental Fig. 1A. Two of the cTnC mutants, S37G and L48Q, caused a significant increase in the basal force during mutant cTnC incubation (Fig. 1D). V44Q showed a trend to increase the basal force; however, this effect was not significant when compared with WT. In addition, these three mutants impaired the ability of the actomyosin ATPase to inhibit at low Ca2+ concentrations. However, A23Q did not increase the basal force compared with WT. The three mutants, E40A, I61Q, and V79Q, with decreased Ca2+ sensitivity of force development in skinned fibers did not cause an increase in basal force (see Fig. 1D). Interestingly, when these experiments were performed with the mutants in the cTnC (+F27W) background, the same trends were seen as with the −F27W; however, all the mutants maintained a much higher base line after incorporation into the skinned fibers (supplemental Fig. 1B). It appeared that the F27W mutation itself altered the basal force in skinned fibers.
Effects of the cTnC Mutations on the Ca2+ Dependence of ATPase Activation
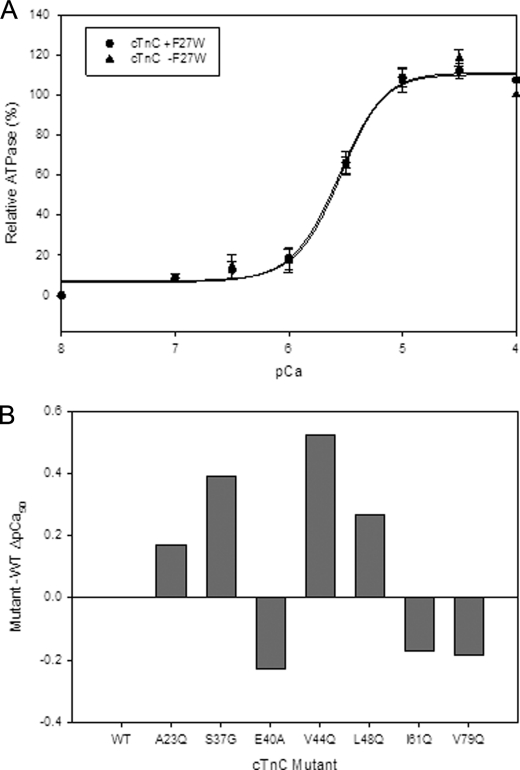
We looked at the effects of the cTnC mutants on Ca2+-dependent ATPase measurements. The Ca2+ sensitivity values obtained for the cTnC mutants were largely consistent with results seen in skinned fibers. However, the actomyosin ATPase pCa curves generally displayed enhanced Ca2+ sensitivity compared with the values seen in skinned fibers (see Fig. 2B and Table 1). There were no significant changes in the WT (plus and minus the F27W mutation) pCa 50 values obtained by actomyosin ATPase (see Fig. 2A). A couple of mutants A23Q and V44Q showed a further and significant increase in Ca2+ sensitivity of the actomyosin ATPase when F27W was absent (see Table 1). On the other hand, the mutant E40A was significantly less sensitive to Ca2+ when F27W was present (see Table 1).
FIGURE 2.
Effect of the cTnC mutants on the Ca2+ dependence of actomyosin ATPase activity. A shows the Ca2+ sensitivity (pCa50) of the ATPase obtained using 1 μm cTn complex containing WT cTnC (+F27W) versus WT cTnC (−F27W). B, comparison of the ΔpCa50 values obtained from complexes containing cTnC mutants (−F27W). Data represent n = 4–8 and are reported as S.E. and shown in Table 1.
Effects of the cTnC Mutations on Maximal and Minimal Actomyosin ATPase Activities
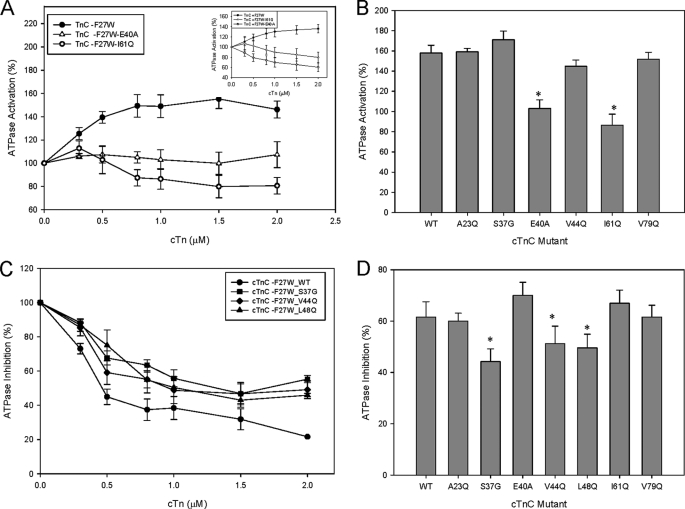
ATPase measurements were performed to examine the ability of the cTn mutants to activate the actomyosin ATPase when Ca2+ is present. The data obtained give a measure of how well cTnI relocates along the actin filament, allowing actin-myosin interactions. It was expected that some of the cTnC mutations studied would be able to fully activate the reconstituted thin filament. However, mutations that followed the trends shown by DCM mutants tended to have impaired activation of the ATPase as well as lowered force recovery in skinned fibers (see Fig. 3, A and B, and Fig. 1C). In general, the cTnC mutants displayed much higher activation levels at 1 μm cTn (which represents the physiological ratio of thin filament proteins) when F27W was absent. However, I61Q was an exception to this trend (Fig. 3A and data not shown). Only two of the cTnC mutants E40A and I61Q showed a statistically significant decrease in % activation at 1 μm cTn complex (see Fig. 3, A and B). These two mutants greatly affected the ability of cTn to activate the thin filament compared with WT (see Fig. 3B). These data are consistent with the impaired ability of skinned fibers reconstituted with cTnC E40A and I61Q to recover force (see Fig. 1C). Activation of reconstituted actomyosin ATPase was the most widely affected parameter with an approximate 20–40% higher activation when F27W was absent. The cTnC mutant S37G showed a slight trend toward increasing the ATPase activation (Fig. 3B).
FIGURE 3.
Effect of the cTnC mutants on activation and inhibition of the actomyosin-ATPase. A, comparison of the ATPase activation values with increasing concentrations of cTn complexes containing WT and DCM-like mutants E40A and I61Q cTnC (−F27W) at pCa 4.0. The inset shows ATPase activation of WT (+F27W) and the DCM-like mutants E40A and I61Q in the (+F27W) background. B, comparison of the relative ATPase activation levels for cTnC (−F27W) WT and mutants. C, inhibition of ATPase activity by increasing concentrations of cTn complexes containing mutant and WT cTnC (−F27W) was assessed at pCa 8.5. D, comparison of the relative ATPase inhibition levels for cTnC (−F27W) WT and mutants. The values were obtained from an average of 4–5 experiments performed in triplicate. Each point represents an average number of experiments performed and is expressed as mean ± S.E. *, p < 0.05 compared with wild type.
The ATPase actomyosin inhibition assays were completed for the cTnC mutants lacking F27W in the absence of Ca2+ and revealed that S37G impaired the inhibitory properties of the actomyosin ATPase (Fig. 3, C and D). This impaired inhibition coupled with increased Ca2+ sensitivity in skinned fibers is consistent with the RCM phenotype shown in a number of studies (10, 12). The “DCM-like” mutant cTnC-E40A showed a statistically significant increase in the ability to inhibit ATPase compared with WT cTnC (see Fig. 3D). Occasionally, DCM mutants may inhibit the ATPase activity better than the corresponding WT. The reduction in ATPase inhibition measurements for the cTnC mutants S37G, V44Q, and L48Q also reached significance at 2 μm cTn complex compared with WT (see Fig. 3C). The mutant A23Q inhibited the ATPase activity to the same degree as WT (see Fig. 3D).
Effect of the cTnC Mutations on the Apparent Ca2+ Affinity in Isolated cTnC
Steady-state tryptophan fluorescence was utilized to determine the relative affinities of the cTnC N terminus for Ca2+. The F27W mutation was introduced along with the cTnC mutations as a method to report the Ca2+-dependent structural changes of the N-domain and the effects of the introduced mutations on Ca2+ affinity. The Ka values reported here did not always directly correspond to the Ca2+ sensitivity of force development in skinned fibers. All of the cTnC mutants were analyzed by tryptophan fluorescence.
The tryptophan fluorescence studies determined the Ca2+ affinity of isolated cTnC-F20Q, A23Q, F24Q, S37G, E40A, K43A, V44Q, L48Q, T53G, E55A, L57Q, I61Q, V64Q, F77Q, V79Q, M80Q, M81Q, and R83A mutants. In general, the mutations showing an increase in the Ca2+ sensitivity of force development also had an increase in the apparent Ca2+ affinity of site II in isolated cTnC. An exception was S37G, which had a similar apparent Ca2+ affinity value as WT (see Table 1) but showed a large increase in Ca2+ sensitivity of skinned cardiac fibers (see Fig. 1B). The mutants A23Q, V44Q, L48Q had increased Ca2+ affinity of cTnC compared with WT, which is consistent with the increased Ca2+ sensitivity seen in skinned cardiac fibers. Most of the mutants that decreased the Ca2+ sensitivity of force development in fibers also decreased the Ca2+ affinity of site II. For the DCM candidate mutants, E40A had a lower affinity for Ca2+ versus WT, whereas the apparent Ca2+ affinity of I61Q was only slightly lower than for WT. The mutants E55A and R83A showed little change in the Ca2+ sensitivity of skinned fibers and Ca2+ affinity of isolated cTnC (data not shown). All of the mutants above showed a strong correlation between their Ca2+ sensitivity in fibers and the Ca2+ affinity of isolated cTnC; however, F20Q, S37G, and L57Q showed no correlation between these measurements. Both F20Q and L57Q increased the Ca2+ affinity of isolated cTnC and decreased the Ca2+ sensitivity at the level of the skinned fiber. I61Q had a significant decrease in the Ca2+ sensitivity of skinned fibers but insignificant changes at the level of Ca2+ binding to isolated cTnC (see Table 1). The cTnC mutants F20Q, L57Q, V79Q, M80Q, and M81Q caused substantial decreases in Ca2+ sensitivity in skinned fibers; however, only M80Q and M81Q showed a slight corresponding decrease in apparent Ca2+ affinity (data not shown, for V79Q see Figs. 1B and 2B). In contrast, the affinity for Ca2+ of this group was only increased for F20Q and L57Q compared with WT (data not shown). Absent from the tryptophan fluorescence data is the mutant F24Q where no measurable increase in fluorescence was detected during Ca2+ titration. It is possible that the F24Q mutation is located too close to F27W and interfered with obtaining a measurable fluorescent signal.
Circular Dichroism Assessed the Secondary Structural Changes That Occurred because of the cTnC Mutations
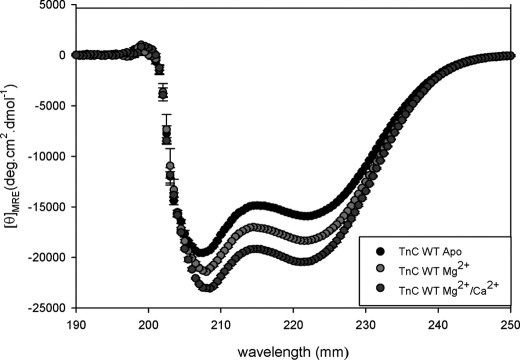
The CD studies showed that some of the mutations caused structural changes in isolated cTnC. These measurements were performed in the following states in WT cTnC: apo (no divalent cations), Mg2+-bound, and Mg2+/Ca2+-saturated conditions. The amount of α-helical content change in the different metal-bound states is shown in Fig. 4. In the apo state, only V79Q did not show a change in secondary structure. The DCM-like mutant I61Q mutant (with decreased apparent Ca2+ affinity and Ca2+ sensitivity of force development) had a decrease in the α-helical content (see Table 2), whereas V44Q, an RCM-like mutant (with increased apparent Ca2+ affinity and Ca2+ sensitivity of force development), had an increased amount of α-helical structure compared with WT as shown in Table 2. Subsequently, the HCM-like mutant A23Q also showed an increase in the amount of α-helical secondary structure to WT.
FIGURE 4.
Determination of secondary structural characteristics of WT-cTnC by CD in various cation-bound states. The spectra compares the changes in α-helical content of WT-cTnC (−F27W) under different conditions as follows: apo, Mg2+-bound, Mg2+/Ca2+-bound states. Far-UV CD was recorded at 195–250 nm with a bandwidth of 1 nm at a speed of 50 nm/min and a resolution of 0.5 nm at room temperature (20 °C). Mean residue ellipticity ([θ]MRE, in degree·cm2/dmol) for the spectra was calculated utilizing the same Jasco system software and the following equation: [θ]MRE = [θ]/(10 × Cr × l), where [θ] is the measured ellipticity in millidegrees; Cr is the mean residue molar concentration, and l is the path length in centimeters.
TABLE 2.
Summary of circular dichroism results of cTnC mutants in the −F27W background
n = number of experiments.
| TnC | [θ]222 nm (degrees·cm2 dmol−1) |
n | ||
|---|---|---|---|---|
| APO | Mg2+ | Ca2+/Mg2+ | ||
| WT | −15928.5 ± 51.2 | −18344.3 ± 105.8 | −20429.0 ± 147.6 | 4 |
| A23Q | −16370.1 ± 50.1a | −18479.2 ± 288.7 | −22041.0 ± 94.4a | 3 |
| S37G | −15136.7 ± 139.8a | −17873.9 ± 91.6a | −20466.7 ± 372.3 | 4 |
| E40A | −14962.6 ± 212.1a | −17049.9 ± 127.9a | −19246.5 ± 363.4a | 4 |
| V44Q | −16876.7 ± 423.3a | −18612.2 ± 375.9 | −21851.8 ± 352.6a | 3 |
| L48Q | −15042.8 ± 179.5a | −18287.2 ± 268.7 | −20557.4 ± 329.7 | 3 |
| I61Q | −14591.8 ± 180.7a | −18282.7 ± 306.9 | −19956.6 ± 425.9 | 3 |
| V79Q | −15590.8 ± 302.1 | −17511.6 ± 206.6a | −19970.2 ± 144.8 | 3 |
a p < 0.05 compared with the respective WT at same condition.
The CD data of the cTnC mutants bound with Mg2+ show that E40A and V79Q had a significant decrease in α-helical content. In the Ca2+/Mg2+-bound structures, E40A caused a similar decrease in the α-helical content of the protein (see Table 2). Also, in the Ca2+/Mg2+ state, A23Q and V44Q displayed significant increases in α-helical content compared with WT. As the CD data indicate, not all the mutants that caused distinct functional changes had noticeable structural changes in isolated cTnC. Additional parameters may be at play in these mutant proteins in that subtle changes may alter important protein-protein contacts in the cTn complex, which may affect thin filament function and ultimately alter cross-bridge recruitment.
DISCUSSION
It was explored whether changing the Ca2+ affinity of cTnC was sufficient to recreate the effects and functional profiles of known cardiomyopathic mutations. It was found that the cTnC mutations displayed phenotypic properties that paralleled known HCM, RCM, and DCM mutations. These mutations were introduced into the regulatory N terminus and involved only the non-Ca2+-liganding residues. Therefore, the changes in Ca2+ affinity were more likely because of conformational changes that altered the dynamics of Ca2+ binding or perturbed thin filament protein-protein interactions and not to direct changes in Ca2+ coordination of the regulatory site II. Previous studies have shown that substitution of glutamic acid for key nonpolar nonliganded residues greatly enhances the Ca2+ affinity of cTnC (39, 48). This study, however characterized mutations that increased or decreased the Ca2+ sensitivity of force development in skinned cardiac fibers. In addition, the Ca2+ affinities of the isolated cTnCs were measured using tryptophan fluorescence; however, these data do not always correlate with the Ca2+ sensitivity of the skinned fiber containing the mutations (16, 49). Some of the mutant cTnCs were identified in a previous study and selected for their effects on Ca2+ affinity (39, 48) and on the basis of mutational modeling. Inclusion of the cTnC mutants in the functional screen was also based on PyMol illustrations. From this information, it was found that V44Q is located in close proximity to the F27W mutation, and Trp substituted at this position may modify the V44Q functional outcomes.
Previously, Chandra et al. (36) reported that the F27W mutation affects activation levels of the myofibrillar ATPase; however, the Ca2+ sensitivity or maximal tension was not altered in fibers. We found that the Ca2+ sensitivity values in cardiac skinned fibers were decreased for cTnC S37G and V44Q when F27W was present (see Table 1). The Ca2+ sensitivity of actomyosin ATPase was also reduced for A23Q and V44Q when F27W was included (see Table 1). The mutant V44Q is located in close proximity to F27W (evident from modeling), which may explain why it altered the functional parameters of this mutant. Force recovery values were further decreased in A23Q and S37G in the presence of F27W.
The cTnC mutants were classified as HCM, DCM or RCM-like mutations based on the results of the functional screen. From the data presented here, potential candidate mutations were selected to represent each type of cardiomyopathy. Mutations that increased the Ca2+ sensitivity of force development were first classified as HCM or RCM-like mutations due to the paradigm that these mutations most often sensitize the myocardium to Ca2+ (17). Conversely, mutations that desensitized the skinned fiber to Ca2+ and decreased force recovery were classified as DCM-like mutations.
In addition to the disease classification, it has been seen that mutations that cause large changes in cooperativity tend to manifest a more severe phenotype (12, 50). Changes in the cooperativity of force activation (nHill) are typically displayed as an altered slope of the force-pCa relationship in skinned fibers (51). It was observed that the A23Q, S37G, V44Q, and L48Q mutations with greatly enhanced Ca2+ affinity of isolated cTnC displayed a loss of cooperativity in the fluorescence spectra and cardiac skinned fibers (see Table 1). This might be partially attributed to the large fluorescence spectral change that occurs when Ca2+ binds the Ca2+/Mg2+ sites (C terminus) and the ability of the N-terminal probe to detect conformational changes occurring when Ca2+ binds to the C-terminal sites. It is possible that these mutations alter the communication of cTnC with cTnT which may be transmitted through the thin filament and ultimately lead to impaired nearest-neighbor cooperative interactions (52).
The activation of the thin filament requires cTnT, which makes several contacts with the cTnC C-domain (53). The N terminus of cTnT, which binds at the Tm-Tm overlap region and helps stabilize the position of Tm on the actin filament, controls the degree of activation obtained (54–56). Mutations in the N terminus that affect cTnC interactions with cTnT may alter the ability of the thin filament to undergo activation and result in increased or decreased force recovery. The process of muscle activation should be considered in the context of the three-state model developed by McKillop and Geeves (57). This model considers the thin filament to exist in dynamic equilibrium between three different states as follows: 1) the blocked state (myosin heads are generally unbound and cTnI lies over the actin-myosin sites and Tm lies along the outer actin domain); 2) the closed state (myosin heads bind weakly to actin and cTnI has moved further allowing cross-bridges to bind, and Tm is present); and 3) the open state (myosin heads isomerize into a strongly bound state where cTnI is in tight association with cTnC and Tm has moved further). It was shown that TnC is able to modulate the rate at which myosin is able to shift from a weakly bound (nonforce generating state) to a force-generating state (58). This reciprocal interplay is believed to control both the steady state of force development and the rate of force generation and relaxation.
The reconstituted actomyosin ATPase data for the WT and mutant cTnC containing cTn complexes all demonstrated increased levels of activation (10–40%) when F27W was not present (Fig. 3B). Those mutants that increased the activation of the thin filament generally increased the force recovery as well (see Figs. 3B and 1C). The two assays are measuring the same parameter, although in the skinned fibers there is higher level of organization of sarcomeric proteins than in the reconstituted ATPase system (59). In addition, force is measured under isometric conditions, although ATPase measurements do not have a tension component. Also, the reconstituted ATPase system has a tendency to exaggerate the Ca2+ sensitivity changes possibly due to the availability of more myosin heads binding to actin than is possible in the skinned fiber system (60). The ATPase actomyosin inhibition assays performed in the absence of Ca2+ revealed that several of the mutations (S37G, V44Q, and L48Q) impaired inhibition by the actomyosin ATPase.
Distinct RCM-like candidates were S37G, V44Q, and L48Q that had increased Ca2+ sensitivity of force development, slightly decreased to normal force recovery, impaired actomyosin ATPase inhibition, and increased basal force. The mutant V44Q in cTnC showed a drastic increase in Ca2+ sensitivity of ATPase activation compared with cTnC WT (see Fig. 2B). These results are consistent with the skinned fiber data for V44Q in cTnC that also resulted in a large increase in the Ca2+ sensitivity of force development. One of the primary reasons that these mutations were classified as RCM-like mutants is that they showed a significant impairment in their ability to inhibit that actomyosin ATPase compared with WT (Fig. 3, C and D). This impaired relaxation stems from an altered ability of cTn to fully inhibit force generation at low Ca2+ concentrations. Another important parameter of muscle regulation is the level of basal force. An increase in the amount of basal force could contribute to the impaired relaxation seen during diastole in RCM patients (11, 12, 61). The difference in basal force in the skinned fibers was measured by examining the increase in the base line during the cTnC incubation. The decreased inhibition seen in the RCM-like mutations correlates well with the impaired relaxation and increased basal force seen in S37G and L48Q. Also, the mutant V44Q impaired ATPase inhibition although it did not cause a significant increase in the basal force (Figs. 1D and 3, C and D).
Increased contractility in cardiac muscle may be the result of a prolonged Ca2+ transient due to higher affinity of the myofilament containing Ca2+-sensitizing mutations (14). This may lead to altered diastolic function at lower [Ca2+] leading toward impaired relaxation. These mutations in cTnC could contribute to an altered dynamic of cTnC-cTnI interactions that may interfere with the three states that occur during myofilament activation. In this scenario, cTnI does not effectively prevent cross-bridge formation (the thin filament maintains a low level of activation) even at low levels of Ca2+ that are not sufficient to stimulate contraction. This is consistent with the RCM mutant phenotype where the ventricular walls become stiffened, providing resistance to filling and maintaining a higher base line (slightly activated) force between heart beats resulting in impaired relaxation.
The candidate mutation that was generated in this study and had the phenotype closest to HCM was cTnC A23Q. This mutant demonstrated ATPase inhibition levels similar to those of WT at 1 μm concentration. Also, the ATPase activation levels and force recovery levels were similar to what was found for WT (see Fig. 3, B and D). The functional phenotype seen with A23Q is consistent with results seen with HCM-linked mutants. A previous study had shown that HCM-cTnC mutants had no effects on ATPase inhibition, which supports the findings for A23Q (16).
The DCM candidates (E40A and I61Q) were identified that had a decreased Ca2+ sensitivity of force development (Fig. 1B), lowered % force recovery (Fig. 1C), unchanged to slightly lower inhibition in actomyosin ATPases (Fig. 3, C and D), and no significant changes in basal force (data not shown). Both E40A and I61Q inhibited the reconstituted thin filament as measured by actomyosin ATPase assays at activating Ca2+ levels (Fig. 3, A and B). These mutations may alter the protein-protein contacts between cTnC and cTnI and interfere with the effective transmission of the Ca2+ binding signal. Even in the presence of Ca2+, the cTnI regulatory region might be less efficient at moving away from its inhibitory position on actin, thus strengthening its association with cTnC. This also may have an effect on the number of cross-bridges that can be recruited. These factors would directly impact the amount of force or degree of activation that is achieved. One result of decreased cross-bridge formation is a diminished cTnC Ca2+ affinity because these two processes are tightly coupled (62).
An inability to recover force is a common outcome of the DCM phenotype, and for most of these mutants it was coupled with decreased Ca2+ sensitivity of force recovery. Actomyosin ATPase inhibition levels in the absence of Ca2+ were similar in value to WT. However, E40A inhibited even better than WT in the absence of Ca2+, which suggests that the cTnI inhibitory region may be blocking the myosin-actin interactions in a more efficient manner. Because cTnT has a fundamental role in ATPase activation and is tightly coupled with force recovery, it may be that these mutations alter crucial TnT interactions that are necessary to produce normal activation levels (31, 63). These two parameters were considered key features of mutations that cause DCM (19, 20). Interestingly, the tryptophan fluorescence data indicated that E40A had lower affinity for Ca2+ versus WT, whereas I61Q had a Ca2+ affinity similar to WT (Table 1). The mutants E40A and I61Q decreased Ca2+ sensitivity in both reconstituted actomyosin pCa curves and in cardiac skinned fibers (see Figs. 1B and 2B). A number of candidates shared these characteristics; therefore, it appeared to be easier to recapitulate the DCM phenotype. Also, it is known that even small changes in Ca2+ sensitivity and a reduction in force recovery may lead to severe DCM disease (23). The mutants E40A and I61Q from this screen were both classified as DCM-like mutations. Other mutations showed drastic changes in these parameters and were not selected as they may have lethal consequences when studied in vivo.
It is anticipated that the cTnC mutants that displayed both a decrease in the Ca2+ sensitivity (in fibers and ATPase) and a decreased Ca2+ affinity of WT cTnC may be altering the Ca2+ sensitivity of force in skinned fibers by a distinct mechanism. The effects of these mutations are expected to be due to direct local conformational changes that directly alter affinity of cTnC for its binding partners (cTnI and cTnT) or long range effects on the other members of the thin filament. For example, only E40A, T53G, and V79Q showed a decrease in the Ca2+ affinity of cTnC by fluorescence, and these effects were transmitted to the thin filament and manifested as decreased Ca2+ sensitivity of force development in skinned fibers (see Table 1 and data not shown). However, a consistent trend was that the mutants A23Q, V44Q, and L48Q that increased the Ca2+ sensitivity of force development in fibers showed ATPase activation levels similar to or greater than WT and also increased Ca2+ affinity of isolated cTnC as shown by fluorescence. This could be explained as follows: the increased Ca2+ affinity of cTnC may play a dominant role in sensitizing the thin filament to Ca2+. However, this inherent property of increased Ca2+ affinity may contribute partially to the pCa50 seen in skinned fibers. In addition, the thin filament proteins or cross-bridges may also heighten the Ca2+ sensitivity to the level seen in the skinned fiber experiments (16).
As structure underlies function, it was important to assess whether the mutations also alter the structure of the cTnC mutants. Cardiac TnC has a high degree of α-helical content, and mutations may alter the structure (increase or decrease the amount of α-helix). The CD measurements are reported as relative changes in α-helix compared with the WT spectrum. As cTnC has three EF-hands, there are short β-sheet strands that may also be altered in the presence of the mutations. In addition, because Mg2+ predominantly binds to the C-domain, Mg2+-saturated conditions provide evidence of whether one of the N-domain mutations can alter the structure of the C-domain.
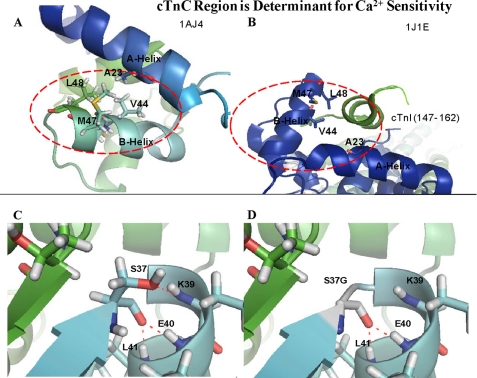
The location of the L48Q mutation is depicted by PyMol in PDB 1AJ4 (Fig. 5, A and B). The mutation was modeled into PDB 1MXL (the cTnC N-domain complexed with cTnI(147–163)) and shows that mutation of Leu to Gln is likely to affect the interaction between cTnC and cTnI (Fig. not shown). This may affect Ca2+ sensitivity by altering the binding of cTnI to cTnC, which increases its affinity for Ca2+ (34). Also, CD did not show any significant structural changes for L48Q in either of the metal bound states. This suggests that the increased apparent Ca2+ affinity of the regulatory site II and increased Ca2+ sensitivity of force development may not begin with detectable alterations in cTnC itself. The mutation A23Q was designed specifically to affect the interactions made between cTnC and the cTnI regulatory domain (40). See the location of the A23Q mutation in Fig. 5, A and B.
FIGURE 5.
Hot spot region in cTnC for increased Ca2+ sensitivity of force development. A, region in cTnC in the absence of TnI binding PDB 1AJ4. B, region is pictured in the presence of the TnI regulatory domain from PDB 1J1E. Modeling of the Ser-37 (C) and the S37G mutation (D) in the PDB file 1AJ4. Potential hydrogen bonds are shown as dotted red lines.
In addition, the mutation V79Q involves a substitution that removes stabilizing hydrophobic interactions between the N- and D-helices. The N-helix has an important role in modulating the Ca2+ affinity of the N-domain (65). The PyMol modeling suggests that V79Q may form a hydrogen bond with the side chain of Asp-75. This is the site of a known DCM mutation D75Y, which is situated at the beginning of the Ca2+-binding loop of the regulatory Ca2+-binding site II (see supplemental Fig. 2) (66, 67). The newly formed hydrogen bond between the side chains of Asp-75 and Gln-79 could perturb the Ca2+-binding loop conformation and subsequently alter the Ca2+ affinity of site II. This decrease in Ca2+ affinity of isolated cTnC may be the first alteration in a series of perturbed contacts and ultimately may result in dysfunction of the myofilament containing V79Q-cTnC. Also, the CD data for V79Q reports a loss of α-helical content in the Mg2+-bound state that is consistent with an effect on binding of this divalent cation (see Table 1).
The V44Q mutation lies between helices A and B in the N-domain. Val-44 appears to be involved in hydrophobic intramolecular interactions that stabilize the interface of these two helices (see Fig. 5A). This mutation is responsible for the largest Ca2+ sensitivity increase in skinned cardiac fibers found in this study. Therefore, the substitution of Gln at Val-44 causes structural changes in cTnC that greatly sensitizes the N terminus for Ca2+. A structural study demonstrates that the relationship between residues Val-44, Leu-48 and Ala-23 are an important determinant that influences the closed conformation of cTnC even when Ca2+ binds site II (68). The NMR data showed that nuclear Overhauser effect contacts (up to 5 Å) were maintained between the A- and B-helices when Ca2+ binds, in sTnC these nuclear Overhauser effect contacts would become weaker as the distance between the A- and B-helices increases. It is anticipated that the insertion of Gln at position 44 would increase the distance between the A- and B-helices. The loss of hydrophobic contacts specifically between the A- and B-helices is important to the mechanism that increases the Ca2+ sensitivity. The CD data indicate that this mutation causes structural changes primarily in the N-domain because the binding of Mg2+ did not cause a significant change in structure (see Table 2). Previously, it was noted that metal binding to the N terminus induces only minor conformational changes in the N terminus of WT cTnC (69). The apo state and Ca2+/Mg2+-bound states both show an increase in α-helical content, suggesting that an intrinsic structural difference exists in apo cTnC-V44Q compared with the WT in the apo state (see Table 2). Also, Ca2+ binds to site II in the V44Q mutant with a much higher affinity than WT, which may mean that an increase in α-helical content correlates with heightened Ca2+ affinity of isolated cTnC (see Table 1). The mutants A23Q, S37G, V44Q, and L48Q with increased Ca2+ sensitivity of force have increased α-helical content in the apo state; therefore, it is possible that the more ordered structure allows a more coordinated response to Ca2+ binding.
The effects of skeletal sTnC-E41A (equivalent to cTnC-E40A in this study) were reported by a number of groups (70–72). Sykes and co-workers (71, 72) found that the N-terminal mutant E41A-sTnC located in the Ca2+-binding loop of site I remains in a closed structure upon binding Ca2+. Normally, the sTnC N-domain opens upon binding of two Ca2+ ions; however, it was shown that Glu-41 lies at a critical hinge region that moves to separate helices B and C from the NAD unit (74). The data obtained with cTnC-E40A parallels the discoveries found in the sTnC. The E40A-cTnC was also modified at site I, and although this site is inactive in the cardiac isoform, this residue plays a critical role in modulating the openness of the N-domain upon Ca2+ binding (71). As the cardiac N-domain does not open in the same sense as sTnC, it might be suggested that this mutation increases the flexibility of the hinge region thus influencing the rate of Ca2+ dissociation from cTnC. The PyMol model showed that the loss of the Glu removes a potential hydrogen bond made with the N-capping residue Ser-37 of helix B, which may contribute to structural stability (see supplemental Fig. 3, A and B). Therefore, it is not surprising that E40A-cTnC has decreased Ca2+ sensitivity of force development in skinned fibers as well as decreased Ca2+ affinity of isolated TnC as seen by tryptophan fluorescence (Table 1). In addition, E40A appeared to cause a global change in cTnC structure as tryptophan fluorescence detected Ca2+-binding events in both the C- and N-domains (data not shown). In reference to the above studies, the CD data revealed a predictable decrease in α-helical content for the cTnC-E40A mutation that may correlate with its decreased Ca2+ sensitivity (see Table 2).
The I61Q mutation decreased the cTnC N-terminal affinity for Ca2+, and studies in skinned fibers have revealed that it greatly decreases force development (see Fig. 1C and Table 1). One study showed that the equivalent residue in skeletal muscle sTnC-I60Q affected the duration of relaxation and was likely to interfere with cooperative interactions between the regulatory units in skinned fibers (64). This mutation is modeled in supplemental Fig. 3, C and D, and suggests that the introduction of Gln causes the side chain of I61Q to make a new potential hydrogen bond with Thr-38 near helix B. These altered interactions within cTnC are likely to affect protein interactions along the thin filament that are important for the transmission of force obtained upon cross-bridge binding.
A study examining the effects of different amino acids at the N-capping position of the C-helix in sTnC revealed that T54G had the greatest attenuation of Ca2+ affinity of the N-terminal domain regulatory sites as seen by tryptophan and stopped-flow fluorescence (73). The N-capping residue may influence the equilibrium that exists between the partially formed (Ca2+-free) and fully formed C-helix in the Ca2+-bound state. The mutation S37G was selected based on modeling that suggested it to be the N-capping residue for the B-helix, and mutation of this residue was designed to disrupt N-capping and destabilize the B-helix. Fig. 5, C and D, shows the loss of hydrogen bonding by the Ser side chain at the N-cap. Destabilizing the B-helix is expected to assist the BC unit in moving faster away from the NAD unit upon Ca2+ binding. This may enhance the opening of the hydrophobic cleft upon Ca2+ binding, thereby enhancing Ca2+ affinity.
The structural data (CD and modeling) assist in our understanding of the crucial intra- and intermolecular interactions within the thin filament, which ultimately may reveal the mechanism of disrupted myofilament function. In general, the cTnC mutants did not have large changes in secondary structure as shown by CD analysis. A higher level of complexity may be necessary to discern the level at which the mutations affect the function of the thin filament. Although there are no detectable changes in secondary α-helical structure, properties of the cTnC mutants may in fact alter important interactions with cTnI and cTnT and manifest the effects to the additional members of the thin filament (16, 49). For example, it was shown that a number of the mutations related to HCM or DCM in Tm, TnT, or TnI and TnC need the presence of all the thin filament proteins (actin-Tm-Tn) to reproduce the Ca2+ sensitivity changes observed in fibers or in reconstituted systems (8, 16). In addition, some cTnC mutations (A8V and D145E) need the addition of myosin because they may influence the number of cross-bridges that are formed and thereby further influence the Ca2+ sensitivity.
This study identified a “hot spot” region of cTnC that when mutated increased the cTnC Ca2+ affinity that could be directly translated into increased Ca2+ sensitivity in skinned fibers (see Fig. 5, A and B). These mutations A23Q, V44Q, M47A, and L48Q all increased the Ca2+ affinity of isolated cTnC, which indicates that they have a direct mechanism of increased Ca2+ sensitivity in skinned fibers, unlike the HCM cTnC mutations A8V and D145E (16). The corresponding mutations in skeletal muscle (V45T, M48A, and L49T) increased the Ca2+ affinity from a pCa50 of 5.7 for WT (F29W) to values varying from 5.8 to 6.2 as measured by tryptophan fluorescence using F29W (38). This region may have promise as a potential drug target in that compounds could be designed to bind in this region and interfere with the interactions between helices A and B. This may sensitize or desensitize the N-domain regulatory Ca2+-binding site to Ca2+.
A future direction would be to determine the effects of these cTnC “HCM, RCM, and DCM candidates,” which demonstrated the in vitro properties of the respective diseases in mice to assess whether altering (increasing or decreasing) the Ca2+ affinity of cTnC is sufficient to re-create complex disease phenotypes of HCM, RCM, and DCM by simply changing the cTnC Ca2+ affinity and fiber Ca2+ sensitivity. The screen of cTnC mutants allowed the selection of mutations that mimic the in vitro consequences of cardiomyopathy mutations as follows: A23Q (HCM), S37G (RCM), and E40A (DCM). It is possible that some of the mutants studied here may be found in patients in the future and be associated with a particular form of cardiomyopathy.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL-42325 (to J. D. P.). This work was also supported by American Heart Postdoctoral Fellowships AHA 0825368E (to J. R. P.) and AHA 09POST2300030 (to M. S. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- HCM
- hypertrophic cardiomyopathy
- DCM
- dilated cardiomyopathy
- RCM
- restrictive cardiomyopathy
- PDB
- Protein Data Bank
- Tn
- troponin
- cTn
- cardiac Tn
- Tm
- tropomyosin
- CDTA
- 1,2-cyclohexylenedinitrilotetraacetic acid.
REFERENCES
- 1.Kurebayashi N., Ogawa Y. (1988) J. Physiol. 403, 407–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollesello P., Ovaska M., Kaivola J., Tilgmann C., Lundström K., Kalkkinen N., Ulmanen I., Nissinen E., Taskinen J. (1994) J. Biol. Chem. 269, 28584–28590 [PubMed] [Google Scholar]
- 3.Sorsa T., Heikkinen S., Abbott M. B., Abusamhadneh E., Laakso T., Tilgmann C., Serimaa R., Annila A., Rosevear P. R., Drakenberg T., Pollesello P., Kilpelainen I. (2001) J. Biol. Chem. 276, 9337–9343 [DOI] [PubMed] [Google Scholar]
- 4.el-Saleh S. C., Solaro R. J. (1987) J. Biol. Chem. 262, 17240–17246 [PubMed] [Google Scholar]
- 5.Chandra M., Rundell V. L., Tardiff J. C., Leinwand L. A., De Tombe P. P., Solaro R. J. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H705–H713 [DOI] [PubMed] [Google Scholar]
- 6.Gomes A. V., Potter J. D. (2004) Ann. N.Y. Acad. Sci. 1015, 214–224 [DOI] [PubMed] [Google Scholar]
- 7.Chang A. N., Harada K., Ackerman M. J., Potter J. D. (2005) J. Biol. Chem. 280, 34343–34349 [DOI] [PubMed] [Google Scholar]
- 8.Robinson P., Griffiths P. J., Watkins H., Redwood C. S. (2007) Circ. Res. 101, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 9.Yumoto F., Lu Q. W., Morimoto S., Tanaka H., Kono N., Nagata K., Ojima T., Takahashi-Yanaga F., Miwa Y., Sasaguri T., Nishita K., Tanokura M., Ohtsuki I. (2005) Biochem. Biophys. Res. Commun. 338, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 10.Gomes A. V., Liang J., Potter J. D. (2005) J. Biol. Chem. 280, 30909–30915 [DOI] [PubMed] [Google Scholar]
- 11.Davis J., Wen H., Edwards T., Metzger J. M. (2007) Circ. Res. 100, 1494–1502 [DOI] [PubMed] [Google Scholar]
- 12.Pinto J. R., Parvatiyar M. S., Jones M. A., Liang J., Potter J. D. (2008) J. Biol. Chem. 283, 2156–2166 [DOI] [PubMed] [Google Scholar]
- 13.Lang R., Gomes A. V., Zhao J., Housmans P. R., Miller T., Potter J. D. (2002) J. Biol. Chem. 277, 11670–11678 [DOI] [PubMed] [Google Scholar]
- 14.Wen Y., Pinto J. R., Gomes A. V., Xu Y., Wang Y., Wang Y., Potter J. D., Kerrick W. G. (2008) J. Biol. Chem. 283, 20484–20494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller T., Szczesna D., Housmans P. R., Zhao J., de Freitas F., Gomes A. V., Culbreath L., McCue J., Wang Y., Xu Y., Kerrick W. G., Potter J. D. (2001) J. Biol. Chem. 276, 3743–3755 [DOI] [PubMed] [Google Scholar]
- 16.Pinto J. R., Parvatiyar M. S., Jones M. A., Liang J., Ackerman M. J., Potter J. D. (2009) J. Biol. Chem. 284, 19090–19100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willott R. H., Gomes A. V., Chang A. N., Parvatiyar M. S., Pinto J. R., Potter J. D. (2010) J. Mol. Cell. Cardiol. 48, 882–892 [DOI] [PubMed] [Google Scholar]
- 18.Huang X. P., Du J. F. (2004) Acta Pharmacol. Sin. 25, 1569–1575 [PubMed] [Google Scholar]
- 19.Venkatraman G., Harada K., Gomes A. V., Kerrick W. G., Potter J. D. (2003) J. Biol. Chem. 278, 41670–41676 [DOI] [PubMed] [Google Scholar]
- 20.Morimoto S., Lu Q. W., Harada K., Takahashi-Yanaga F., Minakami R., Ohta M., Sasaguri T., Ohtsuki I. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershberger R. E., Pinto J. R., Parks S. B., Kushner J. D., Li D., Ludwigsen S., Cowan J., Morales A., Parvatiyar M. S., Potter J. D. (2009) Circ. Cardiovasc. Genet. 2, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dweck D., Reynaldo D. P., Pinto J. R., Potter J. D. (2010) J. Biol. Chem. 285, 17371–17379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burkett E. L., Hershberger R. E. (2005) J. Am. Coll. Cardiol. 45, 969–981 [DOI] [PubMed] [Google Scholar]
- 24.Endoh M. (1998) Jpn. Heart J. 39, 1–44 [DOI] [PubMed] [Google Scholar]
- 25.Mathew L., Katz S. D. (1998) Drugs Aging 12, 191–204 [DOI] [PubMed] [Google Scholar]
- 26.Scholz H., Brückner R., Mügge A., Reupcke C. (1986) J. Mol. Cell. Cardiol. 18, Suppl. 5, 79–87 [DOI] [PubMed] [Google Scholar]
- 27.Baudenbacher F., Schober T., Pinto J. R., Sidorov V. Y., Hilliard F., Solaro R. J., Potter J. D., Knollmann B. C. (2008) J. Clin. Invest. 118, 3893–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzberg O., James M. N. (1985) Nature 313, 653–659 [DOI] [PubMed] [Google Scholar]
- 29.Moews P. C., Kretsinger R. H. (1975) J. Mol. Biol. 91, 201–225 [DOI] [PubMed] [Google Scholar]
- 30.Holroyde M. J., Robertson S. P., Johnson J. D., Solaro R. J., Potter J. D. (1980) J. Biol. Chem. 255, 11688–11693 [PubMed] [Google Scholar]
- 31.Potter J. D., Sheng Z., Pan B. S., Zhao J. (1995) J. Biol. Chem. 270, 2557–2562 [DOI] [PubMed] [Google Scholar]
- 32.Johnson J. D., Collins J. H., Robertson S. P., Potter J. D. (1980) J. Biol. Chem. 255, 9635–9640 [PubMed] [Google Scholar]
- 33.Güth K., Potter J. D. (1987) J. Biol. Chem. 262, 13627–13635 [PubMed] [Google Scholar]
- 34.Potter J. D., Gergely J. (1975) J. Biol. Chem. 250, 4628–4633 [PubMed] [Google Scholar]
- 35.Solaro R. J., Bousquet P., Johnson J. D. (1986) J. Pharmacol. Exp. Ther. 238, 502–507 [PubMed] [Google Scholar]
- 36.Chandra M., da Silva E. F., Sorenson M. M., Ferro J. A., Pearlstone J. R., Nash B. E., Borgford T., Kay C. M., Smillie L. B. (1994) J. Biol. Chem. 269, 14988–14994 [PubMed] [Google Scholar]
- 37.da Silva A. C., de Araujo A. H., Herzberg O., Moult J., Sorenson M., Reinach F. C. (1993) Eur. J. Biochem. 213, 599–604 [DOI] [PubMed] [Google Scholar]
- 38.Pearlstone J. R., Borgford T., Chandra M., Oikawa K., Kay C. M., Herzberg O., Moult J., Herklotz A., Reinach F. C., Smillie L. B. (1992) Biochemistry 31, 6545–6553 [DOI] [PubMed] [Google Scholar]
- 39.Tikunova S. B., Davis J. P. (2004) J. Biol. Chem. 279, 35341–35352 [DOI] [PubMed] [Google Scholar]
- 40.Li M. X., Spyracopoulos L., Sykes B. D. (1999) Biochemistry 38, 8289–8298 [DOI] [PubMed] [Google Scholar]
- 41.Knott A., Purcell I., Marston S. (2002) J. Mol. Cell. Cardiol. 34, 469–482 [DOI] [PubMed] [Google Scholar]
- 42.Chen Z., Higashiyama A., Yaku H., Bell S., Fabian J., Watkins M. W., Schneider D. J., Maughan D. W., LeWinter M. M. (1997) J. Mol. Cell. Cardiol. 29, 2345–2354 [DOI] [PubMed] [Google Scholar]
- 43.Zhang R., Zhao J., Potter J. D. (1995) J. Biol. Chem. 270, 30773–30780 [DOI] [PubMed] [Google Scholar]
- 44.Szczesna D., Zhang R., Zhao J., Jones M., Guzman G., Potter J. D. (2000) J. Biol. Chem. 275, 624–630 [DOI] [PubMed] [Google Scholar]
- 45.Gomes A. V., Guzman G., Zhao J., Potter J. D. (2002) J. Biol. Chem. 277, 35341–35349 [DOI] [PubMed] [Google Scholar]
- 46.Dweck D., Reyes-Alfonso A., Jr., Potter J. D. (2005) Anal. Biochem. 347, 303–315 [DOI] [PubMed] [Google Scholar]
- 47.Fiske C. H., Subbarow Y. (1925) J. Biol. Chem. 66, 375–400 [Google Scholar]
- 48.Tikunova S. B., Rall J. A., Davis J. P. (2002) Biochemistry 41, 6697–6705 [DOI] [PubMed] [Google Scholar]
- 49.Dweck D., Hus N., Potter J. D. (2008) J. Biol. Chem. 283, 33119–33128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandra M., Tschirgi M. L., Tardiff J. C. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H2112–H2119 [DOI] [PubMed] [Google Scholar]
- 51.Olsson M. C., Patel J. R., Fitzsimons D. P., Walker J. W., Moss R. L. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H2712–H2718 [DOI] [PubMed] [Google Scholar]
- 52.Pearlstone J. R., Smillie L. B. (1983) J. Biol. Chem. 258, 2534–2542 [PubMed] [Google Scholar]
- 53.Takeda S., Yamashita A., Maeda K., Maéda Y. (2003) Nature 424, 35–41 [DOI] [PubMed] [Google Scholar]
- 54.Hinkle A., Goranson A., Butters C. A., Tobacman L. S. (1999) J. Biol. Chem. 274, 7157–7164 [DOI] [PubMed] [Google Scholar]
- 55.Lehrer S. S., Geeves M. A. (1998) J. Mol. Biol. 277, 1081–1089 [DOI] [PubMed] [Google Scholar]
- 56.Tobacman L. S., Nihli M., Butters C., Heller M., Hatch V., Craig R., Lehman W., Homsher E. (2002) J. Biol. Chem. 277, 27636–27642 [DOI] [PubMed] [Google Scholar]
- 57.McKillop D. F., Geeves M. A. (1993) Biophys. J. 65, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenner B., Kraft T., Chalovich J. M. (1998) Adv. Exp. Med. Biol. 453, 177–184; discussion 185 [DOI] [PubMed] [Google Scholar]
- 59.Eastwood A. B., Wood D. S., Bock K. L., Sorenson M. M. (1979) Tissue Cell 11, 553–566 [DOI] [PubMed] [Google Scholar]
- 60.Zot A. S., Potter J. D. (1987) J. Biol. Chem. 262, 1966–1969 [PubMed] [Google Scholar]
- 61.Kubo T., Gimeno J. R., Bahl A., Steffensen U., Steffensen M., Osman E., Thaman R., Mogensen J., Elliott P. M., Doi Y., McKenna W. J. (2007) J. Am. Coll. Cardiol. 49, 2419–2426 [DOI] [PubMed] [Google Scholar]
- 62.Zot A. S., Potter J. D. (1989) Biochemistry 28, 6751–6756 [DOI] [PubMed] [Google Scholar]
- 63.Malnic B., Farah C. S., Reinach F. C. (1998) J. Biol. Chem. 273, 10594–10601 [DOI] [PubMed] [Google Scholar]
- 64.Kreutziger K. L., Piroddi N., Scellini B., Tesi C., Poggesi C., Regnier M. (2008) J. Physiol. 586, 3683–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith L., Greenfield N. J., Hitchcock-DeGregori S. E. (1994) J. Biol. Chem. 269, 9857–9863 [PubMed] [Google Scholar]
- 66.Baryshnikova O. K., Li M. X., Sykes B. D. (2008) J. Mol. Biol. 375, 735–751 [DOI] [PubMed] [Google Scholar]
- 67.Lim C. C., Yang H., Yang M., Wang C. K., Shi J., Berg E. A., Pimentel D. R., Gwathmey J. K., Hajjar R. J., Helmes M., Costello C. E., Huo S., Liao R. (2008) Biophys. J. 94, 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sia S. K., Li M. X., Spyracopoulos L., Gagné S. M., Liu W., Putkey J. A., Sykes B. D. (1997) J. Biol. Chem. 272, 18216–18221 [DOI] [PubMed] [Google Scholar]
- 69.Tsalkova T. N., Privalov P. L. (1985) J. Mol. Biol. 181, 533–544 [DOI] [PubMed] [Google Scholar]
- 70.Pearlstone J. R., Chandra M., Sorenson M. M., Smillie L. B. (2000) J. Biol. Chem. 275, 35106–35115 [DOI] [PubMed] [Google Scholar]
- 71.McKay R. T., Saltibus L. F., Li M. X., Sykes B. D. (2000) Biochemistry 39, 12731–12738 [DOI] [PubMed] [Google Scholar]
- 72.Li M. X., Gagné S. M., Spyracopoulos L., Kloks C. P., Audette G., Chandra M., Solaro R. J., Smillie L. B., Sykes B. D. (1997) Biochemistry 36, 12519–12525 [DOI] [PubMed] [Google Scholar]
- 73.Leblanc L., Bennet A., Borgford T. (2000) Arch. Biochem. Biophys. 384, 296–304 [DOI] [PubMed] [Google Scholar]
- 74.Gagné S. M., Li M. X., Sykes B. D. (1997) Biochemistry 36, 4386–4392 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.